Abstract
A conserved, independently folding domain in the large ribosomal subunit consists of 58 nt of rRNA and a single protein, L11. The tertiary structure of an rRNA fragment carrying the E. coli sequence is marginally stable in vitro but can be substantially stabilized by mutations found in other organisms. To distinguish between possible reasons why natural selection has not evolved a more stable rRNA structure in E. coli, mutations affecting the rRNA tertiary structure were assessed for their in vitro effects on rRNA stability and L11 affinity (in the context of an rRNA fragment) or in vivo effects on cell growth rate and L11 content of ribosomes. The rRNA fragment stabilities ranged from −4 to +9 kcal/mol relative to the wild type sequence. Variants in the range of −4 to +5 kcal/mol had almost no observable effect in vivo, while more destabilizing mutations (> 7 kcal/mol) were not tolerated. The data suggest that the in vivo stability of the complex is roughly −6 kcal/mol, and that any single tertiary interaction is dispensable for function as long as a minimum stability of the complex is maintained. Based on these data, it seems that the evolution of this domain has not been constrained by inherent structural or functional limits on stability. The estimated stability corresponds to only a few ribosomes per bacterial cell dissociated from L11 at any time; thus the selective advantage for any further increase in stability may be so small as to be outweighed by other competing selective pressures.
Introduction
The complex between ribosomal protein L11 and its small, compactly folded rRNA binding domain (L11BD1, Figure 1) has become an important focus of ribosome functional studies. The complex appears as a knob protruding from the surface of the ribosome (4, 5) and interacts with elongation factors during the ribosome cycle (6, 7). The functional importance of this complex is underscored by the conservation of key residues across all phylogenetic domains, including bases making tertiary hydrogen bonds in the RNA structure, amino acids supporting the homeodomain-like fold of the L11 C-terminal domain, and residues making protein-RNA contacts (8, 9). The complex is also uniquely suited for studies of folding and stability in the ribosome, since a 58 nt rRNA fragment bound to L11 adopts the same structure as the corresponding sequence in the intact ribosome (4, 5, 9, 10). Here we use the functional importance of the L11BD and its convenience for thermodynamic studies to explore relationships between sequence conservation, thermodynamic stability, and function.
Figure 1.
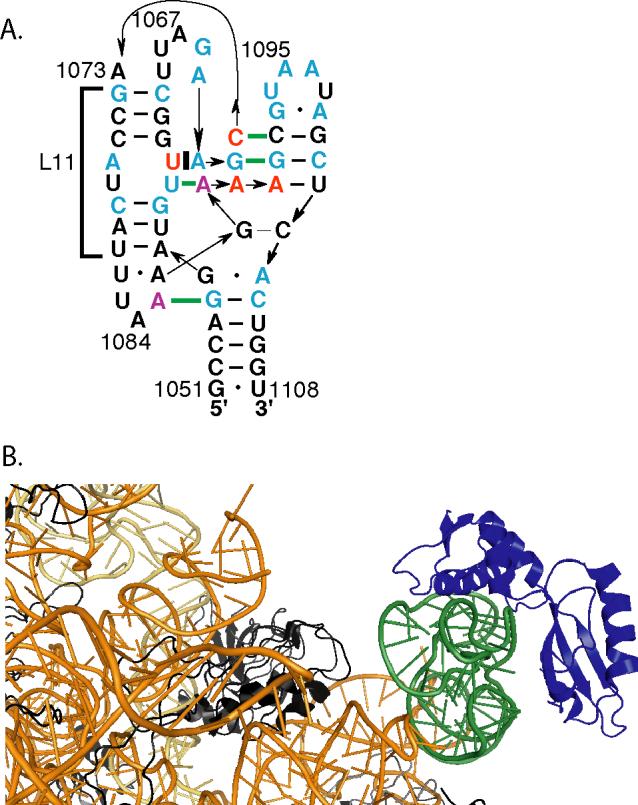
A, Schematic of the L11BD RNA. Nucleotides mutated in this study are indicated in red (U1061 and AA1089−1090, stabilizing) or purple (A1085, A1088, and C1072, destabilizing). Cyan bases are conserved in greater than 97% of bacterial, archaeal, and eukaryotic sequences (15). Of particular importance to this study, at position 1061 only U is found in bacteria, G among the eucarya, but all four bases in archaea (28). The two nucleotide sequence at 1089−1090 is either AA or GU in nearly all organisms. Base-base tertiary interactions are indicated by horizontal thick green lines. The segment of distorted minor groove contacted by L11 is indicated by a bracket. B, View of the Escherichia coli 50S subunit (5) illustrates that L11BDL11 complex is an independent domain isolated from the remainder of the ribosome. L11, blue; other proteins, black; nucleotides 1051−1108 (L11BD), green; 23S RNA, gold.
The present work was stimulated by the observation that the 58 nt L11BD RNA derived from E. coli ribosomes is relatively unstable in vitro (11), adopting its folded structure only when L11 is bound (12, 13). It seemed plausible at the time that the RNA structure might be intrinsically unfoldable in the absence of a stabilizing protein, but later it was found that base changes at two different positions, U1061 or AA1089−1090, yield a very stable RNA (14). Such stabilizing sequence variants are found in other organisms (see Figure 1A). If one or two base changes can yield a more stable and apparently functional rRNA domain, why have the changes not been adopted by E. coli (and many other bacteria)? In more general terms, these observations raise the question of what selective pressures have been at work to define the stability of this protein - RNA domain. Several possibilities are plausible:
(i) U1061 and AA1089−1090 might be required for some functional reason specific to E. coli (or to all bacteria in the case of U1061), and cannot mutate without disrupting a necessary ribosome or regulatory function.
(ii) The function of the L11 - RNA domain could require certain flexible motions or alternative conformations that place a limit on the stability consistent with efficient function.
(iii) The E. coli L11-RNA complex might not be under selective pressure for increased stability simply because it is already well-folded enough in vivo to carry out its functional role.
The last two possibilities pose intriguing questions. A number of studies have suggested that there is a correlation between the adaptation temperature of an organism and thermodynamic stability for series of orthologous proteins or protein-RNA complexes (15, 16). For many protein enzymes, there is evidence that selection favors a balance between stability and flexibility (17): selection has driven stability to an optimum value rather than the maximum possible value. Is stability of the L11-RNA complex also limited by functional considerations, and if not, what is the maximum stability that natural selection can achieve?
In this work we examine a number of mutations in the E. coli 58mer rRNA domain that alter the stability of the RNA in the L11-RNA complex from −4 to +9 kcal/mol relative to wild type. Based on the growth rates of cells dependent on rRNAs containing these same mutations, it appears that specific sequence or functional requirements (possibilities i and ii above) do not limit the stability of the complex. We conclude from our data that natural selection has brought the in vivo stability of the complex to a free energy of roughly −6 kcal/mol. At this stability, only a few ribosomes in a bacterial cell will be dissociated from L11 at any one time, and we suggest that the selective advantage for any further stabilization is so small as to be outweighed by other competing selective pressures.
