Abstract
Background
Previous studies have compared sperm phenotypes between men with partial [1] deletions within the AZFc region of the Y chromosome with non-carriers, with variable results. Here, we have investigated a separate question, the basis of the variation in sperm phenotype within gr/gr deletion carriers, which ranges from normozoospermia to azoospermia. Differences in the genes removed by independent gr/gr deletions, the occurrence of subsequent duplications or the presence of linked modifying variants elsewhere on the chromosome have been suggested as possible causal factors. We set out to test these possibilities in a large sample of gr/gr deletion carriers with known phenotypes spanning the complete range.
Results
We assembled a collection of 169 men diagnosed with gr/gr deletions from six centres in Europe and one in Australia, and characterized the DAZ and CDY1 copies retained, the presence or absence of duplications and the Y-chromosomal haplogroup. Although our study had good power to detect factors that accounted for ≥5.5% of the variation in sperm concentration, no such factor was detected. A negative effect of gr/gr deletions followed by b2/b4 duplication was observed within the normospermic group, which remains to be further explored in a larger study population. Finally, we observed significant geographical differences in the frequency of different subtypes of gr/gr deletions which may have relevance for the interpretation of case control studies dealing with admixed populations.
Conclusions
We conclude that the phenotypic variation of gr/gr carriers in men of European origin is largely independent of the Y-chromosomal background.
Keywords: gr/gr deletions, male infertility, spermatogenesis, genetics, polymorphisms, AZFc
INTRODUCTION
The long arm of the human Y chromosome hosts a number of genes involved in spermatogenesis, and several types of recurrent Yq deletions are firmly associated with spermatogenic failure [2, 3, 4] (Figure 1). Apart from the classical AZF deletions, new types of Yq rearrangements have recently attracted the attention of geneticists and andrologists [5]. The complex duplicated structure of the AZFc region predisposes to a series of rearrangements mediated by non-allelic homologous recombination, including the formation of partial deletions, deletion/duplications and partial duplications [6, 7, 8]. Among them, the most relevant clinically is termed the “gr/gr” deletion [9] and removes half of the AZFc gene content. This deletion is a significant risk factor for spermatogenic failure in some populations but apparently not in others [10]. Contradictory results are likely to derive from methodological differences between studies together with recruitment biases. Concerning the latter only in a minority of studies are the controls normozoospermic men; controls and patients are not matched for ethnic background in highly admixed populations from Paris [11], Southern France [12], Brazil [13] used in some studies, and the proportion of azoospermic/oligozoospermic subjects differs between studies.
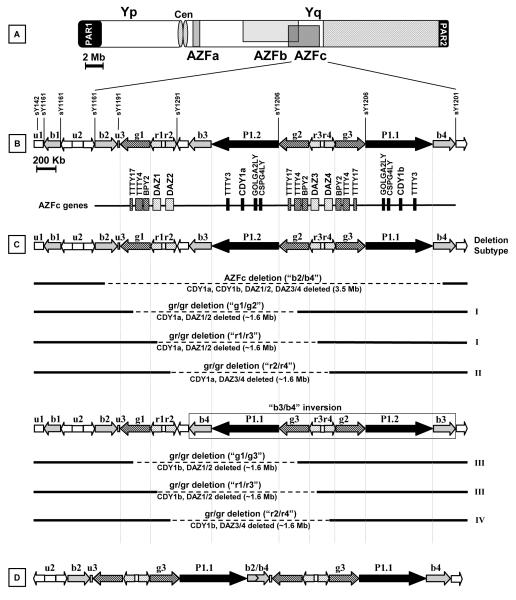
Figure 1.
Schematic representation of the Y chromosome showing the structures, genes, STSs and rearrangements relevant to this study. A) Deletions of the AZFa, AZFb and AZFc regions result in spermatogenic failure. AZFb deletions (two subtypes) overlap with the AZFc region. Yp: short arm of the Y chromosome, cen: centromere, Yq: long arm of the Y chromosome, PAR: pseudoautosomal region. B) The AZFc region is presented in more detail showing the location of multicopy genes and transcription units in the reference sequence published by Skaletsky et al. 2003. This region contains a number of repeated sequences with the same orientation (matching arrows) which through intrachromosomal recombination may lead to deletions. C) The clinically relevant “b2/b4” (top) and “gr/gr” deletions (remainder) are shown. gr/gr deletions remove half of the AZFc gene content, but can vary in breakpoints (deletion subtypes I and II ), or according to the presence of a b3/b4 inversion (deletion subtypes III and IV). D) gr/gr deletion of subtype I or II followed by b2/b4 duplication.
While the debate about the significance of the gr/gr deletion as a risk factor for impaired sperm production is ongoing and unresolved, and is not further investigated here, there is a firm consensus about the heterogeneous phenotype associated with gr/gr deletions. Even in studies in which gr/gr deletion is clearly shown as a risk factor for impaired spermatogenesis, the sperm phenotype ranges from azoospermia to normozoospermia [10]. This phenomenon is in sharp contrast to the classical AZF deletion phenotypes, which are invariably associated with impaired sperm production [14, 15, 16, 17]. Given that the number of genes removed by the gr/gr deletion is half that of the classical AZFc deletion, it is not unexpected that the effect of the deletion is milder and we are dealing with a co-factor for spermatogenic impairment with variable penetrance. Thus it is also possible that other Y-linked or non-Y genetic factors influence the pathogenicity of the deletion. The significance of the presence of polymorphisms or mutations in the autosomal homologue of the DAZ gene, DAZL [18] has been examined in one study [15], but it was concluded that the polymorphic Thr12Ala change (T12A) was unlikely to contribute and no new mutations in the entire coding region of the DAZL gene were identified. No other autosomal factors modulating the deletion phenotype have yet been reported.
For Y-related factors, interpretation is complicated by the lack of recombination in the male-specific part of the chromosome, which results in complete linkage between variants throughout most of the chromosome. But their likely importance is illustrated by certain Y haplogroups (Y chromosome backgrounds defined by Y-SNPs; hgs) which carry fixed gr/gr deletions but are nevertheless present at high frequency in some populations, indicating that compensatory mechanisms may exist on these backgrounds [19, 20, 21]. The specific genes removed can vary between gr/gr deletions, and this has been suggested as the most direct modulating factor. In this regard, the DAZ and CDY1 copies, which lie within the deleted region, have been examined as possible predictors for pathogenicity. The loss of DAZ1/DAZ2 and CDY1a has been reported as more deleterious than the removal of DAZ3/DAZ4 and CDY1b [15, 19, 22]. Concerning the DAZ gene copies, different members of the DAZ gene family have different number of RNA recognition motifs (RRMs) and DAZ repeats, which may confer different functional activity on the four DAZ copies. However, it is also known that both elements show inter-individual variability which in theory might be linked to a particular Y lineage. Similarly, the loss of CDY1a or CDY1b copies might reflect a different Y structure. In fact, in the reference sequence, CDY1b lies outside the region that is predicted to be deleted by gr/gr recombination. This implies that the CDY1b loss may occur only in other Y chromosomes that carry an inversion polymorphism in the AZFc region [12], or must arise independently. Such an inversion polymorphism may affect the transcription of the remaining AZFc genes or may be linked to other Y structural variations. An alternative explanation for the gr/gr deletions observed in normospermic men could be that a compensatory duplication, such as a gr/gr deletion followed by b2/b4 duplication, has restored normal gene number and function. Furthermore, Y variants outside the AZFc region may influence spermatogenesis, and these will be associated with particular hgs, so may be recognized indirectly using a phylogenetic analysis. A well-resolved Y chromosome tree of 311 hgs is now available [23], as well as extensive information about the hgs present in different regions of the world, including Europe [24, 25, 26]. Consequently, a small number of informative Y-SNPs can be tested to identify the most prevalent hgs in Europe.
The majority of studies of gr/gr deletions have used STS plus/minus PCR analysis to identify the deletion and thus have been unable to provide information about the type of missing gene copies or about deletion/duplication events. For a fuller understanding, a methodology such as a combined analysis based on a first step of STS plus/minus screening followed by a confirmatory dosage analysis and gene copy characterization of the samples with suspected deletions is needed, and has been described by Giachini et al. [15].
