Abstract
Background/Aims
The TRPC1 gene on chromosome 3q22–24 resides within the linkage region for diabetic nephropathy (DN) in type 1 (T1D) and type 2 diabetes mellitus (T2D). A recent study has demonstrated that TRPC1 expression is reduced in the kidney of diabetic ZDF- and STZ-treated rats. The present study aimed to evaluate the genetic and functional role of TRPC1 in the development of DN.
Methods
Genetic association study was performed with two independent cohorts, including 1,177 T1D European Americans with or without DN from GoKinD population and 850 African-American subjects with T2D-associated end-stage renal disease (ESRD), or with hypertensive (non-diabetic) ESRD, and nondiabetic controls. Seven tag SNP markers derived from HapMap data (phase II) were genotyped. TRPC1 gene expression was examined using real time RT-PCR.
Results
No significant association of TRPC1 DNA polymorphisms with DN or ERSD was found in GoKinD and African-American populations. TRPC1 gene mRNA expression in kidney was found to be trendily reduced in 12-week and significantly in 26-week-old db/db mice.
Conclusions
TRPC1 genetic polymorphism may not fundamentally contribute to the development of DN, while reduction of the gene expression in kidney may be a late phenomenon of DN as seen in diabetic animal models.
Key Words: TRPC1 gene, Single-nucleotide polymorphism, Diabetic nephropathy, End-stage renal disease, Diabetes types 1 and 2
Introduction
Diabetic nephropathy develops in approximately 40% of individuals with type 1 and type 2 diabetes, and is the most common cause of end-stage renal disease (ESRD) in the western world [1]. The prevalence and course of diabetic nephropathy are similar in patients with type 1 and type 2 diabetes, when they were matched for disease duration [2]. The development of diabetic nephropathy is likely impacted by both environmental and genetic factors. Genetic analysis of positional candidate genes may provide useful information for understanding the pathogenesis of this complex disease [3, 4].
Several genome-wide scans and linkage analyses indicate that a region of chromosome 3q is linked to diabetic nephropathy [5,6,7,8,9,10]. In recent years, we have focused on searching the susceptibility genes of diabetic nephropathy in this chromosomal region. Glomerulosclerosis is the major morphological change in patients with diabetic nephropathy [11]. Patients with type 1 and type 2 diabetes appear to progress through similar stages of nephropathy, while the routes they take may differ [12]. Glomerular mesangial cells (MC), located adjacent to glomerular capillaries, appear to play important roles in physiologic regulation of glomerular hemodynamics. The transient receptor potential channel 1 (TRPC1) molecule has been identified in both glomeruli and glomerular MC [13,14,15,16]. Importantly, the TRPC1 gene is located in chromosome 3q22–24 within the linkage region to diabetic nephropathy. Thus, TRPC1 represents a strong positional and biological candidate for diabetic nephropathy.
A recent study has demonstrated that significant reduction in TRPC1 gene expression in kidney and liver in diabetic ZDF- and STZ-treated rats appears to result from dysfunction in HNF4α. Kidney tissue TRPC1 expression at the protein level is also decreased in patients with diabetic nephropathy. It has been proposed that dysregulation of HNF4α and TRPC1 may underlie susceptibility to diabetic nephropathy [17]. To evaluate the potential role of TRPC1 gene in the development of diabetic nephropathy, in the present study, we have carried out genetic association analyses in two independent cohorts. The first cohort is a collection of 1,286 type 1 diabetes patients with and without diabetic nephropathy from the Genetics of Kidney Diseases in Diabetes (GoKinD) [18]. The second cohort consists of 850 African-American individuals collected from Wake Forest University School of Medicine [19]. They are divided into three groups, i.e. patients with type 2 diabetes-associated ESRD, or with hypertension-associated (nondiabetic) ESRD and nondiabetic subjects (controls). We have also examined TRPC1 gene expression with 12- and 26-week-old db/db mice in order to understand whether the reduction of this gene expression is primary or subsequent defect in the development of diabetic nephropathy.
Material and Methods
Genetic Association Study
Subjects
Clinical material in this study was composed of two independent cohorts. The first cohort consisted of 1,286 type 1 diabetes patients (female 683/male 603) with and without diabetic nephropathy from GoKinD study collected by the Juvenile Diabetes Research Foundation in collaboration with the Joslin Diabetes Centre, George Washington University, and the United States Centres for Diabetes Control and Prevention [18]. Among the GoKinD subjects, 1,177 (91.5%) (622 F/555 M) were of European descent, 109 (8.5%) were African Americans (n = 42), Hispanic Americans (n = 38), Native Indians (n = 7), Asian Americans (n = 4) and 18 unknown. The patients with type 1 diabetes were diagnosed before 31 years of age. Treatment with insulin was instituted within 1 year of diagnosis, and had been uninterrupted since then. Presence of diabetic nephropathy (case) was defined either by persistent proteinuria (urinary albumin excretion rate >300 mg/24 h) in 2 of 3 consecutive measurements (at least 1 month apart), or ESRD (not due to condition other than diabetes). Absence of diabetic nephropathy (control) was considered as persistent normalbuminuria (<30 mg/24 h) despite duration of type 1 diabetes for at least 15 years. The clinical characteristics of the GoKinD subjects are presented in table 1a.
