Abstract
Fusarium head blight caused by Fusarium graminearum is an important disease of wheat and barley. In a previous study, we identified several mutants with reduced virulence by insertional mutagenesis. A transducin beta-like gene named FTL1 was disrupted in one of these nonpathogenic mutants. FTL1 is homologous to Saccharomyces cerevisiae SIF2, which is a component of the Set3 complex involved in late stages of ascospore formation. The Δftl1 mutant was significantly reduced in conidiation and failed to cause typical disease symptoms. It failed to colonize the vascular tissues of rachis or cause necrosis on the rachis of inoculated wheat heads. The Δftl1 mutant also was defective in spreading from infected anthers to ovaries and more sensitive than the wild type to plant defensins MsDef1 and osmotin. However, the activation of two mitogen-activated protein kinases, Mgv1 and Gpmk1, production of deoxynivalenol, and expression of genes known to be important for plant infection in F. graminearum were not affected, indicating that the defect of the Δftl1 mutant in plant infection is unrelated to known virulence factors in this pathogen and may involve novel mechanisms. The Δftl1 deletion mutant was significantly reduced in histone deacetylation, and many members of the yeast Set3 complex are conserved in F. graminearum. FTL1 appears to be a component of this well-conserved protein complex that plays a critical role in the penetration and colonization of wheat tissues.
The filamentous ascomycete Fusarium graminearum (teleomorph Gibberella zeae) is the main causal agent of Fusarium head blight (FHB), or scab, which is an important disease on wheat and barley throughout the world (18). It also causes stalk and ear rots of maize and infects other small grains. In addition to causing yield losses, this pathogen often contaminates infested grains with trichothecene and estrogenic mycotoxins, such as deoxynivalenol (DON) and zearalenone. Unfortunately, complete resistance to F. graminearum is lacking in wheat, and fungicide application is not cost-effective for FHB control in wheat and barley.
F. graminearum overwinters in infected plant debris and produces ascospores in the spring. Ascospores are forcibly discharged from mature perithecia (52) and function as the primary inoculum for FHB. The multicellular conidia or macroconidia are important for spreading the disease in the field and colonizing plant vegetative tissues. Wheat spikes are most susceptible to FHB at anthesis (34a). Although F. graminearum can colonize glumes, anthers are the main site of primary infection on flowering wheat heads (3, 38). Earlier studies indicated that wheat anther extracts stimulate F. graminearum virulence on wheat. Choline and glycine betaine were identified as two major components in anthers that stimulate fungal growth and predispose wheat to F. graminearum infection (50, 51). Under conducive conditions, the fungus can spread from the infected floret along the rachis and cause severe damage. The production of DON, the first virulence factor identified in F. graminearum (11, 42), is not necessary for the initial infection but is important for the spread of FHB on infected wheat heads (2).
In the past few years, genetic and genomic studies of F. graminearum have advanced significantly. The genome of F. graminearum has been sequenced (10) and a whole-genome microarray of this haploid homothallic fungus is commercially available (21). A number of pathogenicity or virulence factors have been identified by insertional mutagenesis or targeted gene deletion approaches. Two mitogen-activated protein (MAP) kinase genes, MGV1 and GPMK1, are essential for pathogenicity in F. graminearum (23, 24). Genes that are important for full virulence in F. graminearum on wheat include FGL1 (54), GzCPS1 (31), FBP1 (22), FSR1 (48), SID1 (19), NPS6 (37), RAS2 (5), GzGPA2 and GzGPB1 (56), and HMR1 (47). These virulence-associated genes encode proteins with various biochemical activities, such as lipase, nonribosomal peptide synthase, Ras protein, and 3-hydroxy 3-methylglutaryl coenzyme A reductase. Several genes involved in the primary metabolism, such as the CBL1, RSY1, GzHIS7, ADE5, and ARG2 genes (29, 44, 46) that are required for methionine, histidine, and arginine syntheses, also have been implicated in plant infection in F. graminearum. Overall, molecular mechanisms underlying F. graminearum pathogenesis appear to be complex and remain to be fully understood.
In a previous study, we identified 11 restriction enzyme-mediated integration (REMI) mutants that are defective in plant infection (46). In one of these mutants, the transforming vector was inserted in a predicted gene named FTL1 (for Fusarium transducin beta-like gene 1). FTL1 is homologous to the mammalian TBL1 or TBLR1 genes (40, 55) and the Saccharomyces cerevisiae SIF2 gene (8). The products of these genes are components of protein complexes involving histone deacetylases (HDACs). In mammalian cells, TBL1 and TBLR1 are parts of the N-CoR/SMRT/HDAC complexes (40). In yeast, SIF2 is a part of the Set3 complex regulating ascospore formation. In F. graminearum, the Δftl1 gene replacement mutant was significantly reduced in conidiation and failed to cause typical head blight symptoms on flowering wheat heads. It failed to colonize vascular tissues or cause necrosis on the rachis of inoculated wheat heads. The Δftl1 mutant also was defective in spreading from infected anthers to ovaries and was more sensitive than the wild type to plant defensins MsDef1 and osmotin. Although it was normal in the production of deoxynivalenol and the expression of known virulence factors, the Δftl1 mutant was significantly reduced in HDAC activities. FTL1 appears to be a component of this well-conserved HDAC complex that plays a critical role in the penetration and colonization of wheat tissues.
MATERIALS AND METHODS
Culture conditions and fungal transformation.
The F. graminearum wild-type strain PH-1 (NRRL 31084) and transformants generated in this study (see Table 3, below) were cultured at 25°C on V8 juice agar or minimal medium (MM) plates (23, 53). Transformant GZT501 expressing the green fluorescent protein (GFP) construct pTEFEGFP (49) was kindly provided by Robert Proctor at USDA Agricultural Research Service. Protoplast preparation and fungal transformation were performed as described previously (42). Complete medium (CM) supplemented with 250 μg/ml hygromycin B (Calbiochem, La Jolla, CA) or 150 μg/ml Geneticin (Sigma, St. Louis, MO) was used for selection of transformants (23). DON and ergosterol production were measured with rice grain cultures as described previously (45). The nit1 mutant 11622 was used for generating outcross perithecia with the Δftl1 mutant and isolation of random ascospore progeny (23). Sensitivities to MsDef1 (43) and osmotin (9) were measured by assaying conidium germination and germ tube growth. MsDef1 was added to conidium suspensions to final concentrations of 0.5, 1, 2, 4, and 10 μM. Osmotin was added to concentrations of 10 μM, 25 μM, and 32 μM.
TABLE 3.
Defects of the Δftl1 mutants in growth, conidiation, and plant infection
| Strain | Growth rate (cm/day) | Conidiation (104 conidia/ml) | Disease indexa |
|---|---|---|---|
| PH-1 (WT) | 1.33 ± 0.04 | 293.3 ± 21.7 | 6.8 ± 1.2 |
| M75 (REMI mutant) | 0.80 ± 0.03 | 0.7 ± 0.2 | 0.0 ± 0 |
| T1 (Δftl1) | 0.83 ± 0.04 | 1.3 ± 0.3 | 0.2 ± 0.1 |
| TDD2 (Δftl1/FTL1ΔLisH) | 0.80 ± 0.04 | 0.5 ± 0.1 | 0.0 ± 0 |
| TC1 (Δftl1/FTL1WT) | 1.32 ± 0.03 | 290.7 ± 12.7 | 7.1 ± 0.8 |
The disease index is reported as the mean (± standard deviation) number of symptomatic spikelets at 14 dpi in four independent infection assays.
Molecular manipulations.
RNA isolated with the TRIzol reagent (Invitrogen, Carlsbad, CA) was used for cDNA synthesis with the ProtoScript first strand cDNA synthesis kit (New England Biolabs, Ipswich, MA). The full-length FTL1 open reading frame (ORF) was amplified with primers TY1F and TY2R2 from first-strand cDNA and cloned into pBD-GAL4 (Stratagene, La Jolla, CA) as pZMH17. For microarray analysis, RNA was isolated from conidia germinated in CM for 18 h as described previously (21). Sequence analysis of pZHH17 revealed that the annotation of the third intron of FGSG_00332.3 was incorrect. Total proteins were extracted from mycelia collected from 2-day-old CM cultures as described previously (7). For Western blot analysis, proteins (approximately 20 μg per sample) were separated on a 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel and transferred to nitrocellulose membranes. The expression and activation of Mgv1 and Gpmk1 MAP kinases were detected with the PhosphoPlus p44/42 MAP kinase antibody kit (Cell Signaling Technology, Beverly, MA) using the ECL Supersignal system (Pierce, Rockford, IL).
For microarray analysis, RNA quality was analyzed with a Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA). One Fusarium GeneChip (21) was used for each of three biological replicates of the wild type and Δftl1 mutant. For each sample, 5 μg of total RNA was labeled with the Affymetrix (Santa Clara, CA) eukaryotic RNA labeling kit. Hybridization and washing followed standard Affymetrix procedures at the Purdue Core Genomics Facility. The hybridization signals were scanned with a GeneChip GCS 3000 scanner (Affymetrix). The resulting CEL files were processed with the software MAS5.0 (Affymetrix) to test signal intensities. The CEL files also were imported to and analyzed by using GeneSpring GX V7.2 (Agilent Technologies).
