Abstract
Edwardsiella tarda is a gram-negative pathogen with a broad host range that includes humans, animals, and fish. Recent studies have shown that the LuxS/autoinducer type 2 (AI-2) quorum sensing system is involved in the virulence of E. tarda. In the present study, it was found that the E. tarda LuxS mutants bearing deletions of the catalytic site (C site) and the tyrosine kinase phosphorylation site, respectively, are functionally inactive and that these dysfunctional mutants can interfere with the activity of the wild-type LuxS. Two small peptides, 5411 and 5906, which share sequence identities with the C site of LuxS, were identified. 5411 and 5906 proved to be inhibitors of AI-2 activity and could vitiate the infectivity of the pathogenic E. tarda strain TX1. The inhibitory effect of 5411 and 5906 on AI-2 activity is exerted on LuxS, with which these peptides specifically interact. The expression of 5411 and 5906 in TX1 has multiple effects (altering biofilm production and the expression of certain virulence-associated genes), which are similar to those caused by interruption of luxS expression. Further study found that it is very likely that 5411 and 5906 can be released from the strains expressing them and, should TX1 be in the vicinity, captured by TX1. Based on this observation, a constitutive 5411 producer (Pseudomonas sp. strain FP3/pT5411) was constructed in the form of a fish commensal isolate that expresses 5411 from a plasmid source. The presence of FP3/pT5411 in fish attenuates the virulence of TX1. Finally, it was demonstrated that fish expressing 5411 directly from tissues exhibit enhanced resistance against TX1 infection.
Quorum sensing is a process of cell-cell communication whereby the population behaviors of bacteria are coordinated to adapt to various environmental situations (15, 17). During quorum sensing, bacteria synthesize and secrete small signaling molecules called autoinducers that can diffuse across cellular membranes and be sensed by neighboring cells. In response to the signal, the cells adjust the expression of certain genes, thus resulting in alterations of community behaviors. For gram-negative bacteria, the classical quorum-sensing system, as represented by the LuxI/LuxR circuit of Vibrio fischeri (12, 13), involves autoinducer type 1 (AI-1). AI-1 molecules are acyl homoserine lactones that are synthesized by the enzyme LuxI and its homologues. Since AI-1 molecules are generally species specific and can only be responded to by the same bacterial species that produced them, AI-1 is considered an intraspecies signaling signal. In contrast, AI-2, which was first discovered in Vibrio harveyi (2) and later found in diverse bacteria, is a universal signaling molecule that communicates between bacteria of different species and genera. The V. harveyi AI-2 is a furanosyl borate diester that is synthesized from S-adenosylhomocysteine (SAH) via the enzymatic steps involving the nucleosidase Pfs, which converts SAH to S-ribosylhomocysteine (SRH), and LuxS, which catalyzes the cleavage of the thioether linkage of SRH to produce 4,5-dihydroxy-2,3-pentanedione, from which AI-2 is derived (29, 43). AI-2 and its synthase, LuxS, have been discovered to exist in both gram-negative and gram-positive bacteria (8, 45), and interruption of LuxS/AI-2-mediated quorum sensing is known to affect multiple aspects of cellular processes, such as bioluminescence, biofilm formation, conjugation, sporulation, and virulence development (9, 16, 19, 26, 27, 30, 33, 36, 40, 44, 48, 51).
LuxS is conserved at the primary structure in many different bacterial species. Sequence comparisons of the known LuxS proteins have revealed the existence in these proteins of a highly conserved motif called the catalytic site (C site), with the sequence feature H-X-X-E-H. In addition, a semiconserved tyrosine kinase phosphorylation site (P site), characterized by K/R-X2-3-D/E-X2-3-Y (8, 14), is found in some of the LuxS proteins. Site-directed mutagenesis analyses have shown that the C site is essential to the catalytic activity of LuxS (58). The potential importance of the P site is not clear. Being a metalloenzyme, the activity of LuxS requires a divalent metal ion, which was initially proposed to be Zn2+ but later demonstrated to be Fe2+. Structural analyses have indicated that LuxS exists as a homodimer with two active sites, each of which contains an Fe2+ ion that is coordinated tetrahedrally by two residues of the C site, a water molecule, and a conserved cysteine residue (7, 20, 30, 37, 41, 58).
Edwardsiella tarda is a gram-negative pathogen with a broad host range that includes both humans and animals. It is considered an important aquaculture pathogen because of its ability to cause edwardsiellosis, a systematic disease that affects a number of farm-reared marine species. Recently, we have cloned and analyzed the luxS gene of E. tarda (55). We found that the E. tarda LuxS is an enzyme of 171 amino acid residues that possesses the conserved C site and P site motifs. Both luxS expression and the AI-2 activity of E. tarda are regulated by the culturing conditions, and the temporal production of LuxS/AI-2 is required for optimal bacterial pathogenicity. In the present study, we investigate the potential for mitigating E. tarda infection by blocking the LuxS/AI-2 signal transduction process. Our results show that small peptides bearing homology to the C site of LuxS can function as specific inhibitors of the LuxS/AI-2 pathway and, as a result, attenuate the virulence of E. tarda.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The Escherichia coli strains DH5α (Takara) and S17-1λpir (Biomedal, Spain), the Vibrio harveyi strain BB170 (ATCC), the Edwardsiella tarda strain TX1 (fish isolate, tetracycline resistant) (55), and the Pseudomonas sp. strain FP3 (fish isolate, ampicillin resistant, 50% lethal dose of >1 × 108 CFU/g with Japanese flounder) were cultured in Luria-Bertani (LB) medium (42) at 37°C (for E. coli) or 28°C (for all others). Appropriate antibiotics were added at the following concentrations: ampicillin, 100 μg/ml; chloramphenicol, 25 μg/ml; kanamycin, 50 μg/ml; and tetracycline, 15 μg/ml.
Plasmid construction.
The plasmids and primers used in this study are listed in Table 1. To construct pH54A, pH58A, pE122K, pDC, pDP, and pDE122, the luxS mutants expressed by these plasmids were generated by using overlap extension PCR (21) as follows: the overlapping PCR amplifications were conducted by using the primer pairs F19/R31 and F33/R8, F19/R32 and F34/R8, F19/R24 and F26/R8, F19/R27 and F29/R8, F19/R25 and F27/R8, and F19/R23 and F25/R8, respectively; the fusion PCR amplifications were performed with the primer pair F19/R8; and the PCR products were purified and inserted into pL1 (46) at the SmaI site. To create pELS, the SwaI fragment of pBTES carrying Ptrc-luxS was inserted into pACYC184 (New England Biolabs) at the EcoRV site. To construct pJDC and pJDP, the SwaI fragments of pDC and pDP carrying the mutant luxS were inserted into pJRA at the EcoRV site. pJR20 was created by inserting the linker ESPB (5′-AATTCATTTAAATGTTTAAACG-3′) into pDN18 (34) between the EcoRI/BamHI sites. To construct p5411, p5906, and p1026, the coding sequences for 5411, 5906, and 1026 (5′-TATGCATACCCTTGAGCACCTATTCGCCGGCTTTATGTAAC-3′, 5′-TATGCTATTCGCCGGCTTTATGTAAC-3′, and 5′-TATGTTCTATATGAGCCTGATCTAAC-3′, respectively) were inserted into pBT3 (56) between the NdeI/XhoI sites, resulting in pBT54, pBT59, and pBT10, respectively; these plasmids were digested with SwaI, and the fragments carrying the Ptrc promoter fused to the coding sequences for the small peptides were inserted into pJRA at the EcoRV site. pT5411 was created by inserting the SwaI fragment of pBT54 carrying Ptrc and the coding sequence for 5411 into pJR20 at the SwaI site. pC5411 was created by inserting the coding sequence for 5411 (5′-AATTCACCACCATGCATACCCTTGAGCACCTATTCGCCGGCTTTATGTAACCC-3′) into pCI between the EcoRI/SmaI sites. pETLS and pETF2 were created by inserting the coding sequences for luxS and fur, respectively, into pET258 (57) between the NdeI/XhoI sites.