Materials and Methods
In vitro experiments and data analysis
Run-off transcription and UV melting experiments (18), filter binding assays with 35S-labeled RNA (19), and purification of Bacillus stearothermophilus L11-C76 (20) were carried out as described in the cited articles. Buffer for protein binding constant measurements was 10 mM MOPS pH 7.5, 3 mM MgCl2, and 175 mM KCl. Thiostrepton (CalBiochem) binding experiments were carried out in the same buffer, substituting NH4Cl for KCl and including 5% DMSO. Melting experiments were carried out in 10 mM HEPES pH 7.5, 5 mM MgSO4, and 100 mM NH4Cl, and are shown as the first derivative of absorbance vs. temperature data normalized to the absorbance observed at 15°C (21).
L11 and L11-C76 recognize only rRNA with intact tertiary structure (12). Therefore rRNA folding and protein binding are coupled reactions, as described by
| (1) |
where I is a partially folded RNA containing only secondary structure and F has the fully folded tertiary structure. KL11 is an intrinsic affinity of L11 for fully folded RNA and KF is the equilibrium constant for RNA folding in the absence of protein. This reaction scheme implies that the measured L11-RNA binding constant, Kapp, may depend on the stability of the RNA tertiary structure:
| (2) |
In the cases of RNA mutations that alter KF without affecting KL11, this equation has been used to estimate KF from measurements of Kapp.
To compare in vitro measurements of KL11 and KF with in vivo experiments, it is convenient to define a free energy that reflects the overall stability of the L11-RNA complex. There are two free energies that could be useful for this purpose. One is the apparent free energy of RNA folding in the presence of L11,
| (3) |
It must be true that KL11[L11] >> 1 under normal in vivo conditions, since ribosomes are nearly saturated with L11. In this case, the standard state free energy ([L11] = 1 M) for folding RNA in the presence of L11 becomes
| (4) |
This is also the free energy that is associated with the reaction I + L11 ⇌ F•L11; compared to equation 1, it assumes that the amount of RNA that is folded but not bound by protein (F) is small. This approximation should apply to all the mutant RNAs discussed here that do not change KL11. The other potentially useful measure of complex stability is the apparent free energy of L11 binding, which is simply the free energy associated with Kapp as expressed in equation (2),
| (5) |
In the event that KF << 1, the last term approaches zero and this equation reduces to equation 4. This approximation is valid for all the destabilizing mutations in Table I (ΔG°F(37°) > 0).
Table 1.
Effects of rRNA variants on RNA tertiary structure stability, L11 binding affinity, and cell growth ratesa
| L11BD RNA 3° structureb | L11-RNA affinitiesc | Relative RNA Stabilityd | Relative growth rates, mutant/wild-typee | ||||
|---|---|---|---|---|---|---|---|
| Mutant | Tm °C | ΔH°F kcal/mol | ΔG°F(37°) kcal/mol | ΔG°L11(0°) kcal/mol | ΔΔG°F(37°) kcal/mol | TA531 | MC1000 |
| Wild-type | 41.5 | −30.5 | −0.4 | −8.8(Eco);−8.7(Bst) | 0 | 1 | 1 |
| U1061A | 65.7 | −57.9 | −4.9 | −8.8(Eco);−9.3(Bst) | −4.4 | 1.1 | 1.0 |
| A1085U | -- | -- | -- | > −7.5(Eco) | 4.6f | 1.0 | ND |
| A1085U/U1061A | 43.3 | −37.2 | −0.74 | −8.6 (Bst) | −0.30 | 1.0 | ND |
| C1072U | -- | -- | -- | > −6.3 (Bst) | 9.9f | Lethal | 0.53 |
| C1072U/U1061A | -- | -- | -- | −7.8 (Bst) | 5.0g | 0.85 | 0.83 |
| A1088U | -- | -- | -- | ND | 4.3f | Lethal | 0.74 |
| A1088U/U1061A | 45.3 | −38.5 | −1.0 | > −6.2 (Bst) | −0.6 | 0.84 | 0.91 |
| AA1090GU | 62.0 | −30.0 | −2.2 | −9.3 (Eco) | −1.8 | 1.0 | ND |
Dashes indicates that values could not be extracted from experimental data; ND means the experiment was not run.
Thermodynamic parameters for RNA tertiary structure unfolding were extracted from melting profiles such as those shown in Figure 2. ΔH° and ΔG° values are for the folding reaction. Values for wild-type, U1061A, A1085U, A1088U, AND AA1090GU RNAs were reported by (14); C1072U/U1061A was reported by (18).
L11 binding affinities were obtained from filter binding assays carried out at 0° C as described in Materials and Methods. “Eco” refers to measurements with the E. coli L11 protein, reported by (45). “Bst” are measurements made with the L11 C-terminal domain from B. stearothermophilus, reported by (38) (wild-type), (18) (C1072U/U1061A), and this work (the remainder). In some cases binding was too weak to measure and only an upper limit on the association constant could be estimated.
Stability of each variant L11BD RNA tertiary structure relative to that of the wild type L11BD RNA was estimated from melting curves and L11 binding affinities as described in the text.
Reference doubling times were 64 ± 5 min (TA531-pNO2680) or 25 ± 5 min (MC1000-pNO2680). In order to account for variability in growth curve measurements, mutant growth rates are reported relative to wild-type growth rates measured in the same experiment. “Lethal” means transformants with the mutant plasmid could not be obtained.
Relative stability estimated from that of the corresponding double mutation by adding 4.9 kcal/mol for the stability contributed by U1061A.
Stability estimated from reduced affinity of L11-C76 relative to wild type RNA, and extrapolation to 37 °C using ΔH°F for wild type RNA (14).
We will use ΔG°tot (equation 4) as a convenient expression for the overall stability of the L11-RNA complex. As long as the large majority of the RNA molecules are either unfolded (I) or folded and bound to L11 (F•L11), the fraction of RNAs that are folded and the fraction of RNAs bound to L11 are essentially the same value and it is valid to refer to ΔG°tot as the stability of the “complex” . Potential exceptions to this approximation are the two rRNAs incorporating A1088U, which decreases the intrinsic L11 binding affinity to an undetectable level. Equation 3 implies that when KL11[L11] < 1, L11 has little effect on the apparent RNA folding free energy and a significant concentration of F could be present. If the in vivo value of KL11[L11] is on the order of 100 (to insure ribosome saturation with L11), then mutations that decrease KL11 can increase ΔG°F,app by no more than ∼3 kcal/mol. This limitation has no effect on the conclusions that will be drawn from measurements with A1088U RNAs.
In comparing stabilities of different mutant L11-RNA complexes, we have assumed that the intracellular concentration of L11 is unaffected by the mutation. Since L11 is not involved in autoregulatory pathways (22) and all the viable mutations maintain a full complement of L11 on the ribosomes, this assumption seems reasonable.