Generation of the Δftl1 mutant.
The FTL1 gene replacement vector pKY56 was constructed by the ligation-PCR approach (58). A 0.96-kb upstream fragment and a 0.92-kb downstream fragment were amplified with primers DL1F/DL2R and primers DL3F/DL4R (Table 1) and digested with FseI and AscI, respectively. After ligating these PCR products with the AscI-FseI hph fragment, a 3.3-kb FTL1 gene replacement construct was amplified with primers DL1F and DL4R and cloned into pGEM-T Easy (Promega, Madison, WI) as pYK56, which was used to transform PH-1 protoplasts. Hygromycin-resistant transformants were screened by PCR with primers TN1F and TN2R (see Fig. 2A, below) and further characterized by Southern blot analysis. For complementation assays, a 4.5-kb fragment containing the FTL1 gene was amplified with primers TC3F and TC4R (Table 1) and cloned into pGEM-T Easy as pZMH7 (see Fig. 2A), which was cotransformed with pSM334 (23) into the Δftl1 mutant T1.
TABLE 1.
Wild-type and mutant strains of Fusarium graminearum used in this study
| Strain | Genotype description | Reference |
|---|---|---|
| PH-1 | Wild type | Trail et al. (53) |
| 11622 | nit1 mutant of the wild-type strain G3639 | Hou et al. (23) |
| GZT501 | Transformant of G3639 expressing GFP constitutively | Skadsen and Hohn (49) |
| M75 | REMI mutant of PH-1 | Seong et al. (46) |
| T1 | Δftl1 mutant | This study |
| T2 | Δftl1 mutant | This study |
| T3 | Δftl1 mutant | This study |
| TC1 | Complementation strain of Δftl1 mutant T1 | This study |
| TC3 | Complementation strain of Δftl1 mutant T1 | This study |
| TDD1 | FTL1ΔLisH (pZHM13) in Δftl1 mutant T1 | This study |
| TDD2 | FTL1ΔLisH (pZHM13) in Δftl1 mutant T1 | This study |
| TDD3 | FTL1ΔLisH (pZHM13) in Δftl1 mutant T1 | This study |
| RM1 | Transformant of Δftl1 mutant expressing EGFP | This study |
| TGN1 | FTL1-GFP (pZMH12) in PH-1 | This study |
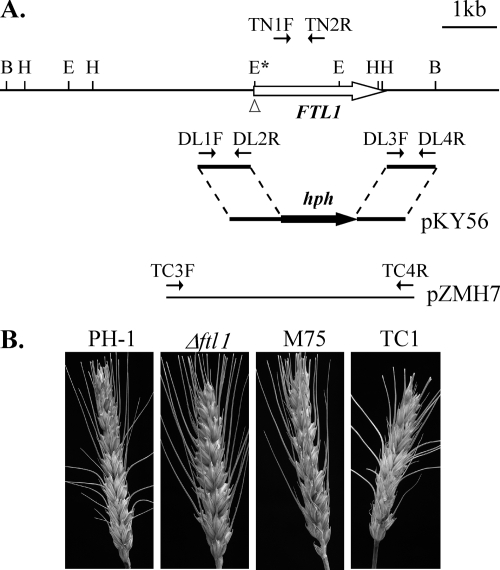
FIG. 2.
The FTL1 gene replacement construct and mutants. (A) The entire FTL1 ORF was replaced by hph in the gene replacement construct pKY56. The position and direction of primers DL1F, DL2R, DL3F, DL4R, TN1F, TN1R, TC3F, and TC4R are marked with small arrows. B, BamHI; E, EcoRI; H, HindIII. (B) Scab disease developed on flowering wheat heads inoculated with conidia of PH-1, Δftl1 mutant T1, REMI mutant M75, and complemented strain TC1. Photos were taken 2 weeks after inoculation.
To construct the GFP reporter strain, the enhanced GFP (EGFP) cassette under the control of the Magnaporthe grisea RP27 promoter was amplified from pKB04 (7) with primers GFP-R and GFP-F (Table 2). The Geneticin resistance gene was amplified from pSM334 with primers NeoF and NeoR. About 10 μl of each of the resulting PCR products was cotransformed into protoplasts of the Δftl1 mutant T1. Geneticin-resistant transformants expressing the EGFP cassette were examined for GFP signals and analyzed by Southern blot analysis.
TABLE 2.
PCR primers used in this study
| Primer | Sequence (5′→3′) |
|---|---|
| DL1F | CCGGAATTCAAAAACAACCACGACCTGCG |
| DL2R | CAGGTTGGCGCGCCACAGATCACTAAGGCTCTCG |
| DL3F | AAGGTTGGCCGGCCGAGGATACGATACGGATGCT |
| DL4R | TTAATTCTGGCACCGGATCC |
| TN1F | GATGAGAGTTTCGCCTCAGC |
| TN2R | AGGATAGACGGGTCTCTTGG |
| TC3F | AGAAGCGATATTCCTCTGTCATC |
| TC4R | CAAAGGAACTCTTCCACCAAAC |
| TDDYC3F | GGCGAATTGGGTACTCAAATTGGTAAGAAGCGATATTCCTCTGTCATC |
| TDD1R | AAAAGCGAACTCTCTATGAGGGAGGAATTCCTTAGCGACCAT |
| TDD2F | ATGGTCGCTAAGGAATTCCTCCCTCATAGAGAGTTCGCTTTT |
| TDDYC2R | ATTCCACCGAGCAGGGTATTGCCTCAAAGGAACTCTTCCACCA |
| NeoF | AATACCCGGGATTAACGCTTACAATT |
| NeoR | ATAACCCGGGAATAGGAACTTCGGAA |
| GFP-F | GTGAGCAAGGGCGAGGAGCTGTT |
| GFP-R | CTAAACAGACATTATCATCATCATGC |
| M27F/eGFP | CGTCACAGAGTTTTAGTAGTTTTTCTG |
| M27R/eGFP | AACAGCTCCTCGCCCTTGCTCATTTTGAAGATTGGGTTCCTAC |
| TY1F | GGCTCGAGGATGGTCGCTAAGGAATTCCTCGACTCGGAC |
| TY2R2 | GGCTGCAGATTGCGTTGGTCCCTTGTCAT |
| TDDYC1F | CTATAGGGCGAATTGGGTACTCAAATTGGTAAGAAGCGATATTCCTCT |
| TGFP2R | GAACAGCTCCTCGCCCTTGCTCACATTGCGTTGGTCCCTTGTCATCT |
| TRT5F | GACCAAGGAGGACGATAGCTT |
| TB-RM-R2 | TTAATTGCGTTGGTCCCTTGTCAT |
| FSNT1F | CTACGAGCCAGCGCAAGACGCCTCGAAGAC |
| FSNT2R | TTGACCTCCACTAGCTCCAGCCAAGCCCTGCAAATTAGGATGGAC |
| FSNT3F | GAATAGAGTAGATGCCGACCGCGGGTTTACATGTTGAGCTTCGTT |
| FSNT4R | ACCACCTTCTCTCCATCAAGCATCCTCATGC |
FTL1-GFP fusion and FTL1ΔLisH constructs.
The FTL1-GFP and FTL1ΔLisH constructs were generated by the yeast gap-repair method (7). A 4.5-kb fragment of the FTL1 gene was amplified with primers TDDYC1F and TGFP2R (Table 1) and cotransformed with XhoI-digested pKB04 into S. cerevisiae XK1-25 (7). Plasmid pZHM12 was rescued from the Trp+ yeast transformant. The FTL1-GFP fusion construct was confirmed by sequence analysis and transformed into PH-1. For constructing the FTL1ΔLisH allele, primers TDDYC3F and TDD1R were used to amplify the first seven codons and 1.6-kb upstream promoter region of FTL1. The rest of FTL1 was amplified with primers TDD2F and TDDYC4R. The resulting PCR products were cotransformed into XK1-25 with XhoI-digested pKB04. Plasmid pZHM13 was rescued from the resulting Trp+ yeast transformant and confirmed by sequence analysis to contain the expected deletion of amino acid residues 8 to 40.
Wheat infection assays.
Conidia were collected from 5-day-old CMC (1.5% carboxymethylcellulose, 0.1% NH4NO3, 0.1% KH2PO4, 0.05% MgSO4 7H2O, 0.1% yeast extract) cultures and resuspended in 0.01% (vol/vol) Tween 20 to a concentration of 106 conidia/ml. Six-week-old plants of wheat cultivar Norm were used for inoculation at flowering as described previously (15, 26). The fifth, full-sized spikelet from the base of the inflorescence was injected with 10 μl of the spore suspension. Inoculated plants were placed in a humidity chamber for 3 days and then transferred to ambient growth conditions in a greenhouse. Discoloration in the rachis was examined by removing wheat florets. For assaying disease indices, symptomatic spikelets in each head were counted 14 days postinoculation (dpi). At least nine inoculated wheat heads per treatment were used for each test and all tests were repeated four times.