TABLE 1.
Plasmids and primers used in this study
| Plasmid or primer | Relevant characteristicsa or sequence (5′ → 3′) | Source or reference |
|---|---|---|
| Plasmids | ||
| p5411 | Apr; expressing 5411 | This study |
| p5906 | Apr; expressing 5906 | This study |
| p1026 | Apr; expressing 1026 | This study |
| pBTES | Apr; expressing luxS | 55 |
| pCI | Apr; eukaryotic expression vector | Promega |
| pC5411 | Apr; pCI expressing 5411 | This study |
| pDC | Apr, expressing luxS bearing C site deletion | This study |
| pDE122 | Apr, expressing luxS bearing E122 deletion | This study |
| pDP | Apr, expressing luxS bearing P site deletion | This study |
| pE122K | Apr; expressing luxS bearing E122K mutation | This study |
| pELS | Cmr; expressing luxS | This study |
| pETF2 | Knr; expressing E. tarda fur | This study |
| pETLS | Knr; expressing luxS | This study |
| pH54A | Apr; expressing luxS bearing H54A mutation | This study |
| pH58A | Apr; expressing luxS bearing H58A mutation | This study |
| pJDC | Apr, expressing luxS bearing C site deletion | This study |
| pJDP | Apr, expressing luxS bearing P site deletion | This study |
| pJR20 | Tcr; broad-host-range vector | This study |
| pJRA | Apr; broad-host-range vector | 55 |
| pT5411 | Tcr; expressing 5411 | This study |
| Primers | ||
| F19 | GATATCGCAGTATGGGAAAAGGT (EcoRV) | |
| F25 | GCATCCCACTGAATGAGTTTCAGTGTG | |
| F26 | GCATCCCAAAGCTGAATGAGTTTCAGTG | |
| F27 | GCGGAGCCAGTGTGGTACCTACA | |
| F29 | CGCGGTATTCTATTCGCCGGCTTTAT | |
| F33 | CGGTATTGCTACCCTTGAGCACCTAT | |
| F34 | CTTGAGGCCCTATTCGCCGGCTT | |
| R8 | GATATCTGTGGGGGCTTTTCTA (EcoRV) | |
| R23 | CATTCAGTGGGATGCTCCGCTG | |
| R24 | TTCAGCTTTGGGATGCTCCGCT | |
| R25 | ACACTGGCTCCGCTGATCGC | |
| R27 | CGAATAGAATACCGCGCTCCGG | |
| R31 | AAGGGTAGCAATACCGCGCTCCG | |
| R32 | CGAATAGGGCCTCAAGGGTATGAATAC |
Ap, ampicillin; Kn, kanamycin; Cm, chloramphenicol; Tc, tetracycline.
bUnderlined nucleotides are restriction sites of the enzymes indicated in parentheses.
Peptide synthesis.
Biotin-labeled and unlabeled peptides were synthesized by Sangon (Shanghai, People's Republic of China). Biotin was labeled at the N termini of the peptides. The peptides were purified by high-performance liquid chromatography to ∼96% purity.
AI-2 assay.
The preparation of cell-free culture supernatant was performed as described previously (47, 55). Briefly, cells were cultured in LB medium with appropriate supplements to an optical density at 600 nm (OD600) of 0.9. To obtain cell-free culture supernatant, cells were subjected to centrifugation, and the supernatant was filtered through a 0.22-μm filter (Millipore). For measurement of bioluminescence induction, the overnight culture of the Vibrio harveyi strain BB170 was diluted 1:5,000 in fresh AB medium (47) supplemented with the cell-free culture supernatant (10%) of the tested strain or with the growth medium (as the control). Where indicated, the synthetic small peptides were added as supplements at a concentration of 0.2 μM. Light production was measured in a Glomax luminometer (Promega).
To examine the effects of DH5α/pDC/pELS and DH5α/pDP/pELS on the AI-2 activity of DH5α/pELS, the cell-free culture supernatants of DH5α/pDC/pELS and DH5α/pDP/pELS (prepared as described above) were each mixed with an equal volume of cell-free culture supernatant of DH5α/pELS. After incubation at 37°C for 0.5 h, the mixtures were assayed for AI-2 activity as described above.
Purification of recombinant protein.
BL21(DE3) (Tiangen, China) was transformed with pETLS and pETF2. The recombinant LuxS and Fur proteins were purified from the transformants by using nickel-nitrilotriacetic acid (Ni-NTA) columns as described previously (57). Et49, an E. tarda antigenic protein, was purified as described previously (23).
Antisera.
Antiserum to the recombinant LuxS was prepared by subcutaneously injecting an adult New Zealand White rabbit with 300 μg of purified recombinant LuxS mixed in complete Freund's adjuvant, followed by a boost with the same amount of LuxS in incomplete Freund's adjuvant 25 days later. A second boost was performed 12 days after the first boost, and a third boost was performed 10 days after the second boost. The rabbit was bled 10 days after the third boost, and the serum was obtained from the collected blood by centrifugation.
Western and immunoblot analysis.
Cells were grown in LB medium to an OD600 of 1 and lysed with lysis buffer (100 mM NaH2PO4, 10 mM Tris-Cl, and 8 M urea, pH 8.0). The lysed cells were centrifuged at 4°C, and the supernatant was subjected to electrophoresis in 0.1% sodium dodecyl sulfate-12% polyacrylamide gels. After electrophoresis, the proteins were transferred to nitrocellulose membranes, which were then blotted with LuxS antibodies as described previously (56).
Pull-down assay.
One-milliliter Ni-NTA columns (GE Healthcare) were incubated with 100 μg of His-tagged recombinant LuxS or 100 μg of His-tagged recombinant Fur in binding buffer (50 mM NaH2PO4, 300 mM NaCl, and 10 mM imidazole, pH 8.0) for 1 h at 4°C. The columns were washed three times with 10 ml of wash buffer (50 mM NaH2PO4, 300 mM NaCl, and 20 mM imidazole, pH 8.0) and incubated with 30 μg of biotin-labeled small peptides or, for competition assays, with 1.5 μg of biotin-labeled small peptides that had been preincubated with 100 μg of LuxS, 300 μg of Et49, or 300 μg of bovine serum albumin in 0.5 ml of binding buffer at 4°C for 1 h. After incubation at 4°C for 1 h, the columns were washed three times with 10 ml of wash buffer and eluted with 10 ml of elution buffer (50 mM NaH2PO4, 300 mM NaCl, and 250 mM imidazole, pH 8.0). The eluates were concentrated by using Amicon ultra centrifugal filter devices (Millipore) and spotted (10 μl per spot) in parallel onto two nitrocellulose membranes; one of the membranes was used for the immunoblotting assay with LuxS antibodies (as described above), and the other membrane was used for the detection of biotin signals with horseradish peroxidase-conjugated streptavidin (Amersham), according to the manufacturer's instructions.
Bacterial conjugation.
pJRA variants were introduced into strain S17-1λpir by transformation. Conjugation between the transformants and strain TX1 or FP3 was performed as described previously (55).
Biofilm assay.
Cells were cultured in LB medium to exponential phase and diluted to 1 × 105 CFU/ml. The diluted cells were transferred into a 96-well polystyrene plate (Nunc, Danmark) and incubated at 28°C for 24 h without agitation. After the incubation, the unattached cells were removed from the wells with a pipette, and the wells were washed rigorously with phosphate-buffered saline (PBS). The attached cells were treated with Bouin fixative for 1 h and stained with 1% crystal violet solution for 20 min. The samples were then measured at A570.
qRT-PCR.