23S rRNA variants in E. coli MC1000 and TA531 cells
23S rRNA mutations were first constructed in pCM10, a pUC18 derivative containing the SacI-SphI fragment of pNO2680 (23) using the QuikChange mutagenesis kit (Stratagene). Mutant SacI-SphI fragments were ligated with pNO2680 that had been cut with the same restriction enzymes. Mutant pNO2680 was transformed into E. coli MC1000 (ATCC 39351, a λ lysogen expressing the λ cI857 temperature sensitive repressor). E. coli MC1000 growth rates were measured in Luria Broth (24) by diluting saturated overnight cultures 1:100 into fresh medium and monitoring optical density at 550 nm. Cultures were grown at 30°C until OD550= 0.2 at which time the cells were transferred to 42°C for the remainder of the growth curve. E. coli TA531 was transformed as described (25), and growth rates were measured in a similar manner at 37°C. Plasmid was isolated and sequenced at the end of growth curves with either MC1000 or TA531 cells, to make sure mutations had been stably maintained . The presence of all ribosomal proteins in mutant TA531- derived ribosomes was confirmed by two-dimensional gel analysis (26) of isolated ribosomes (27).
Growth Competition Experiments
In growth competitions, a single flask of LB medium (24) was inoculated with 1:200 dilutions of saturated cultures of TA531 cells transformed with either wild type or mutant pNO2680. Viability of the stock culture was confirmed by measuring plating efficiencies. The mixed culture was allowed to grow to saturation overnight, and used the next day to inoculate a fresh culture with a 1:100 dilution. This process was repeated 9−10 times. Plasmid DNA was extracted from cells taken from each day's saturated culture and sequenced through the 58 nucleotide region. The fraction of cells with mutant or wild-type ribosomes was estimated from peak heights at the relevant position on the output of an ABI automated sequencer (Johns Hopkins School of Medicine Core Facility).
Results
Characterization of tertiary structure stability in mutant L11BD RNAs
Our object is to modify the stability of the L11BD – L11 protein complex by mutating conserved tertiary interactions, and then correlate relative stabilities measured in vitro with the effects of these mutations on L11-ribosome association in vivo and on cell growth rate. To ask whether the deleterious effect of a mutation is intrinsic to the base change or the result of rRNA destabilization, each destabilizing mutation has also been combined with a compensatory mutation, U1061A, that stabilizes the rRNA by ∼4.3 kcal/mol. This stabilization is primarily due to disruption of a U1061-A1077 base pair which competes for tertiary structure formation (14). Since U1061A destabilizes a competing structure, its effect on the overall free energy of tertiary structure folding is expected to be additive with mutations that disrupt the folded structure.
There are four tertiary base pairings in L11BD RNA (Figure 1A), all of them universally conserved (9, 28). Three of these were mutated. A1085U disrupts a minor groove base triple with G1055-C1104, C1072U breaks a highly conserved base triple with C1092-G1099, and A1088U disrupts an intercalated Hoogsteen base pair, A1088-U1060. The latter mutation also disrupts hydrogen bonds between L11 and the Hoogsteen base pair.
UV melting profiles were used to assess the stability of the tertiary structures of these RNAs. As shown in previous work (14), tertiary unfolding in this RNA is identified by a transition that appears at 260 nm but is absent at 280 nm (Figure 2A). The melting temperature (Tm) and enthalpy (ΔHF°) for the tertiary unfolding transition can be extracted from the two wavelength melting profiles by fitting one tertiary and three secondary structure folding transitions (Table 1) (14, 21). These parameters were used in the van't Hoff equation to calculate ΔGF° for folding the RNA tertiary structure at 37 °C (Table 1).
Figure 2.
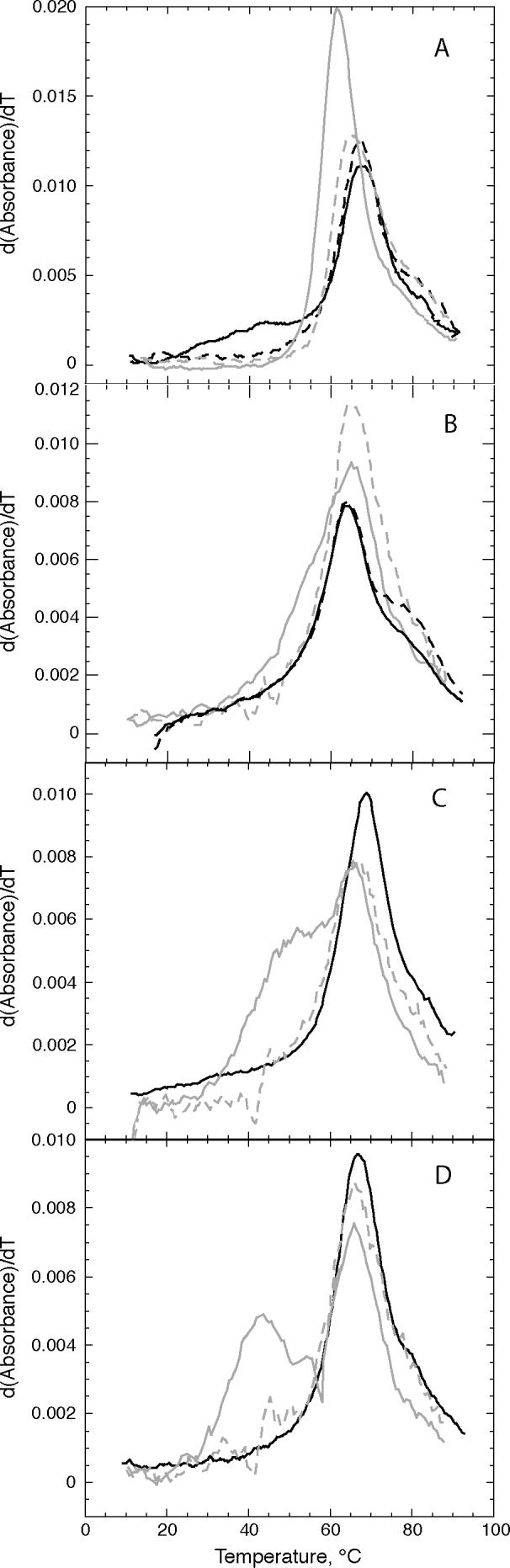
UV melting profiles of wild-type and mutant L11BD RNAs. 260 nm data are solid lines and 280 nm data are dashed; the tertiary unfolding transition is detected at 260 nm but not 280 nm. A, wild-type (black) (14) and U1061A (gray) (14) . B, C1072U (black) and C1072U/U1061A (gray) (18). C, A1088U (black) (14) and A1088U/U1061A (gray). D, A1085U (black) (14) and A1085U/U1061A (gray). Only 260 nm data are shown for A1088U and A1085U RNAs, taken from (14). That paper also examined the effect of Mg2+ and monovalent ions on the shape of the melting profile in concluding that the RNAs have no tertiary unfolding transition.
