Abstract
Cap1p, a transcription factor of the basic region leucine zipper family, regulates the oxidative stress response (OSR) in Candida albicans. Alteration of its C-terminal cysteine-rich domain (CRD) results in Cap1p nuclear retention and transcriptional activation. To better understand the function of Cap1p in C. albicans, we used genome-wide location profiling (chromatin immunoprecipitation-on-chip) to identify its transcriptional targets in vivo. A triple-hemagglutinin (HA3) epitope was introduced at the C terminus of wild-type Cap1p (Cap1p-HA3) or hyperactive Cap1p with an altered CRD (Cap1p-CSE-HA3). Location profiling using whole-genome oligonucleotide tiling microarrays identified 89 targets bound by Cap1p-HA3 or Cap1p-CSE-HA3 (the binding ratio was at least twofold; P ≤ 0.01). Strikingly, Cap1p binding was detected not only at the promoter region of its target genes but also at their 3′ ends and within their open reading frames, suggesting that Cap1p may associate with the transcriptional or chromatin remodeling machinery to exert its activity. Overrepresented functional groups of the Cap1p targets (P ≤ 0.02) included 11 genes involved in the OSR (CAP1, GLR1, TRX1, SOD1, CAT1, and others), 13 genes involved in response to drugs (PDR16, MDR1, FLU1, YCF1, FCR1, and others), 4 genes involved in phospholipid transport (PDR16, GIT1, RTA2, and orf19.932), and 3 genes involved in the regulation of nitrogen utilization (GST3, orf19.2693, and orf19.3121), suggesting that Cap1p has other cellular functions in addition to the OSR. Bioinformatic analyses of the bound sequences suggest that Cap1p recognizes the DNA motif 5′-MTKASTMA. Finally, transcriptome analyses showed that increased expression generally accompanies Cap1p binding at its targets, indicating that Cap1p functions as a transcriptional activator.
Candida albicans is an opportunistic human fungal pathogen that causes superficial infections in healthy patients. However, in patients with impaired immunity C. albicans can cause life-threatening invasive infections, including systemic candidiasis or candidemia. In the United States, candidemia represents the fourth-most-common cause of nosocomial bloodstream infections (7). Several options for the treatment of invasive candidiasis are available to clinicians, including the administration of azole derivatives, amphotericin B preparations, or echinocandin antifungal agents, while for the treatment of mucocutaneous infections, azoles are preferred over other antifungals due to their low toxicity and increased efficacy and availability for both topical and oral use (21, 50).
Azoles, including both imidazoles (e.g., ketoconazole) and triazoles (e.g., fluconazole [FLC]), inhibit the function of the lanosterol demethylase enzyme Erg11p, a component of the ergosterol biosynthesis pathway, leading to methylsterol accumulation, sterol depletion, and consequently to growth arrest (1). This fungistatic property of azoles coupled to their repeated use in the clinic renders the surviving C. albicans cells prone to the selection of mutations conferring azole resistance. The clinical resistance of Candida spp. to azole is a challenging problem for clinicians. Azole resistance develops particularly in human immunodeficiency virus-infected patients with recurrent episodes of oropharyngeal or esophageal candidiases (18). The molecular mechanisms of clinical resistance to azole in C. albicans involve (i) mutations in the target of azoles, Erg11p, resulting in altered drug binding, and/or (ii) the constitutive overexpression of genes responsible for the drug resistance phenotype, including CDR1 and CDR2, which encode ATP binding cassette transporters; MDR1, which encodes a transporter of the major facilitator superfamily; PDR16, which codes for a phospholipid transferase; and/or ERG11 (1, 40, 46, 55, 56). Recent studies revealed the direct involvement in clinical azole resistance of gain-of-function mutations in genes encoding transcription factors of the fungus-specific zinc cluster family (42). It was shown that activating mutations in the transcription factor Tac1p (for transcriptional activator of CDR genes) leads to the constitutive overexpression of its target genes, CDR1, CDR2, and PDR16, in clinical isolates of C. albicans (15-17, 38, 70). Similarly, MDR1 constitutive overexpression is due to gain-of-function mutations in the zinc cluster transcription factor Mrr1p (for multidrug resistance regulator) (19, 47). Finally, an activating mutation in the transcription factor Upc2p (for uptake control) was shown to be responsible for clinical azole resistance and the upregulation of ERG11 and, to a lesser extent, MDR1 (20).
Genome-wide location and/or expression studies have shown that several targets of the transcription factors Tac1p, Upc2p, and Mrr1p have established or predicted roles in the oxidative stress response (OSR) (20, 38, 47, 71), suggesting that multidrug resistance and the OSR are interconnected processes in C. albicans. Studies of the OSR in Saccharomyces cerevisiae have shown that Yap1p, a basic region leucine zipper (bZIP) transcription factor homologous to mammalian activating protein 1 (AP-1), is a key regulator of this process (27, 48). Yap1p binds to Yap1 recognition elements (YRE, for Yap1 recognition elements) TTA(C/G)T(A/C)A, located in the promoter of its target genes (49), and controls the expression of genes encoding the majority of antioxidants and thiol-oxidoreductases, such as the glutathione reductase GLR1 and the thioredoxin reductase TRR1 (27, 48). Yap1p is also essential for the response to cadmium or drug exposures and can be activated by chemicals (e.g., diamide), antifungal agents (e.g., benomyl), ionizing radiation, or toxic endogenous cellular metabolites (e.g., methylglyoxal) (2, 27, 43, 45, 48, 49). Interestingly, Yap1p confers azole resistance in S. cerevisiae by activating the expression of FLR1, the functional homolog of C. albicans MDR1 (2). Yap1p is activated by a mechanism acting on its nuclear export (33, 34). In response to high levels of oxidants, Yap1p undergoes redox conformational changes caused by intramolecular bond formation between cysteine residues within the C-terminal cysteine-rich domain (CRD) of the protein (33, 34). This prevents the interaction of Yap1p with the nuclear exportin Crm1p, leading to nuclear retention and transcriptional activation (27, 35, 48, 67).
The C. albicans CAP1 gene encodes the functional homolog of Yap1p and has been isolated based on its ability to confer azole resistance when expressed in S. cerevisiae (2, 3, 68). The mechanisms whereby Cap1p exerts its function are reminiscent of S. cerevisiae Yap1p, as a truncation of the Cap1p CRD (CAP1-TR) or mutagenesis of the third cysteine residue of the Cap1p CRD (CAP1-C477A) results in enhanced resistance to toxic compounds, including azoles, the heavy metal cadmium, and the oxidative stress-inducing agent 4-nitroquinoline N-oxide (4-NQO), as well as Cap1p constitutive transcriptional activation and nuclear retention (3, 68). Interestingly, the Cap1p-TR protein was shown to constitutively activate MDR1 expression in azole-susceptible C. albicans cells, demonstrating that MDR1 is a direct or indirect target of Cap1p (3). However, deleting CAP1 in an azole-resistant strain overexpressing MDR1 did not decrease MDR1 RNA levels (3), indicating that another transcription factor, possibly Mrr1p, is responsible for the constitutive overexpression of MDR1 in that strain. In addition, Cap1p is involved in protecting C. albicans against the oxidative stress induced by neutrophils during the course of the immune response (13, 24). The OSR in C. albicans involves oxidant sensing and response to oxidative damage via two major pathways that appear to act distinctly, namely, the Cap1p pathway and the high osmolarity glycerol (HOG) mitogen-activated protein kinase pathway (through a mechanism involving Ssk1p) (13, 22). These pathways respond differently to the OSR in a concentration- and/or oxidant-dependent manner, reflecting a complex process. For instance, while CAP1 is required for growth on both low and high concentrations of H2O2, HOG1 is required for growth only on high concentrations of peroxide (22). Also, a CAP1-deficient strain appears to be more susceptible to cadmium but more resistant to menadione than a HOG1-deficient strain (4). Genome-wide expression and proteomic studies showed that Cap1p regulates the expression of many genes involved in the OSR as well as other metabolic pathways, including energy metabolism and substance transport (3, 36, 63, 64); however, it was not determined whether Cap1p regulates these genes directly. In this paper, we used genome-wide location and expression analyses to better characterize the Cap1p regulon as well as Cap1p function in C. albicans.
MATERIALS AND METHODS
Strains and growth media.
The C. albicans strains used in this study are listed in Table 1. The pCaEXP integrants (Table 1) were grown in synthetic complete (SC) medium lacking uracil (SC-ura) (59), in SC medium lacking uracil, methionine, and cysteine (SC-ura-met-cys) to induce the MET3 promoter (PMET3), or in SC-ura supplemented with methionine (2.5 mM) and cysteine (2.5 mM) (SC-ura+met+cys) under PMET3-repressing conditions. Strain CAI4 and its cap1Δ derivatives were grown in YPD broth (1% yeast extract, 2% peptone, and 1% dextrose) supplemented or not with the indicated drug. The Escherichia coli MC1061 strain was used for DNA cloning and maintenance of the plasmid constructs.
TABLE 1.
Strains used in this study
| Strain | Parental strain | Relevant genotype | Reference |
|---|---|---|---|
| SGY243 (parental) | ade2/ade2 Δura3::ADE2/Δura3::ADE2 | 31 | |
| SGY243-CaEXP-A | SGY243 | RP10::(pCaEXP) URA3 PMET3 | This study |
| SGY243-CaEXP-B | SGY243 | RP10::(pCaEXP) URA3 PMET3 | This study |
| SGY243-CaEXP-CAP1-HA-A | SGY243 | RP10::(pCaEXP) URA3 PMET3-CAP1-HA3 sequence | This study |
| SGY243-CaEXP-CAP1-HA-B | SGY243 | RP10::(pCaEXP) URA3 PMET3-CAP1-HA3 sequence | This study |
| SGY243-CaEXP-CAP1-CSE-HA-A | SGY243 | RP10::(pCaEXP) URA3 PMET3-CAP1-CSE-HA3 sequence | This study |
| SGY243-CaEXP-CAP1-CSE-HA-B | SGY243 | RP10::(pCaEXP) URA3 PMET3-CAP1-CSE-HA3 sequence | This study |
| CAI4 (parental) | ura3Δ::imm434/ura3Δ::imm434 | 23 | |
| CJD21/PMK | CJD21 | cap1Δ::hisG/cap1Δ::hisG::(pPMK) | 3 |
| CJD21/PMK-CAP1 | CJD21 | cap1Δ::hisG/cap1Δ::hisG::(pPMK) CAP1 | 3 |
| CJD21/PMK-CAP1-CSE | CJD21 | cap1Δ::hisG/cap1Δ::hisG::(pPMK) CAP1-CSE | This study |
Generation of plasmids and Cap1p-expressing strains.
Plasmid PMK-CAP1-CSE was obtained by site-directed mutagenesis of PMK-CAP1 (3) such that residues C477, S478, and E479 were replaced with alanine residues in the protein sequence of Cap1p. DNA fragments overlapping positions −36 to +1497 (relative to the ATG translation start site) of the C. albicans CAP1 gene and corresponding to the wild-type or the mutated (CAP1-CSE) allele of CAP1 were PCR amplified with PfuTurbo DNA polymerase (Stratagene, La Jolla, CA) from plasmid PMK-CAP1 or PMK-CAP1-CSE, respectively, using primers 5′-ATATGGATCCAACAACCATTTTCAACTATCC (introduces a BamHI site [underlined]) and 5′-TATACTGCAGttaGCGGCCGCCATGTTTTATACTTCGCTCTAG (introduces sequentially a PstI site [underlined], a TAA stop codon [in lowercase letters], and a NotI site [underlined]). The resulting fragments (1,565 bp) were digested with BamHI and PstI and cloned into the corresponding sites of plasmid pCaEXP (9), generating plasmids pCaEXP-CAP1 and pCaEXP-CAP1-CSE. A triple hemagglutinin (HA3)-encoding sequence (111 bp) was released from plasmid pMPY-3×HA (58) by NotI enzymatic digestion and cloned into the NotI site of plasmids pCaEXP-CAP1 and pCaEXP-CAP1-CSE, generating plasmids pCaEXP-CAP1-HA3 and pCaEXP-CAP1-CSE-HA3, respectively. DNA sequencing was performed to ascertain that the fragments were cloned in frame and that no unintended mutations were introduced during the amplification process. The pCaEXP-CAP1-HA3 and pCaEXP-CAP1-CSE-HA3 plasmids were digested with StuI, and the resulting fragments were used to transform strain SGY243 (Table 1). Strain CJD21/PMK-CAP1-CSE (Table 1) was created by introducing plasmid PMK-CAP1-CSE into strain CJD21, as described previously (3).
C. albicans transformations.
C. albicans transformations were conducted as described in MacPherson et al. (41), using a modified standard lithium acetate procedure. The transformed cells were plated on SC-ura plates and incubated for 3 days at 30°C.
Antifungal drugs and susceptibility testing.