Table 1.
Clinical characteristics
| a Clinical characteristics of the GoKinD cohort | ||||||
|---|---|---|---|---|---|---|
| T1D with DN |
T1D without DN |
|||||
| all |
female |
male |
all |
female |
male |
|
| Number | 578 | 265 | 313 | 599 | 357 | 242 |
| Age, years | 44±6 | 44±7 | 45±6 | 40±8 | 40±9 | 40±8 |
| Duration, years | 32±8 | 32±8 | 32±8 | 26±8 | 26±8 | 26±8 |
| HbA1c,% | 7.4±1.9 | 7.4±2.1 | 7.4±1.7 | 7.5±1.1 | 7.5±1.1 | 7.4±1.1 |
| BMI | 25.7±5.3 | 25.2±5.7 | 26.1±4.9 | 26.0±4.4 | 25.7±4.7 | 26.6±3.7 |
| Creatinine, mg/dl | 2.1±1.9 | 1.9±1.6 | 2.3±2.0 | 0.9±0.2 | 0.8±0.1 | 1.0±0.1 |
| Cystatin, mg/l | 2.2±1.7 | 2.2±1.6 | 2.4±1.7 | 0.8±0.1 | 0.8±0.1 | 0.8±0.1 |
| Cholesterol, mg/dl | 186.3±45.9 | 190.3±47.3 | 183.0±44.6 | 185.0±31.3 | 188.7±30.9 | 179.7±31.2 |
| HDL, mg/dl | 53.6±17.3 | 59.3±18.2 | 48.9±15.1 | 59.1±15.9 | 64.8±15.5 | 51.0±12.4 |
| Systolic BP, mm Hg | 131±19 | 130±20 | 133±18 | 118±12 | 116±12 | 122±12 |
| Diastolic BP, mm Hg | 74±11 | 72±11 | 75±11 | 71±8 | 70±7 | 74±8 |
| b Clinical characteristics of the African-American cohort | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| T2D-ESRD |
HTN-ESRD |
ND controls |
|||||||
| all |
female |
male |
all |
female |
male |
all |
female |
male |
|
| Number | 284 | 176 | 108 | 284 | 126 | 158 | 282 | 132 | 150 |
| Age, years | 64±10 | 65±11 | 62±9 | 54±15 | 56±16 | 52±14 | 50±10 | 50±10 | 49±10 |
| Age of onset ESRD | 60±10 | 62±11 | 58±10 | 48±16 | 50±17 | 47±15 | − | − | − |
| Age of onset HTN | − | − | − | 33±12 | 31±12 | 35±11 | − | − | − |
| Age of onset T2D | 43±11 | 43±11 | 43±11 | − | − | − | − | − | − |
| BMI | 29.3±7.3 | 29.9±7.8 | 28.5±6.3 | 26.5±7.3 | 27.5±8.4 | 25.8±6.2 | 29.3±6.8 | 31.2±7.6 | 27.6±5.5 |
All data are means ± SD.
T2D = Type 2 diabetes; ESRD = end-stage renal disease; HTN = hypertension; ND = nondiabetic controls.
The second cohort included 850 African Americans (434 F/ 416 M) collected by the Wake Forest University School of Medicine. This cohort was divided into three groups: (1) 284 (176 F/ 108 M) with type 2 diabetes-associated ESRD, (2) 284 (158 F/ 126 M) with hypertension-associated (nondiabetic) ESRD, and (3) 282 (132 F/150 M) nondiabetic controls. In Wake Forest samples, type 2 diabetes-associated and hypertension-associated ESRD were diagnosed as previously reported [19]. Clinical parameters in each group are summarized in table 1b.
Sample collections in both GoKinD and Wake Forest studies were approved by the local ethics committees. Data and material transfer agreements were completed prior to the study, respectively.
Genotyping of SNPs
Examined SNPs (rs953239, rs7638459, rs17624218, rs7621642, rs2033912, rs3821647, rs7610200) were tag markers in the TRPC1 gene selected from the International HapMap Project database (HapMap data release 21a/phasell Jan 07) [20] and based upon information in the dbSNP database [21]. Haploview (ver 4.0) was employed to visualize linkage disequilibrium (LD) and haplotype block structures between these loci (r2 >0.8, LOD threshold 3.0). Validation of the polymorphisms in 32 subjects randomly selected from GoKinD was performed and at least 0.016 (1/64 chromosomes) of MAF in the polymorphisms was detected. Assays used for allelic discrimination in genotyping experiments were purchased from Applied Biosystems and performed with TaqMan technique (ABI 7300, Foster City, Calif., USA). For quality control, the subjects were distributed randomly across plates with equal numbers of cases and controls on each PCR plate. Negative controls (Universal-mixture blanks) were included on each plate. Genotyping experiments in approximately 20% of samples were performed in duplicate. Successful genotype calls were ∼97% and duplication accuracy ∼99%.