For inoculation assays with detached flowering wheat heads, conidia were resuspended to 105 conidia/ml in 0.25% gelatin. A drop of 1 to 2 μl of the conidium suspension was inoculated onto each exposed anther on detached wheat heads. After incubation at 25°C for 4 to 5 days in a moist chamber, fungal growth and infection of wheat florets were examined under a dissecting microscope. For inoculation assays with unexposed anthers, wheat ovaries with intact stigmas and anthers were separated from glumes and paleae and placed over sterile water agar (2%) plates. Each sample was inoculated by placing a drop of 1 to 2 μl of conidium suspension on the anther and incubated at 25°C. For infection through wounding, wheat ovaries and paleae were punched with a dissecting needle and inoculated with 1 to 2 μl of conidium suspension at the wound site. Fungal growth and discoloration of wheat ovaries were observed 3 to 4 dpi.
Confocal and TEM observations.
Wheat spikelets inoculated with the wild-type or Δftl1 mutant strains were sampled at 6 dpi. Longitudinal sections of wheat rachises were examined under a LSM510 confocal laser scanning microscope (Zeiss, Oberkochem, Germany) with excitation and emission wavelengths of 488 nm and 510 to 530 nm. For transmission electron microscopic (TEM) examinations, rachises were sliced into pieces and fixed in 3% (vol/vol) glutaraldehyde for 3 to 6 h at 4°C. Samples were then rinsed thoroughly with 50 mM phosphate buffer (pH 6.8) and treated with 1% (wt/vol) osmium tetroxide for 2 h at 4°C. After being dehydrated in a graded ethanol series, samples were embedded in LR White (TAAB Laboratories, Munich, Germany) and polymerized at 50°C for 2 days as described previously (26, 27). For electron microscopy, ultrathin sections were prepared with a diamond knife and collected on 200-mesh copper grids. After treatment with uranyl acetate and lead citrate (27), the grids were examined with a JEM-1230 electron microscope (Jeol Ltd., Tokyo, Japan) at 80 kV. Three biological replicates were examined for both the wild-type and mutant strains.
HDAC activity assays.
Protoplasts of PH-1 and the Δftl1 mutant were resuspended to 4 × 108 protoplasts/ml in the lysis buffer (10 mM Tris-HCl, pH 7.5, 10 mM NaCl, 15 mM MgCl2, 250 mM sucrose, 0.5% NP-40, 0.1 mM EGTA, 200 μM phenylmethylsulfonyl fluoride), vortexed for 10 s, and kept on ice for 15 min. One milliliter of the resulting lysate was layered over 4 ml of 30% sucrose in the suspension buffer (10 mM Tris-HCl, pH 7.5, 10 mM NaCl, 3 mM MgCl2) and centrifuged at 1,300 × g for 10 min at 4°C. Nuclei pellets were washed once with the suspension buffer and resuspended in 0.1 ml of the extraction buffer (50 mM HEPES, pH 7.5, 420 mM NaCl, 0.5 mM EDTA, 0.1 mM EGTA, 10% glycerol, 200 μM phenylmethylsulfonyl fluoride). After sonication for 20 s and incubation on ice for 30 min, the nuclear suspension was centrifuged at 10,000 × g for 10 min at 4°C. The supernatant containing crude nuclear extracts was used for assaying HDAC activities with the HDAC activity assay kit (Cayman Chemical Company, Ann Arbor, MI) following the manufacturer's instructions. The deacetylation reaction was carried out at 37°C for 30 min and stopped by addition of 40 μl of the HDAC developer. Fluorescent signals were detected with a plate reader (360 nm excitation and 465 nm emission; Synergy HT; Bio-Tek). For negative controls, 1 μM trichostatin A was added before the deacetylation reaction. The concentration of deacetylated compounds was calculated with the deacetylation standard curve and used to estimate the HDAC activity (in nmol/min/ml) with the formula provided with the HDAC activity assay kit (Cayman Chemical Company).
RESULTS
A transducin beta-like gene FTL1 was disrupted in REMI mutant M75.
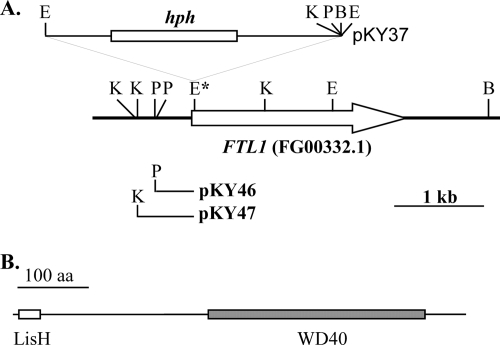
The REMI mutant M75 was generated by transforming EcoRI-digested pKY37 vector DNA into the F. graminearum strain PH-1 (46). It was defective in plant infection and had a slightly reduced growth rate (Table 3). Plasmids pKY46 and pKY47 (Fig. 1A) were recovered by self-ligation of KpnI- and PstI-digested genomic DNA of M75, respectively. Sequence analysis of these two plasmids revealed that pKY37 was inserted at an EcoRI site (Fig. 1A) of a predicted gene, FGSG_00332.3, which was named FTL1 (for F. graminearum TBLR1-like gene 1). We sequenced the reverse transcription-PCR (RT-PCR) product amplified with primers Y2H1F and Y2H2R and found that the third intron of FTL1 was 75 bp longer than predicted by automated annotation. FTL1 encoded a 637-amino-acid protein with an N-terminal LisH domain (residues 8 to 40) and WD40 repeats (Fig. 1B). Its best hit in GenBank with characterized function is the mouse TBLR1 gene, which encodes a subunit of a nuclear receptor corepressor/HDAC3 complex (40).
FIG. 1.
Mutant M75 was disrupted in the FTL1 gene. (A) The transforming vector pKY37 was inserted into the EcoRI site (marked by an asterisk) of the FTL1 gene (empty arrow). Plasmids pKY12 and pKY25 were recovered from M75 by self-ligation with PstI- and KpnI-digested genomic DNA, respectively. B, BamHI; E, EcoRI; K, KpnI; P, PstI. (B) The FTL1 gene contains one N-terminal LisH domain (empty box) and WD40 repeats (solid box) at the C-terminal half of the protein.
Homologs of FTL1 are well conserved among sequenced genomes of filamentous ascomycetes, including F. oxysporum (FO00731), M. grisea (MG03198), and Neurospora crassa (NCU06838). However, none of them has been functionally characterized. Its homolog in the budding yeast, Sif2, is a component of the Set3 complex that regulates sporulation (8, 41). In F. graminearum, FTL1 was not present in about 30,000 expressed sequence tags from three different cDNA libraries (53), indicating that the expression level of FTL1 is relatively low under culture conditions used for library construction. We also failed to detect any GFP signals in conidia and vegetative hyphae in transformants of PH-1 expressing the FTL1-GFP fusion construct under the control of its native promoter (data not shown).
FTL1 plays a critical role in pathogenesis.
The FTL1 gene replacement construct pKY56 (Fig. 2A) was generated by replacing the entire FTL1 ORF with the hygromycin phosphotransferase (hph) gene and transformed into the wild-type strain PH-1. Three Δftl1 mutants, T1, T2, and T3 (Table 2), were identified by PCR with primers TN1F and TN2R and confirmed by Southern blot analysis (data not shown). Similar to REMI mutant M75, the Δftl1 mutants were significantly reduced in conidiation and defective in plant infection (Table 3) but formed conidia of normal morphology. They were normal in DON production in cracked corn cultures and accumulated a reddish pigment in liquid CMC culture (data not shown). Typical WHB symptoms were not observed on spikelets inoculated with the Δftl1 mutants (Fig. 2B). These three Δftl1 mutants had the same phenotype, and only one of them, T1, was selected for further characterization.
To determine whether the observed phenotypes of Δftl1 mutants were caused by deletion of the FTL1 gene, we reintroduced the wild-type allele amplified with primers TC3F and TC4R (Fig. 2A) into mutant T1. The resulting Geneticin-resistant transformant TC1 had a normal vegetative growth rate, conidiation, and virulence on flowering wheat heads (Table 3). F. graminearum is a homothallic fungus, but it can be forced to outcross. On selfing plates, no protoperithecia or perithecia were observed. When crossing with a nit1 mutant, Δftl1 nit1 recombinant progeny were isolated, indicating that the Δftl1 mutant was female sterile but fertile when mated as the male. From outcross perithecia between mutant T1 and a nit1 mutant 11622 (23), we isolated 31 hygromycin-resistant ascospore progeny. All of these progeny displayed characteristic phenotypes of the Δftl1 mutant (data not shown). When scored for growth on CM with 1% potassium chlorate, 14 of them were NIT1+ and 19 were nit1−, indicating that the observed defect of the Δftl1 mutant, but not the nit1 mutation, cosegregated with the hph marker.
The Δftl1 mutant was defective in colonizing the rachis.
