Abstract
Epidemics of Vibrio parahaemolyticus in Chile have occurred since 1998. Direct genome restriction enzyme analysis (DGREA) using conventional gel electrophoresis permitted discrimination of different V. parahaemolyticus isolates obtained from these outbreaks and showed that this species consists of a highly diverse population. A multiple-locus variable-number tandem-repeat (VNTR) analysis (MLVA) approach was developed and applied to 22 clinical and 91 environmental V. parahaemolyticus isolates from Chile to understand their clonal structures. To this end, an advanced molecular technique was developed by applying multiplex PCR, fluorescent primers, and capillary electrophoresis, resulting in a high-resolution and high-throughput (HRHT) genotyping method. The genomic basis of this HRHT method was eight VNTR loci described previously by Kimura et al. (J. Microbiol. Methods 72:313-320, 2008) and two new loci which were identified by a detailed molecular study of 24 potential VNTR loci on both chromosomes. The isolates of V. parahaemolyticus belonging to the same DGREA pattern were distinguishable by the size variations in the indicative 10 VNTRs. This assay showed that these 10 VNTR loci were useful for distinguishing isolates of V. parahaemolyticus that had different DGREA patterns and also isolates that belong to the same group. Isolates that differed in their DGREA patterns showed polymorphism in their VNTR profiles. A total of 81 isolates was associated with 59 MLVA groups, providing fine-scale differentiation, even among very closely related isolates. The developed approach enables rapid and high-resolution analysis of V. parahaemolyticus with pandemic potential and provides a new surveillance tool for food-borne pathogens.
Food-borne infections by Vibrio parahaemolyticus cause gastroenteritis, which is the most common clinical manifestation (38). An increasing number of V. parahaemolyticus infections and outbreaks caused by strains belonging to a pandemic clonal complex have been observed throughout the world since 1996 (2, 6, 9, 12, 13, 31, 32, 36, 40). Epidemics of Vibrio parahaemolyticus in Chile have occurred since the summer of 1998 and were caused by the pandemic clone O3:K6 that had emerged in Southeast Asia in 1996 (12, 13, 15). However, this strain was only a minor component of a highly diverse V. parahaemolyticus population in shellfish, as demonstrated by an improved method for restriction enzyme analysis, using total bacterial DNA, named direct genome restriction enzyme analysis (DGREA), in combination with conventional gel electrophoresis (12). This method has a discrimination index similar to that of restriction fragment length polymorphism-pulsed-field gel electrophoresis (PFGE) (12, 13, 19).
A variety of molecular typing methods have been applied to V. parahaemolyticus, such as ribotyping (3, 10, 14), PFGE (3, 30), group-specific PCR (32), arbitrarily primed PCR (18, 32, 36), and multilocus sequence typing (7, 16). The use of DGREA permitted discrimination of different V. parahaemolyticus Chilean isolates and showed that these bacteria consist of a highly diverse population comprising at least 23 different genotypic groups among the environmental isolates obtained from shellfish and 5 different groups of clinical isolates (19).
Epidemiological analyses of infections caused by pathogenic bacteria depend on the accurate identification of strains, preferably at the clonal level. Variable-number tandem repeats (VNTRs) comprising short sequence repeats constitute a rich source of genetic polymorphism and have been used extensively as markers for discrimination between strains of many different bacterial genera (27, 46). VNTRs have been used to discriminate among individual strains within several food- or waterborne pathogens with little genetic variation, including Escherichia coli O157:H7 (25, 35), Pseudomonas aeruginosa (37), Staphylococcus aureus (41), and Salmonella enterica subsp. enterica serovar Typhimurium (26), and to characterize other important human pathogens, such as Neisseria meningitidis (42), Listeria monocytogenes (28), Legionella pneumophila (34, 39), Leptospira interrogans (43), and Mycobacterium tuberculosis (45). VNTR loci have even been found in genetically highly homogenous pathogens, such as Bacillus anthracis (1, 21, 29). Multiple-locus VNTR analysis (MLVA) is defined as the analysis of a set of loci spread throughout the bacterial genome (23). Individual strains within a bacterial species often maintain the same sequence elements but with different copy numbers due to variations introduced by slipped-strand mispairing during DNA replication (33).
Recently, a study of the polymorphism of tandem repeats in V. parahaemolyticus showed the utility of the MLVA approach for characterizing recently emerged and highly homogeneous pandemic strains of serotype O3:K6 (22). These authors reported a scheme of eight genomic VNTR loci, comparing PFGE results for clinical strains of V. parahaemolyticus serotype O3:K6. The study by Kimura et al. (22) comprised only strains of serogroup O3:K6 and used conventional gel electrophoresis to evaluate VNTRs. In epidemiological studies, a more rapid technique is needed for mass application of MLVA that also provides improved resolution and has been validated for nonserogroup O3:K6 isolates. Capillary electrophoresis has become the preferred technology to improve resolution and accuracy in bacterial VNTR analysis due to the availability of multiple fluorescent labels and better accuracy and reproducibility (27).