Stock solutions of FLC (a gift from Pfizer) and 4-NQO (Sigma) were prepared at concentrations of 5 mg/ml and 500 μM in water or dimethyl sulfoxide (DMSO), respectively. Drug susceptibility testing was performed using spot assays. Cells were grown overnight on SC-ura-met-cys plates and resuspended in water to an optical density at 600 nm (OD600) of 0.1. Serial dilutions (10-fold) of each strain were spotted onto SC-ura-met-cys plates supplemented with 2 μg/ml of FLC and 1.5 μM of 4-NQO or the solvent alone (water or DMSO, respectively). The plates were incubated for 2 days at 30°C.
Total protein preparation and Western blotting.
Total protein extracts were prepared as described for S. cerevisiae (54) from 2 OD units of two independent strains expressing CAP1-HA3 (SGY243-CaEXP-CAP1-HA clones A and B) or CAP1-CSE-HA3 (SGY243-CAP1-CSE-HA clones A and B) (Table 1) grown overnight in SC-ura-met-cys (PMET3-inducing conditions) or SC-ura+met+cys (PMET3-repressing conditions). Extracts were boiled for 1 min, and 25-μl extracts were separated from total extracts of 100 μl by electrophoresis on a sodium dodecyl sulfate-10% polyacrylamide gel. Proteins were transferred to a nitrocellulose membrane with a Trans-Blot SD semidry transfer apparatus (Bio-Rad, Hercules, CA), and the membrane was incubated with a mouse anti-HA monoclonal antibody (12CA5; Roche) at a dilution of 1:1,000, followed by incubation with rabbit anti-mouse immunoglobulin G antibodies coupled to alkaline phosphatase (Bio-Rad). The membrane was then developed with 5-bromo-4-chloro-3-indolylphosphate p-toluidine salt and nitroblue tetrazolium chloride substrates, as recommended by the manufacturer (Bio-Rad).
ChIP-on-chip (ChIP-chip) and data analysis.
Three independent cultures (50-ml each) of strains SGY243-CaEXP-A (untagged; control strain) and SGY243-CAP1-HA-A or SGY243-CAP1-CSE-HA-A (tagged strains) (Table 1) were grown overnight in SC-ura+met+cys diluted to an OD600 of 0.005 in SC-ura-met-cys (to induce PMET3) and grown until the OD600 reached 1.0. The subsequent steps of DNA cross-linking, DNA shearing, chromatin immunoprecipitation (ChIP), DNA labeling with Cy dyes, hybridization to intergenic DNA microarrays, and data analysis were conducted exactly as described in Liu et al. (38). Both pools of labeled DNA from the tagged strain (SGY243-CAP1-HA-A or SGY243-CAP1-CSE-HA-A; Cy5-labeled) and the corresponding untagged control strain (SGY243-CaEXP-A; Cy3-labeled) were mixed and hybridized to a C. albicans whole-genome tiled oligonucleotide DNA microarray described elsewhere (61). Hybridization was performed as recommended by the manufacturer (NimbleGen Systems, Inc). Scanning of the slides (n = 3) was performed using a GenePix 4000B scanner (Molecular Devices). Scanned images were preprocessed using NimbleScan software (version 2.4; NimbleGen Systems, Inc). General feature format reports were created for the Cy5 (tagged strain) and Cy3 (untagged control strain) intensity signals from each independent replicate and were then imported into the Tilescope program (http://tilescope.gersteinlab.org:8080/mosaic/pipeline.html) (69). Quantile normalization was applied to the data (69). The parameters used were as follows: a window size of 400 bp, a maximum genomic distance of 60 bp, and a minimum length of 120 bp. The replicate data were combined, and peak finding (i.e., determining the Cap1p binding sites) was done using a pseudomedian signal threshold of at least twofold and a P value cutoff of 0.01 or less (69).
Q-PCR for confirmation of the ChIP-chip data.
Quantitative real-time PCR (Q-PCR) was performed with three independent SGY243-CaEXP-A and SGY243-CAP1-HA-A or SGY243-CAP1-CSE-HA-A ChIP samples prepared as described above. Quantification of the recovered DNA was performed using a Quant-iT PicoGreen double-stranded DNA assay kit (Molecular Probes-Invitrogen) as previously described (38). The DNA concentration ranged from 0.08 ng/μl to 0.60 ng/μl for the tagged strains and 0.61 ng/μl to 1.22 ng/μl for the untagged strains. Q-PCR assays were conducted using Universal ProbeLibrary (Roche Applied Science) or TaqMan (Integrated DNA Technologies) methodology (38). The different primers and probe combinations used for Q-PCR are listed in Table 2. Optimal specific primer sequences and probes for the CIP1, IFR1 (targets), FUR1 (control for statistical analyses by t test), and SPS4 (orf19.7568; reference for normalization) promoters were obtained using Universal ProbeLibrary Web-based ProbeFinder software (version 2.34; Roche Applied Sciences) as previously described (38). Design of the TaqMan probe and specific forward and reverse primers for the MDR1 target promoter have been described previously (71). Q-PCR mixtures, Q-PCR conditions, and data analyses were performed as described previously (71). Statistical significance was determined using Welsh's two-sample t test. The statistical significance threshold was set at α = 0.05.
TABLE 2.
Primers and probe sequences used for Q-PCR binding assays
| Promoter | Primers/probe sequencesa (5′-3′) | Amplicon locationb |
|---|---|---|
| MDR1 | F: GGCGGATTTACTCCTGATACAACTC | −580 → −401 |
| R: GCGACGGGCTGTTGAGTAAACTAT | ||
| Pc: AGCTCGTTTAGTTGTTCCCATTCGCA | ||
| CIP1 | F: CCAATACAATTTAGTAAGCAGAAACAA | −583 → −457 |
| R: TTTCATAACAAATCAATAACAACAACC | ||
| Pd: CAGCCACA | ||
| IFR1 | F: CGTTTATTCAATAGGATTGAGAAGG | −127 → −50 |
| R: AATGGTGGGCAAGTATCAAAAC | ||
| Pe: TTCCACCA | ||
| FUR1 | F: GGTGCTTTTGGGAGAATGAA | −987 → −913 |
| R: CTTCCTCAAAACAAAACTGCAA | ||
| Pf: GCTGCCTG | ||
| SPS4g | F: TACAGTTGCCCCAGTCAACA | −636 → −574 |
| R: TGTCTTGGAACGGAAACTCA | ||
| Ph: TCCTGCTC |
F, forward; R, reverse; P, probe.
Position according to the ATG start codon.
TaqMan probe (Integrated DNA Technologies).
Probe no. 5 from Universal ProbeLibrary (catalogue no. 04685024001; Roche).
Probe no. 31 from Universal ProbeLibrary (catalogue no. 04687647001).
Probe no. 27 from Universal ProbeLibrary (catalogue no. 04687582001).
orf19.7568.
Probe no. 15 from Universal ProbeLibrary (catalogue no. 04685148001).
Bioinformatic analyses.
Visualization of the ChIP-chip results was conducted using a custom-designed C. albicans genome browser representing the original assembly 19 of the C. albicans genome as described in the Candida Genome Database ([CGD] http://www.candidagenome.org/cgi-bin/gbrowse/candida/). To group the overrepresented functional categories of Cap1p targets, 152 hits out of the 306 Cap1p-HA3- or Cap1p-CSE-HA3-bound targets were removed from the analysis, as they were not clearly associated with specific open reading frames (ORFs) (see Results for details). The orf19 nomenclatures of the genes were then used as input for functional grouping using the CGD Gene Ontology (GO) Term Finder tool (http://www.candidagenome.org/cgi-bin/GO/goTermFinder). Three ontologies, “Biological process,” “Molecular function,” and “Cellular component,” were selected. GO Term Finder calculates a P value for the overrepresented GO terms (relative to the 6,334 annotated C. albicans genes) using a hypergeometric distribution with multiple hypothesis correction (http://www.candidagenome.org/help/goTermFinder.html). If some GO terms contained overlapping gene lists, the GO term with the largest number of genes was selected. The P value cutoff was P ≤ 0.02. For motif discovery analyses, DNA sequences covered by the 189 or 117 peaks identified in Cap1p-HA3 or Cap1p-CSE-HA 3binding data, respectively, were extracted and used as input for motif discovery, using the SCOPE (Suite for Computational Identification of Promoter Elements) program (http://genie.dartmouth.edu/scope/) (10, 12). This program allows accurate determination of potential transcription factor binding sites in a set of sequences using three different motif discovery algorithms (10, 12). To search for the TTASTAA motif within Cap1p-bound sequences, the same sequences analyzed with the SCOPE program were used as input for the DNA pattern matching TTASTAA, using the pattern-matching tool from Regulatory Sequence Analysis Tools ([RSAT] http://rsat.ulb.ac.be/rsat/). As a control, up to 1.0 kb of promoter sequences upstream of the ATG translation start site of the 6,093 promoters of the C. albicans ORFs was retrieved from the RSAT database (http://rsat.ulb.ac.be/rsat/). To search for the MTKASTMA sequence (where M designates A or C, K designates G or T, and S designates C or G) within the promoter region of the genes modulated in CJD21/PMK-CAP1-CSE versus CJD/PMK-CAP1, up to 1.0 kb of the promoter sequence upstream of the ATG translation start site of each gene was retrieved from the assembly 21 genome sequence such that overlap with neighboring ORFs was prevented and was then used as input for the DNA pattern matching MTKASTMA, using the RSAT pattern-matching tool.
RNA isolation.
Strains were grown overnight in 10 ml YPD at 30°C. The next day, an aliquot of the overnight culture was used to inoculate 200 ml YPD broth to a starting OD600 of 0.2. This culture was grown for 3 h as before, cells were collected by centrifugation, and cell pellets were immediately frozen and stored at −80°C until RNA isolation. Benomyl-exposed cultures were treated with 25 μg/ml benomyl (Sigma-Aldrich, St. Louis, MO) for 30 min before cells were harvested. Three independently obtained sets of cell cultures were used. RNA was isolated from frozen cell pellets using the hot-phenol method (57). Briefly, cells were resuspended in 12 ml AE buffer (50 mM sodium acetate [pH 5.2] and 10 mM EDTA) at room temperature, after which 800 μl 25% sodium dodecyl sulfate and 12 ml acid phenol (Fisher Scientific, Houston, TX) were added. The cell lysate was then incubated for 10 min at 65°C with vigorous shaking each minute, cooled on ice for 5 min, and subjected to centrifugation for 15 min at 11,952 × g. The supernatants were transferred to new tubes containing 15 ml chloroform, mixed, and subjected to centrifugation at 200 × g for 10 min. RNA was precipitated from the resulting aqueous layer by mixing that portion in new tubes with 1 volume of isopropanol and 0.1 volume of 2 M sodium acetate [pH 5.0] and subjecting the mixture to centrifugation at 17,211 × g for 35 min at 4°C. The supernatants were removed, the pellet was resuspended in 10 ml 70% ethanol, and the RNA was collected by centrifugation at 17,211 × g for 20 min at 4°C. The supernatants were again removed, and the RNA was resuspended in 50 to 200 μl diethyl pyrocarbonate-treated water. The RNA was stored at −80°C until needed.
cRNA synthesis and microarray hybridization.
Immediately prior to cDNA and subsequent cRNA syntheses, the purity and concentration of RNA samples were determined from A260/A280 readings and RNA integrity was determined by capillary electrophoresis, using an RNA 6000 Nano laboratory-on-a-chip kit and 2100 bioanalyzer (Agilent Technologies) per the manufacturer's instructions. First- and second-strand cDNA was synthesized from 15 μg total RNA, using a SuperScript double-stranded cDNA synthesis kit (Invitrogen) and an oligo-dT24-T7 primer (Proligo) according to the manufacturers' instructions. cRNA was synthesized and labeled with biotinylated UTP and CTP by in vitro transcription, using T7 promoter-coupled double-stranded cDNA as a template and a BioArray HighYield RNA transcript labeling kit (Enzo Diagnostics). Double-stranded cDNA synthesized from the previous steps was washed twice with 70% ethanol and suspended in 22 μl of RNase-free water. The cDNA was incubated as recommended with reaction buffer, biotin-labeled ribonucleotides, dithiothreitol, RNase inhibitor mix, and T7 RNA polymerase for 5 h at 37°C. The labeled cRNA was separated from unincorporated ribonucleotides with a Chroma Spin-100 column (Clontech) and was ethanol precipitated at −20°C overnight.