Gene Expression Study
Animals
A total of 29 male homozygous diabetic (db/db) mice and non-diabetic control littermates of the C57BL/KsJ strain were obtained from colonies at Karolinska University Hospital, Stockholm. They were housed 4 per cage in a 12-/12-hour light/dark cycle, water and standard diet was provided ad libitum. Before death, the mice were tested for blood glucose and body weight. The mean blood glucose levels were 21 ± 6 mmol in db/db and 8 ± 1 mmol in db/m mice. All procedures were approved by the North Stockholm's Ethical Committee for Care and Use of Laboratory Animals.
RNA Extraction and Real-Time RT-PCR
Kidney tissues from 12- and 26-week-old db/db and control mice were harvested, and quickly submerged in RNA/later solution (Ambion, Austin, Tex., USA). Tissue samples were placed at 4°C overnight to allow the solution to permeate the cells. The stabilized RNAs were then stored at −80°C until use. Tissue homogenization was prepared according to a protocol developed in our laboratory. Briefly, 30 mg of whole kidney tissue was placed into a 2-ml microcentrifuge tube containing 0.5 ml of 1-mm diameter glass beads and 0.6 ml of RLT buffer (Qiagen, Hilden, Germany). The tube was then placed in the Mini Beadbeater (BioSpec, Bartlesville, Okla., USA) and shaken twice at 5,000 rpm for 60 s. The supernatant were collected. Total RNAs were extracted by using an RNeasy Mini Kit (Qiagen, Hilden, Germany). The integrity of the RNA was assessed by electrophoresis of 10 μl of each sample through a 1.2% agarose gel. The RNA concentration was determined spectrophotometrically by measuring the A260/ A280 ratio. First-strand cDNA was synthesized from 1 μg total RNA from the kidney of each mouse employing random hexamer oligonucleotide in a final volume of 20 μl using TaqMan reverse transcription reagents (Applied Biosystems), according to the manufacturer's protocol. To perform the real time RT-PCR, a specific TaqMan assay (ID No. Mm00441975_m1) was designed (Applied Biosystems) according to mRNA sequence of the TRPC1 gene in mouse (NM_011643.2). The probe was labeled with 6′-carboxy-fluorescein (FAM) as reporter dye and TAMRA as quencher dye. Amplification was performed using the 5′-nuclease TaqMan method with a two-step PCR protocol (95°C for 10 min, followed by 36 cycles of 95°C for 15 s and 60°C for 1 min) in an ABI 7300 real-time PCR system (Applied Biosystems). Experiments were replicated on at least two occasions.
Statistical Analyses
Clinical data are expressed as mean ± SD. The proportions of genotypes or alleles between the cases and controls were compared using a χ2 test. Additive model and Armitage's trend tests were also performed. Odds ratios (OR) and 95% CI were calculated to test the relative risk for association. The sample sizes in the GoKinD and African-American populations had ∼90% power (at the 5% level) assuming MAFs of 0.15–0.20. Statistical powers were calculated using software of PowerSampleSize (PS version 2.1.31) [22]. Testing for differences in clinical parameters between cases and controls was done, and non-normally distributed traits (plasma cystatin C, serum creatinine, BMI, and cholesterol) were transformed into natural logarithms before analysis. Multivariate logistic regression analysis was also used to examine independent variables associated with diabetic nephropathy between cases and controls in the GoKinD population. The confounders including creatinine, cystatin and systolic blood pressure were applied in the model. LD values between markers were tested using Arlequin programs (ver 2.0) [23] and Haploview (ver 4.0) [24]. Analyses were performed using Statistica (ver 7.1, StatSoft, Tulsa, Okla., USA) and the BioMed Data Program (BMDP ver 1.12, Cork, Ireland). In the animal study, a non-parametric Kruskal-Wallis comparison analysis and/or test for equality of means were done using BMDP (ver 1.12). p < 0.05 was considered significant.