In our study we describe the use of an improved MLVA for discriminating genotypically a diverse collection of clinical and environmental V. parahaemolyticus isolates from Chile. These very closely related isolates have been analyzed and grouped by DGREA previously (12). To this end, we developed and applied multiplex PCR of 10 VNTR loci, tagged with multiple fluorescent dyes, and analyzed the amplicons by capillary electrophoresis. The results demonstrated that MLVA typing is able to distinguish between V. parahaemolyticus isolates that have different DGREA patterns and isolates that belong to the same group, allowing accurate sizing of amplicons by assignment of the fragment size. Validation of this typing method with 113 Chilean isolates demonstrated the utility of this technique also for nonserogroup O3:K6 clinical isolates, thereby providing a new tool for the study of the molecular epidemiology of V. parahaemolyticus.
MATERIALS AND METHODS
Bacterial strains.
A total of 113 Vibrio parahaemolyticus strains, originally isolated from a variety of sources (clinical and shellfish samples) from 2004 to 2007 during Chilean outbreaks, were used in this study (see Table S1 in the supplemental material). For comparative reasons, the reference strains RIMD2210633 (VpKX) and ATCC 17802 (VpD) from the Research Institute for Microbial Diseases and the American Type Culture Collection were included. The genotypic characterization of these isolates using DGREA has been reported previously, including the determination of principal virulence markers (12).
Growth and DNA extraction.
Strains were cultured overnight at 37°C on Luria Bertani broth containing 3% NaCl. DNA was extracted with the Wizard genomic kit (Promega). DNA was quantified by UV absorption using a NanoDrop ND-1000 spectrophotometer (PeqLab Biotechnologie GmbH, Germany). Diluted samples of 1 ng/μl in water were used as DNA template for PCR amplification.
Design of VNTR primers.
The genome sequences of the two chromosomes of V. parahaemolyticus RIMD2210633, deposited in GenBank under accession numbers NC004603 and NC004605, were used to detect the VNTR loci. Analysis using the Tandem Repeats Finder program (http://tandem.bu.edu/trf/trf.html [23]) and the microorganism tandem repeat database (http://minisatellites.u-psud.fr) were used to identify potential VNTR loci. Primer3 (http://biotools.umassmed.edu/bioapps/primer3_www.cgi) was used to design PCR primers for amplifying the loci within the flanking regions. Additionally, dedicated programs (Python language) were also used to help analyze previous results in order to select proper primers. Some VNTR loci used in this analysis have been described previously by Kimura et al. (22). However, the primer for the VPTR7 locus was redesigned and called VPTR7-1 to obtain a longer PCR product (243 instead of 221 bp). Four more primer pairs were designed in this study for loci: VP1-11, VP1-17, VP1-10, and VP2-07.
VNTR PCR amplification.
A total of 12 VNTR loci selected for MLVA were amplified in three multiplex PCRs named MultiA (VPTR7, VPTR5, VP1-11, and VPTR2), MultiB (VP1-17, VPTR1, VPTR8, and VP1-10), and MultiC (VP2-07, VPTR4, VPTR3, and VPTR6). Forward primers were labeled with VIC, PET, NED, and 6-carboxyfluorescein (6-FAM) (Applied Biosystems) at the 5′ end. Reverse primers were synthesized unlabeled (Operon, Germany) (Table 1). The PCRs were performed using the Qiagen multiplex PCR kit (Qiagen, Germany). DNA (2.5 ng) was amplified in a 25-μl reaction mix containing 12.5 μl of 2× multiplex PCR master mix, 2.5 μl of 10× primer mix, and RNase-free water. Primer mix (10×) contained a final concentration of 0.2 μM of each primer, except for VP2-07 in MultiC, for which the final concentration was 0.4 μM. The PCRs for MultiA and MultiB were performed with predenaturing at 95°C for 15 min, 20 cycles of touchdown PCR (30 s at 94°C; 90 s at 63°C, with a 0.2°C drop in temperature during each cycle; and 60 s at 72°C), and 10 cycles of regular PCR (30 s at 94°C, 90 s at 59°C, and 60 s at 72°C), with a final step for 30 min at 60°C (Mastercycler gradient; Eppendorf, Germany). For MultiC, the following program was used: 15 min at 95°C and 30 cycles of regular PCR (30 s at 94°C, 90 s at 61°C, and 60 s at 72°C), with a final 30 min at 60°C (Thermocycler 96 iCycler; Bio-Rad). We amplified the DNA of VpKX as a test control in various test PCRs.
TABLE 1.