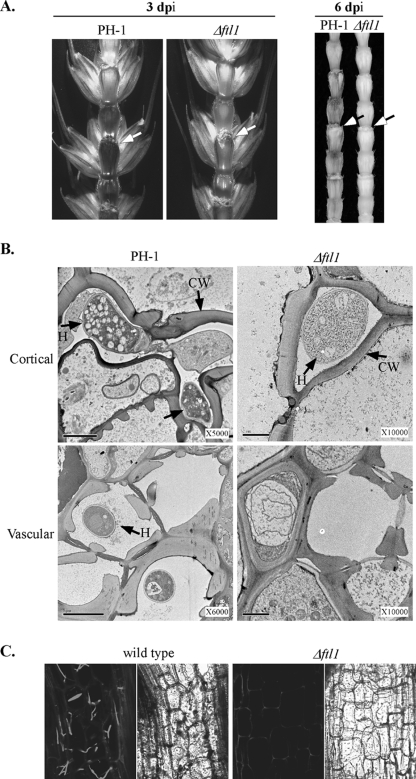
To further characterize the defect of the Δftl1 mutant in plant infection, we removed the spikelets on the same side of drop-inoculated florets 3 or 6 dpi and examined for necrosis in the rachis (Fig. 3A). In wheat heads inoculated with PH-1, discoloration was observed in the rachis both above and below the inoculation site, indicating that the fungus had spread from the inoculated floret through the rachis to other spikelets. In contrast, the Δftl1 mutant caused only limited necrosis at the bases of the inoculated floret. Discoloration was not observed in the rachis (Fig. 3A). These observations indicate that the Δftl1 mutant was defective in spreading from infected florets to the rachis and neighboring florets on wheat heads.
FIG. 3.
The Δftl1 mutant was blocked in colonizing the wheat rachis. (A) Rachises of wheat heads inoculated with the wild type (PH-1) and the Δftl1 mutant strain were examined at 3 (left) or 6 (right) dpi. Wheat kernels on the inoculation side were removed to expose the rachis. The inoculation sites are marked with arrows. (B) TEM images of the cortical (top row) and vascular (bottom row) tissues of the wheat rachis next to the florets inoculated with the wild type (PH-1) or the Δftl1 mutant (Δftl1) at 6 dpi. H, inter- and intracellular hyphae; CW, plant cell wall. Bar, 2 μm. Hyphal growth was only observed in the xylem vessels in wheat heads inoculated with the wild type. (C) Microscopic images of longitudinal sections of rachises and adjacent nodes from wheat heads infected with transformants of the wild type (GZT501) and Δftl1 mutant (RM1) that expressed GFP constitutively. At 6 dpi, intracellular hyphae of the wild type colonized the parenchyma tissue in the rachis. Hyphal growth of the Δftl1 mutant was rare or not observed in the rachis. Left and right panels depict the same areas observed under confocal and differential interference contrast microscopy. A basal level of autofluorescence was visible in the plant cell wall.
When examined at 6 dpi by TEM, intercellular and intracellular hyphae were readily observed in the cortical and vascular tissues of the wheat rachis next to the kernels inoculated with the wild type (Fig. 3B). In wheat heads inoculated with the Δftl1 mutant, intercellular, but not intracellular, hyphae were occasionally observed in the cortical tissues (Fig. 3B). However, no hyphal growth was observed in the vascular tissues of the rachis (Fig. 3B). The Δftl1 mutant appeared to be blocked in intracellular growth and colonization of vascular tissues in the rachis of inoculated wheat heads. This defect may be directly related to the failure of the Δftl1 mutant in causing typical FHB symptoms and spreading from spikelet to spikelet.
To further confirm this observation, we transformed an RP27-EGFP construct into the Δftl1 mutant T1. EGFP was constitutively expressed in the conidia and hyphae of the resulting transformant, RM1 (data not shown). In wheat heads inoculated with RM1, hyphal growth was not or only rarely observed in the rachis by confocal microscopy (Fig. 3C). In the wheat heads inoculated with a GFP-tagged wild-type strain, hyphal growth was readily observed in cortical and vascular tissues (Fig. 3C). These results further confirmed that the Δftl1 mutant is defective in the penetration and colonization of vascular tissues in the rachis.
The Δftl1 mutant failed to infect wheat florets through anthers.
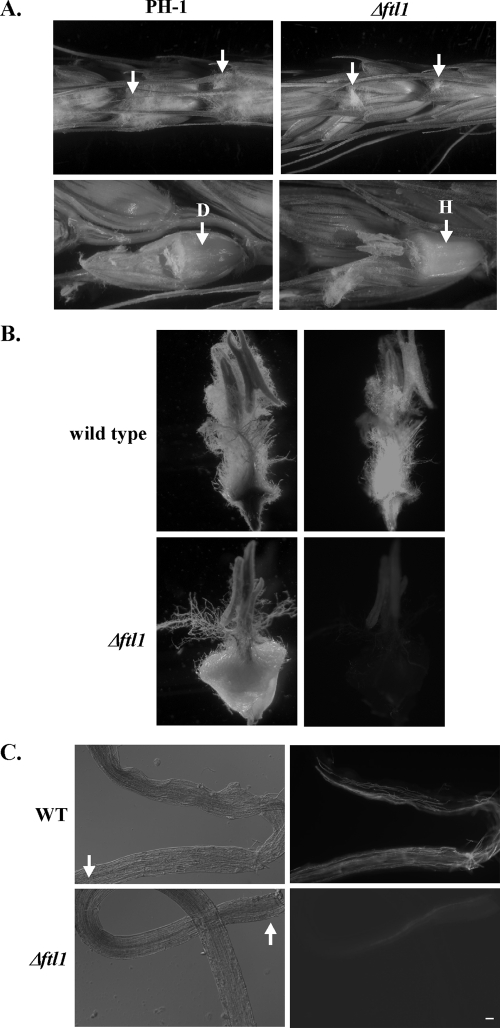
Because anthers are the main site of primary infection, we also inoculated detached wheat heads by placing drops of conidium suspension from PH-1 or the Δftl1 mutant on exposed anthers. After incubating at 25°C for 4 to 5 days, extensive fungal growth was visible on florets inoculated with PH-1 (Fig. 4A). When the glumes were removed, discolored ovaries and fungal hyphae were observed inside wheat florets inoculated with PH-1 (Fig. 4A). Under the same conditions, hyphal growth was limited to the anthers inoculated with the Δftl1 mutant and the ovaries remained healthy (Fig. 4A).
FIG. 4.
Defects of the Δftl1 mutant in colonizing wheat heads through anthers. (A) Flowering wheat heads were inoculated with conidium drops of the wild-type (PH-1) and Δftl1 mutant strains at exposed anthers (marked with arrows). Photos were taken 5 dpi. Glumes were removed (bottom row) to examine hyphal growth and discoloration of the ovaries. The Δftl1 mutant grew on exposed anthers but failed to cause discoloration of the ovaries. D, diseased or discolored ovary; H, healthy wheat ovary. (B) Unexposed anthers were inoculated with conidium drops of GFP-tagged wild-type (GZT501) and Δftl1 mutant (RM1) strains. After incubating for 3 days, the ovaries with inoculated anthers were observed under light (left) and epifluorescence (right) microscopy. (C) After being separated from the anthers inoculated with conidium suspensions at 2 dpi, filaments were observed under differential interference contrast (left) or epifluorescence (right) microscopy. The Δftl1 mutant was defective in colonizing the filaments. Arrows points to the ends connected to the anthers. Bar, 20 μm.
We also isolated ovaries with unexposed anthers with drops of conidium suspensions of the wild-type and Δftl1 mutant strains that constitutively expressed GFP. After incubation at 25°C for 3 days, the wild type colonized the anthers, filaments, and ovaries. Discoloration of the ovaries and abundant hyphal growth (evidenced by strong GFP signals) were observed (Fig. 4B). In contrast, the Δftl1 mutant had limited growth on the inoculated anthers (Fig. 4B). However, the ovaries remained healthy and GFP signals were relatively weak. These observations indicate that the Δftl1 mutant was defective in colonizing wheat ovaries through the anthers and filaments. To further confirm this observation, we examined fungal growth in the filaments connecting inoculated anthers to the ovaries (Fig. 4C). While the wild-type colonized the entire length of the filaments at 2 dpi, the Δftl1 mutant only had limited hyphal growth at the ends next to the anthers (inoculation sites). In most of the samples examined, it failed to spread more than 1/10 of the filaments at 2 dpi.
The Δftl1 mutant is defective in infectious growth in wheat tissues.
To further examine defects of the Δftl1 mutant in plant colonization, we wounded developing seeds with a needle and inoculated with F. graminearum conidia. After incubating for 2 days, hyphae growing on the plant surface were gently removed. When examined by epifluorescence microscopy, the wild type had extensive intracellular and extracellular hyphal growth in seed coat tissues (data not shown). Under the same conditions, conidia of the Δftl1 mutant germinated and grew on plant surfaces. However, intracellular growth of infectious hyphae was rarely observed. While the wild type was able to penetrate and colonize neighboring plant cells, hyphal growth of the Δftl1 mutant was limited. In similar infection assays with wounded wheat paleae, extensive growth of infectious hyphae also was only observed in tissues colonized by the wild type at 2 dpi (data not shown). These observations indicate that the Δftl1 mutant was defective in the penetration and invasive growth in seed coat and palea cells.