Total RNA was extracted from fish tissues and from cells grown in appropriate medium to an OD600 of 0.8 by using an SV total RNA isolation system (Promega). To verify that the RNA was free of DNA contamination, PCR amplifications were performed using the total RNA (at least 10 times the amount used in subsequent quantitative real-time PCR [qRT-PCR]) as the template and the primers intended for all qRT-PCR as primers. The results showed that all PCR amplifications were negative. qRT-PCR was carried out in an ABI 7300 real-time detection system (Applied Biosystems) by using a SYBR ExScript qRT-PCR kit (Takara, Dalian, People's Republic of China). Each assay was performed in triplicate with the 16S rRNA as control. Dissociation analysis of amplification products was performed at the end of each PCR to confirm that only one PCR product was amplified and detected. The comparative cycle threshold method (2−ΔΔCT method) (32) was used to analyze the mRNA level. All data are given in terms of relative quantities of mRNA expressed as the means plus or minus standard errors of the means (SE). Statistical analyses were performed by using the two-tailed t test.
Bacterial dissemination in tissues and blood.
To examine bacterial dissemination, TX1/p5411, TX1/p5906, and TX1/pJRA were cultured to an OD600 of 0.6 in LB medium, washed, and resuspended in PBS to 2 × 106 CFU/ml. Three groups (15 fish/group) of Japanese flounder (Paralichthys olivaceus; ∼11 g each) were injected intraperitoneally (i.p.) with 100 μl of TX1/p5411, TX1/p5906, or TX1/pJRA (as prepared above). The blood and livers of the fish were taken under aseptic conditions at various time points postinfection. The livers were homogenized in PBS. The homogenates and the blood were plated on LB agar plates supplemented with ampicillin (marker for pJRA derivatives) and tetracycline (marker for TX1). After incubation at 28°C for 48 h, the colonies that appeared on the plates were enumerated. The genetic nature of these colonies was verified by PCR analysis using primers specific to TX1 and the plasmids. The PCR products were subsequently analyzed by DNA sequencing. Similarly, to examine the effect of FP3/pT5411 on TX1 infectivity, two groups (15 fish/group) of Japanese flounder were injected i.p. with ∼5 × 107 CFU of FP3/pT5411 or FP3/pJR20 that had been cultured in LB medium to an OD600 of 0.7, washed, and resuspended in PBS; 12 h later, the fish were challenged with 2 × 105 CFU of TX1. The fish were sacrificed at various time points postchallenge, and bacteria recovered from the blood were enumerated as described above. Statistical analyses were performed by using the Student t test.
Analysis of the in vivo survival of FP3/pT5411.
FP3/pT5411 was cultured as described above and resuspended in PBS to 5 × 108 CFU. Twelve Japanese flounder were each injected i.p. with 100 μl of FP3/pT5411 suspension. The fish (three at each time point) were sacrificed at 12, 48, 72, and 96 h after the injection; peritoneal fluids, blood, livers, and spleens of the fish were collected and examined for bacterial recovery as described above and previously (56).
Transient expression of 5411 in fish tissues.
Two groups (15 fish/group) of Japanese flounder (∼11 g each) were administered 12 μg of pC5411 or pCI via muscle injection. Ten days later, the fish were challenged with 100 μl of TX1 that had been cultured to an OD600 of 0.6 in LB medium, washed, and resuspended in PBS to 2 × 106 CFU/ml. The fish were sacrificed at various time points postchallenge and examined for bacterial dissemination in the liver and blood as described above.
RESULTS
Mutational analysis of LuxS.
To determine the functional importance of the C site and P site of E. tarda LuxS (Fig. 1), alanine substitution was performed upon H54 and H58 and lysine substitution was performed upon E122. In addition, deletion mutagenesis was also carried out whereby the C site, P site, and E122 of the P site were each deleted. E. coli strain DH5α, which is defective in AI-2 production (47) and therefore can serve as a reporter of exogenous LuxS/AI-2 activity, was transformed separately with the plasmids pH54A, pH58A, pE122K, pDC, pDP, pDE122, and pBTES and the control plasmid pL1. pH54A, pH58A, and pE122K carry the luxS mutants bearing H54A, H58A, and E122K substitutions, respectively; pDC, pDP, and pDE122 carry the luxS mutants bearing deletions of the C site, P site, and E122, respectively; and pBTES carries the wild-type luxS. The transformants were subjected to AI-2 assay, which showed that except for DH5α/pE122K, which displayed AI-2 activity to a level similar to that of the AI-2 activity displayed by DH5α/pBTES, all other transformants exhibited no detectable AI-2 activity (Table 2). Western immunoblotting analyses showed that the amounts of LuxS detected in DH5α harboring pH54A, pH58A, pDC, pDP, or pDE122 were comparable to the amount detected in DH5α/pBTES (Fig. 2 and data not shown), suggesting that none of these mutations affected LuxS production/stability. Taken together, these results demonstrate that the integrity of both the C site and P site is essential to the functioning of LuxS.
FIG. 1.
The amino acid sequence of E. tarda LuxS. The C site and P site are underlined. The residues contained in 5411, 5906, and 1026 are indicated.
TABLE 2.
AI-2 activities of strains DH5α and TX1 expressing LuxS variants and small peptidesa
| Strain and plasmid(s) | LuxS variant | Level of induction (fold) of AI-2 activityb |
|---|---|---|
| DH5α/pL1 | None | 1 |
| DH5α/pBTES | Wild type | 319 ± 19.14 |
| DH5α/pH54A | H54A | 1 ± 0.02 |
| DH5α/pH58A | H58A | 1 ± 0.04 |
| DH5α/pE122K | E122K | 302 ± 12.08 |
| DH5α/pDC | C site deletion | 1 ± 0.01 |
| DH5α/pDP | P site deletion | 1 ± 0.01 |
| DH5α/pDE122 | E122 deletion | 1 ± 0.02 |
| DH5α/pELS | Wild type | 305 ± 21.35 |
| DH5α/pL1/pELS | Wild type | 308 ± 26.18 |
| DH5α/pH54A/pELS | Wild type + H54A mutant | 289 ± 10.40 |
| DH5α/pDC/pELS | Wild type + C site deletion mutant | 48 ± 1.34 |
| DH5α/pDP/pELS | Wild type + P site deletion mutant | 55 ± 3.16 |
| TX1/pJRA | Wild type | 524 ± 41.92 |
| TX1/pJDC | Wild type + C site deletion mutant | 78 ± 2.63 |
| TX1/pJDP | Wild type + P site deletion mutant | 99 ± 5.78 |
| TX1/p5411 | Wild type + 5411 | 68 ± 4.37 |
| TX1/p5906 | Wild type + 5906 | 104 ± 5.86 |
| TX1/p1026 | Wild type + 1026 | 512 ± 18.94 |
Cell-free culture fluids taken from cultures of plasmid-containing DH5α and TX1 grown in LB medium to an OD600 of 0.9 were used for AI-2 assay.
AI-2 activities are presented as the change in level of induction over that in the DH5α/pL1 culture. Data are representative of at least three independent experiments and are presented as the means ± SE.
FIG. 2.
Western immunoblot analysis of LuxS production in DH5α carrying luxS variants. Whole-cell proteins were prepared from cells grown in LB medium to an OD600 of ∼1 and resolved by electrophoresis in a 0.1% sodium dodecyl sulfate-12% polyacrylamide gel. After electrophoresis, the proteins were transferred to a nitrocellulose membrane and blotted with LuxS antibodies. Lanes 1 to 5, proteins extracted from DH5α harboring the indicated plasmids; lane 6, protein marker.
LuxS mutants can act as dominant-negative inhibitors and interfere with the activity of the wild-type LuxS.