The cRNA pellet was suspended in 10 μl of RNase-free water, and 10 μg was fragmented at 95°C for 35 min in 200 mM Tris-acetate (pH 8.1), 500 mM potassium acetate, and 150 mM magnesium acetate. The fragmented cRNA was hybridized for 16 h at 45°C to either C. albicans Affymetrix GeneChip arrays (CAN04a530004N; manufactured by NimbleExpress) or to an Affymetrix custom expression array (CAN07a520619F; manufactured by Affymetrix) for C. albicans. Arrays were washed at 25°C with 6× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]) and 0.01% Tween 20 followed by a stringent wash at 50°C with 100 mM MES (morpholineethanesulfonic acid), 0.1 M NaCl, and 0.01% Tween 20. Affymetrix Fluidics Station 450 was used for hybridizations and washes according to standard EukGE-WS2v5 protocol. The arrays were then stained with phycoerythrin-conjugated streptavidin (Molecular Probes), and the fluorescence intensities were determined using a GCS 3000 high-resolution confocal laser scanner (Affymetrix). The scanned images were analyzed using software resident in the GeneChip operating system, version 2.0 (Affymetrix). Sample loading and variations in staining were standardized by scaling the average of the fluorescent intensities of all genes on an array to a constant target intensity of 250. The signal intensity for each gene was calculated as the average intensity difference, represented by Σ(PM − MM)/number of probe pairs, where PM and MM denote perfectly matched and mismatched probes, respectively.
Gene expression microarray data analysis.
The scaled gene expression values from GeneChip operating system version 2.0 software were imported into GeneSpring 7.2 software (Agilent Technologies) for preprocessing and data analysis. Probe sets were deleted from subsequent analysis if they were called absent by the Affymetrix criterion and displayed an absolute value below 20 in all experiments. The expression value of each gene was normalized to the median expression of all genes in each chip as well as to the median expression for that gene across all chips in the study. A pairwise comparison of gene expression was performed for each matched experiment.
Q-PCR for expression data.
Real-time PCR was performed in follow-up experiments to validate the microarray results. First-strand cDNAs were synthesized from 2 μg of total RNA in a 21-μl volume of reaction mixture, using a SuperScript first-strand synthesis system for reverse transcription (RT)-PCR (Invitrogen, Carlsbad, CA) in accordance with the manufacturer's instructions. Q-PCRs were performed in triplicate using a 7000 sequence detection system (Applied Biosystems, Foster City, CA). Independent PCRs were performed using the same cDNA for both the gene of interest and the 18S rRNA, using SYBR green PCR master mix (Applied Biosystems). Gene-specific primers were designed for the gene of interest and the 18S rRNA, using Primer Express software (Applied Biosystems) and an oligo analysis and plotting tool (Qiagen, Valencia, CA), and are shown in Table 3. The PCR conditions consisted of AmpliTaq Gold activation at 95°C for 10 min, followed by 40 cycles of denaturation at 95°C for 15 s and annealing/extension at 60°C for 1 min. A dissociation curve was generated at the end of each PCR cycle to verify that a single product was amplified, using software provided with the 7000 sequence detection system. The change in fluorescence of SYBR green I dye in every cycle was monitored by the system software, and the threshold cycle (CT) above the background for each reaction was calculated. The CT value of 18S rRNA was subtracted from that of the gene of interest to obtain a ΔCT value. The ΔCT value of an arbitrary calibrator (e.g., untreated sample) was subtracted from the ΔCT value of each sample to obtain a ΔΔCT value. The gene expression level relative to that of the calibrator was expressed as 2−ΔΔCT. Statistical analysis was performed using R software, version 2.5.0 (www.r-project.org). Relative changes were compared using a two-sample t test. The statistical significance threshold was set at α = 0.05.
TABLE 3.
Primers used for Q-PCR expression analysis
| Gene | Primer paira | Amplicon size (bp) |
|---|---|---|
| 18S | F: 5′-CACGACGGAGTTTCACAAGA-3′ | 135 |
| R: 5′-CGATGGAAGTTTGAGGCAAT-3′ | ||
| MDR1 | F: 5′-ACATAAATACTTTGCCCATCCAGAA-3′ | 82 |
| R: 5′-AAGAGTTGGTTTGTAATCGGCTAAA-3′ | ||
| GLR1 | F: 5′-ATCAACAACAACTATGGCTCCAACT-3′ | 51 |
| R: 5′-CAGATCCACCACCAATGACTAAATA-3′ |
F, forward; R, reverse.
Microarray data accession number.
Microarray data can be found at the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/projects/geo/) under series numbers GSE14258 and GSE15104.
RESULTS
Epitope-tagging of Cap1p.
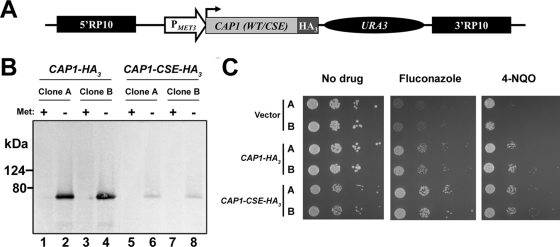
To immunoprecipitate Cap1p, we fused it to a C-terminal HA3 epitope, using the pCaEXP expression system (Fig. 1A; see Materials and Methods). Cap1p-HA3 expression is driven by the MET3 promoter, which is induced in the absence of methionine and repressed in its presence (9). To test the function of a constitutively activated Cap1p, we also constructed a C. albicans strain expressing an HA3-tagged Cap1p carrying the amino acid substitutions C477A, S478A, and E479A (CAP1-CSE-HA3 allele) (Fig. 1A; see Materials and Methods). These substitutions are equivalent to those introduced in S. cerevisiae Yap1p (C629A, S630A, and E631A, respectively), leading to a constitutively activated Yap1p protein (65). Immunoblotting showed that under inducing conditions, the tagged wild-type Cap1p protein was readily detectable, whereas much lower levels of the Cap1p-CSE-HA3 protein were detected (Fig. 1B). It therefore seems that introduction of the CSE mutation is accompanied by a decrease in protein stability, as previously observed for the CAP1-TR allele (3). The strains were also characterized phenotypically by spot assay on PMET3-inducing media containing the azole antifungal agent FLC or the oxidative stress-inducing agent 4-NQO (Fig. 1C). This experiment showed that overexpression of the CAP1-HA3 or CAP1-CSE-HA3 allele conferred resistance to FLC and 4-NQO, with the CAP1-CSE-HA3 allele conferring slightly higher resistance to FLC than the CAP1-HA3 allele (Fig. 1C). Taken together, these results showed that both proteins were properly tagged and functional and that the CSE mutation in CAP1-CSE-HA3 functions as a gain-of-function mutation.
FIG. 1.
Strategy for tagging Cap1p with a HA3 epitope and characterization of the tagged strains. (A) Schematic representation of the CAP1-tagging cassette. The wild-type or mutated version of the CAP1 gene [CAP1 (WT/CSE); light gray box], cloned as an in-frame fusion with a HA3 tag (dark gray box) in plasmid pCaEXP (9), is under the control of the MET3 promoter (PMET3; open arrow) and is followed by the C. albicans URA3 marker (black oval). The 5′ and 3′ fragments of the RP10 gene (5′RP10 and 3′RP10; black boxes) flank the cassette and allow targeted integration at the RP10 locus (9). (B) Western blot analysis of strains expressing HA3-tagged versions of the CAP1 gene (CAP1-HA3 or CAP1-CSE-HA3). Total proteins were extracted from two independent clones of the SGY243 transformants (A and B) grown in the absence (−) or presence (+) of 2.5 mM methionine (Met). Western blotting was performed using the anti-HA antibody 12CA5. Positions of the molecular mass standards are indicated on the left (kDa). (C) Drug resistance profiles of C. albicans strains expressing HA3-tagged CAP1 alleles. Two independent transformants (A and B) for each of the CAP1-HA3- or CAP1-CSE-HA3-expressing strains or the strain carrying the empty vector as the negative control (vector) were analyzed by spot assay for their ability to grow on SC-ura-met plates in the absence or presence of 2 μg/ml of FLC or 1.5 μM of 4-NQO. The plates were incubated for 2 days at 30°C.
Identification of Cap1p binding sites in vivo.
We performed genome-wide location profiling (ChIP-chip) of Cap1p-HA3 or Cap1p-CSE-HA3, using C. albicans whole-genome oligonucleotide tiling microarrays (61) (see Materials and Methods). We identified 189 and 117 hits (i.e., peaks) for Cap1p-HA3 and Cap1p-CSE-HA3, respectively (the log2-transformed pseudomedian signal intensity cutoff was 1 or less; P ≤ 0.01) (see Tables S1 and S2 in the supplemental material).
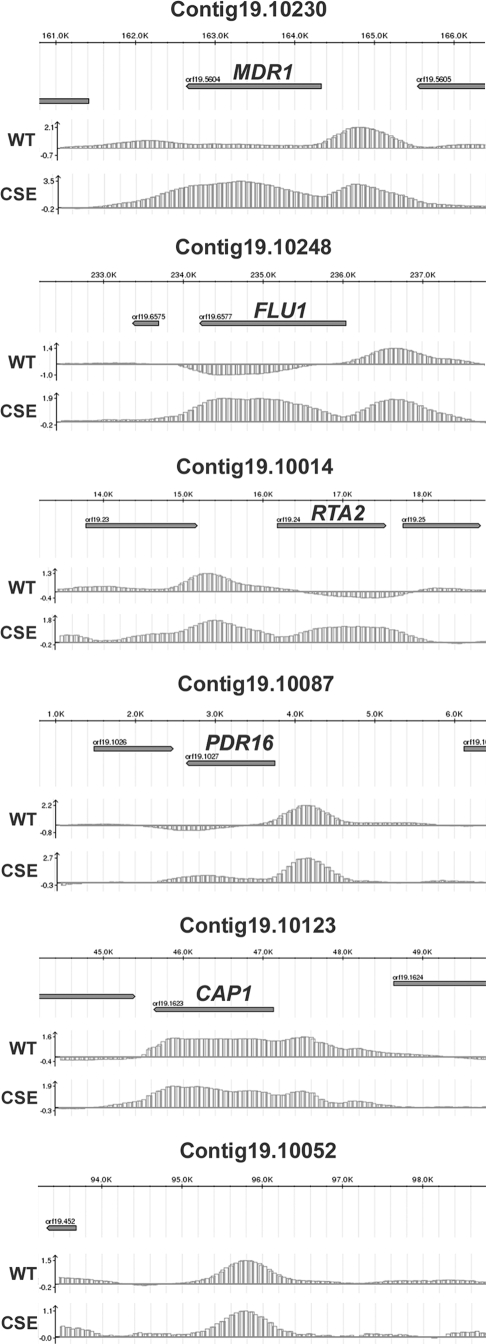
We visualized the ChIP-chip data using a C. albicans genome browser representing the entire assembly 19 (see Materials and Methods). We found that a high proportion of Cap1p binding peaks clearly associated with ORFs, totaling 89 target genes for Cap1p-HA3 or Cap1p-CSE-HA3 (60 of the 89 genes were common to both proteins; 23 additional genes were specific to Cap1p-HA3, while 6 additional genes were specific to Cap1p-CSE-HA3) (Fig. 2; see also Tables S1 and S2 in the supplemental material). In some cases, more than one peak associated with one ORF, while in one occurrence one peak associated with two ORFs (orf19.3121 and orf19.3122) (see Tables S1 and S2 in the supplemental material). Interestingly, we also found that 101 Cap1p-HA 3binding peaks and 35-Cap1p-CSE-HA 3binding peaks did not clearly associate with defined ORFs, including peaks that were located in intergenic regions (see the bottom panel of Fig. 2 for an example), suggesting that these regions may encode unidentified ORFs or small RNAs. Strikingly, Cap1p binding was detected not only at the promoter region of its target genes but also at their 3′ ends and within their ORFs (Fig. 2). This surprising binding profile could be explained by the association of Cap1p with chromatin-associated proteins and/or the transcriptional machinery (see Discussion). Finally, it is noteworthy that the Cap1p-CSE-HA3 signal was often increased relative to that of Cap1p-HA3 at common targets (Fig. 2; see also Tables S1 and S2 in the supplemental material).
FIG. 2.
Cap1p binding at selected C. albicans genomic regions. Plotted are the normalized log2-transformed signal intensities (lower graphs of each panel) of HA3-tagged wild-type (WT) or hyperactive (CSE) Cap1p binding (y-axis) versus the corresponding position of each signal (x-axis) in selected C. albicans genomic regions from assembly 19 (the corresponding contig 19 number is indicated at the top of each panel). Log2-transformed signal intensity values are indicated at the left of the y-axis. The y-axis intercept is the value 0 (i.e., a binding ratio of 1). The location of each selected region from the corresponding contig 19 is shown on the scaled upper axis of each panel in kilobases (K). The spacing is 1.0 kb between each major graduation and 0.2 kb between each minor graduation. The orientation of each ORF is depicted by the arrowed gray rectangle. Negative enrichment values in the FLU1, RTA2, and PDR16 panels may be due to background noise, preferential amplification, or normalization biases inherent to the ChIP-chip technology (6, 51).