Results
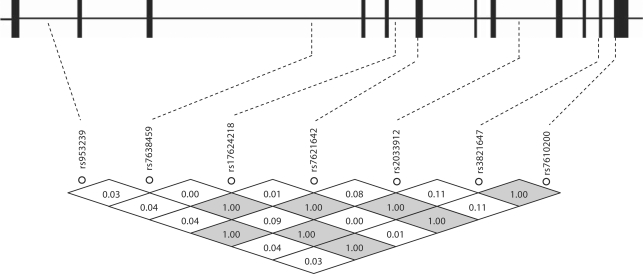
In an attempt to refine the possible TRPC1 association with diabetic nephropathy in type 1 diabetes, seven tag SNPs extending across the TRPC1 gene were genotyped in type 1 diabetes patients with and without diabetic nephropathy from the GoKinD samples. In addition to the 1,177 European American (Caucasian) type 1 diabetes patients, this cohort also included 109 non-Caucasian samples (8.5% of the total). To reduce type 1 or type 2 error resulting from population-specific variants, we analyzed the data in European Americans. All examined SNPs were satisfied in Hardy-Weinberg equilibrium assumption. Figure 1 demonstrates the location and LD matrix of examined SNPs in the TRPC1 gene. Pairwise marker correlations suggested strong LD in GoKinD European Americans, in agreement with the haplotype block structure in European Caucasians in HapMap.
Fig. 1.
Tag SNP markers examined in the TRPC1 gene. Seven examined Tag SNP markers and their linkage disequilibrium (LD) matrix along the TRPC1 gene in the GoKinD population. The coding exons in the gene are marked with the blocks. The LD values (D′) are shown inside the rhombs.
Single marker association analysis was conducted on the genotype distribution and allele frequency in the GoKinD population. Detailed information of examined SNPs and their minor allele frequencies (MAFs) in GoKinD European Americans are summarized in table 2. Genotype distribution and the association of the TRPC1 polymorphisms with diabetic nephropathy in GoKinD European American type 1 diabetes patients are shown in table 3a. The MAF of SNPs rs7621642 and rs2033912 were of borderline significant difference (p = 0.045 and 0.047, respectively) in female European American type 1 diabetes patients with versus without diabetic nephropathy, but not in males (p = 0.661 and 0.694, respectively). The MAF of SNP rs3821647, residing in a haplotype block with rs7610200, trended toward a higher frequency in female type 1 diabetes patients with diabetic nephropathy, compared to those without diabetic nephropathy (0.187 vs. 0.151, p = 0.096). To examine independent variables associated with diabetic nephropathy among female type 1 diabetes patients in the GoKinD population, further multivariate logistic regression analysis was performed. No significant association of SNPs rs7621642 (p = 0.233, OR = 1.457, CI 95% 0.785–2.705), rs2033912 (p = 0.232, OR = 1.458, CI 95% 0.785–2.707) and rs3821647 (p = 0.389, OR = 1.328, CI 95% 0.696–2.532) with diabetic nephropathy among female type 1 diabetic patients with and without diabetic nephropathy was observed.
Table 2.
TRPC1 SNPs and MAFs in GoKinD Caucasian type 1 diabetic subjects with and without diabetic nephropathy
| dbSNP rsID |
SNP type |
Position in chr. 3 |
Genomic/cDNA identity∗ |
Location |
Group |
n |
MAF |
|---|---|---|---|---|---|---|---|
| rs953239 | M= A/C | 143928895 | g.48941351A>C | intron1 | T1DwithDN | 576 | C0.419 |
| T1Dwithout DN |
598 |
C0.409 |
|||||
| rs7638459 | Y= C/T | 143972841 | g.48985297T>C | intron3 | T1DwithDN | 576 | C0.213 |
| T1Dwithout DN |
599 |
C0.191 |
|||||
| rs17624218 | Y= C/T | 143983170 | g.48995626T>C | intron5 | T1DwithDN | 577 | C0.210 |
| T1Dwithout DN |
599 |
C0.217 |
|||||
| rs7621642 | R= A/G | 143986295 | g.48998751G>A | exon6 | T1DwithDN | 576 | A0.213 |
| c.1054G>A(S306S) |
T1D without DN |
596 |
A0.191 |
||||
| rs2033912 | W= A/T | 143999057 | g.49011513A>T | intron8 | T1DwithDN | 578 | T0.213 |
| T1Dwithout DN |
599 |
T 0.191 |
|||||
| rs3821647 | R= A/G | 144006039 | g.49018495G>A | exon11 | T1DwithDN | 578 | A0.189 |
| c.2065G>A(T643T) |
T1D without DN |
599 |
A0.167 |
||||
| rs7610200 | R= A/G | 144007548 | g.49020004G>A | exon12 | T1DwithDN | 578 | A0.192 |
| c.2197G>A(R687R) | T1D without DN | 599 | A0.168 |
T1D = Type 1 diabetes; DN = diabetic nephropathy; n = number of samples genotyped; MAF = minor allele frequency.
M = Adenine or cytosine; Y = cytosine or thymine; R = adenine or guanine; W = adenine or thymine.
References: NT_005612.15, NM_003304 and NP_003295. M, Y, R and W are International Union of Pure and Applied Chemistry (IUPAC) codes used in FASTA sequences at the SNP position.
Table 3.