Increased sensitivity of the Δftl1 mutant to plant defensin MsDef1.
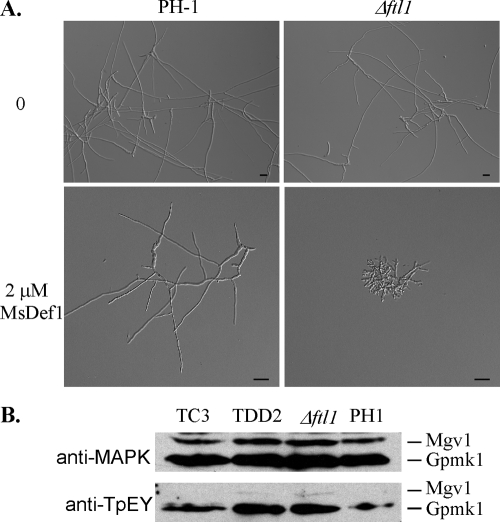
Because the Δftl1 mutant was defective in infectious growth, we tested its sensitivity to plant defensin MsDef1 (43). In the presence of 0.5 μM MsDef1, conidium germination and germ tube growth were not affected in either the wild-type or Δftl1 mutant strains. When the concentration of MsDef1 was increased to 2 μM, conidial germination and germ tube growth were normal in the wild-type strain. Under the same conditions, the Δftl1 mutant produced short germ tubes and displayed a hyperbranching phenotype (Fig. 5A). At 10 μM, MsDef1 completely blocked conidium germination in the Δftl1 mutant but only stunted germ tube growth in PH-1 (data not shown). These results indicated that the Δftl1 mutant was more sensitive to MsDef1 than the wild type in conidium germination and germ tube growth. MsDef1 may interfere with calcium signaling and polarized tip growth in F. graminearum. The Δftl1 mutant also was more sensitive to osmotin, although F. graminearum appeared to be more tolerant to this antifungal protein. In the presence of 32 μM osmotin (9), conidium germination was blocked in the Δftl1 mutant. Under the same conditions, germination was normal for the vast majority of conidia, but germ tube growth was slightly reduced in the wild-type strain PH-1 (data not shown).
FIG. 5.
Enhanced sensitivity of the Δftl1 mutant to MsDef1. (A) Conidia of the wild-type strain PH-1 (upper panels) and the Δftl1 mutant (lower panels) were incubated in CM for 15 h with 0 or 2 μM defensin MsDef1. Bar, 20 μm. (B) Western blot analyses with proteins isolated from vegetative hyphae of PH-1, Δftl1 mutant T1, LisH domain deletion transformant TDD2, or Δftl1-complemented strain TC1. The upper panel shows detection with the anti-TpEY antibody. The lower panel shows detection with an anti-MAPK antibody.
In F. graminearum, Mgv1 and Gpmk1 are two MAP kinases known to be essential for pathogenesis. The defects of the Δftl1 mutant in wheat infection and enhanced sensitivity to MsDef1 were similar to those of the gpmk1 and mgv1 mutants (43). To determinate whether Ftl1 is involved in the activation of Mgv1 or Gpmk1, we assayed the activation of these two MAP kinases. In both the Δftl1 mutant and transformant TDD2, phosphorylation of Gpmk1 and Mgv1 was detectable in total proteins isolated from vegetative hyphae (Fig. 5B). The expression and phosphorylation levels of Mgv1 were lower than those of Gpmk1. However, a similar pattern was observed in PH-1 and the Δftl1 mutant. Therefore, deletion of FTL1 had no obvious effect on the expression or activation of Gpmk1 and Mgv1. Increased sensitivities of the Δftl1 mutant to MsDef1 were unlikely to be related to Gpmk1 or Mgv1 activities.
The LisH domain is essential for FTL1 function.
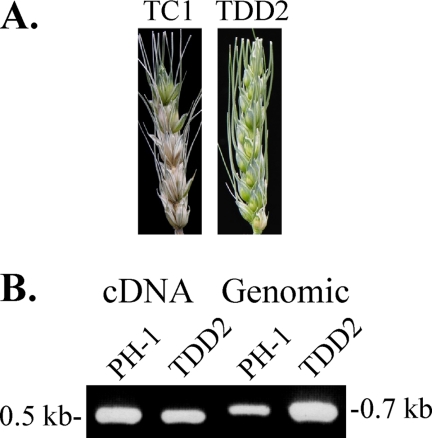
To determine the role of the LisH domain in Ftl1, we generated a FTL1ΔLisH allele and transformed it into the Δftl1 mutant T1. Three resulting transformants expressing the FTL1ΔLisH allele, including TDD2 (Table 1), were isolated and confirmed by Southern blot analysis to contain a single copy of the transforming vector (data not shown). Similar to the Δftl1 mutant, TDD2 was defective in growth, conidiation, and plant infection (Table 3; Fig. 6A). It also accumulated a reddish pigment in liquid CMC cultures (data not shown). A 0.55-kb cDNA band was amplified by RT-PCR in both PH-1 and transformant TDD2 (Fig. 7C) with primers TRT5F and TB-RM-R2 (Table 2). The same primer pairs amplified a 0.7-kb band from genomic DNA. These results indicated that the FTL1ΔLisH allele was expressed in TDD2 but it failed to complement the defects of the Δftl1 mutant. Therefore, the LisH domain is essential for the function of the FTL1 gene in F. graminearum.
FIG. 6.
The LisH domain is essential for the function of FTL1. (A) Wheat heads inoculated with conidia from transformants expressing the wild-type FTL1 gene (TC1) or the FTL1ΔLisH allele (TDD2). Typical Fusarium head blight symptoms were only observed in wheat heads inoculated with TC1 and not in those inoculated with TDD2. (B) PCR products amplified with primers TRT5F and TB-RM-R2 using either the first-strand cDNA or genomic DNA of the wild-type strain (PH-1) and transformant TDD2 as the template.
FIG. 7.
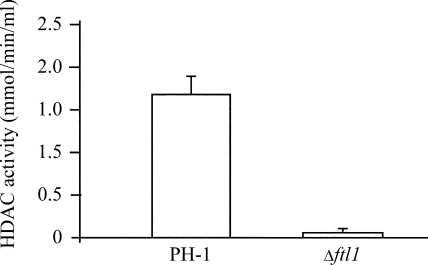
HDAC activity assays. Nuclear extracts of the wild-type and the Δftl1 mutant strains were assayed for HDAC activities with the HDAC activity assay kit (Cayman Chemical Company). The Δftl1 mutant was significantly reduced in HDAC activities.
The Δftl1 mutant has reduced HDAC activities.
Because the FTL1 homologs in yeast and mammalian cells are components of a conserved HDAC complex (41, 55), we assayed the HDAC activities in the Δftl1 mutant with an HDAC activity assay kit (Cayman Chemical Company). In comparison with the wild-type strain, the Δftl1 mutant was significantly reduced in HDAC activity (Fig. 7).
Microarray analysis with the Δftl1 mutant.
To determine what genes are regulated by FTL1, we used the F. graminearum GeneChip (21) to conduct microarray analysis of the Δftl1 mutant. In comparison with the wild type, 448 and 168 genes were down- and upregulated, respectively, over fivefold in the Δftl1 mutants in three independent biological replicates. The majority (over 70%) of those genes with significantly altered expression patterns in the Δftl1 mutant encoded hypothetical proteins with unknown biological functions. Thirty-seven of them were unique to F. graminearum. Fourteen putative hydrolase genes were among the downregulated genes, but none of them was homologous to known fungal virulence factors. The polyketide synthase gene PKS12 (FGSG_02324.3), which is responsible for aurofusarin synthesis, was increased 18-fold in the Δftl1 mutant. Interestingly, several other members of the aurofusarin synthesis gene cluster (28, 33), including FGSG_02321.3 (5.1×) FGSG_02325.3 (8.7×), FGSG_02326.3 (9.6×), FGSG_02327.3 (7.8×), FGSG_02328.3 (11.5×), and FGSG_02329.3 (11.2×), also were expressed at higher levels in the Δftl1 mutant, suggesting that FTL1 negatively regulates aurofusarin biosynthesis.
We also checked the expression levels of known virulence factors, including GPMK1, MGV1, FBP1, SID1, ADE5, ARG2, NPS6, RSY1, CBL1, FSR1, RAS2, ZIF1, TRI5, FGL1, GzHIS7, GzCPS1, GzGPA2, GzGPB1, and HMR1. None of these genes had more than twofold changes in their expression levels in the Δftl1 mutant. Therefore, the defect of the Δftl1 mutant in plant infection may involve novel mechanisms. However, with the number of genes affected in the Δftl1 mutant, it was impossible to determine which genes are directly involved in plant infection. To verify the microarray data, we selected eight genes for quantitative RT-PCR (qRT-PCR) analysis. Six of them, FGSG_08347.3, FGSG_01767.3, FGSG_05622.3, FGSG_05597.3, FGSG_07582.3, and FGSG_11080.3, encoding a putative choline permease, cytochrome P450, trehalase, hexose transporter, triacylglycerol lipase, and membrane receptor, respectively, were confirmed by qRT-PCR to be significantly downregulated in the Δftl1 mutant (data not shown). Some of these genes may be involved in plant infection processes.