We used the GO Term Finder tool from the CGD (http://www.candidagenome.org/cgi-bin/GO/goTermFinder) to identify functional groupings among the 89 Cap1p-HA3 or Cap1p-CSE-HA3 target genes that were significantly overrepresented relative to the annotated C. albicans genome (Table 4; see Materials and Methods). As expected, Cap1p binding was significantly enriched at the loci of target genes involved or predicted to be involved in the OSR, including CIP1, orf19.2262, CCP1, SOD1, orf19.3537, GLR1, GCS1, MDR1, CAT1, and TRX1, which were grouped into the functional category “Response to oxidative stress,” which was among the most overrepresented categories of the GO terms identified (P = 4.1 × 10−6). In line with this finding, the functional grouping “Oxidoreductase activity” was also among the overrepresented GO terms (P = 1.1 × 10−6) and included 19 genes, such as the superoxide dismutase-encoding gene SOD1, the predicted old yellow enzyme family-encoding genes OYE2, OYE23, and OYE32, and the peroxidase-encoding genes CAT1 and CCP1. Genes grouped into the GO term “Response to chemical stimulus,” the most overrepresented functional category (P = 5.0 × 10−9), were also found in the parent overrepresented category “Response to stimulus” (P = 5.9 × 10−6) in addition to four genes involved or predicted to be involved in the OSR (GRE2 and orf19.6757), transcriptional regulation of morphogenesis (EFG1), and the response to DNA damage (YIM1). Interestingly, Cap1p also bound to genes previously shown to be involved in azole resistance, including PDR16 (55), RTA2 (29), MDR1 (66), and FLU1 (8), which were grouped into the overrepresented functional category “Response to drug” (P = 2.0 × 10−6) (Fig. 2). Other overrepresented functional categories included “Hyphal cell wall,” “Response to cadmium ion,” “Phospholipid transport,” and “Regulation of nitrogen utilization”.
TABLE 4.
Overrepresented functional categories in Cap1p ChIP-chip data
| GO terma | CGD accession no. (ontology classification)b | % Frequencyc (no. of genes) | % Genome frequencyd (no. of genes) | P valuee | Genesf |
|---|---|---|---|---|---|
| Response to chemical stimulusg | GO:0042221 (P) | 27 (24) | 5.4 (342) | 5.0 × 10−09 | PDR16, SSA2, CIP1, CAP1, orf19.2262, CCP1, RTA2, SOD1, RIB1, orf19.3537, SHA3, GLR1, GCS1, CDR4, HXK2, RHR2, MDR1, CAT1, CYS3, YCF1, FLU1, FCR1, ALS6, TRX1 |
| Oxidoreductase activity | GO:0016491 (F) | 21.3 (19) | 4.6 (293) | 1.1 × 10−06 | IFD6, EBP1, RNR22, orf19.2262, CCP1, SOD1, OYE32, GRE2, orf19.3234, OYE23, OYE2, orf19.3537, ADH1, GLR1, GRP2, ERO1, orf19.5517, CAT1, orf19.6757 |
| Response to oxidative stress | GO:0006979 (P) | 12.4 (11) | 1.3 (84) | 4.1 × 10−06 | CIP1, CAP1, orf19.2262, CCP1, SOD1, orf19.3537, GLR1, GCS1, MDR1, CAT1, TRX1 |
| Response to stimulusg | GO:0050896 (P) | 31.5 (28) | 10.2 (647) | 5.9 × 10−06 | PDR16, SSA2, CIP1, CAP1, orf19.2262, CCP1, RTA2, SOD1, RIB1, GRE2, orf19.3537, SHA3, GLR1, GCS1, CDR4, HXK2, RHR2, MDR1, EFG1, CAT1, CYS3, YCF1, FLU1, orf19.6757, FCR1, ALS6, TRX1, YIM1 |
| Response to drug | GO:0042493 (P) | 14.6 (13) | 2.3 (145) | 2.0 × 10−05 | PDR16, CAP1, RTA2, SOD1, RIB1, GCS1, CDR4, RHR2, MDR1, CYS3, YCF1, FLU1, FCR1 |
| Cell fraction | GO:000267 (C) | 15.7 (14) | 3.3 (212) | 6.5 × 10−05 | PDR16, SSA2, EBP1, orf19.251, orf19.2693, PDC11, orf19.3121, PGI1, ADH1, GRP2, RHR2, orf19.5517, CCC1, GST3 |
| Hyphal cell wall | GO:0030446 (C) | 5.6 (5) | 0.6 (35) | 6.2 × 10−03 | SSA2, EBP1, orf19.251, PDC11, ADH1 |
| Response to cadmium ion | GO:0046686 (P) | 3.4 (3) | 0.1 (6) | 1.2 × 10−02 | CIP1, CAP1, GCS1 |
| Phospholipid transport | GO:0015914 (P) | 4.5 (4) | 0.3 (17) | 1.7 × 10−02 | PDR16, GIT1, RTA2, orf19.932 |
| Regulation of nitrogen utilization | GO:0006808 (P) | 3.4 (3) | 0.1 (7) | 2.0 × 10−02 | orf19.2693, orf19.3121, GST3 |
Grouping of the Cap1p (Cap1p-HA3 or Cap1p-CSE-HA3) targets identified in ChIP-chip data according to GO terminology determined by using the online CGD GO Term Finder tool (http://www.candidagenome.org/cgi-bin/GO/goTermFinder). Analysis conducted in September 2008.
Ontology classification: P, biological process; C, cellular component; F, molecular function.
Percentages were calculated based on the number of genes in each GO category divided by the total number (89 genes).
Percentages were calculated based on the number of genes in each category divided by the total number of annotated genes of the C. albicans genome, according to CGD (6,334 genes).
P values for the overrepresented categories were calculated using a hypergeometric distribution with multiple hypothesis correction (i.e., Bonferroni's correction) as described in the GO Term Finder tool website (http://www.candidagenome.org/help/goTermFinder.shtml). The P value cutoff used was ≤0.02.
Gene name or orf19 nomenclature according to CGD. Some genes were attributed to more than one GO term.
The selection criteria for GO term groups with overlapping gene lists (see Materials and Methods for details) was not applied to these two groups, in order to show that “Response to chemical stimulus” was the most significantly overrepresented functional category.
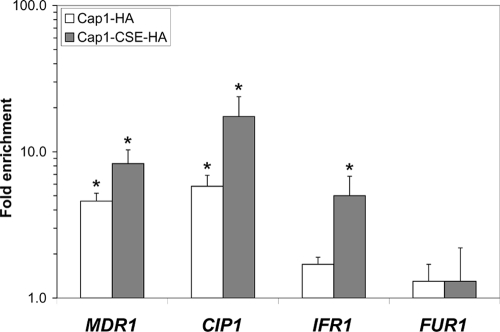
Using Q-PCR, we confirmed the binding of Cap1p to the MDR1, CIP1, and IFR1 promoters, with enrichment ratios (± standard deviations) of 4.6 ± 0.6, 5.8 ± 1.1, and 1.7 ± 0.2, respectively, for Cap1p-HA3 binding and 8.3 ± 2.0, 17.4 ± 6.4, and 5.0 ± 1.8, respectively, for Cap1p-CSE-HA3 binding (Fig. 3). As a control, we investigated the binding of Cap1p to the promoter of the FUR1 gene, which was not enriched in the ChIP-chip experiments, and found no significant enrichment of that promoter by Q-PCR (1.3 ± 0.4 for Cap1p-HA3 and 1.3 ± 0.9 for Cap1p-CSE-HA3) (Fig. 3), confirming the validity of the data obtained in the ChIP-chip experiments.
FIG. 3.
Quantification of the in vivo enrichment of Cap1p-HA3 (Cap1-HA) and Cap1p-CSE-HA3 (Cap1-CSE-HA) binding at the MDR1, CIP1, IFR1, and FUR1 targets using Q-PCR. SGY243-CaEXP-A (control strain; untagged) and SGY243-CAP1-HA-A or SGY243-CAP1-CSE-HA-A (tagged strains) were submitted to ChIP (three biological replicates), and the recovered DNA samples were analyzed by Q-PCR, using Universal ProbeLibrary probes (Roche Diagnostics) for the CIP1, IFR1, FUR1, and SPS4 (reference for normalization) targets and a TaqMan probe for the MDR1 target. Relative enrichment values (n-fold) are presented in logarithmic scale. Error bars denote standard deviations. Asterisks denote statistical significance determined by using Welsh's two-sample t test (P ≤ 0.05).
Identification of potential Cap1p binding motifs.
Recent studies with C. albicans characterized the cis-acting elements controlling MDR1 expression upon treatment with the antifungal agent benomyl or the oxidative stress inducer H2O2 and suggested that YRE-like sequences found in the promoter of MDR1 were responsible for its induction upon treatment with H2O2 (26, 28, 53). Furthermore, it was shown that an H2O 2response element (HRE), including two YRE-like sequences (TTASTAA) located between positions −561 and −520 relative to the ATG translation start site of MDR1, were required for this induction in a Cap1p-dependent manner (53). These data suggest that Cap1p binds to the promoter regions of its target genes via this YRE-like element. Our data show that the Cap1p-HA3 binding peak reached its maximal intensity within a region roughly encompassing positions −250 to −600 of the MDR1 promoter (Fig. 2; see also Table S1 in the supplemental material), consistent with published observations (53). To determine if the TTASTAA motif was significantly enriched in our ChIP-chip data, we looked at its occurrence in the 189 Cap1p-HA3- or the 117 Cap1p-CSE-HA3-bound sequences (see Materials and Methods) and identified a total of 93 and 61 TTASTAA-containing sequences, respectively. As a control, we searched for the TTASTAA motif in the promoter regions from the 6,093 ORFs of the C. albicans genome, using up to 1,000 bp of a promoter sequence upstream of the ATG translation start site of each ORF, and found an average of 49 (per 189) and 30 (per 117) promoters containing this motif, yielding 1.9- and 2.0-fold enrichments for the presence of TTASTAA in Cap1p-HA3 and Cap1p-CSE-HA3 targets, respectively. The fact that 96 (out of 189) and 56 (out of 117) sequences do not contain the TTASTAA motif suggests that Cap1p might recognize different sequence motifs. We thus conducted a motif search using the SCOPE program (http://genie.dartmouth.edu/scope/), which allows the accurate detection of conserved putative transcription factor binding sites among a given set of promoter sequences, using three independent motif discovery algorithms (10, 12) (see Materials and Methods). This analysis identified the MTKASTMA motif (Fig. 4), which includes the palindrome sequence TKASTMA, in both the Cap1p-HA3 (significance value, 340.6; coverage, 66.1%)- and Cap1p-CSE-HA3 (significance value, 196.0; coverage, 64.1%)-bound sequences, suggesting that Cap1p can bind to a more degenerate recognition sequence in C. albicans.
FIG. 4.
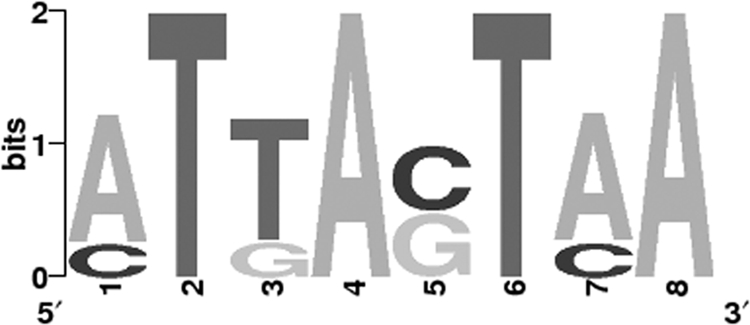
Motif logo of a conserved sequence in Cap1p-HA3- or Cap1p-CSE-HA3-enriched DNA fragments. The 189 Cap1p-HA3- or 117 Cap1p-CSE-HA3-bound sequences were used as input for motif discovery, using the SCOPE program (http://genie.dartmouth.edu/scope/) (10, 12). The highest scoring motif MTKASTMA common to both conditions (Cap1p-HA3 and Cap1p-CSE-HA3) is shown.
Global gene expression profile.