Genotype distributions and minor allele frequencies of TRPC1 polymorphisms: American Caucasians (a) and African Americans (b)
| a American Caucasians in GoKinD | ||||||||
|---|---|---|---|---|---|---|---|---|
| dbSNPrsID |
Group |
Gender |
Genotype |
MAF |
pvalue |
OR(95%CI) |
||
| rs7621642 | GG | GA | AA | |||||
| T1Dwith DN | female | 161 (60.8) | 93 (35.1) | 11(4.1) | A 0.217 | 0.045∗ | 1.336(1.005–1.774) | |
| male |
195 (62.7) |
102 (32.8) |
14 (4.5) |
A 0.209 |
0.661∗ |
|||
| T1D without DN | female | 241 (67.9) | 106 (29.9) | 8 (2.2) | A 0.172 | |||
| male |
147 (61.0) |
82 (34.0) |
12 (5.0) |
A 0.220 |
||||
| rs2033912 | AA | AT | TT | |||||
| T1Dwith DN | female | 161 (60.8) | 93 (35.1) | 11(4.1) | T 0.217 | 0.047∗ | 1.331(1.003–1.768) | |
| male |
196 (62.6) |
103 (32.9) |
14 (4.5) |
T 0.209 |
0.694∗ |
|||
| T1D without DN | female | 242 (67.8) | 107 (30.0) | 8 (2.2) | T 0.173 | |||
| male |
148 (61.1) |
82 (33.9) |
12 (5.0) |
T 0.219 |
||||
| rs3821647 | GG | GA | AA | |||||
| T1D with DN | female | 172 (64.9) | 87 (32.8) | 6 (2.3) | A 0.187 | 0.096 | ||
| male |
200 (63.9) |
106 (33.9) |
7 (2.2) |
A 0.192 |
0.946 |
|||
| T1D without DN | female | 256 (71.7) | 94 (26.3) | 7 (2.0) | A 0.151 | |||
| male | 157 (64.9) | 78 (32.2) | 7 (2.9) | A 0.190 | ||||
| b African Americans from Wake Forest | ||||||||
| rs7621642 | GG | GA | AA | |||||
| T2D with ESRD | female | 16 (9.2) | 74 (42.5) | 84 (48.3) | G 0.305 | 0.847∗∗ | ||
| male |
13 (12.6) |
40 (38.8) |
55 (50.9) |
G 0.306 |
0.382∗∗ |
|||
| HTN with ESRD | female | 18 (14.4) | 42 (33.6) | 65 (52.0) | G 0.312 | 0.204∗∗∗ | ||
| male |
11(7.0) |
63 (40.1) |
83 (52.9) |
G 0.271 |
0.392∗∗∗ |
|||
| ND | female | 9 (6.8) | 51 (38.6) | 72 (54.6) | G 0.261 | |||
| male |
15 (10.0) |
60 (40.3) |
74 (49.7) |
G 0.302 |
||||
| rs2033912 | AA | AT | TT | |||||
| T2D with ESRD | female | 14 (8.1) | 71 (41.0) | 88 (50.9) | A 0.286 | 0.416∗∗ | ||
| male |
12 (11.2) |
36 (33.7) |
59 (55.1) |
A 0.280 |
0.365∗∗ |
|||
| HTNwith ESRD | female | 11(8.8) | 42 (33.6) | 72 (57.6) | A 0.256 | 0.722∗∗∗ | ||
| male |
11(6.9) |
56 (35.2) |
92 (57.9) |
A 0.245 |
0.261∗∗∗ |
|||
| ND | female | 9 (6.8) | 46 (34.9) | 77 (58.3) | A 0.242 | |||
| male |
14 (9.4) |
57 (38.3) |
78 (52.3) |
A 0.285 |
||||
| rs3821647 | AA | AG | GG | |||||
| T2Dwith ESRD | female | 11(6.3) | 50 (28.8) | 113 (64.9) | A 0.207 | 0.142∗∗ | ||
| male |
9 (8.3) |
38 (35.2) |
61 (56.5) |
A 0.259 |
0.070∗∗ |
|||
| HTN with ESRD | female | 10 (7.9) | 45 (35.7) | 71 (56.4) | A 0.258 | 0.612∗∗∗ | ||
| male |
7 (4.4) |
47 (29.8) |
104 (65.8) |
A 0.193 |
0.033∗∗∗ |
0.663(0.454–0.969) |
||
| ND | female | 6 (4.6) | 51 (38.6) | 75 (56.8) | A 0.239 | |||
| male | 7 (4.7) | 65 (43.6) | 77 (51.7) | A 0.265 | ||||
T1D = Type 1 diabetes; DN = diabetic nephropathy; T2D = type 2 diabetes; ESRD = end-stage renal disease; HTN = hypertension; ND = non-diabetic subjects; MAF = minor allele frequency. Comparison analyses of ∗ T1D with DN vs. T1D without DN, ∗∗ T2D with ESRD vs. ND, and ∗∗∗ T2D with DN vs. HTN with ESRD. Figures in parentheses are percentages.