Since LuxS functions as a dimer, it is possible that in an environment in which the wild type and a mutant LuxS coexisted, the mutant could block the activity of the wild type by forming inactive heterodimers with the wild type or by interacting negatively with the wild type in other ways. To examine this possibility, DH5α/pH54A, DH5α/pDC, DH5α/pDP, and DH5α/pL1 were transformed with the plasmid pELS, which carries the wild-type luxS. Subsequent AI-2 assays showed that DH5α/pL1/pELS and DH5α/pH54A/pELS exhibited AI-2 activities to levels comparable to the level of AI-2 activity exhibited by DH5α/pELS; in contrast, DH5α/pDC/pELS and DH5α/pDP/pELS exhibited, respectively, 6.5- and 5.5-fold less AI-2 activity than DH5α/pELS (Table 2). Hence, it appeared that the presence of pDC and pDP inhibited the AI-2 activity of DH5α/pELS. Incubation of the cell-free culture supernatants of DH5α/pDC/pELS and DH5α/pDP/pELS with the cell-free culture supernatant of DH5α/pELS had no effect on the AI-2 activity of the latter, which ruled out the possibility that DH5α/pDC/pELS and DH5α/pDP/pELS may produce certain elements that inactivate the AI-2 molecules directly. Taken together, these results suggest that the LuxS mutants bearing C site and P site deletions very likely reduce AI-2 activity by interfering with the activity of the wild-type LuxS.
To examine whether the inhibitory effect of the mutant LuxS could be observed in the native genetic background of E. tarda, the conjugative plasmids pJDC and pJDP, which carry the luxS mutants bearing C site and P site deletions, respectively, as well as the control plasmid pJRA, were introduced separately into the pathogenic E. tarda strain TX1 via conjugation. The transconjugants were assayed for AI-2 activity, which showed that the AI-2 activities of TX1/pDC and TX1/pDP were lower than those of TX1/pJRA during the entire growth phase and were, respectively, 6.7- and 5.3-fold less than that of TX1/pJRA at an OD600 of 0.9 (Table 2). Hence, the mutant LuxS bearing C site and P site deletions probably can interfere with the activity of LuxS in TX1.
Selection of small peptides that can interfere with the AI-2 activity of E. tarda.
With the above results, we speculated that the activity of LuxS might be interfered with by, in addition to mutant LuxS, certain specific sequence/structure motifs in the form of peptide elements that could interact specifically with LuxS in such a manner as to prevent the proper function of LuxS. To investigate this idea, we set forth to select small peptides that could mitigate AI-2 production through negative interaction with LuxS. For this purpose, three small peptides were created: (i) 5411 (MHTLEHLFAGFM), which is a 12-residue peptide containing the C site of LuxS; (ii) 5906 (MLFAGFM), which contains the six residues located immediately downstream of the C site; and (iii) 1026 (MFYMSLI), which contains the six residues between F87 and I92 of LuxS (Fig. 1). Plasmids p5411, p5906, and p1026, which carry the genetic elements encoding 5411, 5906, and 1026, respectively, were each introduced into TX1 via conjugation. The transconjugants were assayed for AI-2 production, which showed that, compared to the AI-2 activity of TX1 harboring the control plasmid pJRA, the AI-2 activities of TX1/p5411 and TX1/p5906 were lower during the entire growth phase and were, respectively, 7.7-, and 5-fold less than that of TX1/pJRA at an OD600 of 0.9 (Table 2). In contrast, the AI-2 activity of TX1/p1026 was similar to that of TX1/pJRA. These results indicate that the presence of the small peptides 5411 and 5906, but not 1026, causes a drastic decrease in the AI-2 activity of TX1.
5411 and 5906 can bind specifically to LuxS.
Since our previous study (55) has demonstrated that in TX1, the AI-2 activity is controlled by LuxS, it was likely that the effects of 5411 and 5906 on AI-2 activity described above were exerted on LuxS. To investigate whether or not this was the case, pull-down assays were performed to examine whether there was any physical interaction between the small peptides and LuxS. For this purpose, the biotin-labeled 5411 was passed through a Ni-NTA column saturated with His-tagged LuxS (the bait). As negative controls, the biotin-labeled 5411 was also passed through Ni-NTA columns that were without proteins or saturated with the His-tagged ferric uptake regulator of E. tarda (Fur), a global regulatory protein that is involved in the regulation of iron assimilation and many other cellular processes (39, 49). After washing, the bound proteins/peptides were eluted from the columns. Subsequent detection assays showed that biotin signals were found only in the eluate of the LuxS-saturated column (Fig. 3). Immunoblotting analyses using LuxS antibodies indicated that LuxS appeared in the eluate containing biotin signals (Fig. 3), suggesting that LuxS was coeluted with 5411. Competition assays showed that when the biotin-labeled 5411 was passed through the LuxS-bound Ni-NTA column that had been presaturated with unlabeled 5411, no biotin signals could be detected from the eluate (Fig. 3). Preincubation of biotin-labeled 5411 with LuxS, but not with bovine serum albumin or Et49 (23), abolished the ability of 5411 to be trapped subsequently by LuxS bound to Ni-NTA columns (data not shown). Taken together, these results suggest that 5411 can interact specifically with LuxS. Similar pull-down assay results were also obtained with 5906 (data not shown).
FIG. 3.
Analysis of the interaction between LuxS and 5411 by pull-down assay. Ni-NTA columns were preincubated with His-tagged recombinant E. tarda LuxS (LuxSEt; lanes 1 and 4), E. tarda Fur (FurEt; lane 2), or no protein (lane 3) in the binding buffer for 1 h. Lanes 1 to 3, the columns were then incubated with biotin-labeled 5411 for 1 h; lane 4, the column was first incubated with unlabeled 5411 for 1 h and then incubated with biotin-labeled 5411 for 1 h. After washing, the columns were eluted, and the eluates were spotted in parallel onto two nitrocellulose membranes (A and B). Membrane A was used for the detection of biotin signals with horseradish peroxidase-conjugated streptavidin, and membrane B was blotted with LuxS antibodies. +, present; −, absent.
Interaction between 5411 and LuxS requires the integrity of the P site.
To further examine the specificity of the interaction between 5411 and LuxS, we wanted to determine whether 5411 could bind to the mutant LuxS bearing C site and P site deletions. However, the C site deletion mutant, when expressed in the E. coli strain BL21(DE3), existed mainly in the form of insoluble inclusion bodies and thus could not be purified under native conditions. We therefore only examined the potential interaction between 5411 and the mutant LuxS bearing the P site deletion, which could be purified under native conditions. To this end, the biotin-labeled 5411 was passed through Ni-NTA columns bound with the mutant LuxS. Subsequent detection assays showed that no biotin signals could be found in the eluates (data not shown), suggesting that deletion of the P site disables LuxS for interaction with 5411.
Expression of 5411 and 5906 in TX1 has multiple effects. (i) Effect on biofilm formation and the expression of virulence-associated genes.
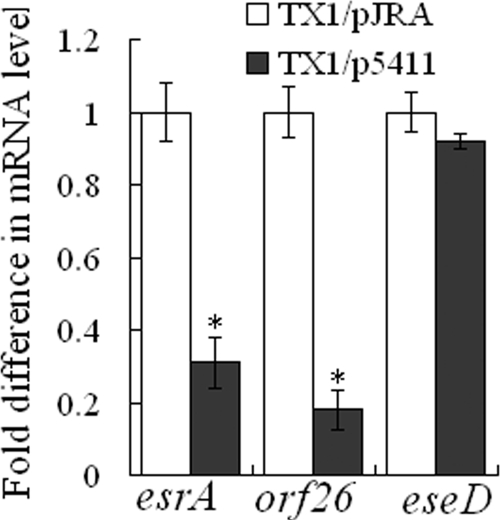
Since previous studies have shown that interference with luxS expression, which results in a reduction of AI-2 activity, has a negative effect on biofilm formation and on the expression of the type III secretion system genes esrA and orf26 but has no effect on the expression of the type III secretion system gene eseD (55), we examined whether the expression of 5411 and 5906 would produce the same effect on TX1. The results showed that the growth profiles of TX1/p5411 and TX1/p5906 in standard LB medium were similar to that of TX1/pJRA (data not shown), but the biofilm growths of TX1/p5411 and TX1/p5906 on a polystyrene surface were, respectively, 2.5- and 2-fold less than that of TX1/pJRA. qRT-PCR analyses indicated that esrA and orf26 expression levels in TX1/p5411 were, respectively, 3.2- and 5.8-fold less than those in TX1/pJRA (Fig. 4), whereas eseD expression in TX1/p5411 was comparable to that in TX1/pJRA. Similar levels of reduction in esrA and orf26 expression were also observed for TX1/p5906 (data not shown). Hence, the expression of 5411 and 5906 in TX1 produces effects that are similar to those caused by interruption of luxS expression.