The aim of the present study was to determine whether any of the postulated Y-related factors can account for the different semen phenotypes observed in gr/gr deletion carriers, rather than to re-investigate possible differences between carriers and non-carriers. For this purpose, we collected a total of 169 DNA samples from seven countries that were previously defined by STS plus/minus analysis as gr/gr deleted men. All subjects were further characterized by gene dosage analysis, gene copy type and Y hg, and these factors were related to spermatogenic phenotype in order to search for a genetic profile specific for spermatogenic failure.
MATERIALS AND METHODS
Subjects analyzed
A total of 169 DNA samples were analyzed for AZF deletions (classical) and for partial AZFc region deletions in the participating laboratories from the following countries: Australia (n=50), Denmark (n=25), France (n=13), Germany (n=35), Hungary (n=4), Italy (n=24) and Spain (n=18). All 169 samples showed a lack of amplification for sY1291 but the presence of the other specific AZFc markers sY142, sY1258, sY1161, sY1197, sY1191, sY1206, and sY1201 (see GenBank for PCR primers) indicating the presence of a gr/gr deletion. Some of the patients/controls with gr/gr deletions (based on STS plus/minus analysis) have been included in previous publications [14, 15, 16, 17].
The composition of the study population (n=169) was as follows: 152 infertile and 17 fertile or normospermic men sent as “controls” (Supplementary Table 1). All subjects were of European ancestry either based on surname or on direct questioning during recruitment about the origin of the father and mother. Of the 152 infertile patients where semen parameters were available for 134 patients. In an additional 7 patients, only the semen phenotype (azoospermia, oligozoospermia) was given by the laboratories, whereas for the rest of patients (n=11) the only information given was that they were infertile. In the 141 patients with known semen phenotype there were 27 azoospermic, 18 cryptozoospermic (<1 million spzoa/ml), 82 oligozoospermic and 14 asthenoteratozoospermic subjects. The mean values of the three principal sperm parameters in the infertile group in which semen parameters were provided were as follows: sperm concentration: 6.9 ± 14.7 × 106 /ml (n=134); total sperm count: 24.8 ± 60.6 × 106 (n=114); % progressive motility: 16.2 ± 16.8 (n=122); % normal morphology: 8.9 ± 8.1 (n=82). Motility and/or morphology were not determined in azoospermic and severely oligozoospermic men (< 1 × 106 spermatozoa/ml). The mean sperm concentration are reported for each country in supplementary table 2.
Semen parameters were available for all 17 controls (except two cases in which morphology was not provided (D255; B11) and one case in which motility was not provided (D255). The control population consisted of: i) 10 subjects presenting all the three major sperm parameters (concentration, motility and morphology) above the normal range [27]; ii) 7 subjects presenting normal sperm concentration but with motility and morphology below the normal range. The mean values of the sperm parameters in the control group were: sperm concentration: 76.8 ± 44.9 × 106 /ml (n=17); total sperm count: 328.4 ± 396.7 × 106 (n=13); % progressive motility: 49.2 ± 13.3 (n=16); % normal morphology: 20.8 ± 11.8 (n=15).
The 169 samples were analysed further at the Andrology Unit (Department of Clinical Physiopathology, University of Florence) where a fine-scale molecular characterization of the deletions using AZFc gene dosage and gene copy analysis was carried out as described by Giachini et al. (2007). Y chromosome hg analysis was performed using the multiplexed primers previously described [5] adapted for SNaPshot single base extension (Applied Biosystems) at the Wellcome Trust Sanger Institute (Cambridge, UK) for a total of 153 subjects. The markers RPS4Y711, M145, M96, M89, M9 and M45 were typed on all samples, and M123, M78, V6, M35 and M81 (hg E – derived for M96), M201, M170, M52 and 12f2 (hg F – derived for M89), M106, M61, M147, M214, M27, M76 and M70 (hg K – derived for M9), or M17, M343, M369 and M18 (hg P – derived for M45) on appropriate subsamples, but data were combined into the major hgs E, F*(xK), K*(xP) and P to provide numbers suitable for most statistical analyses. Full data are available for 150 samples (Supplementary Table 1).
Gene dosage
Quantitative analysis of CDY1 and DAZ, in order to measure the copy number of these genes, was performed using a modified PCR-based method [12]: we simultaneously amplified the AZFc locus to be quantified (CDY1 or DAZ) and a homologous locus outside the AZFc interval (CDY2 and DAZL, respectively) – as an internal standard with a known number of copies – using a single primer pair in a PCR reaction with a maximum of 24 cycles (end point of the exponential phase). The primers flank an insertion/deletion difference of 3-5 bp, which allowed the products amplified from the AZFc loci and the control loci to be separated by polyacrylamide gel electrophoresis. One of the primers was labelled at its 5′ end with a fluorochrome (FAM). Then, the reaction was mixed with formamide, denatured at 95°C for 5 min and the different sized loci separated on an automatic sequencer (ABI PRISM 310 Genetic Analyzer PE). Quantification was performed by comparing the peak area of the AZFc locus with its homolog.
- CDY1 vs CDY2 (primers: oMY953a/o1023): there are two identical copies each of CDY1 and CDY2, which themselves share 98% nucleotide identity. We amplified CDY1 and CDY2 across a 3 bp indel difference in the coding region, to give fragments of 134 bp for CDY1 and 137 bp for CDY2.
- DAZ vs DAZL (primers: o1130/01313): we co-amplified a fragment of intron 10 from DAZ (214 bp) and DAZL (217 bp). This intron is present in one copy per DAZ gene (in the reference 46,XY man there are four copies of DAZ and two copies of DAZL). Some samples show a 40 bp insertion polymorphism in the DAZL intron 10, resulting in an extra band at 260 bp, which could be heterozygous or, more rarely, homozygous.
Gene copy type
Qualitative analysis for CDY1 and DAZ, in order to determine which copies of these genes have been removed by the gr/gr deletion, was performed according to Machev et al. [12]. For DAZ, we chose the sequence family variant (SFV) at STS sY587 in intron 10, which discriminates DAZ1/2 from DAZ3/4. For CDY1, we used a C/A SFV situated 7750 bp 5′ of the CDY1 translation start codon (CDY7750), which distinguishes CDY1a from CDY1b. SFVs were scored by PCR followed by restriction enzyme digestion (5 U for at least 4 hours): DAZ sY587, DraI (DAZ1/2 cut); CDY1-7750, PvuII (CDY1b cut). Digestion products were then analyzed by electrophoresis at 100 V on 4% agarose gels containing ethidium bromide and visualized under ultraviolet light. Primer pairs: sY587, o912/o913; CDY1-7750, o1025/o1026 [12].
Statistical analyses
Median values between groups were compared using a non-parametric Mann-Whitney U test, or Student’s T-test after normalization of the distribution by a log transformation, as shown by a one-sample Kolmogorov-Smirnov test; p = 0.023. We also performed a regression analysis on the log-transformed sperm counts to determine whether variation in sperm count (the dependent variable) could be explained by any of the other variables. Analyses were carried out using SPSS 14.0, except for power calculations which were performed on the website http://www.danielsoper.com/statcalc/calc09.aspx.
RESULTS
Molecular analysis of the AZFc region:
Definition of different gr/gr rearrangements
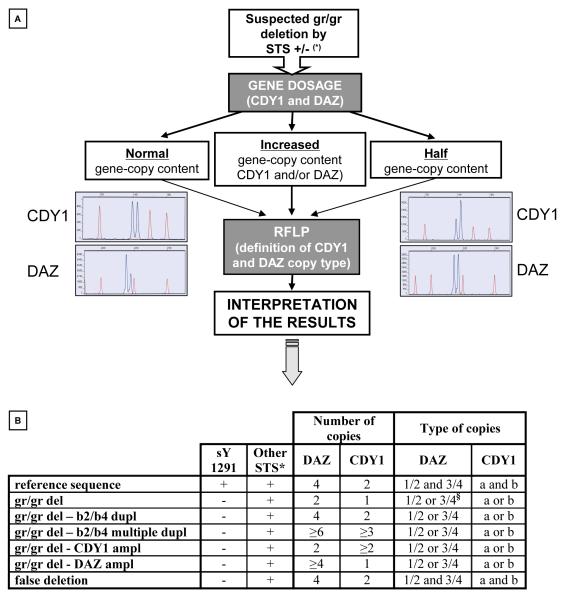
The molecular characterization of samples sharing the absence of the sY1291 was necessary in order to distinguish between different gr/gr rearrangements which may not be detected by the STS plus/minus analysis used by the participating laboratories and to confirm the effective loss of genetic material, i.e. exclude “false” deletions (Figure 2).