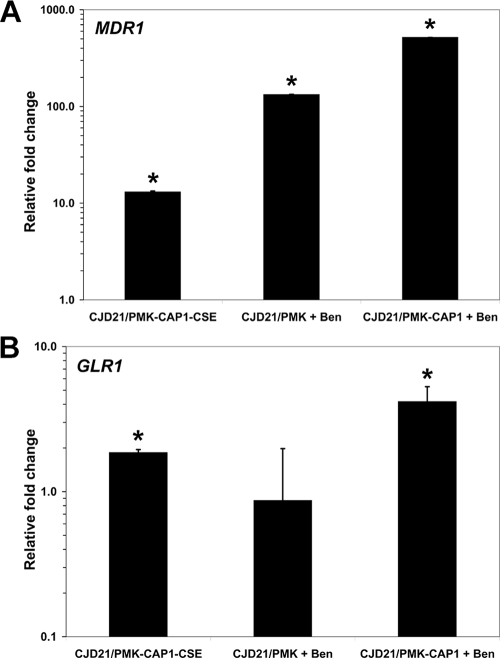
To test whether the expression of the Cap1p target genes identified by ChIP-chip was modulated by the CSE mutation, we compared the gene expression profiles of strains CJD21/PMK-CAP1-CSE with those of CJD21/PMK-CAP1 (Table 5) . Three independent RNA samples per strain were hybridized to custom-designed Affymetrix C. albicans microarrays, and the data were analyzed as described in Materials and Methods. Genes were considered as differentially expressed if (i) their average change (n-fold) in expression was ≥2.0, (ii) their expression changed by at least 2.0-fold in each experiment, and (iii) the change (n-fold) was considered statistically significant by Student's t test. Based on these criteria, we found 51 differentially expressed genes (Table 5), including GLR1, GRE2, OYE32, CIP1, MDR1, orf19.2262, orf19.3537, orf19.5060, orf19.1167, orf19.1340, orf19.5517, and orf19.6757, all of which are involved in responses to oxidative stress or coding for oxidoreductases. Indeed, using the GO term Finder tool at the CGD, we found that “Response to oxidative stress” and “Oxidoreductase activity” were the most significantly overrepresented GO terms among the 51 modulated genes (P = 0.004 and P = 0.01, respectively). We performed real-time RT-PCR assays to validate the expression array results (see Materials and Methods) and confirmed the differential expression of MDR1 and GLR1 with relative expression changes (± standard errors) of 13.1 ± 0.3 and 1.9 ± 0.1, respectively (Fig. 5).
TABLE 5.
List of genes whose expression is modulated in CJD21/PMK-CAP1-CSE versus CJD21/PMK-CAP1
| Systematic namea | CGD nameb | Molecular functionc | Relative level of gene expression (n-fold)d | Cap1p-CSE-HA3 binding ratioe | Cap1p-HA3 binding ratiof | MTKASTMA motifg | Starting positionh | Ending positionh | Strandi |
|---|---|---|---|---|---|---|---|---|---|
| orf19.3121 | Transcription corepressor activity | 145.3 | 1.9 | 1.2 | ATTACTAA | −177 | −170 | D | |
| ATTAGTAA | −39 | −32 | D | ||||||
| orf19.3131 | OYE32 | NADPH dehydrogenase activity | 79.2 | 3.3 | 1.6 | CTTAGTAA | −744 | −737 | D |
| orf19.113 | CIP1 | 67.5 | 2.0 | 1.2 | CTTACTAA | −573 | −566 | R | |
| orf19.5604 | MDR1 | Fluconazole transporter activity | 57.9 | 2.5 | 1.8 | ATGACTCA | −737 | −730 | D |
| ATTAGTAA | −533 | −526 | D | ||||||
| orf19.1763 | IFR1 | 38.2 | 2.5 | 1.2 | ATTAGTAA | −304 | −297 | D | |
| ATTAGTAA | −161 | −154 | D | ||||||
| orf19.2285 | 27.2 | 1.2 | 1.7 | ATTACTAA | −120 | −113 | D | ||
| orf19.3150 | GRE2 | Oxidoreductase activity | 25.9 | 1.8 | 1.7 | CTTACTAA | −390 | −383 | D |
| orf19.1149 | MRF1 | DNA binding | 18.1 | 1.3 | 1.4 | ATGAGTCA | −561 | −554 | D |
| orf19.2262 | NADPH:quinone reductase activity | 17.7 | 1.8 | ||||||
| orf19.2693 | Transcription corepressor activity | 12.7 | 1.4 | 1.7 | CTTAGTAA | −162 | −155 | D | |
| orf19.1167 | Sulfonate dioxygenase activity | 11.9 | — | — | CTTACTAA | −239 | −232 | D | |
| orf19.3122 | ARR3 | Arsenite transmembrane transporter activity | 11.2 | 1.9 | 1.2 | ATTACTAA | −329 | −322 | D |
| ATTAGTCA | −178 | −171 | D | ||||||
| CTTAGTCA | −968 | −961 | R | ||||||
| orf19.7531 | 10.4 | — | — | ATTACTAA | −79 | −72 | D | ||
| orf19.7042 | 9.9 | 1.9 | 1.9 | ATTACTAA | −709 | −702 | D | ||
| ATGAGTAA | −339 | −332 | D | ||||||
| ATTACTAA | −214 | −207 | D | ||||||
| orf19.1237 | ARO9 | Aromatic amino acid transaminase activity | 7.7 | — | — | ATTAGTAA | −153 | −146 | D |
| orf19.6898 | 7.0 | — | 1.4 | ATTAGTCA | −426 | −419 | D | ||
| ATTAGTAA | −340 | −333 | D | ||||||
| CTGACTAA | −516 | −509 | R | ||||||
| orf19.251 | 7.0 | 2.0 | 1.5 | ATTAGTAA | −939 | −932 | D | ||
| ATTACTAA | −725 | −718 | D | ||||||
| orf19.847 | YIM1 | Endopeptidase activity | 5.4 | 1.4 | 1.3 | ATTAGTAA | −169 | −162 | D |
| orf19.5770 | OPT8 | Oligopeptide transporter activity | 5.3 | — | 1.4 | CTGACTAA | −243 | −236 | D |
| orf19.1340 | Aldehyde reductase activity | 5.2 | — | 1.2 | ATTAGTCA | −156 | −149 | D | |
| orf19.4757 | NAR1 | 4.9 | — | — | ATTACTAA | −113 | −106 | D | |
| orf19.5259 | 4.9 | — | — | ||||||
| orf19.344 | 4.9 | 1.4 | 1.4 | ATTACTAA | −505 | −498 | D | ||
| orf19.747 | NBP35 | 4-Iron, 4-sulfur cluster binding | 4.8 | — | — | ||||
| orf19.5060 | GCS1 | Glutamate-cysteine ligase activity | 4.5 | — | — | ||||
| orf19.2862 | RIB1 | Cyclohydrolase activity | 4.0 | 1.3 | 1.3 | ATTACTAA | −913 | −906 | D |
| ATTACTAA | −882 | −875 | D | ||||||
| ATTAGTAA | −808 | −801 | D | ||||||
| CTTACTAA | −612 | −605 | R | ||||||
| CTTAGTAA | −139 | −132 | R | ||||||
| orf19.4449 | Superoxide dismutase copper chaperone activity | 3.6 | 1.2 | — | ATTACTAA | −114 | −107 | D | |
| orf19.6757 | GCY1 | Aldehyde reductase activity | 3.1 | 1.3 | 1.5 | ||||
| orf19.2396 | IFR2 | 3.1 | 1.5 | 1.2 | CTTACTCA | −33 | −26 | D | |
| orf19.1162 | 3.1 | — | — | ||||||
| orf19.3537 | Sulfiredoxin activity | 3.0 | — | 1.2 | ATGACTAA | −257 | −250 | D | |
| ATGAGTAA | −56 | −49 | R | ||||||
| orf19.5517 | Alcohol dehydrogenase (NADP+) activity | 2.9 | 2.0 | — | ATTACTAA | −180 | −173 | D | |
| orf19.4337 | Monocarboxylic acid transmembrane transporter activity | 2.8 | — | — | ATTAGTCA | −538 | −531 | D | |
| ATGACTCA | −601 | −594 | R | ||||||
| ATGAGTAA | −389 | −382 | R | ||||||
| orf19.3668 | HGT2 | Glucose transmembrane transporter activity | 2.6 | — | — | ATTACTAA | −239 | −232 | D |
| orf19.5133 | ZCF29 | Specific RNA polymerase II transcription factor activity | 2.5 | 1.3 | 1.2 | ATTACTAA | −64 | −57 | D |
| orf19.3130 | 2.5 | — | — | CTTACTAA | −144 | −137 | D | ||
| orf19.4147 | GLR1 | Glutathione-disulfide reductase activity | 2.5 | 1.2 | 1.4 | ATTAGTAA | −230 | −223 | D |
| orf19.3139 | 2.4 | — | — | ATTACTAA | −307 | −300 | D | ||
| orf19.6610 | Microtubule binding | 2.4 | — | — | |||||
| orf19.6464 | 2.2 | — | — | ATGACTAA | −697 | −690 | D | ||
| CTGACTCA | −463 | −456 | D | ||||||
| ATTACTAA | −90 | −83 | D | ||||||
| ATTACTCA | −167 | −160 | R | ||||||
| orf19.5257 | d-erythro-sphingosine kinase activity | 2.2 | — | — | ATTAGTAA | −681 | −674 | D | |
| orf19.105 | HAL22 | 3′(2′),5′-bisphosphate nucleotidase activity | 2.2 | — | — | CTTACTCA | −359 | −352 | D |
| orf19.4906 | 2.1 | — | — | CTTACTCA | −791 | −784 | D | ||
| ATTAGTAA | −333 | −326 | D | ||||||
| orf19.5811 | MET1 | Uroporphyrin III C-methyltransferase activity | 2.0 | — | — | ||||
| orf19.7589 | 0.5 | — | — | ||||||
| orf19.2256 | 0.5 | — | — | ||||||
| orf19.4900 | Alpha-1,3-mannosyltransferase activity | 0.5 | — | — | ATTAGTAA | −404 | −397 | R | |
| orf19.1866 | Hydrogen ion-transporting ATPase activity; rotational mechanism | 0.5 | — | — | |||||
| orf19.3523 | CRK1 | Mitogen-activated protein kinase activity | 0.5 | — | — | ||||
| orf19.1996 | CHA1 | Ammonia-lyase activity | 0.5 | — | — | ATTACTCA | −922 | −915 | R |
| ATTACTAA | −201 | −194 | R | ||||||
| orf19.5485 | MEC3 | DNA binding | 0.3 | — | — |
orf19 nomenclature according to the assembly 19 version.
Gene name according to the CGD (http://www.candidagenome.org/).
Descriptions using GO terminology according to the CGD.
Expression (n-fold) of the gene in strain CJD21/PMK-CAP1-CSE relative to that in strain CJD21/PMK-CAP1.
Corresponding log2-transformed pseudomedian binding ratio in Cap1p-CSE-HA3 ChIP-chip data (see Table S1 in the supplemental material). If more than one peak was associated with a target gene, the average log2-transformed pseudomedian binding ratio is shown. A dash indicates that binding peaks around the locus of the corresponding gene were not detected using our criteria (see Materials and Methods).
Corresponding log2-transformed pseudomedian binding ratio in Cap1p-HA3 ChIP-chip data (see Table S2 in the supplemental material). If more than one peak was associated with a target gene, the average log2-transformed pseudomedian binding ratio is shown. A dash indicates that binding peaks around the locus of the corresponding gene were not detected using our criteria (see Materials and Methods).
MTKASTMA sequence within the promoter region of the corresponding gene (see Materials and Methods).
Limits (starting and ending) of the MTKASTMA sequence relative to the ATG translation start site.
D, sense strand; R, antisense strand.
FIG. 5.
Quantitative real-time RT-PCR analysis of (A) MDR1 and (B) GLR1 genes differentially expressed in the microarray experiments. Bars in each graph indicate log-transformed relative changes in RNA expression of the samples indicated compared to their controls: CJD21/PMK-CAP1-CSE versus CJD21/PMK-CAP1, CJD21/PMK plus benomyl (Ben) versus CJD21/PMK plus DMSO and CJD21/PMK-CAP1 plus Ben versus CJD21/PMK-CAP1 plus DMSO. Asterisks denote statistical significance by the t test (P ≤ 0.05). Error bars denote standard errors.
When combining the expression and location data, we found that among the 51 differentially expressed genes, 26 also showed increased Cap1p-HA3 or Cap1-CSE-HA3 binding at their promoters in our ChIP-chip data (Table 5). Interestingly, all of these genes were upregulated, including GLR1, GRE2, OYE32, orf19.1340, orf19.2262, orf19.3537, orf19.5517, and orf19.6757, which code for oxidoreductases; ARR3, MDR1, and OPT8, which code for transporters; and orf19.3121, ZCF29, and orf19.2693, which code for transcriptional regulators (Table 4). Taken together, these data show that increased expression of several Cap1p targets accompanies Cap1p binding at these targets, indicating that Cap1p is a transcriptional activator. Our results also suggest that many genes differentially expressed in response to the CSE mutation, including all the downregulated genes (Table 5), are indirect Cap1p targets.
Transcriptional response of Cap1p targets to benomyl treatment.