FIG. 4.
Effect of 5411 on the expression of esrA, orf26, and eseD. Total RNA was extracted from TX1/pJRA and TX1/p5411 grown in LB medium to an OD600 of 0.8 and used for qRT-PCR. The mRNA level of each gene was normalized to that of 16S rRNA. Data are the means of the results of three independent assays and are presented as the means ± SE. *, P < 0.001.
(ii) Effect on bacterial dissemination and survival in host tissue.
Since in TX1, the temporal production of LuxS and AI-2 activity is important to pathogenicity, we wondered whether the presence of 5411 and 5906 would have any effect on bacterial virulence. To investigate this question, Japanese flounder were infected with the same dose of TX1/p5411, TX1/p5906, or TX1/pJRA. The infections were terminated after 24 h, and the amounts of bacteria recoverable from the livers of the infected fish were determined. The results showed that the average amounts of bacteria recovered from the livers of TX1/p5411- and TX1/p5906-infected fish were, respectively, 212- and 580-fold less than the amount recovered from the livers of TX1/pJRA-infected fish.
To examine whether the differences in bacterial recovery observed above were due to loss of p5411 and p5906 as a result of selective pressures, the loss rates of p5411 and p5906 were determined. For this purpose, the livers were taken from five TX1/p5411- and five TX1/p5906-infected fish and homogenized in PBS; the homogenates were plated on LB plates supplemented with either tetracycline alone (for the selection of TX1 harboring and not harboring plasmids) or tetracycline plus ampicillin (for the selection of TX1 harboring plasmids). The results showed that, for the TX1/p5411- and TX1/p5906-infected fish, the average amounts of bacteria that appeared on the tetracycline-plus-ampicillin plates were, respectively, 11.8 and 12.8% lower than those that appeared on the tetracycline plates; PCR analyses showed that 90% (72/80) of the colonies on the tetracycline plates derived from the TX1/p5411-infected fish were TX1/p5411 and 86.3% (69/80) of the colonies on the tetracycline plates derived from the TX1/p5906-infected fish were TX1/p5906. Hence, the plasmid loss rates of p5411 and p5906 (11.8 and 13.7% at most, respectively) were comparable to that of pJRA (10%) (55). Taken together, these results demonstrate that the expression of 5411 and 5906 in TX1 severely vitiates the tissue dissemination and survival ability of the pathogen.
The presence of 5411 and 5906 in the culture medium can reduce the AI-2 activity of TX1.
Since 5411 and 5906 are small peptides, there was the possibility that they could pass through cell membranes through mechanisms such as passive diffusion. We reasoned that if 5411 and 5906 could, regardless of the mechanism (passive diffusion or active transport), cross the membrane barriers of DH5α and TX1, then TX1 incubated in the presence of the culture supernatants of DH5α/p5411 and DH5α/p5906 would capture the exogenous peptides released from DH5α/p5411 and DH5α/p5906 and, as a result, display reduced levels of AI-2 activity. To examine this hypothesis, TX1 was grown in LB medium supplemented with the cell-free culture fluids of DH5α/p5411, DH5α/p5906, or, as a control, DH5α/pJRA. Subsequent AI-2 assays showed that the AI-2 activities of TX1 grown in the presence of the culture supernatants of DH5α/p5411 and DH5α/p5906 were approximately 2.2-fold less than that of TX1 grown in the presence of the supernatant of DH5α/pJRA. These results supported the notion that (i) 5411 and 5906 were present in the culture supernatants of DH5α/p5411 and DH5α/p5906 and (ii) TX1 could absorb free 5411 and 5906 from the environmental milieu. To further examine the latter part of this notion, TX1 was cultured in LB medium alone or in LB medium supplemented with synthetic 5411 or 1026. Subsequent AI-2 assays showed that the AI-2 activity of TX1 cultured in the presence of synthetic 5411 was fivefold lower than that of TX1 cultured in standard LB medium; in contrast, the AI-2 activity of TX1 cultured in the presence of 1026 (which, as described above, has no effect on AI-2 activity) was comparable to that of TX1 cultured in LB medium. Further analyses showed that the effect of synthetic 5411 on the AI-2 activity of TX1 was apparent only when the concentration of 5411 was above 1 nM and reached a maximum when the concentration of 5411 was 5 nM.
To further examine whether the reduced AI-2 activity observed with TX1 cultured in the presence of synthetic 5411 was due to inactivation of the AI-2 molecules (which are secreted into the culture supernatant) of TX1 directly by 5411, the supernatant of TX1 cultured in LB medium was incubated at 28°C for 5 h with synthetic 5411 and then assayed for AI-2 activity. The results showed that incubation with 5411 caused no apparent change in the AI-2 activity of the TX1 culture supernatant. Taken together, these results suggest that 5411 and 5906 affect AI-2 activity not directly by acting on AI-2 but indirectly by, probably, entering into TX1 and interacting with LuxS.
Construction of a recombinant commensal strain that produces 5411.
The above-described results, i.e., that TX1 could obtain 5411 from the culturing environment and the presence of 5411 in TX1 had a vitiating effect on bacterial infection, led us to speculate that the production of 5411 in fish from a stable source may attenuate the virulence potential of TX1. To examine this idea, the plasmid pT5411, which expresses 5411, and the control plasmid pJR20 were introduced, via conjugation, into FP3, a commensal strain of Pseudomonas sp. isolated from a healthy Japanese flounder. To examine whether FP3/pT5411 could secrete active 5411, the effect of the culture supernatant of FP3/pT5411 on the AI-2 activity of TX1 was determined. For this purpose, TX1 was grown in LB medium supplemented with the cell-free culturing fluids of either FP3/pT5411 or FP3/pJR20. Subsequent AI-2 assays showed that the AI-2 activity of TX1 cultured in the presence of the culture supernatant of FP3/pT5411 was 4.1-fold lower than that of TX1 cultured in the presence of the supernatant of FP3/pJR20, suggesting that TX1 probably recruited the 5411 produced by FP3/pT5411 and secreted into the culturing environment.
To examine the survival ability of FP3/pT5411 in vivo in the animal host, 5 × 107 CFU of the cells were injected i.p. into Japanese flounder, and the numbers of FP3/pT5411 cells that could be recovered from the peritoneal fluids, blood, liver, and spleen of the fish were determined at various times over a period of 96 h. The results showed that the numbers of FP3/pT5411 cells recovered from the peritoneal fluids at 12, 48, 72, and 96 h postinjection were, respectively, 20, 5, 1.6, and 0.1% of the input. No bacterial cells were recovered from the blood and tissues.
Effect of FP3/pT5411 on the infectivity of TX1.
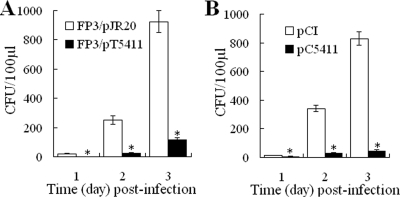
To examine whether FP3/pT5411 had any effect on the infectivity of TX1, Japanese flounder were administered the same dose of FP3/pT5411 or the control strain FP3/pJR20 and, 12 h later, challenged with TX1. The fish were sacrificed at 1, 2, and 3 days postchallenge, and the amounts of TX1 bacteria recoverable from the blood of the fish were determined. The results showed that, during the entire infection period examined, the amounts of TX1 bacteria recovered from the blood of the fish that had FP3/pT5411 administered were significantly lower than those recovered from the fish that had FP3/pJR20 administered (Fig. 5A).