Tests in the entire GoKinD population were then performed. When African Americans from the GoKinD population were combined with European Americans, the difference in MAFs of these three polymorphisms were statistically significant in type 1 diabetes patients with versus without diabetic nephropathy (p = 0.003, 0.003 and 0.004, respectively; Pc = 0.042, 0.042 and 0.056, respectively, after Bonferroni correction). In order to understand whether TRPC1 polymorphisms are the population-specific variants or confer the susceptibility to diabetic nephropathy in African Americans, we further genotyped the three SNPs, rs7621642, rs2033912 and rs3821647, in a second cohort of African Americans, including 284 with type 2 diabetes-associated ESRD, 284 with hypertension-associated ESRD and 282 nondiabetic subjects [19]. The genotype distribution and MAF of these polymorphisms in the population of Wake Forest African Americans are listed in table 3b. The genotype distributions of these SNPs were reversed in African Americans, compared to what had been detected in European Americans (table 3a); major alleles of the SNPs in European Americans were minor alleles in African Americans. No significant associations for these three polymorphisms were detected with type 2 diabetes-ESRD or hypertensive-ESRD in African Americans.
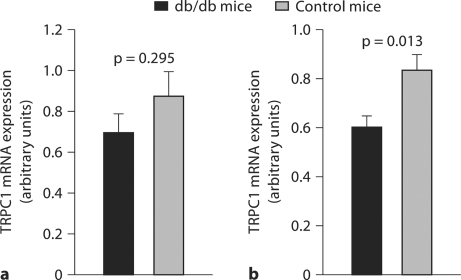
We further investigated TRPC1 gene expression at mRNA levels with a mouse model of diabetic nephropathy to determine whether reduced TRPC1 gene expression was a primary or subsequent defect in the development of diabetic nephropathy. TRPC1 gene expression in kidney tissue from db/db mice at 12 and 26 weeks of age, respectively, was examined using a TaqMan real time RT-PCR protocol. Data indicated that TRPC1 mRNA expression levels kidney tissue from db/db mice at the age of 12 weeks compared to the controls were tendency to reduce (p = 0.295, fig. 2a), while the expression levels of this gene in 26-week-old db/db mice were significantly decreased (p = 0.013, fig. 2b).
Fig. 2.
TRPC1 gene mRNA expression in kidney tissues of db/db mice. TRPC1 gene mRNA expression was examined using real-time RT-PCR in kidney tissue from db/db mice at 12 and 26 weeks of age, respectively. TRPC1 mRNA expression levels between db/db and db/m (control) mice at the age of 12 weeks were not significantly different (mean values 0.874 ± 0.117 SE vs. 0.697 ± 0.061, p = 0.295, a). The expression levels of TRPC1 in kidney tissue from 26-week-old db/db mice, however, were significantly decreased compared with the db/m controls (0.836 ± 0.062 vs. 0.601 ± 0.047, p = 0.013, b).
Discussion
In the present study, we have conducted a genetic association study to detect TRPC1 genetic susceptibility for diabetic nephropathy in the GoKinD type 1 diabetes subjects. Among this population, a large proportion of subjects are American Caucasian while the rest are African American, Asian, Hispanic and Indian. Evidence has demonstrated that race and ethnicity are related to the progression of kidney dysfunction and clinical outcomes in the patients with diabetic nephropathy. In the United States, the incidence of diabetic nephropathy is higher for African Americans, Native Americans and Asians than for American Caucasians [25, 26]. We thus performed the analyses for association of TRPC1 genetic polymorphisms and diabetic nephropathy, separately, in American Caucasians and other American ethnic groups. Although single marker association analysis has showed a borderline association between the TRPC1 genetic polymorphisms and diabetic nephropathy in female type 1 diabetes patients of American Caucasians, multivariate logistic regression analysis negates the evidence for the association.
We have tested whether TRPC1 genetic polymorphisms are associated with diabetic nephropathy among African-American diabetic patients or are the ethical specific variants with the second cohort of African Americans. Data indicate that genotype distribution of TRPC1 polymorphisms in African Americans with type 2 diabetes-ESRD or hypertension and nondiabetic subjects are reversed in comparison with the GoKinD American Caucasians. No significant association of TRPC1 polymorphisms with ESRD in those type 2 diabetes patients is found. The first cohort of GoKinD population is a collection of type 1 diabetes patients with and without diabetic nephropathy, and 70.6% of the patients with diabetic nephropathy have ESRD. In the second cohort of African Americans, the type 2 diabetes patients are associated with EASD. We did not combine the data from American Caucasians’ type 1 diabetes with ESRD and African Americans’ type 2 diabetes with ESRD together for analysis, because the genotype distributions of TRPC1 polymorphisms between these two cohorts are reversely different. The combined analysis in these two populations may simply cause type 1 or type 2 error. Since there are only 42 African-American type 1 diabetes patients with and without diabetic nephropathy enrolled in the present study, we are not able to exclude the possibility of association between TRPC1 genetic polymorphisms and diabetic nephropathy in African-American type 1 diabetes patients.