Figure 2.
Schematic representation of the multi-step gr/gr deletion screening procedure and categories of gr/gr subtypes defined. A) Gene dosage and RFLP analysis was used to i) distinguish between “simple” gr/gr deletions, false deletions and deletion/duplications events and ii) to define the missing gene copies, i.e. gr/gr deletion subtypes (examples of electropherograms shown; see Materials and Methods for further details). (*) STS: Sequence Tagged Site-based PCR method, according to Repping et al. [9]. B) Molecular characterization of the different gr/gr rearrangement types. Gene dosage of CDY1 and DAZ and the possible combinations on the basis of missing DAZ (DAZ1/DAZ2/DAZ3/DAZ4) and CDY1 (CDY1a/CDY1b) copy types are shown for each rearrangement. “+” and “-” represent positive and negative results, respectively. * in the AZFc region: sY142, sY1258, sY1161, sY1197, sY1191, sY1206, sY1201. § 6 cases showed a combination of DAZ copies from each DAZ duplex, possibly DAZ1/3 or DAZ3/4. Abreviations: del=deletion; dupl=duplication; ampl=amplification.
We defined the gene dosage of CDY1 and DAZ and the type of missing DAZ (DAZ1/DAZ2/DAZ3/DAZ4) and CDY1 (CDY1a/CDY1b) copies. The gene dosage and gene copy types are reported in Figure 2 for each gr/gr rearrangement which could be identified by this combined, two-step method.
In 9 subjects (1 normospermic for all three parameters and 8 infertile with reduced sperm count) we observed a pattern compatible with “false” deletions, showing normal DAZ and CDY1 gene dosage and the presence of both CDY1 copies (CDY1a/CDY1b) and all four DAZ copies (DAZ1/DAZ2/DAZ3/DAZ4). False deletions using STS plus/minus analysis can be due either to polymorphisms/rearrangements at the annealing site of the primers or to PCR artefacts. The 9 samples were reanalyzed in simplex PCR using sY1291 at different annealing temperatures which confirmed they were “not deleted” in all cases, indicating that the failure of amplification was due to non-optimized PCR conditions for those specific samples (DNA quality may be responsible for such an amplification failure) and the false deletion rate was 5.3% (9/169). The 9 samples originated from 4 different laboratories and were excluded from further analyses.
In the remaining 160 subjects we found a heterogeneous situation showing both “true” simple gr/gr deletions (n=128), gr/gr deletion followed by b2/b4 duplication (n=23), gr/gr deletion followed by multiple b2/b4 duplications (n=3), four cases with confirmed gr/gr deletion but a higher copy number of CDY genes than the reference sequence (gr/gr deletion – CDY1 amplification) and two cases with confirmed gr/gr deletion but a higher copy number of DAZ genes than the reference sequence (gr/gr deletion - DAZ amplification) (Figure 2, Table 1).
Table 1.
Frequency of each gr/gr rearrangement in the whole study population.
| NORMAL SPERM COUNT (n) |
ABNORMAL SPERM COUNT (n) |
|||||||
|---|---|---|---|---|---|---|---|---|
| Rearrangement type |
TOT (n) |
TOT (n) with semen phenotypes |
Normo | AT | TOT 1 |
AZ | OZ | TOT2 |
| gr/gr del | 128 | 122 | 9 | 16 | 25 | 18 | 79 | 97 |
| gr/gr del – b2/b4 dupl | 23 | 22 | - | 4 | 4 | 7 | 11 | 18 |
| gr/gr del – b2/b4 multiple dupl | 3 | 3 | - | 1 | 1 | - | 2 | 2 |
| gr/gr del - CDY1 ampl | 4 | 1 | - | - | - | - | 1 | 1 |
| gr/gr del - DAZ ampl | 2 | 1 | - | - | - | 1 | - | 1 |
| TOT (n) | 160 * | 149 § | 9 | 21 | 30 | 26 | 93 | 119 |
false deletions were excluded (n=9).
in 11 cases no information about the semen phenotype was given by the laboratories. Subjects with known semen phenotypes were divided into two major categories (normal or abnormal sperm) and further divided in subcategories.
Abbreviations: del=deletion; dupl=duplication; ampl=amplification ; AZ=azoospermic ; OZ=oligozoospermic ; normo=normospermic; AT=Astheno and /or Teratospermic; TOT1=normo plus pure AT; TOT2= AZ plus OZ.
Geographical distribution of gr/gr rearrangements
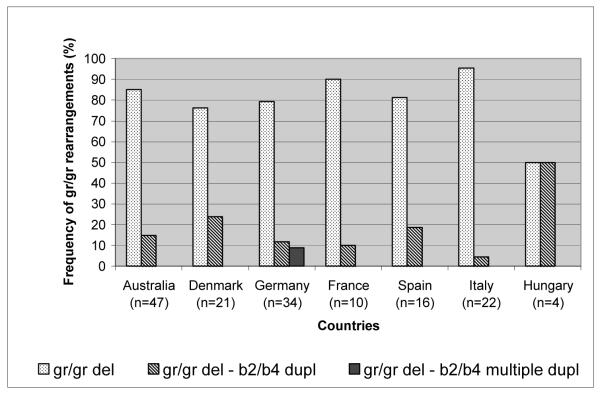
The most frequent gr/gr rearrangement type in all countries was the simple gr/gr deletion which ranged from 50% (Hungary) to 95% (Italy). The number of Hungarian gr/gr deletion carriers was low (n=4) and 50% presented gr/gr deletion – b2/b4 duplication. The frequency of gr/gr deletion –b2/b4 duplication was variable between countries, being otherwise highest in Denmark (24%) and lowest in Italy (4.5%) with a comparable number of total subjects analyzed from the two countries (21 and 22, respectively). Interestingly, we found gr/gr deletion – b2/b4 multiple duplication only in the German study population (8.8%) (Figure 3). Deletions with amplified DAZ or CDY copies are not included in the figure because of their low number (n=6). Three out of 4 subjects with gr/gr deletion –CDY1 amplification were French and 1 was Australian. All 2 subjects with gr/gr deletion –DAZ amplification were of Danish origin. Given the relatively low number of subjects in each group, differences in the distribution of gr/gr rearrangement types are not significant.
Figure 3.
Comparison of the frequencies of the different gr/gr rearrangement types in each participating country. False deletions and gr/gr deletion – DAZ or CDY1 amplification are excluded from this histogram.
Definition of gr/gr deletion patterns (subtypes) based on the missing copies of DAZ and CDY1 genes
Our RFLP method is based on the use of a SFV at sY587 which differentiates DAZ1 and DAZ2 from DAZ3 and DAZ4. In the vast majority of cases (n=154), we found deletion of either the DAZ1/DAZ2 (n=106, 69%) or the DAZ3/DAZ4 (n=48, 31%) gene pairs. In 6 cases belonging either to the group of “true” gr/gr deletions (n=5) or to gr/gr deletion – b2/b4 duplications (n=1) the RFLP analysis identified a peculiar pattern characterized by the combination of one DAZ gene from each gene pair. It can be hypothesized that the recombination site was between the two red amplicons removing either DAZ1 with DAZ3 or DAZ2 with DAZ4. According to previous publications [12, 15, 28] gr/gr deletions may present either the removal of CDY1a or CDY1b (see figure 1). The position of the CDY1b copy on the AZFc reference sequence implies that in those cases in which CDY1b is deleted, an inversion polymorphism in the DAZ3/4 palindrome P1 may have occurred. The alternative hypothesis of gene conversion of CDY1a by CDY1b prior to deletion was considered highly unlikely by Machev et al. [12] because CDY1b deletions were observed at high frequency in the gr/gr deleted subjects (48%) whereas CDY1b gene conversion events were observed only at low frequency ( <1%). In our study population (n=159), we found the deletion of CDY1a in 107 (67%) subjects and of CDY1b in 52 (33%). In one case, there was insufficient DNA for RFLP analysis of CDY1.