FIG. 5.
Effect of 5411 on bacterial dissemination in the blood of fish infected with TX1. (A) Japanese flounder were injected i.p. with FP3/pJR20 or FP3/pT5411 and challenged with TX1 12 h later. The blood of the fish was taken at 1, 2, and 3 days postinfection and plated on selective LB plates. After incubation, the colonies that appeared on the plates were enumerated. (B) Japanese flounder were administered pCI or pC5411 via muscle injection and challenged with TX1 10 days later. Amounts of bacteria recovered from the blood at 1, 2, and 3 days postchallenge were determined as described above. Data are the means of the results of five independent assays and are presented as the means ± SE. *, P < 0.001.
Transient expression of 5411 in fish mitigates TX1 infection.
Given the results described above, we wondered whether 5411 expressed directly by fish tissues would have any effect on bacterial infection. To investigate this question, the mammalian expression plasmid pC5411 was constructed, which constitutively expresses 5411 under the human cytomegalovirus immediate-early promoter. Japanese flounder were administered pC5411 or the control plasmid pCI. The expression of 5411 in fish tissues was examined by qRT-PCR at 1, 5, and 10 days after plasmid administration. The results showed that 5411 expression was detected in the liver, kidney, and muscle at all time points examined and that, in all the tissues examined, the expression levels of 5411 increased with time, reaching the highest level at 10 days after plasmid administration. Based on this observation, the fish were challenged with TX1 10 days after the administration of pC5411 or pCI. The fish were sacrificed at 1, 2, and 3 days postchallenge and examined for bacterial recovery from the blood and liver. The results showed that the amounts of bacteria recovered from the blood of fish administered pC5411 at 1, 2, and 3 days postchallenge were, respectively, 2-, 12-, and 19-fold lower than the amounts recovered from the blood of fish administered pCI (Fig. 5B). Similar differences in bacterial recovery from the liver were also observed for fish administered pC5411 and pCI (data not shown).
DISCUSSION
As a metalloprotein, LuxS requires cognate metal ions for catalytic activity. Structural studies of the LuxS proteins of several bacterial species (Bacillus subtilis, Helicobacter pylori, and Deinococcus radiodurans) have indicated that the metal ion-binding sites of these enzymes are formed similarly by a cysteine residue, a water molecule, and residues corresponding to H54 and H58 of the B. subtilis LuxS (20, 29, 41). Consistent with their essential role in ion coordination, H54 and H58 are highly conserved among the LuxS proteins of diverse bacterial genera. In our study, we found that E. tarda LuxS possesses an active site that is identical to that of B. subtilis LuxS and that alanine replacement of H54 or H58 within this site completely inactivated the enzyme. These results favor the notion that H54 and H58 of E. tarda LuxS may play an essential role similar to that played by their counterparts in B. subtilis LuxS. Since H54 and H58 are known to interact with the coordinated metal ion via the Nɛ atom (20), alanine substitutions at these positions may inactivate the protein as a result of ineffective ionic interactions with the metal ion. It is interesting that LuxS mutants bearing C site and P site deletions, but not the mutant bearing the H54A substitution, could act as effective inhibitors of the wild-type LuxS. It is possible that the failure of the H54A mutant to attenuate the activity of the wild-type LuxS is due to the inability of the mutant protein to form dysfunctional heterodimers or oligomers with the wild type. It is also possible that the H54A mutant could form a heterodimer with the wild-type LuxS but that such heterodimers still possess most of the enzyme activity and therefore act indistinguishably from the wild-type LuxS. If this hypothesis is true, then the defect caused by H54A substitution in the mutant subunit can be rescued by dimerization with the wild-type subunit.
Since quorum sensing is a mechanism by which bacteria regulate the processes of intra-/interspecies communication, interaction with the environment, and, for some pathogens, response to host immune reactions (5, 6, 11, 26, 31, 50, 59), these systems represent the ideal targets for the development of therapeutic procedures aimed at the control of bacterial infections. A large number of studies have been carried out to investigate the effects of quorum sensing inhibitors and their potential applications in disease control (3, 25). For example, for the opportunistic human pathogen Pseudomonas aeruginosa, various quorum sensing inhibitors have been identified, some of which are found to be effective in mitigating P. aeruginosa infection in murine models (4, 18, 22, 24, 38, 53). For the gram-positive bacterial pathogens Staphylococcus aureus and Staphylococcus epidermidis, several quorum sensing antagonists have been reported which interfere with the expression of the virulence-associated agr regulon by blocking autoinducer AIP-mediated signal transduction (1, 10, 28, 35, 52). Also in the study of S. aureus, a small peptide has been identified that can interrupt the RAP/TRAP quorum sensing pathway by binding to RPA (54). In our study, we found that the LuxS-derived small peptides 5411 and 5906 can effectively mitigate the AI-2 activity of TX1. 5411 and 5906 appear to interact directly with LuxS, possibly via hydrophobic interactions since these peptides are constituted mainly of nonpolar amino acids. The observation that 5411 failed to bind the LuxS mutant bearing a P site deletion indicates, on one hand, that the interaction between 5411 and LuxS is indeed specific and, on the other hand, that the P site is either directly involved in the binding of 5411 or required for the formation of certain topological structures that are essential to the binding of 5411. Given the fact that the C site participates in iron binding and is vital to catalytic activity, it is possible that 5411 and 5906, which are derived from the C cite and its immediate surrounding region, may interfere with LuxS activity by hindering the process of iron coordination and/or substrate binding. Alternatively, 5411 and 5906 may block AI-2 activity via other mechanisms, such as by affecting the production/activity of factors that control LuxS activity or the AI-2 level. In the cases of flounder infection studies, it is also possible that the small peptides may have induced a certain response in fish that is disadvantageous to TX1 infection.
Of the quorum sensing inhibitors identified in gram-negative bacteria, most are natural or artificial biochemical elements that cannot be easily synthesized in vivo through genetic manipulations. In our study, the quorum sensing inhibitors, being of the nature of protein, can be expressed and produced in vivo in the animal host from biological sources. Three agents were used to produce the inhibitory peptide in our study: the pathogen itself, a commensal bacterial strain, and the animal host. We found that, as far as the impact on bacterial dissemination and survival is concerned, the small peptide produced in the pathogen (i.e., TX1/p5411) is more effective than those produced by the commensal strain (FP3/pT5411) and the fish (pC5411) This is probably due to the facts that (i) the peptides produced by FP3/pT5411 and pC5411 have to be secreted out of the producer strain/tissues and captured by TX1, during which process large amounts of the secreted peptides may diffuse away and thus be lost; (ii) there may exist certain competition between TX1 and the microfloral strains for capture of the secreted peptides; and (iii) the peptide-expressing plasmids (i.e., pT5411 and pC5411) in FP3 and the fish tissues may be lost with time and the production of the small peptide gradually dwindle to a low level as a result. In spite of these problems, our results indicate that LuxS can serve as a promising target against bacterial infections and that LuxS inhibitors in the form of secretable small peptides possess promising potential as antimicrobial agents. It is reasonable to speculate that in the future, the development of genetic systems (such as transgenic animals) that can stably and effectively express LuxS inhibitors may provide a new approach for the control of diseases caused by bacterial infection.
Acknowledgments
This work was supported by grant 40576071 from the National Natural Science Foundation of China (NSFC), grant 2006CB101807 from the National Basic Research Program of China, and grant 2008 AA092501 from the 863 High Technology Project.
Footnotes
Published ahead of print on 1 May 2009.