None of the L11BD RNAs with single mutations (A1085U, A1088U, or C1072U RNA) showed evidence of tertiary structure unfolding in melting experiments (Figure 2B-D; (14)). The U1061A RNA variant is very stable (Figure 2A) and compensated for the destabilizing effects of A1088U and A1085U to the extent that the double mutants are slightly more stable than the wild-type E.coli sequence (Figure 2C,D and Table 1). There is evidence for some tertiary structure unfolding in the melting profile of the C1072U/U1061A RNA, but the transition is too broad for reliable calculation of its ΔGF° (Figure 2B). On the basis that the stabilizing effect of U1061A is additive with the destabilizing effects of these tertiary structure mutations, the estimated relative stabilities of A1085U and A1088U RNA variants are shown in Table 1 (ΔΔGF(37°)).
Effects of the mutations on L11-binding to the L11BD RNA
For each of the single and double mutations of the L11BD RNA, association constants were determined by filter binding assays for interaction with either E. coli L11 or B. stearothermophilus L11-C76, the C-terminal RNA binding domain of L11 (20). These two proteins bind the L11BD RNA with slightly different affinities; as this study is only concerned with relative affinities for wild type and mutant RNAs, this difference is inconsequential. ΔG°L11(0°), the standard binding free energy change at the assay temperature of 0 °C, is listed for each mutation in Table 1.
Since L11 recognizes the rRNA tertiary structure, destabilization of the RNA fold must correspondingly weaken L11 binding (12, 20). This is apparently the case for A1085U, which weakens L11 binding by >1.3 kcal/mol at 0°C and destabilizes rRNA tertiary structure by about 4.2 kcal/mol at 37°C (Table 1). The two measurements are in agreement if the RNA unfolding enthalpy (ΔH°F) is more negative than −21 kcal/mol. This is consistent with a previous estimate of ΔH°F ≈ −30 kcal/mol for tertiary folding of the E.coli L11BD RNA (14). Since A1085 does not directly contact L11 (9), it is reasonable that the effect of A1085U on RNA tertiary structure stability entirely explains its effect on L11 binding affinity. In A1085U/U1061A RNA, both the tertiary structure stability and the L11-C76 binding affinity are restored to wild-type levels (Table 1). Thus we argue that the A1085U mutation weakens L11 binding because of its effect on tertiary structure stability, not because 1085U affects L11 contacts with the folded RNA.
The C1072U mutation also weakens both tertiary structure stability and L11 binding affinity without directly altering any L11-RNA contacts. The mutation results in no detectable binding of L11-C76 to the RNA by a filter binding assay; since we estimate the measurable lower limit of KL11 as ∼0.1 μM-1, we deduce that L11 binding to C1072U RNA is at least 2.4 kcal/mol weaker than its binding to wild-type RNA. A very broad tertiary unfolding transition appeared when C1072U RNA was melted in the presence of 5 μM L11-C76 (Tm ≈ 40 °C; data not shown). This suggests that L11 can promote folding of this RNA, but it was not possible to deduce a binding constant from these data. However, the double mutation C1072U/U1061A restores detectable L11 affinity for the RNA (Table 1). This affinity was used to estimate that the RNA is 0.9 kcal/mol less stable than wild type RNA at 0 °C, which extrapolates to 5.0 kcal/mol less stable at 37 °C (using ΔH°F = −30 kcal/mol and assuming that C1072U does not affect the intrinsic protein-RNA affinity).
The A1088U mutation is especially interesting in respect to L11 binding, as it disrupts both RNA tertiary structure and L11-RNA hydrogen bonds (9). No L11 binding to either A1088U or A1088U/U1061A RNAs could be detected (Table 1 and Figure 3), even though melting curves suggest that tertiary structure is restored in the double mutant. As further evidence that A1088U/U1061A RNA is properly folded, the antibiotic thiostrepton bound mutant RNA (K ≈ 0.31 μM-1) with about the same affinity as it bound U1061A RNA (K ≈ 0.48 μM-1) (Figure 3). Thiostrepton binds and stabilizes the same RNA tertiary structure as L11 (20, 29, 30).
Figure 3.
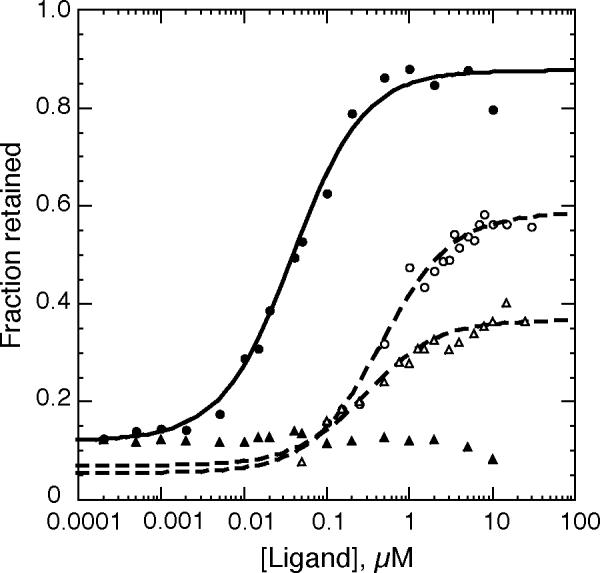
Filter binding assays of L11-C76 and thiostrepton binding to U1061A and A1088U/U1061A RNAs. ●, L11 binding to U1061A RNA; K = 30 μM-1. ○, thiostrepton binding to U1061A RNA; K = 2.1 μM-1. ▴: L11 binding to A1088U/U1061A RNA. ▵: thiostrepton binding to A1088U/U1061A RNA; ; K = 3.2 μM-1. Assay conditions are given in Materials and Methods.
Effects of rRNA mutations in vivo
The same series of ribosomal RNA mutations as described above were introduced into pNO2680, a plasmid which carries the rrnB operon under control of a λ PL promoter (23). The growth rates of two different E.coli strains transformed with the variant pNO2680 plasmids were measured. The first strain, MC1000, has a temperature sensitive λ repressor which controls transcription of the pNO2680 rRNA genes. However, the plasmid genes are always expressed with a 20−50% background of wild-type ribosomes present (23, 31). The second strain, TA531, lacks all seven chromosomal copies of the ribosomal RNA operon; a homogenous population of ribosomes is synthesized from a plasmid (25). Relative growth rates are summarized in Table 1.
TA531 transformants were not obtained with two of the RNA destabilizing mutants, C1072U and A1088U. In addition, growth of these mutants in MC1000 was significantly slower (Table 1). Thus, C1072U and A1088U are deleterious when carried by a fraction of the ribosomes in a cell and lethal when present in a homogenous population of ribosomes. (Here “lethal” means that transformants with the mutated plasmid cannot be found; we estimate that transformants growing 10 fold slower than wild type would be very hard to detect.)