The combination of the different copies of the DAZ and CDY genes gave four major gr/gr deletion patterns characterized by the loss of: i) DAZ1/DAZ2+CDY1a (subtype 1); ii) DAZ3/DAZ4+CDY1a (subtype 2); iii) DAZ1/DAZ2+CDY1b (subtype 3); iv) DAZ3/DAZ4+CDY1b (subtype 4). The frequency of the 4 subtypes in the whole study population (n=153, excluding the 6 cases showing the combination of two DAZ gene for each gene pair and the case with inconclusive CDY1-RFLP results) was: 65/153 (42.5%) of subtype 1; 37/153 (24.2%) of subtype 2; 39/153 (25.5%) of subtype 3; 12/153 (7.8%) of subtype 4.
Geographical distribution of gr/gr subtypes
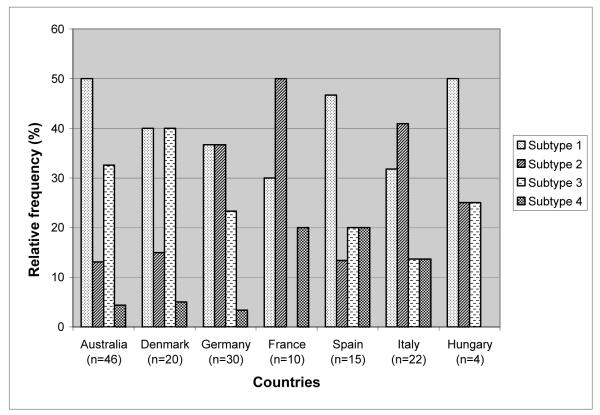
The frequency of gr/gr subtypes 2 and 3 showed significant differences among countries whereas subtype 1 and 4 were more similarly distributed (Figure 4). The highest frequency of subtype 1 was observed in Australia (50%), whereas the lowest was in France (30%). Subtype 4 was more frequent in France, Spain and Italy (13-20%) and less frequent in Australia, Germany and Denmark (3.3-5%). This subtype was absent in the 4 Hungarian gr/gr deletion carriers. Significant differences were observed between the frequencies of subtype 2: Australia with the lowest value (13%) versus Germany (36.7%), France (50%) and Italy (41%) with p =0.016, 0.008 and 0.01, respectively. The difference was also significant between France (the highest with 50%) versus Spain (the second lowest, 13.3%) with p=0.05. Subtype 3 was completely absent from the French samples and differences were significant between France (0%) and Australia (32.6%) and Denmark (40%), with p values 0.036 and 0.022, respectively.
Figure 4.
Comparison of the frequencies of the different gr/gr subtypes in each participating country. False deletions and gr/gr deletion – DAZ or CDY1 amplification are excluded from this histogram. Deletion subtypes remove: 1) DAZ1/DAZ2+CDY1a; 2) DAZ3/DAZ4+CDY1a; 3) DAZ1/DAZ2+CDY1b; 4) DAZ3/DAZ4+CDY1b.
Genotype/phenotype correlations
In order to evaluate the effect of different gr/gr rearrangements and gr/gr deletion subtypes on sperm production, we divided the study population into two groups on the basis of the sperm count: group 1 with abnormal sperm count (azoospermia or oligozoospermia) and group 2 with normal sperm count (including those subjects who presented either all three sperm parameters above the normal range or only normal sperm concentration independently from their fertility status). This subdivision meant that in the group of subjects with “normal sperm count” we also included 13 originating from the patient group with normal sperm concentration but reduced sperm motility and/or morphology (astheno and/or teratozoospermic men). The mean of the sperm parameters in this combined control group was: sperm concentration 59 ± 41.4 × 106 spzoa/ml; % progressive motility 37.2 ± 20; % normal morphology 14.6 ± 10.7.
gr/gr rearrangements and semen phenotype categories
Given that different gr/gr rearrangements are associated with different gene dosage, we tested the hypothesis that differences in gene dosage might explain the heterogeneous semen phenotype observed in gr/gr deletion carriers. However, the comparison of the two groups defined on the basis of the sperm count showed a similar distribution of the different gr/gr rearrangements. The most prevalent rearrangement type was the gr/gr deletion in both group 1 and 2 (81.5% versus 83.3%, respectively). The restoration of the original gene dosage, or higher, by single or multiple duplications did not seem to influence the phenotype since 17% of oligo/azoospermic subjects carried deletion/duplication events and this rearrangement was present at a similar frequency in the normospermic group (17 %). Data are reported in Table 1. The 9 “fully” normospermic controls who presented all three sperm parameters above the normal range were all carriers of a simple gr/gr deletion without duplications.
gr/gr rearrangements and mean sperm concentration
In those subjects for whom sperm parameters were available (n=142), we compared the mean sperm concentration within each rearrangement type. There was no significant difference between subjects with half AZFc gene dosage (gr/gr deletion) compared to normal (deletion/duplication) and higher copy number (deletion/multiple duplication): 16 ± 32.2 × 106 /ml (n=117) versus 9.3 ± 13.7 × 106 /ml (n=24). The corresponding comparison was also performed separately in subjects with abnormal sperm count (n=111) and in subjects with normal sperm concentrations (n=30). The comparison of gr/gr deleted and deleted/duplicated subjects in the first group showed no difference, 2.9 ± 3.8 × 106 /ml (n=92) versus 3.1 ± 3.9 × 106 /ml (n=19). Even after the removal of azoospermic men (given that gr/gr deletions are highly unlikely to be a definitive cause of azoospermia), the mean values remained similar.
In contrast, the group of subjects with normal sperm concentration showed nominally significant differences in gr/gr deletion versus deletion/duplication and multiple duplication: 64.2 ± 43.4 × 106 /ml (n=25) versus 32.8 ±11.8 × 106 /ml (n=5), respectively (t test of log n spermatozoa/ml p=0.042). Since all 9 fully normospermic controls were carriers of gr/gr deletions, such a comparison cannot be performed in this group. If we consider separately only those 16 subjects who have normal sperm concentration and were sent as “controls”, we found a similar effect of duplications in this subgroup: 81.6 ±12.1 × 106 /ml (pure gr/gr deletions in 14 subjects) and 29.7 ± 6.7 × 106 /ml (deletion-b2/b4 duplication and deletion –multiple duplications in 2 subjects). This interesting observation could have a number of explanations, including pure chance since multiple tests have been carried out, and replication in an independent sample is needed as a next step.
These data suggest that gene dosage differences are not a critical determinant of sperm output in the group of oligo/azoospermic men (although the mean sperm concentration was lower when duplications occurred), whereas it may reduce the spermatogenic potential in subjects with >20 millions spermatozoa/ml.
Deletion subtypes and semen phenotype categories
The most frequently deleted DAZ copy was DAZ1/2, whereas the predominant missing CDY1 copy was CDY1a, in both subjects with normal and abnormal sperm concentration (Table 2). We and others have previously hypothesized that certain deletion subtypes based on the definition of the missing DAZ (1/2 or 3/4) and CDY1 (a or b) copies (4 possible combinations) are enriched in patients and thus could be more pathogenic than others. In order to test this hypothesis, we compared the frequency of the 4 different gr/gr subtypes between subjects with abnormal and normal sperm concentration. As stated above, in 6 cases the RFLP analysis of the DAZ gene showed a pattern which was different from the 4 classic combinations and in one case the RFLP analysis of the CDY1 gene gave inconclusive result. These 7 cases, together with those presenting gr/gr deletion and amplified DAZ or CDY1 copy number, were excluded from the statistical analysis. Therefore, for this analysis we selected only those subjects who had either gr/gr deletion or gr/gr deletion – b2/b4 duplication and multiple duplication (Table 2).