REFERENCES
- 1.Balaban, N., O. Cirioni, A. Giacometti, R. Ghiselli, J. B. Braunstein, C. Silvestri, F. Mocchegiani, V. Saba, and G. Scalise. 2007. Treatment of Staphylococcus aureus biofilm infection by the quorum-sensing inhibitor RIP. Antimicrob. Agents Chemother. 51:2226-2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bassler, B. L., M. Wright, R. E. Showalter, and M. R. Silverman. 1993. Intercellular signalling in Vibrio harveyi: sequence and function of genes regulating expression of luminescence. Mol. Microbiol. 9:773-786. [DOI] [PubMed] [Google Scholar]
- 3.Bjarnsholt, T., and M. Givskov. 2008. Quorum sensing inhibitory drugs as next generation antimicrobials: worth the effort? Curr. Infect. Dis. Rep. 10:22-28. [DOI] [PubMed] [Google Scholar]
- 4.Bjarnsholt, T., P. O. Jensen, T. B. Rasmussen, L. Christophersen, H. Calum, M. Hentzer, H. Hougen, J. Rygaard, C. Moser, L. Eberl, N. Hoiby, and M. Givskov. 2005. Garlic blocks quorum sensing and promotes rapid clearing of pulmonary Pseudomonas aeruginosa infections. Microbiology 151:3873-3880. [DOI] [PubMed] [Google Scholar]
- 5.Coulthurst, S. J., S. Clare, T. J. Evans, I. J. Foulds, K. J. Roberts, M. Welch, M. Dougan, and G. P. C. Salmond. 2007. Quorum sensing has an unexpected role in virulence in the model pathogen Citrobacter rodentium. EMBO J. 8:698-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coulthurst, S. J., C. L. Kurz, and G. P. C. Salmond. 2004. luxS mutants of Serratia defective in autoinducer-2-dependent “quorum sensing” show strain-dependent impacts on virulence and production of carbapenem and prodigiosin. Microbiology 150:1901-1910. [DOI] [PubMed] [Google Scholar]
- 7.Das, S. K., S. E. Sedelnikova, P. J. Baker, S. N. Ruzheinikov, S. Foster, A. Hartley, M. J. Horsburgh, and D. W. Rice. 2001. Cloning, purification, crystallization and preliminary crystallographic analysis of Bacillus subtilis LuxS. Acta Crystallogr. D 57:1324-1325. [DOI] [PubMed] [Google Scholar]
- 8.De Keersmaecker, S. C. J., K. Sonck, and J. Vanderleyden. 2006. Let LuxS speak up in AI-2 signaling. Trends Microbiol. 14:114-119. [DOI] [PubMed] [Google Scholar]
- 9.DeLisa, M. P., C. F. Wu, L. Wang, J. J. Valdes, and W. E. Bentley. 2001. DNA microarray-based identification of genes controlled by autoinducer 2-stimulated quorum sensing in Escherichia coli. J. Bacteriol. 183:5239-5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dell'Acqua, G., A. Giacometti, O. Cirioni, R. Ghiselli, V. Saba, G. Scalise, Y. Gov, and N. Balaban. 2004. Suppression of drug-resistant staphylococcal infections by the quorum-sensing inhibitor RNAIII-inhibiting peptide. J. Infect. Dis. 190:318-320. [DOI] [PubMed] [Google Scholar]
- 11.Duan, K. M., and M. G. Surette. 2007. Environmental regulation of Pseudomonas aeruginosa PAO1 Las and Rhl quorum-sensing systems. J. Bacteriol. 189:4827-4836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Engebrecht, J., K. Nealson, and M. Silverman. 1983. Bacterial bioluminescence: isolation and genetic analysis of functions from Vibrio fischeri. Cell 32:773-781. [DOI] [PubMed] [Google Scholar]
- 13.Fuqua, W. C., and S. C. Winans. 1994. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J. Bacteriol. 176:269-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gattiker, A., E. Gasteiger, and A. Bairoch. 2002. ScanProsite: a reference implementation of a PROSITE scanning tool. Appl. Bioinformatics 1:107-108. [PubMed] [Google Scholar]
- 15.Gonzalez, J. E., and N. D. Keshavan. 2006. Messing with bacterial quorum sensing. Microbiol. Mol. Biol. Rev. 70:859-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gonzalez Barrios, A. F., R. Zuo, Y. Hashimoto, L. Yang, W. E. Bentley, and T. K. Wood. 2006. Autoinducer 2 controls biofilm formation in Escherichia coli through a novel motility quorum-sensing regulator (MqsR, B3022). J. Bacteriol. 188:305-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henke, J. M., and B. L. Bassler. 2004. Bacterial social engagements. Trends Cell Biol. 14:648-656. [DOI] [PubMed] [Google Scholar]
- 18.Hentzer, M., H. Wu, J. B. Andersen, K. Riedel, T. B. Rasmussen, N. Bagge, N. Kumar, M. A. Schembri, Z. Song, P. Kristoffersen, M. Manefield, J. W. Costerton, S. Molin, L. Eberl, P. Steinberg, S. Kjelleberg, N. Høiby, and M. Givskov. 2003. Attenuation of Pseudomonas aeruginosa virulence by quorum sensing inhibitors. EMBO J. 22:3803-3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herzberg, M., I. K. Kaye, W. Peti, and T. K. Wood. 2006. YdgG (TqsA) controls biofilm formation in Escherichia coli K-12 through autoinducer 2 transport. J. Bacteriol. 188:587-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hilgers, M. T., and M. L. Ludwig. 2001. Crystal structure of the quorum sensing protein LuxS reveals a catalytic metal site. Proc. Natl. Acad. Sci. USA 98:11169-11174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 22.Hoffmann, N., B. Lee, M. Hentzer, T. B. Rasmussen, Z. Song, H. K. Johansen, M. Givskov, and N. Høiby. 2007. Azithromycin blocks quorum sensing and alginate polymer formation and increases the sensitivity to serum and stationary-growth-phase killing of Pseudomonas aeruginosa and attenuates chronic P. aeruginosa lung infection in Cftr−/− mice. Antimicrob. Agents Chemother. 51:3677-3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hou, J., W. Zhang, and L. Sun. 2009. Immunoprotective analysis of two Edwardsiella tarda antigens. J. Gen. Appl. Microbiol. 55:57-61. [DOI] [PubMed] [Google Scholar]
- 24.Ishida, T., T. Ikeda, N. Takiguchi, A. Kuroda, H. Ohtake, and J. Kato. 2007. Inhibition of quorum sensing in Pseudomonas aeruginosa by N-acyl cyclopentylamides. Appl. Environ. Microbiol. 73:3183-3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Janssens, J. C. A., S. C. J. De Keersmaecker, D. E. De Vos, and J. Vanderleyden. 2008. Small molecules for interference with cell-cell-communication systems in Gram-negative bacteria. Curr. Med. Chem. 15:2144-2156. [DOI] [PubMed] [Google Scholar]
- 26.Joyce, E. A., A. Kawale, S. Censini, C. C. Kim, A. Covacci, and S. Falkow. 2004. LuxS is required for persistent pneumococcal carriage and expression of virulence and biosynthesis genes. Infect. Immun. 72:2964-2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kendall, M. M., D. A. Rasko, and V. Sperandio. 2007. Global effects of the cell-to-cell signaling molecules autoinducer-2, autoinducer-3, and epinephrine in a luxS mutant of enterohemorrhagic Escherichia coli. Infect. Immun. 75:4875-4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kiran, M. D., A. Giacometti, O. Cirioni, and N. Balaban. 2008. Suppression of biofilm related, device-associated infections by staphylococcal quorum sensing inhibitors. Int. J. Artif. Organs 31:761-770. [DOI] [PubMed] [Google Scholar]
- 29.Lewis, H. A., E. Furlong, B. Laubert, G. Eroshkina, Y. Batiyenko, J. Adams, M. Bergseid, C. Marsh, T. Peat, and W. Sanderson. 2001. A structural genomics approach to the study of quorum sensing: crystal structures of three LuxS orthologs. Structure 9:527-537. [DOI] [PubMed] [Google Scholar]