Both the C1072U and A1088U mutations were complemented by U1061A. MC1000 cells carrying either double mutation grew faster than cells with the single mutation but slower than wild-type (Table 1). The combination of either mutation with U1061A also resulted in viable TA531 cells, though the double mutants did not grow at wild-type rates. In two-dimensional gel analysis (26) of the protein complement of ribosomes extracted from TA531 cells containing A1088U/U1061A or C1072U/U1061A mutations, L11 is present on the ribosome in approximately the same stoichiometry (estimated relative to neighboring proteins on the gel) as in wild type ribosomes (data not shown).
Lastly, TA531 cells expressing A1085U rRNA grew at a wild-type rate, despite the substantial RNA destabilization caused by this mutation. The double mutation, A1085U/U1061A, also had a wild-type growth rate (Table 1).
Growth competition experiments
TA531 cells containing the U1061A mutation appeared to grow faster than wild type cells by about 10%, a difference which is close to the confidence level of the measurements (Table 1). To see if this unusual result is reliable, cultures initially containing approximately equal numbers of TA531 cells with wild type pNO2680 or pNO2680-U1061A were repeatedly grown to saturation, diluted 100 fold, and re-grown (see Materials and Methods). Cells containing U1061A consistently overtook the culture of cells with wild type rRNA in a number of generations consistent with approximately 15% faster growth. To see if other mutations stabilizing the rRNA tertiary structure had a similar effect, the variants AA1090GU and U1061G were introduced into pNO2680 and examined in growth competitions against TA531 cells with wild type pNO2680. Neither of these mutations altered the growth rate relative to wild type cells. TA531 cells expressing A1085U were also tested in growth competition experiments. The mutant unexpectedly outgrew wild-type cells with 3−7% faster growth rate.
Discussion
Stability of the L11 – rRNA complex
In this work we have examined the destabilizing effects of mutations at each of three conserved tertiary interactions in a small, independently folding rRNA domain, as well as two mutations that stabilize the same tertiary structure. From melting experiments and measurements of L11 binding affinity for RNAs carrying combinations of stabilizing and destabilizing mutations, the relative stabilities of all the variants could be estimated (ΔΔG°F(37°) in Table 1). We first discuss some general implications of these data, and then comment on the three selective pressures mentioned in the Introduction as potentially having influenced the sequence and stability of this domain.
Our first conclusion from Table 1 data is that the L11-rRNA domain is critical, if not essential, for efficient ribosome function in vivo: mutations that destabilize the rRNA tertiary structure sufficiently are also lethal. This result was expected, since deletion of L11 from the E. coli chromosome slows growth dramatically, by about seven-fold (32). Mutations that destabilize the rRNA may slow growth by an even larger factor, since unfolding of the RNA may disrupt both rRNA and L11 interactions with elongation factors (7, 33).
We also argue from Table 1 data that it is the stability of the L11-RNA complex, rather than any specific RNA tertiary contact, which has been selected during evolution: none of the tertiary interactions is in itself essential for function as long as the overall stability of the L11-RNA complex is maintained above a certain level. Figure 4 illustrates this argument by showing a correlation between L11-RNA stability and cell growth rate. The ordinate in this graph is the overall stability of the complex, ΔG°tot = ΔG°F + ΔG°L11, relative to the wild-type complex (see equation 4 and its derivation in Materials & Methods for a discussion of how this quantity reflects the stability of the complex.) For most of the RNAs, the intrinsic affinity of L11 for folded RNA is unchanged and values of ΔΔG°F(37°) from Table 1 are plotted as horizontal bars (gray for A1088U variants, black for the others). Among the mutations examined, A1088U is the only one that affects the intrinsic L11 binding affinity as well as the stability of the RNA tertiary fold. The gray arrows in Figure 4 account for the additional destabilization (ΔΔG°L11) caused by the disruption of protein contacts in RNAs containing A1088U. (Ranges of free energies, indicated by boxes, are shown because of uncertainties in the extent to which the L11 binding constant is affected and the maximum possible effect of the decrease on the stability of the complex in vivo; see Materials & Methods for further discussion.) Thus the black bars (or boxes) at highest free energy for each of the RNA variants in Figure 4 represent the relative stabilities of the L11 – RNA complexes. (This comparison is strictly valid only when L11 concentrations are saturating, but this condition holds for all the viable mutants; see Materials and Methods.) The dashed horizontal line divides the rRNA variants into two groups based on the growth rates of TA531 cells expressing them: L11-rRNA complexes with stabilities differing by −4 to +5 kcal/mol from wild type grow at 85 − 100% of the normal rate, while destabilization in excess of ∼7 kcal/mol is lethal. Lethality cannot be the consequence of a specific base change per se, since each deleterious mutation is benign when combined with a stabilizing mutation that brings the overall relative stability of the complex back below ∼5 kcal/mol.
Figure 4.
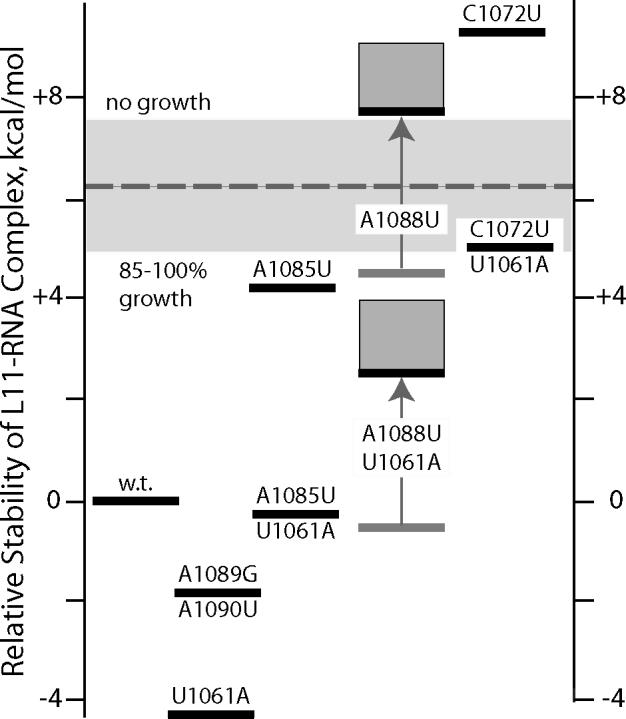
Energy level diagram for rRNA domains and L11-rRNA complexes for all the rRNA variants studied. The relative free energy plotted on the ordinate is ΔΔG°tot = ΔΔG°F + ΔΔG°L11 where the free energies are reported relative to ΔG°tot for wild type rRNA (equation 4, Materials and Methods). ΔΔG°F is taken from Table 1. It is assumed that L11 stabilizes all the rRNA variants by the same amount as wild type (ΔΔG°L11 = 0), with the exception of RNAs containing the A1088U variant; weaker binding of these RNAs is indicated by the gray arrows (shaded boxes indicate uncertainty in the amount of destabilization; see text). The dashed horizontal line divides the variants into two groups, based on the growth rates of cells expressing rRNA with the mutations; the same line represents a hypothetical mutant whose growth rate is 50% of wild type and has 50% of ribosomes with an unfolded L11BD. The wide gray bar is the stability range over which growth rate and the extent to which the L11BD is folded are predicted to vary from 90% (lower edge) to 10% of wild type.