Table 2.
The frequency distribution of different subcategories defined on the basis of the missing DAZ and CDY1 copies. The percentages are calculated from the totals (TOT) in a vertical manner.
| NORMAL SPERM COUNT | ABNORMAL SPERM COUNT | TOT1 +TOT 2(%) |
||||||
|---|---|---|---|---|---|---|---|---|
| gr/gr del (%) |
gr/gr del – b2/b4 dupl and md (%) |
TOT1 (%) |
gr/gr del (%) |
gr/gr del – b2/b4 dupl and md (%) |
TOT2 (%) |
|||
| Type of missing DAZ copies |
DAZ1/2 | 14 (58.3) |
4 (80) |
18 (62.1) |
62 (66.7) |
15 (79) |
77 (68.8) |
95 (67.4) |
| DAZ3/4 | 10 (41.7) |
1 (20) |
11 (37.9) |
31 (33.3) |
4 (21) |
35 (31.2) |
46 (32.6) |
|
| TOT | 24 | 5 | 29 | 93 | 19 | 112 | 141 * | |
| Type of missing CDY1 copy |
CDY1a | 15 (60) |
3 (60) |
18 (60) |
66 (68.8) |
15 (75) |
81 (69.8) |
99 (67.8) |
| CDY1b | 10 (40) |
2 (40) |
12 (40) |
30 (31.2) |
5 (25) |
35 (30.2) |
47 (32.2) |
|
| TOT | 25 | 5 | 30 | 96 | 20 | 116 | 146 * | |
| Deletion subtypes | 1 | 8 (33.3) |
2 (40) |
10 (34.5) |
37 (40.2) |
11 (57.9) |
48 (43.2) |
58 (41.4) |
| 2 | 6 (25) |
1 (20) |
7 (24.1) |
25 (27.2) |
4 (21.05) |
29 (26.1) |
36 (25.7) |
|
| 3 | 6 (25) |
2 (40) |
8 (27.6) |
23 (25) |
4 (21.05) |
27 (24.3) |
35 (25) |
|
| 4 | 4 (16.7) |
- | 4 (13.8) |
7 (7.6) |
- | 7 (6.3) |
11 (7.8) |
|
| TOT | 24 | 5 | 29 | 92 | 19 | 111 | 140 * | |
differences in total numbers (TOT1, TOT2 and TOT1+TOT2) are due to 6 cases with unclassified DAZ deletions (1 normal and 5 abnormal) and 1 case with an undefined CDY1 copy deletion (one from the abnormal sperm count group). The 2 cases with gr/gr del-CDY1 ampl and DAZ ampl were excluded from the table. Deletion subtypes are: 1) DAZ1/DAZ2+CDY1a; 2) DAZ3/DAZ4+CDY1a ; 3) DAZ1/DAZ2+CDY1b; 4) DAZ3/DAZ4+CDY1b.
Abbreviations: del=deletion; dupl=duplication; md=multiple duplication; TOT1=n. gr/gr del plus n. gr/gr del – b2/b4 dupl and md among subject with normal sperm count; TOT2= n. gr/gr del plus n. gr/gr del – b2/b4 dupl and md among subject with abnormal sperm count
According to our previous data [15], the most frequent subtype in oligoazoospermic men was subtype 1, whereas the least frequent was subtype 4. However, men with normal sperm concentration also showed a similar frequency distribution for the four subtypes. It is nevertheless worth noting that, in the present study, subtype 4 – the only gr/gr subtype found in normospermic controls in the Italian sample (Giachini et al 2005, and unpublished data) – is more frequent in the normospermic control group (16.7%) than in the oligo/azoospermic group (7.6%), or 13.8% versus 6.3% if we consider both gr/gr deletions and gr/gr deletion – b2/b4 duplication (single or multiple). The phenotype associated with subtype 4 therefore requires further investigation. The semen phenotype of the 6 patients with the unusual DAZ deletion pattern was heterogeneous including azoospermia (n=1), cryptozoospermia (n=2), severe oligoasthenoteratozoospermia (n=2) and asthenoteratozoospermia (n=1). These data indicate that this rare pattern is mainly associated with abnormal spermatogenesis.
Deletion subtypes and mean sperm concentration
We compared the mean sperm concentration in subjects with CDY1a deletion versus CDY1b deletion in the whole study population and then separately in the group of subjects with normal and abnormal sperm count. Data are reported in Table 3; in which we again excluded the 6 cases with unclassified DAZ copies and gr/gr deletions with DAZ and CDY1 copy amplification. Therefore, for this analysis we selected only those subjects who had either gr/gr deletion or gr/gr deletion – b2/b4 duplication or multiple duplications, and with known sperm concentrations. Although in both groups with normal and abnormal sperm count, the mean values of sperm concentration were lower in cases of loss of DAZ1/DAZ2 or CDY1a copy loss, the differences did not reach statistical significance.
Table 3.
Means (and standard deviations) of sperm concentration of gr/gr deleted and gr/gr deleted – b2/b4 duplicated (single or multiple) men, classified on the basis of the type of missing DAZ and CDY1 copies. The means of sperm concentration in these gr/gr rearrangements, further divided on the basis of the deletion subtype, are also shown. Deletion subtypes remove: 1) DAZ1/DAZ2+CDY1a; 2) DAZ3/DAZ4+CDY1a; 3) DAZ1/DAZ2+CDY1b; 4) DAZ3/DAZ4+CDY1b.
| NORMAL SPERM COUNT (mean sperm concentration ×106/ml) |
ABNORMAL SPERM COUNT (mean sperm concentration ×106/ml) |
TOT (mean sperm concentration ×106/ml) |
||||||
|---|---|---|---|---|---|---|---|---|
| gr/gr del | gr/gr del – b2/b4 dupl and md |
gr/gr del and gr/gr del – b2/b4 dupl and md |
gr/gr del | gr/gr del – b2/b4 dupl and md |
gr/gr del and gr/gr del – b2/b4 dupl and md |
|||
|
Type of missing
DAZcopies |
DAZ1/2 | 62.2±42.0 (n=14) |
31.9±13.4 (n=4) |
55.5±39.4 (=18) | 3.1±3.9 (n=60) |
3.3±3.6 (n=14) |
3.2±3.8 (n=74) | 13.4±27.1 (n=92) |
| DAZ3/4 | 71.4±46.7 (n=10) |
36.5 (n=1) | 68.2±45.5 (n=11) |
2.8±3.5 (n=28) |
2.9±5.7 (n=4) | 2.8±3.7 (n=32) | 19.6±36.6 (n=43) |
|
| TOT | 66.0±43.3 (n=24) |
32.8±11.8 (n=5) |
3.0±3.8 (n=88) |
3.2±3.9 (n=18) |
||||
|
missing
CDY1 |
CDY1a | 52.7±34.2 (n=15) |
40.2 ±8.6 (n=3) |
50.6±31.6 (n=18) |
2.6±3.4 (n=61) |
3.4±3.9 (n=14) |
2.8±3.5 (n=75) | 12.0±23.6 (n=93) |
| CDY1b | 81.38±51.4 (n=10) |
21.7±1.8 (n=2) |
71.4±52.0 (n=12) |
3.5±4.4 (n=30) |
2.0±4.1 (n=5) | 3.3±4.4 (n=35) | 20.7±39.5 (n=47) |
|
| TOT | 64.20±43.3 (n=25) |
32.8±11.8 (n=5) |
2.9±3.8 (n=91) |
3.1±3.9 (n=19) |
||||
| Deletion subtypes | 1 | 48.3±28.2 (n=8) |
42.0±11.2 (n=2) |
47.0±25.3 (n=10) |
2.6±3.5 (n=35) |
3.7±3.3 (n=10) |
2.8±3.5 (n=45) | 10.9±20.3 (n=55) |
| 2 | 64.2±42.0 (n=6) |
36.5 (n=1) | 60.2±39.7 (n=7) | 3.1±3.4 (n=22) |
2.9±5.7 (n=4) | 3.1±3.7 (n=26) | 15.2±29.5 (n=33) |
|
| 3 | 80.8±52.5 (n=6) |
21.7±1.8 (n=2) |
66.0±52.1 (n=8) | 4.1±4.5 (n=23) |
2.5±4.6 (n=4) | 3.9±4.5 (n=27) | 18.1±35.7 (n=35) |
|
| 4 | 82.2±57.8 (n=4) |
(n=0) | 82.2±57.8 (n=4) | 1.5±3.7 (n=7) | (n=0) | 1.5±3.7 (n=7) | 30.9±51.7 (n=11) |
|
| TOT | 66.0±43.3 (n=24) |
32.8±11.8 (n=5) |
3.0±3.8 (n=87) |
3.2±3.9 (n=18) |
Abbreviations: del=deletion; dupl=duplication; md=multiple duplication
Y haplogroups and their relationship to AZFc rearrangements and to the phenotypic expression of gr/gr deletions and deletions/duplications
Y haplogroup analysis showed that hgs E, I, R1a and R1b, here combined with related chromosomes into the broader categories E, F*(xK), K*(xP) and P, made up most of the sample, as expected for a mixed predominantly Western European sample (Supplementary Table 1). Hg P was the most common hg in men with both normal and abnormal sperm counts, and the frequencies of the other hgs followed the same order P > F*(xK) > E > K*(xP) in the two groups (Table 4). Similarly, within each of the normal sperm concentration, abnormal sperm concentration or total classes, there were no significant differences in mean sperm concentration between the haplogroups (Table 5). Duplications (including multiple duplications) following deletion were more rare in hg P chromosomes than in other hgs and more common in hg E chromosomes, but these differences were not statistically significant (p = 0.08, p = 0.08 respectively; Fisher exact test). These duplications were more common in Denmark and Germany than in the other countries examined, but this enrichment could not be accounted for by the hg distributions since hg E is more frequent in south-eastern European countries [25].