- 30.Li, J., C. Attila, L. Wang, T. K. Wood, J. J. Valdes, and W. E. Bentley. 2007. Quorum sensing in Escherichia coli is signaled by AI-2/LsrR: effects on small RNA and biofilm architecture. J. Bacteriol. 189:6011-6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li, L., D. Hooi, S. R. Chhabra, D. Pritchard, and P. E. Shaw. 2004. Bacterial N-acylhomoserine lactone-induced apoptosis in breast carcinoma cells correlated with down-modulation of STAT3. Oncogene 23:4894-4902. [DOI] [PubMed] [Google Scholar]
- 32.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 33.Lyon, W. R., J. C. Madden, J. C. Levin, J. L. Stein, and M. G. Caparon. 2001. Mutation of luxS affects growth and virulence factor expression in Streptococcus pyogenes. Mol. Microbiol. 42:145-157. [DOI] [PubMed] [Google Scholar]
- 34.Marx, C. J., and M. E. Lidstrom. 2001. Development of improved versatile broad-host-range vectors for use in methylotrophs and other Gram-negative bacteria. Microbiology 147:2065-2075. [DOI] [PubMed] [Google Scholar]
- 35.Mayville, P., G. Ji, R. Beavis, H. Yang, M. Goger, R. P. Novick, and T. W. Muir. 1999. Structure-activity analysis of synthetic autoinducing thiolactone peptides from Staphylococcus aureus responsible for virulence. Proc. Natl. Acad. Sci. USA 96:1218-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McNab, R., S. K. Ford, A. El-Sabaeny, B. Barbieri, G. S. Cook, and R. J. Lamont. 2003. LuxS-based signaling in Streptococcus gordonii: autoinducer 2 controls carbohydrate metabolism and biofilm formation with Porphyromonas gingivalis. J. Bacteriol. 185:274-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rajan, R., J. Zhu, X. Hu, D. Pei, and C. E. Bell. 2005. Crystal structure of S-ribosylhomocysteinase (LuxS) in complex with a catalytic 2-ketone intermediate. Biochemistry 44:3745-3753. [DOI] [PubMed] [Google Scholar]
- 38.Rasmussen, T. B., T. Bjarnsholt, M. E. Skindersoe, M. Hentzer, P. Kristoffersen, M. Kote, J. Nielsen, L. Eberl, and M. Givskov. 2005. Screening for quorum-sensing inhibitors (QSI) by use of a novel genetic system, the QSI selector. J. Bacteriol. 187:1799-1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ratledge, C., and L. G. Dover. 2000. Iron metabolism in pathogenic bacteria. Annu. Rev. Microbiol. 54:881-941. [DOI] [PubMed] [Google Scholar]
- 40.Rickard, A. H., R. J. Palmer, D. S. Blehert, S. R. Campagna, M. F. Semmelhack, P. G. Egland, B. L. Bassler, and P. E. Kolenbrander. 2006. Autoinducer 2: a concentration-dependent signal for mutualistic bacterial biofilm growth. Mol. Microbiol. 60:1446-1456. [DOI] [PubMed] [Google Scholar]
- 41.Ruzheinikov, S. N., S. K. Das, S. E. Sedelnikova, A. Hartley, S. J. Foster, M. J. Horsburgh, A. G. Cox, C. W. McCleod, A. Mekhalfia, G. M. Blackburn, D. W. Rice, and P. J. Baker. 2001. The 1.2 Å structure of a novel quorum-sensing protein, Bacillus subtilis LuxS. J. Mol. Biol. 313:111-122. [DOI] [PubMed] [Google Scholar]
- 42.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 43.Schauder, S., K. Shokat, M. G. Surette, and B. L. Bassler. 2001. The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol. Microbiol. 41:463-476. [DOI] [PubMed] [Google Scholar]
- 44.Stroeher, U. H., A. W. Paton, A. D. Ogunniyi, and J. C. Paton. 2003. Mutation of luxS of Streptococcus pneumoniae affects virulence in a mouse model. Infect. Immun. 71:3206-3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun, J., R. Daniel, I. Wagner-Döbler, and A. Zeng. 2004. Is autoinducer-2 a universal signal for interspecies communication: a comparative genomic and phylogenetic analysis of the synthesis and signal transduction pathways. BMC Evol. Biol. 4:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun, K., S. Cheng, M. Zhang, F. Wang, and L. Sun. 2008. Cys-92, Cys-95, and the C-terminal 12 residues of the Vibrio harveyi ferric uptake regulator (Fur) are functionally inessential. J. Microbiol. 46:670-680. [DOI] [PubMed] [Google Scholar]
- 47.Surette, M. G., and B. L. Bassler. 1998. Quorum sensing in Escherichia coli and Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 95:7046-7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Surette, M. G., and B. L. Bassler. 1999. Regulation of autoinducer production in Salmonella typhimurium. Mol. Microbiol. 31:585-595. [DOI] [PubMed] [Google Scholar]
- 49.Wang, F., S. Cheng, K. Sun, and L. Sun. 2008. Molecular analysis of the fur (ferric uptake regulator) gene of a pathogenic Edwardsiella tarda strain. J. Microbiol. 46:350-355. [DOI] [PubMed] [Google Scholar]
- 50.Williams, S. C., E. K. Patterson, N. L. Carty, J. A. Griswold, A. N. Hamood, and K. P. Rumbaugh. 2004. Pseudomonas aeruginosa autoinducer enters and functions in mammalian cells. J. Bacteriol. 186:2281-2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Winzer, K., Y. H. Sun, A. Green, M. Delory, D. Blackley, K. R. Hardie, T. J. Baldwin, and C. M. Tang. 2002. Role of Neisseria meningitidis luxS in cell-to-cell signaling and bacteremic infection. Infect. Immun. 70:2245-2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wright, J. S., R. Jin, and R. P. Novick. 2005. Transient interference with staphylococcal quorum sensing blocks abscess formation. Proc. Natl. Acad. Sci. USA 102:1691-1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu, H., Z. Song, M. Hentzer, J. B. Andersen, S. Molin, M. Givskov, and N. Høiby. 2004. Synthetic furanones inhibit quorum-sensing and enhance bacterial clearance in Pseudomonas aeruginosa lung infection in mice. J. Antimicrob. Chemother. 53:1054-1061. [DOI] [PubMed] [Google Scholar]
- 54.Yang, G., H. Cheng, C. Liu, Y. Xue, Y. Gao, N. Liu, B. Gao, D. Wang, S. Li, B. Shen, and N. Shao. 2003. Inhibition of Staphylococcus aureus pathogenesis in vitro and in vivo by RAP-binding peptides. Peptides 24:1823-1828. [DOI] [PubMed] [Google Scholar]
- 55.Zhang, M., K. Sun, and L. Sun. 2008. Regulation of autoinducer 2 production and luxS expression in a pathogenic Edwardsiella tarda strain. Microbiology 154:2060-2069. [DOI] [PubMed] [Google Scholar]
- 56.Zhang, W., K. Sun, S. Cheng, and L. Sun. 2008. Characterization of DegQVh, a serine protease and a protective immunogen from a pathogenic Vibrio harveyi strain. Appl. Environ. Microbiol. 74:6254-6262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang, W.-W., and L. Sun. 2007. Cloning, characterization, and molecular application of a beta-agarase gene from Vibrio sp. strain V134. Appl. Environ. Microbiol. 73:2825-2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhu, J., E. Dizin, X. Hu, A. S. Wavreille, J. Park, and D. Pei. 2003. S-Ribosylhomocysteinase (LuxS) is a mononuclear iron protein. Biochemistry 42:4717-4726. [DOI] [PubMed] [Google Scholar]
- 59.Zhu, J., M. B. Miller, R. E. Vance, M. Dziejman, B. L. Bassler, and J. J. Mekalanos. 2002. Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 99:3129-3134. [DOI] [PMC free article] [PubMed] [Google Scholar]