The way rRNA mutations switch from benign to lethal over a range of a few kcal/mol in complex stability is consistent with a simple model which assumes that (i) L11 associates and dissociates from ribosomes in an equilibrium manner in vivo, (ii) ribosomes lacking L11 are essentially inactive (32), and (iii) cell growth is roughly proportional to the fraction of active ribosomes.2 In this model, the dashed line dividing viable from lethal mutations in Figure 4 corresponds to the free energy needed to disrupt half of the L11-ribosome complexes in vivo. From this half-dissociation free energy, increasing or decreasing the L11BD RNA stability by 1.3 kcal/mol should vary the equilibrium extent of L11 association with ribosomes from 10% to 90% (see equation 2 in Materials & Methods). This free energy range, shown in Figure 4 as a gray box centered on the 50% dissociation line, corresponds to the gap between the stabilities of C1072U/U1061A (viable) and A1088U (lethal) mutations. Thus the effective in vivo stability of the complex (the free energy needed to move from the 50% dissociation line to the free energy of the wild-type complex in Figure 4) is on the order of −6 kcal/mol. The implications of this value are considered in the following section.
Selective pressures operating on the L11-rRNA complex
The finding that increased stability of the rRNA domain is not deleterious (U1061A and AA1090GU RNAs; Figure 4) argues against the first two explanations suggested in the Introduction for the instability of the E. coli rRNA domain. One possibility was that sequence variants conferring stability might also disrupt a regulatory or functional interaction specific to bacteria maintaining U1061 and AA1089−1090. The unaltered growth rate of cells with these mutations does not support this idea, though it is impossible to rule out the possibility that these sequences become essential under some set of environmental conditions not duplicated in the laboratory. The maintenance of U1061 in all bacteria, while archaea have all four possible bases at this position (28), could in fact be interpreted to mean that bacteria have evolved some functional requirement for U1061 that is missing in archaea. However, the widespread occurrence of both AA and GU1089−1090 variants in bacteria and archaea is more difficult to explain in terms of a specific functional requirement. Another possibility was that a very stable rRNA structure could slow translation by inhibiting an essential conformational switch. Although “open” and “closed” forms of this domain were once postulated to account for the effects of thiazole antibiotics on the function of the domain (34), six independent crystal structures of the domain do not hint at any alternative RNA conformations within the domain despite varying orientations of the domain with respect to the rest of the large subunit (4, 5, 9, 10, 35). This observation, and the lack of any detrimental effect of stabilizing mutations on cell growth rate, argue that this region cannot be made too stable for ribosome function.
We are then left with the third possibility, that the L11-rRNA complex is already so stable in vivo that there is little or no selective pressure to adopt further stabilizing mutations. A stability of −6 kcal/mol means that only 1 − 3 ribosomes in an exponentially growing E. coli cell (out of a total of 3 − 7 × 104 ribosomes; (36)) will be dissociated from L11 at any one time.3 If growth rate is proportional to the fraction of active ribosomes, then selection for further increases in stability can result in a maximum ∼0.01% increase in growth rate. On this basis, it might seem reasonable to argue that the E. coli L11BD is subject to a vanishingly small selective pressure for increased stability.
The above argument begs the question of how small a selective advantage needs to be before it is inconsequential: over the extraordinary time scale of evolution, sequence changes conferring selective advantages of much less than 0.01% should have become widespread. However, even though we have argued that growth rate is roughly proportional to the saturation of ribosomes by L11, it is simplistic to think that this relationship will hold for mutations causing very small changes in L11-ribosome association. Selection is ultimately a multidimensional optimization problem. Most mutations probably affect cell growth in more than one way; rRNA mutations could, for instance, affect the efficiency of ribosome assembly or the overall regulation of ribosome synthesis. There is good evidence that rRNA mutations have pleiotropic effects: both U1061A and A1085U variants grow a few percent faster than cells with wild type ribosomes, despite the opposite effects of these mutations on RNA stability; others have found rRNA mutations that confer a slight growth advantage in rich growth medium but are disadvantageous in minimal medium (37). We therefore argue that changes in L11-rRNA stability that might be expected to enhance protein synthesis rates by 1% may also have subtle deleterious consequences of the same magnitude. The selective pressure for enhanced stability continues to operate even when the advantage is much less than 1%, but the effects become essentially undetectable against the background of competing factors that optimize cell growth.
The view of L11BD RNA evolution presented here has several implications for evolution of the ribosome and the general question of evolution of stability in macromolecules: (i) If overall stability of the L11-rRNA complex is subject to selective pressure, then compensation between the intrinsic L11-rRNA binding affinity and the rRNA stability might have taken place during evolution. A possible instance of this has been seen in a comparison of bacterial and chloroplast L11BDs (38). Chloroplast ribosomes have compensated for a change in the L11 sequence by a base pair substitution; though the base pair is destabilizing it has in turn been compensated by the stabilizing GU1089−1090 substitution. In another instance, we have found that eukaryotic L11 orthologs have much weaker intrinsic affinity for their cognate rRNAs than found in the E. coli system, but the eukaryotic rRNA sequences are correspondingly more stable (E. Poliakova & DED, ms. in preparation). Thus compensatory changes at several positions in the rRNA and protein have maintained about the same overall stability of the complex in diverse mesophilic organisms.
(ii) A surprising result of our studies is that mutations at universally conserved residues may have no apparent phenotype. Our rationalization for this observation is that the L11-rRNA complex has been selected for such a high degree of stability that it has a high tolerance for disruptive mutations. Since the ribosome has been evolving for a very long time, one might expect that many rRNA sequences that confer relatively small advantages to the organism have become universally adopted. In fact, mutations at highly conserved rRNA residues often have no apparent defect (39-41).
(iii) The same question about natural selection and the stability of the L11-rRNA complex that has been posed here has also been asked about globular proteins: if protein stability can be enhanced by directed mutagenesis (15), why has natural selection not generated more stable proteins? There may be strong functional limitations on the stabilities of protein enzymes; for instance, the energy required to maintain an active site (e.g., a pK shift in a catalytic residue) detracts from the overall folding free energy (42), and the conformational flexibility needed for catalysis may be inconsistent with a very stable structure (17). Thus it is generally thought that protein enzymes have been selected for an optimum stability consistent with function, rather than a maximum stability. But most proteins have similar or even greater stabilities than the −6 kcal/mol estimated for the L11-rRNA complex: protein enzymes commonly denature at temperatures 30 − 50 deg above their adaptive temperatures (15, 43) and mesophilic globular proteins tend to have stabilities in the range of −5 to −15 kcal/mol at 37 °C (44). We have argued that natural selection is unable to push the L11-rRNA complex to greater stability because the advantages of any further stability increases are outweighed by other factors not directly related to the function of the complex. It is worth considering whether this conclusion might apply to proteins too. Even though folding free energies of −5 to −15 kcal/mol may seem marginally stable to a physical chemist, they are large enough that if a bacterium had 1000 copies of a protein, only 1 in ∼104 bacteria would have a single unfolded protein at any one time. The selective advantage for further stability is extremely small and perhaps comparable to other subtle effects a stabilizing mutation might have on cell growth.