Previous studies have shown that the genes responding to benomyl overlap significantly with those responding to H2O2 and thus to oxidative stress (30, 36), suggesting that benomyl is an inducer of Cap1p activity. To identify genes whose expression is induced by benomyl in a CAP1-dependent manner, we examined the gene expression profiles of the CAP1-expressing C. albicans strain CJD21/PMK-CAP1 treated with benomyl to those of CJD21/PMK-CAP1 treated with DMSO and those of the CAP1-deficient related strain CJD21/PMK also treated with benomyl to those of CJD21/PMK treated with DMSO (see Materials and Methods). When the gene expression profiles of benomyl-treated and diluent-treated CJD21/PMK-CAP1 were compared using our criteria (see Materials and Methods and Table S3 in the supplemental material), 432 genes were found to be differentially expressed. Similarly, when benomyl-treated and diluent-treated CAP1-deficient C. albicans gene expression profiles were compared using our criteria (see Materials and Methods and Table S4 in the supplemental material), 232 genes were found to be differentially expressed. By considering only those genes differentially expressed in the CAP1-expressing strain in response to benomyl compared to those differentially expressed in CJD21/PMK in response to benomyl, we found a set of 115 genes whose expression fit these criteria, qualifying them as genes induced by benomyl in a CAP1-dependent manner (see Table S5 in the supplemental material). We validated the expression microarray data by confirming the CAP1-dependent benomyl induction of MDR1 and GLR1, using real-time PCR (Fig. 5). This experiment showed that strain CJD21/PMK-CAP1 displayed a 517-fold induction of MDR1 expression in response to benomyl treatment, while the CAP1-deficient mutant CJD21/PMK displayed a 133-fold increase in MDR1 RNA upon treatment with benomyl (Fig. 5A), most likely due to the contribution of another transcriptional regulator (see Discussion). In addition, benomyl-treated CJD21/PMK-CAP1 displayed a 4.2-fold change in GLR1 expression, whereas CJD21/PMK treated with benomyl showed no significant change in GLR1 expression (Fig. 5B), demonstrating that Cap1p is the major regulator of GLR1 expression in response to benomyl.
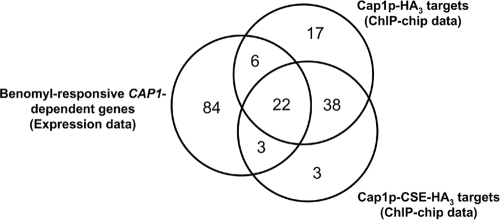
Interestingly, among the 115 genes responding to benomyl in a CAP1-dependent manner, 31 were also identified as target genes of Cap1p-HA3 or Cap1p-CSE-HA3 in our ChIP-chip data, as illustrated in the Venn diagram representing the overlap between the ChIP-chip data set and the benomyl-induced CAP1-dependent gene data set (Fig. 6). These 31 genes include CAP1 itself, MDR1, EBP1, OYE32, OYE23, and GLR1 (Table 6), all of which are involved in the OSR. Taken together, these results indicate that benomyl induces Cap1p activity and suggest that it activates the OSR via Cap1p in C. albicans (see Discussion).
FIG. 6.
Venn diagram of the overlap between the benomyl-responsive CAP1-dependent gene data set (left circle) and the Cap1p-HA3 (right upper circle) and Cap1p-CSE-HA3 (right lower circle) ChIP-chip datasets. The numbers in the Venn diagram indicate the numbers of genes.
TABLE 6.
List of Cap1p-bound targets whose expression is modulated upon benomyl treatment in a CAP1-dependent manner
| Systematic namea | CGD nameb | Molecular functionc | Relative change in gene expression (n-fold)d | Cap1p-CSE-HA3 binding ratioe | Cap1p-HA3 binding ratiof |
|---|---|---|---|---|---|
| orf19.3121 | Transcription corepressor activity | 574.6 | 1.9 | 1.2 | |
| orf19.1623 | CAP1 | Transcription factor activity | 453.7 | 1.5 | 1.5 |
| orf19.2285 | 228.3 | 1.2 | 1.7 | ||
| orf19.125 | EBP1 | NADPH dehydrogenase activity | 127.8 | 3.1 | 2.1 |
| orf19.1763 | IFR1 | 97.1 | 2.5 | 1.2 | |
| orf19.3433 | OYE23 | NADPH dehydrogenase activity | 90.6 | 2.7 | 2.2 |
| orf19.3234 | NADPH dehydrogenase activity | 58.2 | 1.3 | 1.2 | |
| orf19.720 | GST3 | Transcription corepressor activity | 48.5 | 1.5 | 1.4 |
| orf19.6898 | 33.7 | — | 1.4 | ||
| orf19.2262 | 25.7 | 1.8 | — | ||
| orf19.3131 | OYE32 | NADPH dehydrogenase activity | 25.7 | 3.3 | 1.6 |
| orf19.2396 | IFR2 | 18.9 | 1.5 | 1.2 | |
| orf19.3122 | ARR3 | Arsenite transmembrane transporter activity | 17.3 | 1.9 | 1.2 |
| orf19.3395 | 15.7 | — | 1.2 | ||
| orf19.3537 | 10.1 | — | 1.2 | ||
| orf19.5770 | OPT8 | Oligopeptide transporter activity | 8.7 | — | 1.4 |
| orf19.7042 | 6.1 | 1.9 | 1.9 | ||
| orf19.5517 | Alcohol dehydrogenase (NADP+) activity | 5.3 | 2.0 | — | |
| orf19.6586 | 5.1 | 1.4 | 1.6 | ||
| orf19.847 | YIM1 | Peptidase activity | 4.7 | 1.4 | 1.3 |
| orf19.5604 | MDR1 | Multidrug transporter activity | 4.4 | 2.5 | 1.8 |
| orf19.932 | Phospholipid-translocating ATPase activity | 4.3 | 1.2 | 1.2 | |
| orf19.4449 | Superoxide dismutase copper chaperone activity | 3.9 | 1.2 | — | |
| orf19.4147 | GLR1 | Glutathione-disulfide reductase activity | 3.8 | 1.2 | 1.4 |
| orf19.2862 | RIB1 | Cyclohydrolase activity | 3.7 | 1.3 | 1.3 |
| orf19.4907 | 3.5 | 1.4 | 1.7 | ||
| orf19.344 | 3.3 | 1.4 | 1.2 | ||
| orf19.6554 | 2.6 | — | 1.3 | ||
| orf19.5133 | ZCF29 | Specific RNA polymerase II transcription factor activity | 2.4 | 1.3 | 1.2 |
| orf19.6402 | CYS3 | Cystathionine gamma-lyase activity | 2.3 | — | 1.2 |
| orf19.251 | 2.0 | 2 | 1.5 |
orf19 nomenclature according to the assembly 19 version.
Gene name according to the CGD (http://www.candidagenome.org/).
Described using GO terminology according to the CGD.
Average (n = 3) relative change in gene expression of CJD21/PMK-CAP1 treated with benomyl versus CJD21/PMK treated with benomyl.
Corresponding log2-transformed pseudomedian binding ratio in Cap1p-CSE-HA3 ChIP-chip data (see Table S1 in the supplemental material). If more than one peak was associated with a target gene, the average log2-transformed pseudomedian binding ratio is shown. A dash indicates that binding-peaks around the locus of the corresponding gene were not detected using our criteria (see Materials and Methods).
Corresponding log2-transformed pseudomedian binding ratio in Cap1p-HA3 ChIP-chip data (see Table S2 in the supplemental material). If more than one peak was associated with a target gene, the average log2-transformed pseudomedian binding ratio is shown. A dash indicates that binding-peaks around the locus of the corresponding gene were not detected using our criteria (see Materials and Methods).
DISCUSSION
The bZIP transcription factor Cap1p undergoes nuclear retention upon activation by oxidative stress, while in cells grown in the absence of Cap1p-activating conditions, Cap1p shows diffuse cytoplasmic localization (68). It was also shown that Cap1p nuclear localization is constitutive when the activating mutation C477A is introduced in Cap1p (68). In this study, we introduced a similar gain-of-function mutation in Cap1p to identify the Cap1p regulon, hypothesizing that this alteration, by shifting Cap1p cellular localization to the nucleus, would result in increased Cap1p binding to its targets. We used the methionine-regulatable promoter (pCaEXP expression system) to drive high expression of the CAP1-HA3 or CAP1-CSE-HA3 alleles. We found that upon this forced overexpression, both Cap1p-HA3 and Cap1p-CSE-HA3 bound to DNA in vivo. It is likely that forced overexpression of CAP1-HA3 leads to an accumulation of the protein in the nucleus by overcoming Crm1p export activity. However, our finding that in most cases Cap1p-CSE-HA3 binding was increased relative to that of Cap1p-HA3 at common targets (Fig. 2; see also Tables S1 and S2 in the supplemental material) is consistent with the hypothesis that Cap1p-CSE-HA3 levels in the nucleus were higher than those of Cap1p-HA3 as a result of the CSE mutation. It is also consistent with our observation that the Cap1p-CSE-HA3 cells are more resistant to FLC than the Cap1p-HA3 cells (Fig. 1C). Other explanations could be that Cap1p-CSE-HA3 affinity to DNA is higher than that of Cap1p-HA3 or that the introduction of the HA3 tag in Cap1p has affected its nuclear localization. Previous studies used transcription factor overexpression approaches to study genome-wide transcription factor function (14, 60, 71). This strategy appears to mimic transcription factor physiological activation, presumably by increasing promoter occupancy of target genes as a consequence of supraphysiological levels of the protein (14). Importantly, this approach increases both the sensitivity and relevance of the data (14, 60, 62) as reflected in our present study, since several known potential direct Cap1p targets were bound by Cap1p (Fig. 2; see also Tables S1 and S2 in the supplemental material). However, we cannot rule out the possibility that overexpression of Cap1p and/or the introduction of the HA3 epitope tag in Cap1p drives nonphysiological binding in vivo, resulting in spurious binding, which may explain the identification of peaks in genome regions with no obvious ORFs (see the example in Fig. 2, lower panel). Also, transcription factor overexpression often causes cell growth inhibition (14, 71). In the present study, a growth inhibition effect was also detected, as colonies overexpressing CAP1-CSE-HA3 were clearly smaller than the control colonies, whereas a slight growth inhibition was detected in CAP1-HA3-expressing colonies (Fig. 2C). Thus, it is possible that an additive growth inhibition effect was caused by Cap1p hyperactivity.
A previous study was performed to compare genes coregulated with MDR1 in azole-resistant C. albicans strains with those induced in response to benomyl (30). Several genes that responded to benomyl treatment in a CAP1-dependent manner (see Table S5 in the supplemental material) were shown to be benomyl responsive by Karababa et al., including the stress genes orf19.3121, OYE23, EBP1, orf19.2262, OYE32, IFR2, orf19.251, orf19.5517, and TTR1 and the transporter genes MDR1, ARR3, and SNQ2 (30). In addition, genes of unknown function that were defined as CAP1 dependent upon benomyl treatment in our study were also found to be benomyl responsive by Karababa et al., for example, orf19.2285, orf19.6898, PRN1, orf19.7042, orf19.6586, YIM1, orf19.2043, and orf19.1162. Using our criteria (P ≤ 0.01; binding ratio, ≥2), we found a majority of these genes to be bound in vivo by Cap1p (except TTR1, SNQ2, PRN1, orf19.2043, and orf19.1162) (see Tables S1 and S2 in the supplemental material), indicating a direct transcriptional regulation by Cap1p. Although using a microarray with only partial coverage of the C. albicans genome, another previous study examined the C. albicans response to hydrogen peroxide, an inducer of oxidative stress (64). As expected, several genes responsive to benomyl treatment in a CAP1-dependent manner overlapped with those responsive to H2O2 treatment, including the stress genes EBP1, OYE23, orf19.2262, OYE32, orf19.5517, TRR1, orf19.251, IFR2, and GLR1. Another report by Wang et al. aimed at identifying genes differentially expressed under untreated conditions, also termed basal or stress-absent conditions, in a wild-type C. albicans strain versus those in a cap1Δ/cap1Δ mutant (63). This study identified 48 downregulated genes in the cap1 mutant relative to those in the wild-type strain, among which only EBP1, OYE23, OYE32, orf19.2262, ARR3, ZRT2, and orf19.868 were bound by Cap1p in the present study (see Tables S1 and S2 in the supplemental material), suggesting that many genes found by Wang et al. that were differentially expressed under untreated conditions are indirect Cap1p targets.
A striking finding was that Cap1p binds not only to the promoter region of its target genes but also within the ORF and the 3′ region, including the transcriptional termination region (Fig. 2; see also Tables S1 and S2 in the supplemental material). Interestingly, binding of Cap1p-CSE-HA3 within the ORFs was more frequent than that of Cap1p-HA3 (Fig. 2; see also Tables S1 and S2 in the supplemental material), suggesting that activation of Cap1p enhances its propensity to occupy intragenic regions. To our knowledge, this is the first report of such a transcription factor binding profile in C. albicans. Previous studies reported similar associations of transcription factors to both promoter and coding regions of their targets, including the tumor suppressor p53 and the estrogen receptor (5, 11). Interestingly, binding of both transcription factors to intragenic regions overlapped with that of RNA polymerase II binding sites (5, 11). Moreover, it was shown that p53 physically associated with RNA polymerase II, with which it travels within target gene loci (5). The intragenic binding profile of Cap1p as well as its binding at the 3′ end of target genes is consistent with a model in which Cap1p interacts with the transcriptional machinery and travels with the RNA polymerase II across the transcribed region of the target locus in a manner similar to that of p53. Interestingly, it was shown that Yap1p physically interacts with the general transcription factor TFIIA in S. cerevisiae and that TFIIA mutants exhibited an enhanced susceptibility to oxidative stress (32), highlighting a direct link between Yap1p and the RNA polymerase II complex. Another possibility could be that Cap1p interacts with chromatin-associated proteins.