Based upon the data from our genetic association study and the recent report [17], we hypothesized that the reduction of TRPC1 gene expression may be the subsequent defect in the development of diabetic nephropathy. To test this hypothesis, we have examined TRPC1 gene expression in kidney tissue from db/db mice at 12 and 26 weeks of age. This animal model has been used for study of the progression of diabetic nephropathy [27]. We found that TRPC1 mRNA expression levels are trendily decreased in 12-week-old and significantly in 26-week-old db/db mice compared to the controls. The results are consistent with the recent report [17] and provide further evidence that TRPC1 expression was not reduced until the later stage of diabetic nephropathy. Moreover, the reduction of TRPC1 expression has been indicated as a result of HNF4α dysfunction [17]. To detect the susceptibility of the HNF4α in diabetic nephropathy, a genetic association study of this gene has been taken into our consideration.
In conclusion, the present study provides the first evidence that TRPC1 genetic polymorphisms are lack of association with diabetic nephropathy in either European American subjects with type 1 diabetes from GoKinD or African Americans with type 2 diabetes-associated ESRD. Genetic variation in this gene may not fundamentally contribute to the development of diabetic nephropathy. However, a functional analysis with db/db mice, in consistence with the recent report using STZ-treated rats, indicates that reduction of TRPC1 gene expression is the subsequent effect in the late stage of diabetic nephropathy.
Acknowledgements
The authors thank all GoKinD and Wake Forest University study subjects for their participation. GoKinD study sample collection was supported by the Juvenile Diabetes Research Foundation in collaboration with the Joslin Diabetes Center and George Washington University, and by the United States Centers for Diabetes Control and Prevention. Wake Forest University samples were collected by Ms. Joyce Byers and Mitzie Spainhour, supported by NIH grants RO1 DK 070942 (B.I.F.) and RO1 DK53591 (D.W.B.). This work was supported by Novo Nordic Consortium (S.E.), Swedish Research Council 04224 (K.B.), Family Erling-Persson Foundation (K.B.), Loo and Hans Osterman Foundation (H.F.G.) and the Swedish Diabetes Association (H.F.G.). Dongying Zhang is originally from Chongqing Medical University, China.
References
- 1.US Renal Data System, USRDS 2007 Annual Data Report: Atlas of Chronic Kidney Disease and End-Stage Renal Disease in the United States. Bethesda, National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases, 2007.
- 2.Hasslacher C, Ritz E, Wahl P, Michael C. Similar risks of nephropathy in patients with type I or type II diabetes mellitus. Nephrol Dial Transplant. 1989;4:859–863. doi: 10.1093/ndt/4.10.859. [DOI] [PubMed] [Google Scholar]
- 3.Yamagishi S, Fukami K, Ueda S, Okuda S. Molecular mechanisms of diabetic nephropathy and its therapeutic intervention. Curr Drug Targets. 2007;8:952–959. doi: 10.2174/138945007781386884. [DOI] [PubMed] [Google Scholar]
- 4.Freedman BI, Bostrom M, Daeihagh P, Bowden DW. Genetic factors in diabetic nephropathy. Clin J Am Soc Nephrol. 2007;2:1306–1316. doi: 10.2215/CJN.02560607. [DOI] [PubMed] [Google Scholar]
- 5.Moczulski DK, Rogus JJ, Antonellis A, Warram JH, Krolewski AS. Major susceptibility locus for nephropathy in type 1 diabetes on chromosome 3q: results of novel discordant sib-pair analysis. Diabetes. 1998;47:1164–1169. doi: 10.2337/diabetes.47.7.1164. [DOI] [PubMed] [Google Scholar]
- 6.Chistiakov DA, Savost'anov KV, Shestakova MV, Chugunova LA, Samkhalova MSh, Dedov II, Nosikov VV. Confirmation of a susceptibility locus for diabetic nephropathy on chromosome 3q23-q24 by association study in Russian type 1 diabetic patients. Diabetes Res Clin Pract. 2004;66:79–86. doi: 10.1016/j.diabres.2004.02.011. [DOI] [PubMed] [Google Scholar]
- 7.Bowden DW, Colicigno CJ, Langefeld CD, Sale MM, Williams A, Anderson PJ, Rich SS, Freedman BI. A genome scan for diabetic nephropathy in African Americans. Kidney Int. 2004;66:1517–1526. doi: 10.1111/j.1523-1755.2004.00915.x. [DOI] [PubMed] [Google Scholar]