Acknowledgements
We thank Drs. Tamara Hendrickson and Dan Grilley for helpful comments and Dr. Rachel Green for help with two-dimensional gels.
Footnotes
This work was supported by NIH grant R37 GM29048.
Abbreviation used: L11BD, L11-binding domain (nucleotides 1051−1108 of E. coli 23S rRNA).
Cell growth rate might be unaffected by mutations causing a decrease in ribosome efficiency if cells have excess protein synthesis capacity. However, a number of careful studies have shown that protein synthesis is limiting for E. coli growth (1-3).
Macromolecular crowding and osmolytes undoubtedly cause free energy values relevant in vivo to differ from those measured in vitro. However, the estimate of the fraction of L11BD complexes that are folded is based on the difference in free energy between wild type ribosomes and a hypothetical L11BD mutant with 50% growth rate (+6.3 kcal/mol in Figure 4). Therefore the assumption being made in this estimate is that in vivo conditions have affected wild type and mutant ribosomes to the same degree, which seems reasonable.
References
- 1.Jensen KF, Pedersen S. Metabolic growth rate control in Escherichia coli may be a consequence of subsaturation of the macromolecular biosynthetic apparatus with substrates and catalytic components. Microbiol Rev. 1990;54:89–100. doi: 10.1128/mr.54.2.89-100.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pedersen S. Escherichia coli ribosomes translate in vivo with variable rate. Embo J. 1984;3:2895–8. doi: 10.1002/j.1460-2075.1984.tb02227.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sørensen MA, Kurland CG, Pedersen S. Codon usage determines the translation rate in Escherichia coli. J. Mol. Biol. 1989;207:365–377. doi: 10.1016/0022-2836(89)90260-x. [DOI] [PubMed] [Google Scholar]
- 4.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000;289:905–20. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 5.Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH. Structures of the bacterial ribosome at 3.5 A resolution. Science. 2005;310:827–34. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 6.Agrawal RK, Linde J, Sengupta J, Nierhaus KH, Frank J. Localization of L11 protein on the ribosome and elucidation of its involvement in EF-G-dependent translocation. J Mol Biol. 2001;311:777–87. doi: 10.1006/jmbi.2001.4907. [DOI] [PubMed] [Google Scholar]
- 7.Valle M, Zavialov A, Li W, Stagg SM, Sengupta J, Nielsen RC, Nissen P, Harvey SC, Ehrenberg M, Frank J. Incorporation of aminoacyl-tRNA into the ribosome as seen by cryo-electron microscopy. Nat Struct Biol. 2003;10:899–906. doi: 10.1038/nsb1003. [DOI] [PubMed] [Google Scholar]
- 8.Xing Y, GuhaThakurta D, Draper DE. The RNA binding domain of ribosomal protein L11 is structurally similar to homeodomains. Nat Struct Biol. 1997;4:24–27. doi: 10.1038/nsb0197-24. [DOI] [PubMed] [Google Scholar]
- 9.Conn GL, Draper DE, Lattman EE, Gittis AG. Crystal Structure of a Conserved Ribosomal Protein - RNA Complex. Science. 1999;284:1171–1174. doi: 10.1126/science.284.5417.1171. [DOI] [PubMed] [Google Scholar]
- 10.Wimberly BT, Guymon R, McCutcheon JP, White SW, Ramakrishnan V. A detailed view of a ribosomal active site: the structure of the L11- RNA complex. Cell. 1999;97:491–502. doi: 10.1016/s0092-8674(00)80759-x. [DOI] [PubMed] [Google Scholar]
- 11.Laing LG, Draper DE. Thermodynamics of RNA Folding in a Highly Conserved Ribosomal RNA Domain. J. Mol. Biol. 1994;237:560–576. doi: 10.1006/jmbi.1994.1255. [DOI] [PubMed] [Google Scholar]
- 12.Xing Y, Draper DE. Stabilization of a ribosomal RNA tertiary structure by ribosomal protein L11. J Mol Biol. 1995;249:319–31. doi: 10.1006/jmbi.1995.0299. [DOI] [PubMed] [Google Scholar]
- 13.Rosendahl G, Douthwaite S. Ribosomal proteins L11 and L10.(L12)4 and the antibiotic thiostrepton interact with overlapping regions of the 23 S rRNA backbone in the ribosomal GTPase centre. J Mol Biol. 1993;234:1013–20. doi: 10.1006/jmbi.1993.1655. [DOI] [PubMed] [Google Scholar]
- 14.Lu M, Draper DE. Bases Defining an Ammonium and Magnesium Ion-Dependent Tertiary Structure Within the Large Subunit Ribosomal RNA. J. Mol. Biol. 1994;244:572–585. doi: 10.1006/jmbi.1994.1753. [DOI] [PubMed] [Google Scholar]
- 15.Somero GN. Proteins and temperature. Annu Rev Physiol. 1995;57:43–68. doi: 10.1146/annurev.ph.57.030195.000355. [DOI] [PubMed] [Google Scholar]
- 16.Gruber T, Kohrer C, Lung B, Shcherbakov D, Piendl W. Affinity of ribosomal protein S8 from mesophilic and (hyper)thermophilic archaea and bacteria for 16S rRNA correlates with the growth temperatures of the organisms. FEBS Lett. 2003;549:123–8. doi: 10.1016/s0014-5793(03)00760-9. [DOI] [PubMed] [Google Scholar]
- 17.Fields PA. Review: Protein function at thermal extremes: balancing stability and flexibility. Comp Biochem Physiol A Mol Integr Physiol. 2001;129:417–31. doi: 10.1016/s1095-6433(00)00359-7. [DOI] [PubMed] [Google Scholar]
- 18.Conn GL, Gutell RR, Draper DE. A Functional Ribosomal RNA Tertiary Structure Involves a Base Triple Interaction. Biochemistry. 1998;37:11980–11988. doi: 10.1021/bi980825+. [DOI] [PubMed] [Google Scholar]
- 19.Draper DE, Deckman IC, Vartikar JV. Physical Studies of Ribosomal Protein - RNA Interactions. Meth. Enzymology. 1988;164:203–220. doi: 10.1016/s0076-6879(88)64044-4. [DOI] [PubMed] [Google Scholar]
- 20.Xing Y, Draper DE. Cooperative interactions of RNA and thiostrepton antibiotic with two domains of ribosomal protein L11. Biochemistry. 1996;35:1581–8. doi: 10.1021/bi952132o. [DOI] [PubMed] [Google Scholar]
- 21.Draper DE, Bukhman YV, Gluick TC. Thermal Methods for the Analysis of RNA Folding Pathways. In: Beaucage SL, Bergstrom DE, Glick GD, Jones RA, editors. Current Protocols in Nucleic Acid Chemistry. John Wiley & Sons; New York: 2000. p. section 11.3. [DOI] [PubMed] [Google Scholar]