Previous studies have implicated the zinc cluster transcription factors Mrr1p and Upc2p in regulating MDR1 gene expression in C. albicans (19, 20, 47, 71). While Mrr1p appears to act as a potent transcriptional activator of MDR1 in C. albicans azole-resistant clinical isolates overexpressing MDR1 (47), Upc2p appeared to act as a moderate activator or a repressor of MDR1, depending upon the activating signal (71). Consistently, a gain-of-function mutation in UPC2 from a C. albicans azole-resistant strain was shown to cause a moderate upregulation of MDR1 (20). We previously showed, using ChIP experiments, that Upc2p binds to the MDR1 promoter (71), whereas it is still unknown whether Mrr1p associates directly with the promoter region of MDR1. Based on previous studies and the present study, Cap1p appears to be a potent activator of MDR1 expression, as reflected by Northern blot and luciferase reporter analyses (3, 53) and by our expression microarray data (Table 5; see also Table S5 in the supplemental material). Another regulator that was shown to participate in the regulation of MDR1 expression is Mcm1p, which binds directly to an Mcm1p binding motif found in the promoter of MDR1 (52), a finding that was recently confirmed in vivo by genome-wide location analyses (37). Taken together, these observations suggest that a complex network of transcriptional regulators, including Cap1p, Mrr1p, Upc2p, and Mcm1p, is involved in the regulation of MDR1 in C. albicans. Studies are ongoing to determine whether Cap1p interacts physically with these other factors to regulate MDR1 expression.
Among the overrepresented functional categories of genes bound in vivo by Cap1p were “Response to drug,” “Hyphal cell wall,” “Phospholipid transport,” and “Regulation of nitrogen utilization” (Table 3), suggesting other roles for Cap1p besides the OSR. Of particular interest was the grouping of PDR16, RTA2, MDR1, and FLU1 into the functional category “Response to drug.” These genes were previously reported to participate in azole resistance in C. albicans (8, 29, 55, 66). While PDR16 and MDR1 are involved in clinical azole resistance (55, 66), FLU1 and RTA2 were shown to modulate cell susceptibility to azoles in C. albicans (8, 29), suggesting that Cap1p could confer azole resistance via PDR16, FLU1, RTA2, and/or MDR1 in C. albicans. Although no evidence of Cap1p gain-of-function mutations in azole-resistant clinical isolates of C. albicans has been reported to date, a recent study has reported the correlation of constitutive overexpression of FLU1 together with MDR1 with the development of clinical azole resistance (25). Whether upregulation of these two major facilitator-encoding genes is mediated by Cap1p remains to be determined. Another overrepresented functional category was “Hyphal cell wall,” which included SSA2, EBP1, orf19.251, PDC11, and ADH1. As Cap1p appears to be important for protecting C. albicans against the oxidative stress induced by neutrophils during the course of the immune response (24), it might regulate processes contributing to virulence, including hyphal growth. Neutrophils induce oxidative stress through the release of nitric oxide anions; thus, it would not be surprising if Cap1p regulates processes involved in nitrogen metabolism, as reflected by the enrichment of the functional category “Regulation of nitrogen utilization,” which included orf19.2693, orf19.3121, and GST3. Finally, “Phospholipid transport” was also among the overrepresented functional categories and might reflect a role for Cap1p in enhancing the dynamics of phospholipid metabolism, as these molecules are highly sensitive to oxidative stress.
Benomyl is an aneuploidogen, a toxic antimitotic drug also used as an antifungal agent that is thought to exert its effect by binding to tubulin and inhibiting tubulin assembly (44). Based on previous studies as well as the present study, it appears that benomyl induces the OSR in C. albicans. First, genes differentially expressed upon exposure to H2O2 overlap with those responsive to benomyl (30, 36). Second, we showed here that benomyl induces Cap1p activity, reflected by the upregulation of genes involved in oxidative stress in a CAP1-dependent manner. Indeed, the GO term “Response to oxidative stress” was among the most significantly enriched functional categories (P = 0.026) of the CAP1-dependent genes responding to benomyl (data not shown). A study with S. cerevisiae also showed that a subset of OSR genes was upregulated rapidly after the addition of benomyl (39). Interestingly, this transcriptional response involved Yap1p as the central regulator of the early response to benomyl treatment (39). Lucau-Danila et al. suggested that benomyl potentially acts at the level of Yap1p nuclear localization rather than at the level of DNA binding (39). Thus, it is possible that benomyl induces conformational changes within the Cap1p CRD by directly binding to the C-terminal domain of Cap1p, leading to nuclear retention of Cap1p and transcriptional activation. However, an effect on intracellular redox balance is not excluded, as the benomyl metabolite n-butylisocyanate, a cleavage product of benomyl, results in inhibitory effects on dehydrogenases or glutathione reductases (44), which in turn may activate the OSR via Cap1p.
Supplementary Material
Acknowledgments
We are indebted to Raphaëlle Lambert, Pierre Chagnon, and Anne-Sophie Guenier from the IRIC Genomics platform for their support with the Q-PCR and ChIP-chip experiments; Patrick Gendron from the IRIC Bioinformatics platform for the C. albicans genome browser; Ramin Homayouni for assistance with expression data analysis; Mike Snyder for the design of the C. albicans tiling arrays; and the Candida Genome Database for sequence information. We also thank Steve Trosok for critically reading the manuscript and Brian Wilhelm, Marie-Pier Scott-Boyer, Simon Drouin, François Robert, and Sébastien Lemieux for stimulating discussions.
This work was supported by research grants from the Canadian Institutes of Health Research (CIHR) to M.R. (grants MT-15679 and HOP-67260) and from the National Institutes of Health (R01 AI058145) to P.D.R. S.Z. is supported by a doctoral studentship from the Fonds de la Recherche en Santé du Québec (FRSQ). IRIC is supported by the Canadian Center of Excellence in Commercialization and Research, the Canadian Foundation for Innovation, and the Fonds de Recherche en Santé du Québec.
Footnotes
Published ahead of print on 24 April 2009.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Akins, R. A. 2005. An update on antifungal targets and mechanisms of resistance in Candida albicans. Med. Mycol. 43285-318. [DOI] [PubMed] [Google Scholar]
- 2.Alarco, A.-M., I. Balan, D. Talibi, N. Mainville, and M. Raymond. 1997. AP1-mediated multidrug resistance in Saccharomyces cerevisiae requires FLR1 encoding a transporter of the major facilitator superfamily. J. Biol. Chem. 27219304-19313. [DOI] [PubMed] [Google Scholar]
- 3.Alarco, A.-M., and M. Raymond. 1999. The bZip transcription factor Cap1p is involved in multidrug resistance and oxidative stress response in Candida albicans. J. Bacteriol. 181700-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alonso-Monge, R., F. Navarro-Garcia, E. Roman, A. I. Negredo, B. Eisman, C. Nombela, and J. Pla. 2003. The Hog1 mitogen-activated protein kinase is essential in the oxidative stress response and chlamydospore formation in Candida albicans. Eukaryot. Cell 2351-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balakrishnan, S. K., and D. S. Gross. 2008. The tumor suppressor p53 associates with gene coding regions and co-traverses with elongating RNA polymerase II in an in vivo model. Oncogene 272661-2672. [DOI] [PubMed] [Google Scholar]
- 6.Buck, M. J., and J. D. Lieb. 2004. ChIP-chip: considerations for the design, analysis, and application of genome-wide chromatin immunoprecipitation experiments. Genomics 83349-360. [DOI] [PubMed] [Google Scholar]
- 7.Bustamante, C. I. 2005. Treatment of Candida infection: a view from the trenches! Curr. Opin. Infect. Dis. 18490-495. [DOI] [PubMed] [Google Scholar]
- 8.Calabrese, D., J. Bille, and D. Sanglard. 2000. A novel multidrug efflux transporter gene of the major facilitator superfamily from Candida albicans (FLU1) conferring resistance to fluconazole. Microbiology 1462743-2754. [DOI] [PubMed] [Google Scholar]
- 9.Care, R. S., J. Trevethick, K. M. Binley, and P. E. Sudbery. 1999. The MET3 promoter: a new tool for Candida albicans molecular genetics. Mol. Microbiol. 34792-798. [DOI] [PubMed] [Google Scholar]
- 10.Carlson, J. M., A. Chakravarty, C. E. DeZiel, and R. H. Gross. 2007. SCOPE: a web server for practical de novo motif discovery. Nucleic Acids Res. 35W259-W264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carroll, J. S., C. A. Meyer, J. Song, W. Li, T. R. Geistlinger, J. Eeckhoute, A. S. Brodsky, E. K. Keeton, K. C. Fertuck, G. F. Hall, Q. Wang, S. Bekiranov, V. Sementchenko, E. A. Fox, P. A. Silver, T. R. Gingeras, X. S. Liu, and M. Brown. 2006. Genome-wide analysis of estrogen receptor binding sites. Nat. Genet. 381289-1297. [DOI] [PubMed] [Google Scholar]
- 12.Chakravarty, A., J. M. Carlson, R. S. Khetani, and R. H. Gross. 2007. A novel ensemble learning method for de novo computational identification of DNA binding sites. BMC Bioinform. 8249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chauhan, N., J. P. Latge, and R. Calderone. 2006. Signalling and oxidant adaptation in Candida albicans and Aspergillus fumigatus. Nat. Rev. Microbiol. 4435-444. [DOI] [PubMed] [Google Scholar]
- 14.Chua, G., Q. D. Morris, R. Sopko, M. D. Robinson, O. Ryan, E. T. Chan, B. J. Frey, B. J. Andrews, C. Boone, and T. R. Hughes. 2006. Identifying transcription factor functions and targets by phenotypic activation. Proc. Natl. Acad. Sci. USA 10312045-12050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coste, A., A. Selmecki, A. Forche, D. Diogo, M. E. Bougnoux, C. d'Enfert, J. Berman, and D. Sanglard. 2007. Genotypic evolution of azole resistance mechanisms in sequential Candida albicans isolates. Eukaryot. Cell 61889-1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coste, A., V. Turner, F. Ischer, J. Morschhauser, A. Forche, A. Selmecki, J. Berman, J. Bille, and D. Sanglard. 2006. A mutation in Tac1p, a transcription factor regulating CDR1 and CDR2, is coupled with loss of heterozygosity at chromosome 5 to mediate antifungal resistance in Candida albicans. Genetics 1722139-2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coste, A. T., M. Karababa, F. Ischer, J. Bille, and D. Sanglard. 2004. TAC1, transcriptional activator of CDR genes, is a new transcription factor involved in the regulation of Candida albicans ABC transporters CDR1 and CDR2. Eukaryot. Cell 31639-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Repentigny, L., D. Lewandowski, and P. Jolicoeur. 2004. Immunopathogenesis of oropharyngeal candidiasis in human immunodeficiency virus infection. Clin. Microbiol. Rev. 17729-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dunkel, N., J. Blass, P. D. Rogers, and J. Morschhauser. 2008. Mutations in the multi-drug resistance regulator MRR1, followed by loss of heterozygosity, are the main cause of MDR1 overexpression in fluconazole-resistant Candida albicans strains. Mol. Microbiol. 69827-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dunkel, N., T. T. Liu, K. S. Barker, R. Homayouni, J. Morschhauser, and P. D. Rogers. 2008. A gain-of-function mutation in the transcription factor Upc2p causes upregulation of ergosterol biosynthesis genes and increased fluconazole resistance in a clinical Candida albicans isolate. Eukaryot. Cell 71180-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eggimann, P., J. Garbino, and D. Pittet. 2003. Management of Candida species infections in critically ill patients. Lancet Infect. Dis. 3772-785. [DOI] [PubMed] [Google Scholar]