Table 4.
Comparison of the frequency of gr/gr deletion and gr/gr deletion – b2/b4 duplication (single or multiple) in men with normal and abnormal sperm count, divided on the basis of their Y haplogroup. The percentages are calculated from the total (TOT) in a vertical manner.
| NORMAL SPERM COUNT | ABNORMAL SPERM COUNT | TOT1+ TOT2 (%) |
||||||
|---|---|---|---|---|---|---|---|---|
| gr/gr del (%) |
gr/gr del – b2/b4 dupl and md (%) |
TOT1 (%) |
gr/gr del (%) |
Gr/gr del – b2/b4 dupl and md (%) |
TOT2 (%) |
|||
| Y haplogroup |
F*(x
K) |
6 (25) | 1 (20) | 7 (24.1) |
34 (37.4) | 8 (42.1) | 42 (38.2) |
49 (35.2) |
| P | 14 (58.3) | 2 (40) | 16 (55.2) |
42 (46.2) | 6 (31.6) | 48 (43.6) |
64 (46) | |
|
K*(x
P) |
2 (8.3) | 1 (20) | 3 (10.3) |
7 (7.7) | 1 (5.3) | 8 (7.3) | 11 (7.9) | |
| E | 2 (8.3) | 1 (20) | 3 (10.3) |
8 (8.8) | 4 (21.1) | 12 (10.9) |
15 (10.8) |
|
| TOT | 24 | 5 | 29 | 91 | 19 | 110 | 139 | |
Abbreviations: del=deletion; dupl=duplication; md=multiple duplication; TOT1=n. gr/gr del plus n. gr/gr del – b2/b4 dupl and md among subject with normal sperm count; TOT2= n. gr/gr del plus n. gr/gr del – b2/b4 dupl and md among subject with abnormal sperm count
Table 5.
Means (and standard deviations) of sperm concentration of gr/gr deleted and gr/gr deleted – b2/b4 duplicated (single or multiple) men, classified on the basis of their Y haplogroup.
| NORMAL SPERM COUNT | ABNORMAL SPERM COUNT | TOT1+ TOT2 (%) |
||||||
|---|---|---|---|---|---|---|---|---|
| gr/gr del (%) |
gr/gr del – b2/b4 dupl and md (%) |
TOT1 (%) | gr/gr del (%) |
gr/gr del – b2/b4 dupl and md (%) |
TOT2 (%) |
|||
| Y haplogroup |
F*(x
K) |
65.2±49.1 (n=6) |
23.0 (n=1) |
59.2±47.6 (n=7) |
4.3±4.7 (n=34) |
3.7±4.5 (n=8) |
4.2±4.6 (n=42) |
12.1±26.1 (n=49) |
| P | 45.8±27.6 (n=14) |
43.2±9.5 (n=2) |
45.5±25.8 (n=16) |
2.3±3.0 (n=40) |
2.6±3.6 (n=5) |
2.3±3.0 (n=45) |
13.7±23.2 (n=61) |
|
|
K*(x
P) |
94.0±55.1 (n=2) |
20.5 (n=1) |
69.5±57.6 (n=3) |
1.8±3.0 (n=5) |
0.0 (n=1) | 1.5±2.8 (n=6) |
24.2±44.6 (n=9) |
|
| E | 126.5±37.5 (n=2) |
34.1 (n=1) |
95.7±59.6 (n=3) |
1.1±1.9 (n=8) |
3.8±4.2 (n=4) |
2.0±3.0 (n=12) |
20.7±44.9 (n=15) |
|
| TOT | 61.4±42.0 (n=24) |
32.8±11.8 (n=5) |
3.0±3.8 (n=87) |
3.2±3.9 (n=18) |
||||
Abbreviations: del=deletion; dupl=duplication; md=multiple duplication.
Since the removal of both CDY1b and sY1291 as a single event implies the presence of an inversion polymorphism with respect to the reference sequence, it was of interest to analyse the hg distribution in subjects with CDY1a versus CDY1b deletion. We found that hg P is significantly more frequent when there has been CDY1a copy loss (56.7%) compared to CDY1b copy loss (14.6%) p<0.001. On the other hand, 73% of subjects with CDY1b copy loss have hg F*(xP) or E (56.3% and 16.7%), whereas in CDY1a loss carriers the cumulative frequency of the same hgs was lower at 33% (hg F*(xP) = 25% and hg E=7.7%). Within hg F*(xP), there was a highly significant difference within hg I which made up 81% of the CDY1b loss chromosomes but only 19% of the CDY1a loss chromosomes (p=0.00001; data in Supplementary Table 1). Finally, we used a linear regression analysis on the same samples to investigate whether the variation in sperm concentration of oligo- or normospermic men, considered as a continuous variable, could be explained by any of the factors measured: gr/gr versus deletion/duplication, CDY1 deletion, DAZ deletion or Y haplogroup. None of these factors explained a significant proportion of the variation in sperm count (all p > 0.05).
DISCUSSION
We have carried out the largest study thus far to investigate the contribution of Y-chromosomal factors to the extensive and puzzling phenotypic variation exhibited by gr/gr deletion carriers, which ranges from normal spermatogenesis to azoospermia. The factors examined included both the known AZFc structural variants associated with this deletion – removal of different DAZ and CDY1 gene copies, deletion followed by duplication – and the more general Y chromosome background, measured as the Y-SNP-defined hg, which could reveal the influence of factors located anywhere on the male-specific part of the chromosome. We found that none of these factors accounted for a significant proportion of the spermatogenic variation associated with gr/gr deletions. We now consider i) whether any aspects of our study design and execution could have led us to a false negative conclusion, ii) the power of the study to detect the effects we were seeking and iii) the implications of the conclusion for biological understanding and clinical practice.
The study was designed to investigate a relatively homogeneous group of subjects with Western European descent. Genetic ancestry was confirmed by the presence of the expected European Y hgs. All molecular characterizations were conducted in a single centre (eliminating ~5% false positives from the original screen in the process), ensuring that uniform standards of genotype calling were applied to the entire set of samples. Our study was well-powered. For example, with a sample size of 134 and the standard requirement for a p value of 0.05, we would have 80% power to detect a variable that explained ≥5.5% of the variation in sperm count. We therefore conclude that Y-chromosomal factors do not explain any substantial proportion of the variation in sperm phenotype of Western European gr/gr deletion carriers.