- 8.Freedman BI, Bowden DW, Sale MM, Langefeld CD, Rich SS. Genetic susceptibility contributes to renal and cardiovascular complications of type 2 diabetes mellitus. Hypertension. 2006;48:8–13. doi: 10.1161/01.HYP.0000227047.26988.3e. [DOI] [PubMed] [Google Scholar]
- 9.Osterholm AM, He B, Pitkaniemi J, Albinsson L, Berg T, Sarti C, Tuomilehto J, Tryggvason K. Genome-wide scan for type 1 diabetic nephropathy in the Finnish population reveals suggestive linkage to a single locus on chromosome 3q. Kidney Int. 2007;71:140–145. doi: 10.1038/sj.ki.5001933. [DOI] [PubMed] [Google Scholar]
- 10.McKnight AJ, Maxwell AP, Sawcer S, Compston A, Setakis E, Patterson CC, Brady HR, Savage DA. A genome-wide diabetic nephropathy: a microsatellite association screen to identify chromosomal regions harboring candidate genes in diabetic nephropathy. J Am Soc Nephrol. 2006;17:831–836. doi: 10.1681/ASN.2005050493. [DOI] [PubMed] [Google Scholar]
- 11.Kanwar YS, Wada J, Sun L, Xie P, Wallner EI, Chen S, Chugh S, Danesh FR. Diabetic nephropathy: mechanisms of renal disease progression. Exp Biol Med (Maywood) 2008;233:4–11. doi: 10.3181/0705-MR-134. [DOI] [PubMed] [Google Scholar]
- 12.White KE, Marshall SM, Bilous RW. Are glomerular volume differences between type 1 and type 2 diabetic patients pathologically significant? Diabetologia. 2007;50:906–912. doi: 10.1007/s00125-007-0624-1. [DOI] [PubMed] [Google Scholar]
- 13.Wes PD, Chevesich J, Jeromin A, Rosenberg C, Stetten G, Montell C. TRPC1, a human homolog of a Drosophila store-operated channel. Proc Natl Acad Sci USA. 1995;92:9652–9656. doi: 10.1073/pnas.92.21.9652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu SZ, Beech DJ. TrpC1 is a membrane-spanning subunit of store-operated Ca(2+) channels in native vascular smooth muscle cells. Circ Res. 2001;88:84–87. doi: 10.1161/01.res.88.1.84. [DOI] [PubMed] [Google Scholar]
- 15.Goel M, Sinkins WG, Zuo CD, Estacion M, Schilling WP. Identification and localization of TRPC channels in the rat kidney. Am J Physiol Renal Physiol. 2006;290:F1241–F1252. doi: 10.1152/ajprenal.00376.2005. [DOI] [PubMed] [Google Scholar]
- 16.Sours S, Du J, Chu S, Ding M, Zhou XJ, Ma R. Expression of canonical transient receptor potential (TRPC) proteins in human glomerular mesangial cells. Am J Physiol Renal Physiol. 2006;290:F1507–F1515. doi: 10.1152/ajprenal.00268.2005. [DOI] [PubMed] [Google Scholar]
- 17.Niehof M, Borlak J. HNF4 alpha and the Ca-channel TRPC1 are novel disease candidate genes in diabetic nephropathy. Diabetes. 2008;57:1069–1077. doi: 10.2337/db07-1065. [DOI] [PubMed] [Google Scholar]
- 18.Mueller PW, Rogus JJ, Cleary PA, Zhao Y, Smiles AM, Steffes MW, Bucksa J, Gibson TB, Cordovado SK, Krolewski AS, Nierras CR, Warram JH. Genetics of Kidneys in Diabetes (GoKinD) study: a genetics collection available for identifying genetic susceptibility factors for diabetic nephropathy in type 1 diabetes. J Am Soc Nephrol. 2006;17:1782–1790. doi: 10.1681/ASN.2005080822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Freedman BI, Yu H, Spray BJ, Rich SS, Rothschild CB, Bowden DW. Genetic linkage analysis of growth factor loci and end-stage renal disease in African Americans. Kidney Int. 1997;51:819–825. doi: 10.1038/ki.1997.115. [DOI] [PubMed] [Google Scholar]
- 20.http://www.hapmap.org/cgi-perl/gbrowse/hapmap_B35/
- 21.http://www.ncbi.nlm.nih.gov/projects/SNP/
- 22.http://biostat.mc.vanderbilt.edu/twiki/bin/view/Main/PowerSampleSize
- 23.http://lgb.unige.ch/arlequin/
- 24.http://www.ncbi.nlm.nih.gov/projects/SNP/
- 25.Burrows NR, Li Y, Williams DE. Racial and ethnic differences in trends of end-stage renal disease: United States, 1995 to 2005. Adv Chron Kid Dis. 2008;15:147–152. doi: 10.1053/j.ackd.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 26.Lopes AA. Relationships of race and ethnicity to progression of kidney dysfunction and clinical outcomes in patients with chronic kidney failure. Adv Ren Replace Ther. 2004;11:14–23. doi: 10.1053/j.arrt.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 27.Sharma K, McCue P, Dunn SR. Diabetic kidney disease in the db/db mouse. Am J Physiol Renal Physiol. 2003;284:F1138–F1144. doi: 10.1152/ajprenal.00315.2002. [DOI] [PubMed] [Google Scholar]