DISCUSSION
In the REMI mutant M75, the transforming vector was inserted at the N terminus of the FTL1 gene. In this study, we generated the Δftl1 gene replacement mutant. Phenotypically, M75 and the Δftl1 mutant were identical, indicating that the original insertional event completely eliminated the function of the FTL1 gene. FTL1 homologs are well conserved in filamentous ascomycetes. The Fusarium oxysporum (FO00731) and Fusarium verticillioides (FV00784) homologs share 79% and 78% identity with FTL1, respectively. However, the functions of this well-conserved gene in hyphal growth and plant infection are not clear, because none of the FTL1 homologs has been functionally characterized in filamentous fungi.
FTL1 homologs also are conserved in yeast and mammals. FTL1 is homologous to the mammalian TBL1 (transducin beta-like) and TBLR1 (TBL1-related) genes. TBL1 and TBL1R are two paralogous genes with overlapping functions, and they both interact with N-CoR and SMRT (55, 57). N-CoR and SMRT were initially identified as corepressors for the retinoic acid receptor and thyroid hormone receptor, but they have also been implicated, in association with HDACs, in repression of other transcription factors (17, 20, 30). In S. cerevisiae, the Set3 complex contains Sif2, which is a homolog of TBL1 (8). Interestingly, FTL1 shares higher homology with mouse TBL1R than with yeast SIF2. The Set3 complex is the yeast analog of the mammalian HDAC/SMRT complex (55). It represses early and middle sporulation genes, including key meiotic regulators such as the IME2 protein kinase and the NDT80 transcription factor (41). In F. graminearum, the Δftl1 mutant was female sterile but was able to mate as a male and produced ascospore progeny. We were able to isolate normal Δftl1 ascospore progeny from a Δftl1 NIT1 × FTL1 nit1 cross. Yeast has no distinction in male or female fertility. Defects in ascospore formation could be observed in the sif2 × sif2 cross. Because of the loss of female fertility, we could not test the fertility of the Δftl1 × Δftl1 cross in this study. Therefore, it remains possible that FTL1 is important for sexual reproduction in F. graminearum. Conidiation is an asexual reproduction process that is absent in yeast. In F. graminearum, conidiation was significantly reduced in the Δftl1 mutant. There is a distinct homolog of Set3 in F. graminearum, M. grisea, and Neurospora crassa. Other members of the yeast Set3 complex include an NAD-dependent HDAC, Hst1, and an NAD-independent HDAC, Hos2. In mammalian cells, the HDAC gene HDAC3 is a member of the N-CoR complex (20). Our data indicated that the Δftl1 mutant was significantly reduced in HDAC activities. The F. graminearum genome has one putative Hst1 homolog and two putative Hos2 homologs. These genes also are well conserved in M. grisea and other filamentous ascomycetes. To date, there are only limited studies on HDACs in phytopathogenic fungi. In Cochliobolus carbonum, the yeast HOS2 homolog HDC1 is required for full virulence (4).
Another member of the Set3 complex is the SNT1 gene, which encodes a putative DNA-binding protein and directly interacts with SIF2 (8). The equivalent of Snt1 in the mammalian N-CoR/SMRT/HDAC3 complex is N-CoR or SMRT (57). All Sif2 homologs, including FgSnt1 in F. graminearum and a predicted M. grisea protein (MG09174), have a SANT DNA-binding domain (SM00717) that functions as a unique histone tail-binding module coupling histone binding to enzyme catalysis (6). In F. graminearum, our preliminary data have indicated that deletion of FgSNT1 resulted in similar defects as the Δftl1 mutant (S. Ding and J.-R. Xu, unpublished data). Because these genes are conserved from yeast to human, it is likely that FTL1 and FgSNT1 are components of this well-conserved complex, which may have been adapted to regulate plant infection processes, such as infectious or in planta growth in F. graminearum and other plant pathogenic fungi.
The Δftl1 mutant was normal in DON production, which is the first virulence factor that has been characterized in F. graminearum. The activation of Gpmk1 and Mgv1 was not affected in the Δftl1 mutant either. In the microarray analysis, the expression levels of all the F. graminearum genes that are known to be required for pathogenicity or full virulence were normal in the Δftl1 mutant. Therefore, FTL1 is likely involved in novel mechanisms regulating plant infection processes. However, the Δftl1 mutant had pleiotropic defects. It is difficult to determine which genes are responsible for observed defects of the Δftl1 mutant in plant infection based on transcript profiling. In addition, microarray analysis was conducted with RNA isolated from vegetative hyphae, which may not reflect in planta growth conditions.
We noticed that 14 hydrolytic enzyme genes had reduced expression levels in the Δftl1 mutant, including three putative triacylglycerol lipase genes that are unrelated to FGL1 (54). No putative hydrolase genes were upregulated in the Δftl1 mutant. Among eight genes selected for verification by qRT-PCR, FGSG_08347.3 encodes a choline permease that may be involved in the uptake of choline, a compound abundant in wheat anthers (51). FGSG_01767.3 encodes a P450 monooxygenase that is highly homologous to pisatin demethylase (32). FGSG_05622.3 shares strong similarity to a trehalase precursor. In M. grisea and Candida albicans, trehalase plays an important role in pathogenesis (14, 39). FGSG_07582.3 encodes a putative low-affinity hexose transporter. In Sclerotinia sclerotiorum, two hexose transporter genes are upregulated during plant infection (25). FGSG_05597.3 is a putative triacylglycerol lipase gene. FGL1 and other lipase genes are important for full virulence in F. graminearum (35, 54). FGSG_11080.3 is homologous to PTH11, encoding a putative membrane receptor that is important for plant infection in M. grisea (12). Changes in their expression levels in the Δftl1 mutant were confirmed by qRT-PCR for all six of these candidate virulence factors. However, none of them has been functionally characterized, and their functions in the virulence of F. graminearum remain to be analyzed.
Interestingly, PKS12 and six other members of the aurofusarin synthesis gene cluster (28, 33) were upregulated in the Δftl1 mutant. The production of aurofusarin is likely to be elevated in the Δftl1 mutant. We noticed that deletion of the entire FTL1 or the LisH domain resulted in the accumulation of a reddish pigment in the CMC cultures. However, not all genes in the aurofusarin synthesis cluster were expressed at higher levels in the Δftl1 mutant, and the chemical identity of this reddish pigment was not clear. The LisH domain is known to be involved in protein-protein interactions and the regulation of microtubule dynamics and nuclear migration (13, 16, 34). In F. graminearum, the LisH domain is essential for the function of FTL1 (Fig. 6). In the original REMI mutant M75, the transforming vector was inserted in the LisH domain (Fig. 1). The F. graminearum genome appears to have six additional genes containing the putative LisH domain (IPR006594). To date, the Aspergillus nidulans nudF gene, which is known to be involved in nuclear migration (1), is the only LisH domain-containing gene that has been functionally characterized in filamentous fungi.
It is possible that a reduction in the growth rate may contribute to the defects of the Δftl1 mutant in plant infection. However, while the growth rate was only reduced about 40% in the Δftl1 mutant (Table 3), its disease index was near zero. Deletion of FTL1 must have adversely affected other pathogenicity factors in F. graminearum. Based on microscopic examination, loss of full virulence in the Δftl1 mutant was related to its defects in penetration and infectious growth in wheat tissues. In nature, anthers are the main site of primary infection on flowering wheat heads (3, 38). The Δftl1 mutant was defective in the colonization of filaments and spreading from infected anthers to ovaries. In Claviceps purpurea, deletion of two endopolygalacturonase genes (cppg1 and cppg2) also resulted in a near loss of pathogenicity and the mutant was blocked for the colonization of oat ovaries (36). However, the cppg1 cppg2 mutant was normal in growth and conidiation, and C. purpurea is a biotrophic pathogen. Our TEM examination indicated that rare intercellular hyphae developed for the Δflt1 mutant in the cortical layer of wheat rachises. However, the Δftl1 mutant failed to penetrate and grow intracellularly in the cortical layer and was defective in the colonization of palea or seed coat cells. Because colonization of vascular tissues of the rachis may be a critical factor in the development and spread of wheat head blight, it will be important to determine the function of FTL1 in regulating the penetration of plant cells (intracellular growth) and colonization of vascular tissues.
Acknowledgments
We thank Larry Dunkle and Charles Woloshuk for critical reading of the manuscript, Yan-Hong Dong for DON and ergosterol measurements, and Zhanming Hou for assistance with complementation and LisH domain analyses. We also thank Dilip Shah at Danforth Plant Science Center and Paul M. Kasegawa at Purdue University for providing MsDef1 and osmotin proteins.
This work was supported by a grant from the U.S. Wheat and Barley Scab Initiative to H.C.K. and J.-R.X. and grants to J.-R.X. from the National Research Initiative of the USDA CSREES (numbers 2003-35319-13829 and 2007-35319-102681).