The presence of certain Y hgs with constitutive deletions, both gr/gr and b2/b3, at high frequency in populations such as the Japanese and Finns, respectively, led to the hypothesis that these Y chromosomes might carry constitutive duplications as well which would counteract the deleterious effects of the gene dosage reduction. According to the hypothesis that gene dosage differences generally underlie the phenotypic variability, we would expect to have seen a higher rate of del/dupl events in the normospermic group examined here. Our results excluded this possibility and, on the contrary, suggested that deletion followed by duplication may negatively affect spermatogenic efficiency. Subjects with deletion/duplications showed lower sperm concentration in both normospermic and infertile groups, although the finding reached significance only in the normospermic group. A possible explanation could be that deletion followed by duplications may indicate a higher propensity to genomic instability which may not be restricted to the AZFc region.
The definition of different DAZ and CDY1 copies in our study has two implications. A number of studies have indicated that DAZ1/DAZ2 deletions are restricted to gr/gr carriers with impaired sperm production [15, 19, 22] and it was therefore proposed that these two copies may be biologically more important than the others. We observed no significant phenotypic difference according to the deletion of different DAZ and CDY1 gene copies implying that these genes are either irrelevant to sperm output – which is unlikely to be true for both – or functionally equivalent. The second implication is that while the definition of the Y hgs provides information about the Y chromosome structure as a whole, the removal of different DAZ and CDY1 copies may reflect a particular AZFc structure on which gr/gr deletion took place, which can be present in several lineages, albeit at different frequency. Our data indicate that special rearrangements such as inversion polymorphism that should precede, for example, the removal of CDY1b copy in case of sY1291 loss, are probably not relevant for the phenotypic expression of deletions. On the other hand we observed significant differences in the hg distribution between carriers with the loss of CDY1a and CDY1b; this result shows some structural rearrangements in the AZFc region may be associated with particular haplogroups (e.g. CDY1b deletion in hg I).
This study has important implications also for clinical practice. Although we did not find an evident correlation between Y hgs and the phenotypic expression of gr/gr rearrangements, it is worth noting that geographic differences were detected. It is therefore likely that a genetic factor with a relatively mild phenotypic effect may be more penetrant in certain populations than in others and this may contribute to bias in admixed populations. Although we were unable to distinguish between “pathogenic” and “neutral” deletions, gr/gr deletions as a group are considered a significant risk factor for spermatogenic failure [29]. This conclusion might encourage some laboratories to screen for this genetic anomaly prior to assisted reproductive techniques, since this genetic risk factor will be obligatorily transmitted to the male offspring. In this regard it is important to note that in the populations examined here, the false deletion rate of 5.3% in the sY1291-based test as it is routinely used is a cause for concern. Care should be taken to distinguish amplification failures from true deletions. Suspected deletions should be verified through the use of different PCR conditions or by redesigning the assay into a multiplex format with internal positive control fragments or performing a gene dosage analysis as used in this study. Second, there is at present no justification for subtyping of gr/gr deletions in order to inform counselling of carriers of European decent. We emphasize, however, that this last conclusion is only relevant to carriers with ancestry restricted to Western Europe: in other areas of the world the matter remains open, and in a population with mixed Western European/Japanese ancestry, identification of the low-risk hg D gr/gr deletion chromosomes would be positively indicated.
In conclusion, our study has provided evidence that neither the definition of Y hgs nor the definition of gr/gr subtypes on the basis of the dosage or the missing DAZ and CDY copies are able to provide an explanation for the heterogeneous phenotype observed in gr/gr deletion carriers. However, our study provides evidence for significant geographic differences in the distribution of deletion subtypes which may affect the outcome of case control association studies in different geographic areas.
Supplementary Material
Acknowledgements
We would like to thank: the Italian Ministry of Education and Research (MIUR) for the grant PRIN 2005-2007 recipient C.K and the University of Florence; the Wellcome Trust (Y.X. and C.T.-S.); Ministerio de Ciencia y Tecnologia BFU2006-03479/BMC (R.O),
We thank for clinical input and useful discussions: Mario Maggi, Ivo Noci, David M de Kretser, HW Gordon Baker, Elisabeth Carlsen and Niels E. Skakkebaek
We thank for technical assistance and useful discussions: Selene Degl’Innocenti, Francesca Nuti, Elena Guarducci, Anne Reilly, David Cram, Michael Lynch, Sabine Borchert, Inger D. Garn.
We thank Laura Gambera and Francesca Serafini, Center for Couple Sterility, Dept. Pediatrics, Obstetrics and Reproductive Medicine, University of Siena for basic semen analysis.
The Corresponding Author has the right to grant on behalf of all authors and does grant on behalf of all authors, an exclusive licence (or non exclusive for government employees) on a worldwide basis to the BMJ Publishing Group Ltd and its Licensees to permit this article (if accepted) to be published in JMG and any other BMJPGL products to exploit all subsidiary rights, as set out in our licence (http://jmg.bmj.com/ifora/licence.pdf).
REFERENCES
- 1.Choe JH, Kim JW, Lee JS, Seo JT. Routine screening for classical azoospermia factor deletions of the Y chromosome in azoospermic patients with Klinefelter syndrome. Asian J Androl. 2007;9:815–20. doi: 10.1111/j.1745-7262.2007.00315.x. [DOI] [PubMed] [Google Scholar]
- 2.Krausz C, Degl’Innocenti S. Y chromosome and male infertility: update, 2006. Front Biosci. 2006;11:3049–61. doi: 10.2741/2032. [DOI] [PubMed] [Google Scholar]
- 3.McElreavey K, Krausz C. Sex Chromosome Genetics ’99. Male infertility and the Y chromosome. Am J Hum Genet. 1999;64:928–33. doi: 10.1086/302351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Skaletsky H, Kuroda-Kawaguchi T, Minx PJ, Cordum HS, Hillier L, Brown LG, Repping S, Pyntikova T, Ali J, Bieri T, Chinwalla A, Delehaunty A, Delehaunty K, Du H, Fewell G, Fulton L, Fulton R, Graves T, Hou SF, Latrielle P, Leonard S, Mardis E, Maupin R, McPherson J, Miner T, Nash W, Nguyen C, Ozersky P, Pepin K, Rock S, Rohlfing T, Scott K, Schultz B, Strong C, Tin-Wollam A, Yang SP, Waterston RH, Wilson RK, Rozen S, Page DC. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature. 2003;423:825–37. doi: 10.1038/nature01722. [DOI] [PubMed] [Google Scholar]
- 5.Noordam MJ, Repping S. The human Y chromosome: a masculine chromosome. Curr Opin Genet Dev. 2006;16:225–32. doi: 10.1016/j.gde.2006.04.018. [DOI] [PubMed] [Google Scholar]