- 22.Nomura M, Gourse R, Baughman G. Regulation of the Synthesis of Ribosomes and Ribosomal Components. Ann. Rev. Biochem. 1984;53:75–117. doi: 10.1146/annurev.bi.53.070184.000451. [DOI] [PubMed] [Google Scholar]
- 23.Gourse RL, Takebe Y, Sharrock RA, Nomura M. Feedback regulation of rRNA and tRNA synthesis and accumulation of free ribosomes after conditional expression of rRNA genes. Proc Natl Acad Sci U S A. 1985;82:1069–73. doi: 10.1073/pnas.82.4.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Press; 1989. [Google Scholar]
- 25.Asai T, Zaporojets D, Squires C, Squires CL. An escherichia coli strain with all chromosomal rRNA operons inactivated: complete exchange of rRNA genes between bacteria. Proc Natl Acad Sci U S A. 1999;96:1971–6. doi: 10.1073/pnas.96.5.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Agafonov DE, Kolb VA, Nazimov IV, Spirin AS. A protein residing at the subunit interface of the bacterial ribosome. Proc Natl Acad Sci U S A. 1999;96:12345–9. doi: 10.1073/pnas.96.22.12345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spedding G. In: Ribosomes and Protein Synthesis: A Practical Approach. Spedding G, editor. Oxford University Press; Oxford: 1990. pp. 1–27. [Google Scholar]
- 28.Cannone JJ, Subramanian S, Schnare MN, Collett JR, D'Souza LM, Du Y, Feng B, Lin N, Madabusi LV, Muller KM, Pande N, Shang Z, Yu N, Gutell RR. The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics. 2002;3:2. doi: 10.1186/1471-2105-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Draper DE, Xing Y, Laing LG. Thermodynamics of RNA unfolding: stabilization of a ribosomal RNA tertiary structure by thiostrepton and ammonium ion. J Mol Biol. 1995;249:231–8. doi: 10.1006/jmbi.1995.0291. [DOI] [PubMed] [Google Scholar]
- 30.Blyn LB, Risen LM, Griffey RH, Draper DE. The RNA-binding domain of ribosomal protein L11 recognizes an rRNA tertiary structure stabilized by both thiostrepton and magnesium ion. Nucleic Acids Res. 2000;28:1778–84. doi: 10.1093/nar/28.8.1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thompson J, Kim DF, O'Connor M, Lieberman KR, Bayfield MA, Gregory ST, Green R, Noller HF, Dahlberg AE. Analysis of mutations at residues A2451 and G2447 of 23S rRNA in the peptidyltransferase active site of the 50S ribosomal subunit. Proc Natl Acad Sci U S A. 2001;98:9002–7. doi: 10.1073/pnas.151257098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Dyke N, Murgola EJ. Site of functional interaction of release factor 1 with the ribosome. J Mol Biol. 2003;330:9–13. doi: 10.1016/s0022-2836(03)00537-0. [DOI] [PubMed] [Google Scholar]
- 33.Agrawal RK, Penczek P, Grassucci RA, Frank J. Visualization of elongation factor G on the Escherichia coli 70S ribosome: the mechanism of translocation. Proc Natl Acad Sci U S A. 1998;95:6134–8. doi: 10.1073/pnas.95.11.6134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cundliffe E. Involvement of Specific Portions of Ribosomal RNA. In: Hardesty B, Kramer G, editors. Defined Ribosomal Functions: A Study Utilizing Antibiotics, in Structure, Function, and Genetics of Ribosomes. Springer-Verlag; New York: 1986. pp. 586–604. [Google Scholar]
- 35.Harms J, Schluenzen F, Zarivach R, Bashan A, Gat S, Agmon I, Bartels H, Franceschi F, Yonath A. High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell. 2001;107:679–88. doi: 10.1016/s0092-8674(01)00546-3. [DOI] [PubMed] [Google Scholar]
- 36.Dennis PP, Bremer H. Macromolecular composition during steady-state growth of Escherichia coli B-r. J Bacteriol. 1974;119:270–81. doi: 10.1128/jb.119.1.270-281.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moine H, Squires CL, Ehresmann B, Ehresmann C. In vivo selection of functional ribosomes with variations in the rRNA-binding site of Escherichia coli ribosomal protein S8: evolutionary implications. Proc Natl Acad Sci U S A. 2000;97:605–10. doi: 10.1073/pnas.97.2.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.GuhaThakurta D, Draper DE. Protein- RNA Sequence Covariation in a Ribosomal Protein-rRNA Complex. Biochemistry. 1999;38:3633–3640. doi: 10.1021/bi9826411. [DOI] [PubMed] [Google Scholar]
- 39.Dragon F, Spickler C, Pinard R, Carriere J, Brakier-Gringas L. Mutations of non-canonical base-pairs in the 3' major domain of Escherichia coli 16 S ribosomal RNA affect the initiation and elongation of protein synthesis. J Mol Biol. 1996;259:207–15. doi: 10.1006/jmbi.1996.0313. [DOI] [PubMed] [Google Scholar]
- 40.O'Connor M, Lee WM, Mankad A, Squires CL, Dahlberg AE. Mutagenesis of the peptidyltransferase center of 23S rRNA: the invariant U2449 is dispensable. Nucleic Acids Res. 2001;29:710–5. doi: 10.1093/nar/29.3.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marchant A, Hartley MR. Mutational studies on the alpha-sarcin loop of Escherichia coli 23S ribosomal RNA. Eur J Biochem. 1994;226:141–7. doi: 10.1111/j.1432-1033.1994.tb20035.x. [DOI] [PubMed] [Google Scholar]
- 42.Shoichet BK, Baase WA, Kuroki R, Matthews BW. A relationship between protein stability and protein function. Proc Natl Acad Sci U S A. 1995;92:452–6. doi: 10.1073/pnas.92.2.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fields PA, Wahlstrand BD, Somero GN. Intrinsic versus extrinsic stabilization of enzymes: the interaction of solutes and temperature on A4-lactate dehydrogenase orthologs from warm-adapted and cold-adapted marine fishes. Eur J Biochem. 2001;268:4497–505. doi: 10.1046/j.1432-1327.2001.02374.x. [DOI] [PubMed] [Google Scholar]
- 44.Privalov PL, Gill SJ. Stability of protein structure and hydrophobic interaction. Adv Protein Chem. 1988;39:191–234. doi: 10.1016/s0065-3233(08)60377-0. [DOI] [PubMed] [Google Scholar]
- 45.Ryan PC, Lu M, Draper DE. Recognition of the Highly Conserved GTPase Center of 23 S Ribosomal RNA by Ribosomal Protein L11 and the Antibiotic Thiostrepton. J. Mol. Biol. 1991;221:1257–1268. doi: 10.1016/0022-2836(91)90932-v. [DOI] [PubMed] [Google Scholar]