Footnotes
Published ahead of print on 17 April 2009.
REFERENCES
- 1.Ahn, C., and N. R. Morris. 2001. NudF, a fungal homolog of the human Lis1 protein, functions as a dimer in vivo. J. Biol. Chem. 2769903-9909. [DOI] [PubMed] [Google Scholar]
- 2.Bai, G. H., A. E. Desjardins, and R. D. Plattner. 2002. Deoxynivalenol-nonproducing Fusarium graminearum causes initial infection, but does not cause disease spread in wheat spikes. Mycopathologia 15391-98. [DOI] [PubMed] [Google Scholar]
- 3.Bai, G. H., and G. Shaner. 2004. Management and resistance in wheat and barley to Fusarium head blight. Annu. Rev. Phytopathol. 42135-161. [DOI] [PubMed] [Google Scholar]
- 4.Baidyaroy, D., G. Brosch, J. H. Ahn, S. Graessle, S. Wegener, N. J. Tonukari, O. Caballero, P. Loidl, and J. D. Walton. 2001. A gene related to yeast HOS2 histone deacetylase affects extracellular depolymerase expression and virulence in a plant pathogenic fungus. Plant Cell 131609-1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bluhm, B. H., X. Zhao, J. E. Flaherty, J. R. Xu, and L. D. Dunkle. 2007. RAS2 regulates growth and pathogenesis in Fusarium graminearum. Mol. Plant-Microbe Interact. 20627-636. [DOI] [PubMed] [Google Scholar]
- 6.Boyer, L. A., R. R. Latek, and C. G. Peterson. 2004. The SANT domain: a unique histone-tail-binding module. Nat. Rev. Mol. Cell Biol. 5158-163. [DOI] [PubMed] [Google Scholar]
- 7.Bruno, K. S., F. Tenjo, L. Li, J. E. Hamer, and J. R. Xu. 2004. Cellular localization and role of kinase activity of PMK1 in Magnaporthe grisea. Eukaryot. Cell 31525-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cerna, D., and D. K. Wilson. 2005. The structure of sif2p, a WD repeat protein functioning in the SET3 corepressor complex. J. Mol. Biol. 351923-935. [DOI] [PubMed] [Google Scholar]
- 9.Coca, M. A., B. Damsz, D. J. Yun, P. M. Hasegawa, R. A. Bressan, and M. L. Narasimhan. 2000. Heterotrimeric G-proteins of a filamentous fungus regulate cell wall composition and susceptibility to a plant PR-5 protein. Plant J. 2261-69. [DOI] [PubMed] [Google Scholar]
- 10.Cuomo, C. A., U. Gueldener, J. R. Xu, F. Trail, B. G. Turgeon, A. Di Pietro, J. D. Walton, L. J. Ma, S. E. Baker, M. Rep, G. Adam, J. Antoniw, T. Baldwin, S. Calvo, Y. L. Chang, D. DeCaprio, L. R. Gale, S. Gnerre, R. S. Goswami, K. Hammond-Kosack, L. J. Harris, K. Hilburn, J. C. Kennell, S. Kroken, J. K. Magnuson, G. Mannhaupt, E. Mauceli, H. W. Mewes, R. Mitterbauer, G. Muehlbauer, M. Munsterkotter, D. Nelson, K. O'Donnell, T. Ouellet, W. H. Qi, H. Quesneville, M. I. G. Roncero, K. Y. Seong, I. V. Tetko, M. Urban, C. Waalwijk, T. J. Ward, J. Q. Yao, B. W. Birren, and H. C. Kistler. 2007. The Fusarium graminearum genome reveals a link between localized polymorphism and pathogen specialization. Science 3171400-1402. [DOI] [PubMed] [Google Scholar]
- 11.Desjardins, A. E., R. H. Proctor, G. H. Bai, S. P. McCormick, G. Shaner, G. Buechley, and T. M. Hohn. 1996. Reduced virulence of trichothecene-nonproducing mutants of Gibberella zeae in wheat field tests. Mol. Plant-Microbe Interact. 9775-781. [Google Scholar]
- 12.DeZwaan, T. M., A. M. Carroll, B. Valent, and J. A. Sweigard. 1999. Magnaporthe grisea Pth11p is a novel plasma membrane protein that mediates appressorium differentiation in response to inductive substrate cues. Plant Cell 112013-2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Emes, R. D., and C. P. Ponting. 2001. A new sequence motif linking lissencephaly, Treacher Collins and oral-facial-digital type 1 syndromes, microtubule dynamics and cell migration. Hum. Mol. Genet. 102813-2820. [DOI] [PubMed] [Google Scholar]
- 14.Foster, A. J., J. M. Jenkinson, and N. J. Talbot. 2003. Trehalose synthesis and metabolism are required at different stages of plant infection by Magnaporthe grisea. EMBO J. 22225-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gale, L. R., L. F. Chen, C. A. Hernick, K. Takamura, and H. C. Kistler. 2002. Population analysis of Fusarium graminearum from wheat fields in eastern China. Phytopathology 921315-1322. [DOI] [PubMed] [Google Scholar]
- 16.Gerlitz, G., E. Darhin, G. Giorgio, B. Franco, and O. Reiner. 2005. Novel functional features of the LisH domain: role in protein dimerization, half-life and cellular localization. Cell Cycle 41632-1640. [DOI] [PubMed] [Google Scholar]
- 17.Glass, C. K., and M. G. Rosenfeld. 2000. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 14121-141. [PubMed] [Google Scholar]
- 18.Goswami, R. S., and H. C. Kistler. 2004. Heading for disaster: Fusarium graminearum on cereal crops. Mol. Plant Pathol. 5515-525. [DOI] [PubMed] [Google Scholar]
- 19.Greenshields, D. L., G. S. Liu, J. Feng, G. Selvaraj, and Y. D. Wei. 2007. The siderophore biosynthetic gene SID1, but not the ferroxidase gene FET3, is required for full Fusarium graminearum virulence. Mol. Plant Pathol. 8411-421. [DOI] [PubMed] [Google Scholar]
- 20.Guenther, M. G., W. S. Lane, W. Fischle, E. Verdin, M. A. Lazar, and R. Shiekhattar. 2000. A core SMRT corepressor complex containing HDAC3 and TBL1, a WD40-repeat protein linked to deafness. Genes Dev. 141048-1057. [PMC free article] [PubMed] [Google Scholar]
- 21.Guldener, U., K. Y. Seong, J. Boddu, S. H. Cho, F. Trail, J. R. Xu, G. Adam, H. W. Mewes, G. J. Muehlbauer, and H. C. Kistler. 2006. Development of a Fusarium graminearum Affymetrix GeneChip for profiling fungal gene expression in vitro and in planta. Fungal Genet. Biol. 43316-325. [DOI] [PubMed] [Google Scholar]
- 22.Han, Y. K., M. D. Kim, S. H. Lee, S. H. Yun, and Y. W. Lee. 2007. A novel F-box protein involved in sexual development and pathogenesis in Gibberella zeae. Mol. Microbiol. 63768-779. [DOI] [PubMed] [Google Scholar]
- 23.Hou, Z. M., C. Y. Xue, Y. L. Peng, T. Katan, H. C. Kistler, and J. R. Xu. 2002. A mitogen-activated protein kinase gene (MGV1) in Fusarium graminearum is required for female fertility, heterokaryon formation, and plant infection. Mol. Plant-Microbe Interact. 151119-1127. [DOI] [PubMed] [Google Scholar]
- 24.Jenczmionka, N. J., F. J. Maier, A. P. Losch, and W. Schafer. 2003. Mating, conidiation and pathogenicity of Fusarium graminearum, the main causal agent of the head-blight disease of wheat, are regulated by the MAP kinase gpmk1. Curr. Genet. 4387-95. [DOI] [PubMed] [Google Scholar]
- 25.Jobic, C., A. M. Boisson, E. Gout, C. Rascle, M. Fevre, P. Cotton, and R. Bligny. 2007. Metabolic processes and carbon nutrient exchanges between host and pathogen sustain the disease development during sunflower infection by Sclerotinia sclerotiorum. Planta 226251-265. [DOI] [PubMed] [Google Scholar]
- 26.Kang, Z., and H. Buchenauer. 1999. Immunocytochemical localization of Fusarium toxins in infected wheat spikes by Fusarium culmorum. Physiol. Mol. Plant Pathol. 55275-288. [Google Scholar]
- 27.Kang, Z. S., H. Buchenauer, L. L. Huang, Q. M. Han, and H. C. Zhang. 2008. Cytological and immunocytochemical studies on responses of wheat spikes of the resistant Chinese cv. Sumai 3 and the susceptible cv. Xiaoyan 22 to infection by Fusarium graminearum. Eur. J. Plant Pathol. 120383-396. [Google Scholar]
- 28.Kim, J. E., J. M. Jin, H. Kim, J. C. Kim, S. H. Yun, and Y. W. Lee. 2006. GIP2, a putative transcription factor that regulates the aurofusarin biosynthetic gene cluster in Gibberella zeae. Appl. Environ. Microbiol. 721645-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim, J. E., K. Myong, W. B. Shim, S. H. Yun, and Y. W. Lee. 2007. Functional characterization of acetylglutamate synthase and phosphoribosylamine-glycine ligase genes in Gibberella zeae. Curr. Genet. 5199-108. [DOI] [PubMed] [Google Scholar]