- 22.Enjalbert, B., D. A. Smith, M. J. Cornell, I. Alam, S. Nicholls, A. J. Brown, and J. Quinn. 2006. Role of the Hog1 stress-activated protein kinase in the global transcriptional response to stress in the fungal pathogen Candida albicans. Mol. Biol. Cell 171018-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fonzi, W. A., and M. Y. Irwin. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134717-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fradin, C., P. De Groot, D. MacCallum, M. Schaller, F. Klis, F. C. Odds, and B. Hube. 2005. Granulocytes govern the transcriptional response, morphology and proliferation of Candida albicans in human blood. Mol. Microbiol. 56397-415. [DOI] [PubMed] [Google Scholar]
- 25.Goldman, G. H., M. E. Silva Ferreira, M. E. dos Reis, M. Savoldi, D. Perlin, S. Park, P. C. Godoy Martinez, M. H. Goldman, and A. L. Colombo. 2004. Evaluation of fluconazole resistance mechanisms in Candida albicans clinical isolates from HIV-infected patients in Brazil. Diagn. Microbiol. Infect. Dis. 5025-32. [DOI] [PubMed] [Google Scholar]
- 26.Harry, J. B., B. G. Oliver, J. L. Song, P. M. Silver, J. T. Little, J. Choiniere, and T. C. White. 2005. Drug-induced regulation of the MDR1 promoter in Candida albicans. Antimicrob. Agents Chemother. 492785-2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herrero, E., J. Ros, G. Belli, and E. Cabiscol. 2008. Redox control and oxidative stress in yeast cells. Biochim. Biophys. Acta 17801217-1235. [DOI] [PubMed] [Google Scholar]
- 28.Hiller, D., S. Stahl, and J. Morschhauser. 2006. Multiple cis-acting sequences mediate upregulation of the MDR1 efflux pump in a fluconazole-resistant clinical Candida albicans isolate. Antimicrob. Agents Chemother. 502300-2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jia, X. M., Z. P. Ma, Y. Jia, P. H. Gao, J. D. Zhang, Y. Wang, Y. G. Xu, L. Wang, Y. Y. Cao, Y. B. Cao, L. X. Zhang, and Y. Y. Jiang. 2008. RTA2, a novel gene involved in azole resistance in Candida albicans. Biochem. Biophys. Res. Commun. 373631-636. [DOI] [PubMed] [Google Scholar]
- 30.Karababa, M., A. T. Coste, B. Rognon, J. Bille, and D. Sanglard. 2004. Comparison of gene expression profiles of Candida albicans azole-resistant clinical isolates and laboratory strains exposed to drugs inducing multidrug transporters. Antimicrob. Agents Chemother. 483064-3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kelly, R., S. M. Miller, M. B. Kurtz, and D. R. Kirsch. 1987. Directed mutagenesis in Candida albicans: one-step gene disruption to isolate ura3 mutants. Mol. Cell. Biol. 7199-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kraemer, S. M., D. A. Goldstrohm, A. Berger, S. Hankey, S. A. Rovinsky, W. Scott Moye-Rowley, and L. A. Stargell. 2006. TFIIA plays a role in the response to oxidative stress. Eukaryot. Cell 51081-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuge, S., M. Arita, A. Murayama, K. Maeta, S. Izawa, Y. Inoue, and A. Nomoto. 2001. Regulation of the yeast Yap1p nuclear export signal is mediated by redox signal-induced reversible disulfide bond formation. Mol. Cell. Biol. 216139-6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuge, S., N. Jones, and A. Nomoto. 1997. Regulation of yAP-1 nuclear localization in response to oxidative stress. EMBO J. 161710-1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuge, S., T. Toda, N. Iizuka, and A. Nomoto. 1998. Crm1 (XpoI) dependent nuclear export of the budding yeast transcription factor yAP-1 is sensitive to oxidative stress. Genes Cells 3521-532. [DOI] [PubMed] [Google Scholar]
- 36.Kusch, H., S. Engelmann, D. Albrecht, J. Morschhauser, and M. Hecker. 2007. Proteomic analysis of the oxidative stress response in Candida albicans. Proteomics 7686-697. [DOI] [PubMed] [Google Scholar]
- 37.Lavoie, H., A. Sellam, C. Askew, A. Nantel, and M. Whiteway. 2008. A toolbox for epitope-tagging and genome-wide location analysis in Candida albicans. BMC Genomics 9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu, T. T., S. Znaidi, K. S. Barker, L. Xu, R. Homayouni, S. Saidane, J. Morschhauser, A. Nantel, M. Raymond, and P. D. Rogers. 2007. Genome-wide expression and location analyses of the Candida albicans Tac1p regulon. Eukaryot. Cell 62122-2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lucau-Danila, A., G. Lelandais, Z. Kozovska, V. Tanty, T. Delaveau, F. Devaux, and C. Jacq. 2005. Early expression of yeast genes affected by chemical stress. Mol. Cell. Biol. 251860-1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lupetti, A., R. Danesi, M. Campa, M. Del Tacca, and S. Kelly. 2002. Molecular basis of resistance to azole antifungals. Trends Mol. Med. 876-81. [DOI] [PubMed] [Google Scholar]
- 41.MacPherson, S., B. Akache, S. Weber, X. De Deken, M. Raymond, and B. Turcotte. 2005. Candida albicans zinc cluster protein Upc2p confers resistance to antifungal drugs and is an activator of ergosterol biosynthetic genes. Antimicrob. Agents Chemother. 491745-1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.MacPherson, S., M. Larochelle, and B. Turcotte. 2006. A fungal family of transcriptional regulators: the zinc cluster proteins. Microbiol. Mol. Biol. Rev. 70583-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maeta, K., S. Izawa, S. Okazaki, S. Kuge, and Y. Inoue. 2004. Activity of the Yap1 transcription factor in Saccharomyces cerevisiae is modulated by methylglyoxal, a metabolite derived from glycolysis. Mol. Cell. Biol. 248753-8764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McCarroll, N. E., A. Protzel, Y. Ioannou, H. F. Frank Stack, M. A. Jackson, M. D. Waters, and K. L. Dearfield. 2002. A survey of EPA/OPP and open literature on selected pesticide chemicals. III. Mutagenicity and carcinogenicity of benomyl and carbendazim. Mutat. Res. 5121-35. [DOI] [PubMed] [Google Scholar]
- 45.Molin, M., J. P. Renault, G. Lagniel, S. Pin, M. Toledano, and J. Labarre. 2007. Ionizing radiation induces a Yap1-dependent peroxide stress response in yeast. Free Radic. Biol. Med. 43136-144. [DOI] [PubMed] [Google Scholar]
- 46.Morschhäuser, J. 2002. The genetic basis of fluconazole resistance development in Candida albicans. Biochim. Biophys. Acta 1587240-248. [DOI] [PubMed] [Google Scholar]
- 47.Morschhäuser, J., K. S. Barker, T. T. Liu, J. Blass-Warmuth, R. Homayouni, and P. D. Rogers. 2007. The transcription factor Mrr1p controls expression of the MDR1 efflux pump and mediates multidrug resistance in Candida albicans. PLoS Pathog. 3e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moye-Rowley, W. S. 2003. Regulation of the transcriptional response to oxidative stress in fungi: similarities and differences. Eukaryot. Cell 2381-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nguyên, D. T., A. M. Alarco, and M. Raymond. 2001. Multiple Yap1p-binding sites mediate induction of the yeast major facilitator FLR1 gene in response to drugs, oxidants, and alkylating agents. J. Biol. Chem. 2761138-1145. [DOI] [PubMed] [Google Scholar]
- 50.Pappas, P. G., J. H. Rex, J. D. Sobel, S. G. Filler, W. E. Dismukes, T. J. Walsh, and J. E. Edwards. 2004. Guidelines for treatment of candidiasis. Clin. Infect. Dis. 38161-189. [DOI] [PubMed] [Google Scholar]
- 51.Peng, S., A. A. Alekseyenko, E. Larschan, M. I. Kuroda, and P. J. Park. 2007. Normalization and experimental design for ChIP-chip data. BMC Bioinform. 8219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Riggle, P. J., and C. A. Kumamoto. 2006. Transcriptional regulation of MDR1, encoding a drug efflux determinant, in fluconazole-resistant Candida albicans strains through an Mcm1p binding site. Eukaryot. Cell 51957-1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rognon, B., Z. Kozovska, A. T. Coste, G. Pardini, and D. Sanglard. 2006. Identification of promoter elements responsible for the regulation of MDR1 from Candida albicans, a major facilitator transporter involved in azole resistance. Microbiology 1523701-3722. [DOI] [PubMed] [Google Scholar]
- 54.Rose, M. D., F. Winston, and P. Hieter. 1990. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 55.Saidane, S., S. Weber, X. De Deken, G. St-Germain, and M. Raymond. 2006. PDR16-mediated azole resistance in Candida albicans. Mol. Microbiol. 601546-1562. [DOI] [PubMed] [Google Scholar]
- 56.Sanglard, D., and F. C. Odds. 2002. Resistance of Candida species to antifungal agents: molecular mechanisms and clinical consequences. Lancet Infect. Dis. 273-85. [DOI] [PubMed] [Google Scholar]
- 57.Schmitt, M. E., T. A. Brown, and B. L. Trumpower. 1990. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 183091-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schneider, B. L., W. Seufert, B. Steiner, Q. H. Yang, and A. B. Futcher. 1995. Use of polymerase chain reaction epitope tagging for protein tagging in Saccharomyces cerevisiae. Yeast 111265-1274. [DOI] [PubMed] [Google Scholar]
- 59.Sherman, F. 1991. Getting started with yeast. Methods Enzymol. 1943-21. [DOI] [PubMed] [Google Scholar]
- 60.Sopko, R., D. Huang, N. Preston, G. Chua, B. Papp, K. Kafadar, M. Snyder, S. G. Oliver, M. Cyert, T. R. Hughes, C. Boone, and B. Andrews. 2006. Mapping pathways and phenotypes by systematic gene overexpression. Mol. Cell 21319-330. [DOI] [PubMed] [Google Scholar]
- 61.Srikantha, T., A. R. Borneman, K. J. Daniels, C. Pujol, W. Wu, M. R. Seringhaus, M. Gerstein, S. Yi, M. Snyder, and D. R. Soll. 2006. TOS9 regulates white-opaque switching in Candida albicans. Eukaryot. Cell 51674-1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tang, L., X. Liu, and N. D. Clarke. 2006. Inferring direct regulatory targets from expression and genome location analyses: a comparison of transcription factor deletion and overexpression. BMC Genomics 7215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang, Y., Y. Y. Cao, Y. B. Cao, D. J. Wang, X. M. Jia, X. P. Fu, J. D. Zhang, Z. Xu, K. Ying, W. S. Chen, and Y. Y. Jiang. 2007. Cap1p plays regulation roles in redox, energy metabolism and substance transport: an investigation on Candida albicans under normal culture condition. Front. Biosci. 12145-153. [DOI] [PubMed] [Google Scholar]
- 64.Wang, Y., Y. Y. Cao, X. M. Jia, Y. B. Cao, P. H. Gao, X. P. Fu, K. Ying, W. S. Chen, and Y. Y. Jiang. 2006. Cap1p is involved in multiple pathways of oxidative stress response in Candida albicans. Free Radic. Biol. Med. 401201-1209. [DOI] [PubMed] [Google Scholar]
- 65.Wemmie, J. A., S. M. Steggerda, and W. S. Moye-Rowley. 1997. The Saccharomyces cerevisiae AP-1 protein discriminates between oxidative stress elicited by the oxidants H2O2 and diamide. J. Biol. Chem. 2727908-7914. [DOI] [PubMed] [Google Scholar]
- 66.Wirsching, S., S. Michel, and J. Morschhauser. 2000. Targeted gene disruption in Candida albicans wild-type strains: the role of the MDR1 gene in fluconazole resistance of clinical Candida albicans isolates. Mol. Microbiol. 36856-865. [DOI] [PubMed] [Google Scholar]
- 67.Yan, C., L. H. Lee, and L. I. Davis. 1998. Crm1p mediates regulated nuclear export of a yeast AP-1-like transcription factor. EMBO J. 177416-7429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang, X., M. de Micheli, S. T. Coleman, D. Sanglard, and W. S. Moye-Rowley. 2000. Analysis of the oxidative stress regulation of the Candida albicans transcription factor, Cap1p. Mol. Microbiol. 36618-629. [DOI] [PubMed] [Google Scholar]
- 69.Zhang, Z. D., J. Rozowsky, H. Y. Lam, J. Du, M. Snyder, and M. Gerstein. 2007. Tilescope: online analysis pipeline for high-density tiling microarray data. Genome Biol. 8R81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Znaidi, S., X. De Deken, S. Weber, T. Rigby, A. Nantel, and M. Raymond. 2007. The zinc cluster transcription factor Tac1p regulates PDR16 expression in Candida albicans. Mol. Microbiol. 66440-452. [DOI] [PubMed] [Google Scholar]
- 71.Znaidi, S., S. Weber, O. Z. Al Abdin, P. Bomme, S. Saidane, S. Drouin, S. Lemieux, X. De Deken, F. Robert, and M. Raymond. 2008. Genomewide location analysis of Candida albicans Upc2p, a regulator of sterol metabolism and azole drug resistance. Eukaryot. Cell 7836-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.