- 6.Lin YW, Hsu LC, Kuo PL, Huang WJ, Chiang HS, Yeh SD, Hsu TY, Yu YH, Hsiao KN, Cantor RM, Yen PH. Partial duplication at AZFc on the Y chromosome is a risk factor for impaired spermatogenesis in Han Chinese in Taiwan. Hum Mutat. 2007;28:486–94. doi: 10.1002/humu.20473. [DOI] [PubMed] [Google Scholar]
- 7.Vogt PH. AZF deletions and Y chromosomal haplogroups: history and update based on sequence. Hum Reprod Update. 2005;11:319–36. doi: 10.1093/humupd/dmi017. [DOI] [PubMed] [Google Scholar]
- 8.Yen P. The fragility of fertility. Nat Genet. 2001;29:243–4. doi: 10.1038/ng1101-243. [DOI] [PubMed] [Google Scholar]
- 9.Repping S, van Daalen SK, Brown LG, Korver CM, Lange J, Marszalek JD, Pyntikova T, van der Veen F, Skaletsky H, Page DC, Rozen S. Polymorphism for a 1.6-Mb deletion of the human Y chromosome persists through balance between recurrent mutation and haploid selection. Nat Genet. 2003;35:247–51. doi: 10.1038/ng1250. [DOI] [PubMed] [Google Scholar]
- 10.Krausz C, Giachini C. Genetic risk factors in male infertility. Arch Androl. 2007;53:125–33. doi: 10.1080/01485010701271786. [DOI] [PubMed] [Google Scholar]
- 11.Ravel C, Chantot-Bastaraud S, El Houate B, Mandelbaum J, Siffroi JP, McElreavey K. GR/GR deletions within the azoospermia factor c region on the Y chromosome might not be associated with spermatogenic failure. Fertil Steril. 2006;85:229–31. doi: 10.1016/j.fertnstert.2005.07.1278. [DOI] [PubMed] [Google Scholar]
- 12.Machev N, Saut N, Longepied G, Terriou P, Navarro A, Levy N, Guichaoua M, Metzler-Guillemain C, Collignon P, Frances AM, Belougne J, Clemente E, Chiaroni J, Chevillard C, Durand C, Ducourneau A, Pech N, McElreavey K, Mattei MG, Mitchell MJ. Sequence family variant loss from the AZFc interval of the human Y chromosome, but not gene copy loss, is strongly associated with male infertility. J Med Genet. 2004;41:814–25. doi: 10.1136/jmg.2004.022111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carvalho CM, Zuccherato LW, Bastos-Rodrigues L, Santos FR, Pena SD. No association found between gr/gr deletions and infertility in Brazilian males. Mol Hum Reprod. 2006;12:269–73. doi: 10.1093/molehr/gal029. [DOI] [PubMed] [Google Scholar]
- 14.de Llanos M, Ballescà JL, Gázquez C, Margarit E, Oliva R. High frequency of gr/gr chromosome Y deletions in consecutive oligospermic ICSI candidates. Hum Reprod. 2005;20:216–20. doi: 10.1093/humrep/deh582. [DOI] [PubMed] [Google Scholar]
- 15.Giachini C, Guarducci E, Longepied G, Degl’Innocenti S, Becherini L, Forti G, Mitchell MJ, Krausz C. The gr/gr deletion(s): a new genetic test in male infertility? J Med Genet. 2005;42:497–502. doi: 10.1136/jmg.2004.028191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hucklenbroich K, Gromoll J, Heinrich M, Hohoff C, Nieschlag E, Simoni M. Partial deletions in the AZFc region of the Y chromosome occur in men with impaired as well as normal spermatogenesis. Hum Reprod. 2005;20:191–7. doi: 10.1093/humrep/deh558. [DOI] [PubMed] [Google Scholar]
- 17.Lynch M, Cram DS, Reilly A, O’Bryan MK, Baker HW, de Kretser DM, McLachlan RI. The Y chromosome gr/gr subdeletion is associated with male infertility. Mol Hum Reprod. 2005;11:507–12. doi: 10.1093/molehr/gah191. [DOI] [PubMed] [Google Scholar]
- 18.Becherini L, Guarducci E, Degl’Innocenti S, Rotondi M, Forti G, Krausz C. DAZL polymorphisms and susceptibility to spermatogenic failure: an example of remarkable ethnic differences. Int J Androl. 2004;27:375–81. doi: 10.1111/j.1365-2605.2004.00520.x. [DOI] [PubMed] [Google Scholar]
- 19.Fernandes S, Huellen K, Goncalves J, Dukal H, Zeisler J, Rajpert De Meyts E, Skakkebaek NE, Habermann B, Krause W, Sousa M, Barros A, Vogt PH. High frequency of DAZ1/DAZ2 gene deletions in patients with severe oligozoospermia. Mol Hum Reprod. 2002;8:286–98. doi: 10.1093/molehr/8.3.286. [DOI] [PubMed] [Google Scholar]
- 20.Jobling MA, Samara V, Pandya A, Fretwell N, Bernasconi B, Mitchell RJ, Gerelsaikhan T, Dashnyam B, Sajantila A, Salo PJ, Nakahori Y, Disteche CM, Thangaraj K, Singh L, Crawford MH, Tyler-Smith C. Recurrent duplication and deletion polymorphisms on the long arm of the Y chromosome in normal males. Hum Mol Genet. 1996;5:1767–75. doi: 10.1093/hmg/5.11.1767. [DOI] [PubMed] [Google Scholar]
- 21.Repping S, van Daalen SK, Brown LG, Korver CM, Lange J, Marszalek JD, Pyntikova T, van der Veen F, Skaletsky H, Page DC, Rozen S. High mutation rates have driven extensive structural polymorphism among human Y chromosomes. Nat Genet. 2006;38:463–7. doi: 10.1038/ng1754. [DOI] [PubMed] [Google Scholar]
- 22.Ferlin A, Tessari A, Ganz F, Marchina E, Barlati S, Garolla A, Engl B, Foresta C. Association of partial AZFc region deletions with spermatogenic impairment and male infertility. J Med Genet. 2005;42:209–13. doi: 10.1136/jmg.2004.025833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karafet TM, Mendez FL, Meilerman MB, Underhill PA, Zegura SL, Hammer MF. New binary polymorphisms reshape and increase resolution of the human Y chromosomal haplogroup tree. Genome Res. 2008;18:830–8. doi: 10.1101/gr.7172008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paracchini S, Arredi B, Chalk R, Tyler-Smith C. Hierarchical high-throughput SNP genotyping of the human Y chromosome using MALDI-TOF mass spectrometry. Nucleic Acids Res. 2002;30:e27. doi: 10.1093/nar/30.6.e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosser ZH, Zerjal T, Hurles ME, Adojaan M, Alavantic D, Amorim A, Amos W, Armenteros M, Arroyo E, Barbujani G, Beckman G, Beckman L, Bertranpetit J, Bosch E, Bradley DG, Brede G, Cooper G, Côrte-Real HB, de Knijff P, Decorte R, Dubrova YE, Evgrafov O, Gilissen A, Glisic S, Gölge M, Hill EW, Jeziorowska A, Kalaydjieva L, Kayser M, Kivisild T, Kravchenko SA, Krumina A, Kucinskas V, Lavinha J, Livshits LA, Malaspina P, Maria S, McElreavey K, Meitinger TA, Mikelsaar AV, Mitchell RJ, Nafa K, Nicholson J, Nørby S, Pandya A, Parik J, Patsalis PC, Pereira L, Peterlin B, Pielberg G, Prata MJ, Previderé C, Roewer L, Rootsi S, Rubinsztein DC, Saillard J, Santos FR, Stefanescu G, Sykes BC, Tolun A, Villems R, Tyler-Smith C, Jobling MA. Y-chromosomal diversity in Europe is clinal and influenced primarily by geography, rather than by language. Am J Hum Genet. 2000;67:1526–43. doi: 10.1086/316890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Semino O, Passarino G, Oefner PJ, Lin AA, Arbuzova S, Beckman LE, De Benedictis G, Francalacci P, Kouvatsi A, Limborska S, Marcikiae M, Mika A, Mika B, Primorac D, Santachiara-Benerecetti AS, Cavalli-Sforza LL, Underhill PA. The genetic legacy of Paleolithic Homo sapiens sapiens in extant Europeans: a Y chromosome perspective. Science. 2000;290:1155–9. doi: 10.1126/science.290.5494.1155. [DOI] [PubMed] [Google Scholar]
- 27.Organization WH. WHO Laboratory Manual for the Examination of Human Semen and Sperm-Cervical Mucus Interaction 3rd. Cambridge University Press; Cambridge, UK: 1999. pp. 4–33. [Google Scholar]
- 28.Giachini C, Nuti F, Marinari E, Forti G, Krausz C. Partial AZFc deletions in infertile men with cryptorchidism. Hum Reprod. 2007;22:2398–403. doi: 10.1093/humrep/dem186. [DOI] [PubMed] [Google Scholar]
- 29.Tüttelmann F, Rajpert-De Meyts E, Nieschlag E, Simoni M. Gene polymorphisms and male infertility--a meta-analysis and literature review. Reprod Biomed Online. 2007;15:643–58. doi: 10.1016/s1472-6483(10)60531-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.