- 30.Kristen, J., A. Gleiberman, C. Shi, D. I. Simon, and M. G. Rosenfeld. 2008. Cooperative regulation in development by SMRT and FOXP1. Genes Dev. 22740-745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu, S. W., S. Kroken, B. N. Lee, B. Robbertse, A. C. L. Churchill, O. C. Yoder, and B. G. Turgeon. 2003. A novel class of gene controlling virulence in plant pathogenic ascomycete fungi. Proc. Natl. Acad. Sci. USA 1005980-5985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maloney, A. P., and H. D. Vanetten. 1994. A gene from the fungal plant pathogen Nectria haematococca that encodes the phytoalexin-detoxifying enzyme pisatin demethylase defines a new cytochrome-p450 family. Mol. Gen. Genet. 243506-514. [DOI] [PubMed] [Google Scholar]
- 33.Malz, S., M. N. Grell, C. Thrane, F. J. Maier, P. Rosager, A. Felk, K. S. Albertsen, S. Salomon, L. Bohn, W. Schafer, and H. Giese. 2005. Identification of a gene cluster responsible for the biosynthesis of aurofusarin in the Fusarium graminearum species complex. Fungal Genet. Biol. 42420-433. [DOI] [PubMed] [Google Scholar]
- 34.Mateja, A., T. Cierpicki, M. Paduch, Z. S. Derewenda, and J. Otlewski. 2006. The dimerization mechanism of LIS1 and its implication for proteins containing the LisH motif. J. Mol. Biol. 357621-631. [DOI] [PubMed] [Google Scholar]
- 34a.McMullen, M., et al. 1997. Scab of wheat and barley: a re-emerging disease of devastating impact. Plant Dis. 811340-1348. [DOI] [PubMed] [Google Scholar]
- 35.Nguyen, N. L., M. Fehrmann, S. Salomon, and W. Schäfer. 2007. Different secreted lipases are virulence factors of Fusarium graminearum. Fungal Genet. Newslett. 54A486. [Google Scholar]
- 36.Oeser, B., P. M. Heidrich, U. Muller, P. Tudzynski, and K. B. Tenberge. 2002. Polygalacturonase is a pathogenicity factor in the Claviceps purpurea-rye interaction. Fungal Genet. Biol. 36176-186. [DOI] [PubMed] [Google Scholar]
- 37.Oide, S., W. Moeder, S. Krasnoff, D. Gibson, H. Haas, K. Yoshioka, and B. G. Turgeon. 2006. NPS6, encoding a nonribosomal peptide synthetase involved in siderophore-mediated iron metabolism, is a conserved virulence determinant of plant pathogenic ascomycetes. Plant Cell 182836-2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parry, D. W., P. Jenkinson, and L. McLeod. 1995. Fusarium ear blight (scab) in small grain cereals: a review. Plant Pathol. 44207-238. [Google Scholar]
- 39.Pedreno, Y., P. Gonzalez-Parraga, M. Martinez-Esparza, R. Sentandreu, E. Valentin, and J. C. Arguelles. 2007. Disruption of the Candida albicans ATC1 gene encoding a cell-linked acid trehalase decreases hypha formation and infectivity without affecting resistance to oxidative stress. Microbiology 1531372-1381. [DOI] [PubMed] [Google Scholar]
- 40.Perissi, V., A. Aggarwal, C. K. Glass, D. W. Rose, and M. G. Rosenfeld. 2004. A corepressor/coactivator exchange complex required for transcriptional activation by nuclear receptors and other regulated transcription factors. Cell 116511-526. [DOI] [PubMed] [Google Scholar]
- 41.Pijnappel, W., D. Schaft, A. Roguev, A. Shevchenko, H. Tekotte, M. Wilm, G. Rigaut, B. Seraphin, R. Aasland, and A. F. Stewart. 2001. The Saccharomyces cerevisiae SET3 complex includes two histone deacetylases, Hos2 and Hst1, and is a meiotic-specific repressor of the sporulation gene program. Genes Dev. 152991-3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Proctor, R. H., T. M. Hohn, and S. P. McCormick. 1995. Reduced virulence of Gibberella zeae caused by disruption of a trichothecene toxin biosynthetic gene. Mol. Plant-Microbe Interact. 8593-601. [DOI] [PubMed] [Google Scholar]
- 43.Ramamoorthy, V., X. H. Zhao, A. K. Snyder, J. R. Xu, and D. M. Shah. 2007. Two mitogen-activated protein kinase signalling cascades mediate basal resistance to antifungal plant defensins in Fusarium graminearum. Cell. Microbiol. 91491-1506. [DOI] [PubMed] [Google Scholar]
- 44.Seo, B. W., H. K. Kim, Y. W. Lee, and S. H. Yun. 2007. Functional analysis of a histidine auxotrophic mutation in Gibberella zeae. Plant Pathol. J. 2351-56. [Google Scholar]
- 45.Seo, J. A., J. C. Kim, D. H. Lee, and Y. W. Lee. 1996. Variation in 8-ketotrichothecenes and zearalenone production by Fusarium graminearum isolates from corn and barley in Korea. Mycopathologia 13431-37. [DOI] [PubMed] [Google Scholar]
- 46.Seong, K., Z. M. Hou, M. Tracy, H. C. Kistler, and J. R. Xu. 2005. Random insertional mutagenesis identifies genes associated with virulence in the wheat scab fungus Fusarium graminearum. Phytopathology 95744-750. [DOI] [PubMed] [Google Scholar]
- 47.Seong, K., L. Li, Z. M. Hou, M. Tracy, H. C. Kistler, and J. R. Xu. 2006. Cryptic promoter activity in the coding region of the HMG-CoA rediactase gene in Fusarium graminearum. Fungal Genet. Biol. 4334-41. [DOI] [PubMed] [Google Scholar]
- 48.Shim, W. B., U. S. Sagaram, Y. E. Choi, J. So, H. H. Wilkinson, and Y. W. Lee. 2006. FSR1 is essential for virulence and female fertility in Fusarium verticillioides and F. graminearum. Mol. Plant-Microbe Interact. 19725-733. [DOI] [PubMed] [Google Scholar]
- 49.Skadsen, R. W., and T. A. Hohn. 2004. Use of Fusarium graminearum transformed with GFP to follow infection patterns in barley and Arabidopsis. Physiol. Mol. Plant Pathol. 6445-53. [Google Scholar]
- 50.Strange, R. N., and H. Smith. 1978. Specificity of choline and betaine as stimulants of Fusarium graminearum. Trans. Br. Mycol. Soc. 70187-192. [Google Scholar]
- 51.Strange, R. N., H. Smith, and J. R. Majer. 1972. Choline, one of two fungal growth stimulants in anthers responsible for susceptibility of wheat to Fusarium graminearum. Nature 238103-105. [Google Scholar]
- 52.Trail, F., I. Gaffoor, and S. Vogel. 2005. Ejection mechanics and trajectory of the ascospores of Gibberella zeae (anamorph Fusarium graminearum). Fungal Genet. Biol. 42528-533. [DOI] [PubMed] [Google Scholar]
- 53.Trail, F., J. R. Xu, P. San Miguel, R. G. Halgren, and H. C. Kistler. 2003. Analysis of expressed sequence tags from Gibberella zeae (anamorph Fusarium graminearum). Fungal Genet. Biol. 38187-197. [DOI] [PubMed] [Google Scholar]
- 54.Voigt, C. A., W. Schafer, and S. Salomon. 2005. A secreted lipase of Fusarium graminearum is a virulence factor required for infection of cereals. Plant J. 42364-375. [DOI] [PubMed] [Google Scholar]
- 55.Yoon, H. G., D. W. Chan, Z. Q. Huang, J. W. Li, J. D. Fondell, J. Qin, and J. M. Wong. 2003. Purification and functional characterization of the human N-CoR complex: the roles of HDAC3, TBL1 and TBLR1. EMBO J. 221336-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yu, H. Y., J. A. Seo, J. E. Kim, K. H. Han, W. B. Shim, S. H. Yun, and Y. W. Lee. 2008. Functional analyses of heterotrimeric G protein G alpha and G beta subunits in Gibberella zeae. Microbiology 154392-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang, J. S., M. Kalkum, B. T. Chait, and R. G. Roeder. 2002. The N-CoR-HDAC3 nuclear receptor corepressor complex inhibits the JNK pathway through the integral subunit GPS2. Mol. Cell 9611-623. [DOI] [PubMed] [Google Scholar]
- 58.Zhao, X. H., C. Xue, Y. Kim, and J. R. Xu. 2004. A ligation-PCR approach for generating gene replacement constructs in Magnaporthe grisea. Fungal Genet. Newslett. 5117-18. [Google Scholar]