Specific primers used for amplifying the VNTR loci in V. parahaemolyticus isolates
| Locus | Primer | Fluorescence tag-sequencec | DNA sequence length (bp)a
|
Reference | |
|---|---|---|---|---|---|
| Theoretical | CE | ||||
| VP2-03 | VP2-03 F | VIC-CATAAACGAGCGACACGAGA | 168 | 164 | This study |
| VP2-03 R | GCGCAAAAATTCATTGTGATT | ||||
| VP1-17 | VP1-17 F | VIC-TCAACACGAGCTTGATCACC | 206 | 201 | This study |
| VP1-17 R | GAAATCCGGAGTACCTGCAA | ||||
| VPTR7 | VPTR7-1F | VIC-TATCTACAAAGGTGGCGGAGAT | 200 | 239 | This studyb |
| VPTR7-1R | AAGGTGTTACTTGTTCCAGACG | ||||
| VPTR5 | VPTR5 F | NED-GCTGGATTGCTGCGAGTAAGA | 204 | 195 | Kimura et al. (22) |
| VPTR5 R | AACTCAAGGGCTGCTTCGG | ||||
| VP1-11 | VP1-11 F | PET-CTGCCTGGAGAATTGGCTTA | 854 | 835 | This study |
| VP1-11 R | TGAGCCTGAAGCTGAAAACA | ||||
| VPTR1 | VPTR1 F | NED-TAACAACGCAAGCTTGCAACG | 253 | 253 | Kimura et al. (22) |
| VPTR1 R | TCATTCTCGCCACATAACTCAGC | ||||
| VPTR8 | VPTR8 F | PET-ACATCGGCAATGAGCAGTTG | 301 | 307 | Kimura et al. (22) |
| VPTR8 R | AAGAGGTTGCTGAGCAAGCG | ||||
| VP1-10 | VP1-10 F | 6-FAM-CGTCTTGCTCGTGAACGTAA | 955 | 964 | This study |
| VP1-10 R | TCATTAAGTCAGGCGTGCTG | ||||
| VP2-07 | VP2-07 F | VIC-TGATTTTGAAGCAGCGAAGA | 296 | 317 | This study |
| VP2-07 R | TTTGTGACTGCTGTCCTTGC | ||||
| VPTR4 | VPTR4 F | NED-AAACGTCTCGACATCTGGATCA | 227 | 227 | Kimura et al. (22) |
| VPTR4 R | TGTTTGGCTATGTAACCGCTCA | ||||
| VPTR3 | VPTR3 F | PET-CGCCAGTAATTCGACTCATGC | 331 | 332 | Kimura et al. (22) |
| VPTR3 R | AAGACTGTTCCCGTCGCTGA | ||||
| VPTR6 | VPTR6 F | 6-FAM-TGTCGATGGTGTTCTGTTCCA | 316 | 309 | Kimura et al. (22) |
| VPTR6 R | CTTGACTTGCTCGCTCAGGAG | ||||
Sizes according to the genome sequence strain RIMD2210633. CE, capillary electrophoresis.
Primer modified from the work of Kimura et al. (22).
Fluorescence labels appear in bold. VIC, green; NED, yellow; PET, red; 6-FAM, blue.
DNA sequencing of PCR products.
The PCR products from four strains, including the reference strain RIMD2210633, were sequenced using the same primers used to amplify the VNTRs to verify the genetic basis of the results from MLVA. Sequencing reactions were performed using the BigDye Terminator technology according to the manufacturer's protocol (Applied Biosystems), and products were analyzed in an ABI 3130xl capillary electrophoresis system (Applied Biosystems). Data obtained with forward and reverse sequencing primers were combined, and the alignments were obtained using BioEdit (17).
Capillary electrophoresis of fluorescently labeled PCR products.
The PCR products were purified using MinElute PCR purification kit (Qiagen), and DNA was eluted in buffer EB (10 mM Tris-HCl, pH 8.5; Qiagen). Prior to capillary electrophoresis, PCR products from the three multiplex reactions were diluted in distilled water 1:100 for MultiA and MultiB and 1:200 for MultiC. Diluted PCR products (1 μl) were mixed with 8.85 μl HiDi formamide (Applied Biosystems, Foster City, CA) and 0.15 μl GeneScan LIZ1200 size standard (Applied Biosystems), containing 68 single-stranded labeled fragments in the range of 20 to 1,200 bp. Samples were pipetted into a 96-well microtiter plate, denatured for 3 min at 95°C, and cooled on ice for at least 1 min, and the microtiter plate was centrifuged briefly at 500 rpm in a Multifuge 1 centrifuge (Heraeus, Germany). Electrophoresis was performed on a 3130xl sequencer (Applied Biosystems) equipped with 50-cm capillaries, using POP-7 polymer, with the recommended running parameters for the GeneScan LIZ1200 size standard, for 2.5 h, with a running voltage of 8.5 kV and an injection voltage of 15 kV. Each VNTR locus was identified by color and assigned a size by the GeneMapper software version 3.7 (Applied Biosystems) using settings for microsatellite analysis.
Data analysis and statistics.
The number of repeats in the alleles was estimated by subtracting the invariable flanking region from the amplicon size divided by the repeat unit length, as determined for reference strain RIMD2210633. Allele sizes obtained by GeneMapper were converted into repeat numbers using Microsoft Excel and the following equation: number of repeats (bp) = fragment size (bp) − flanking regions (bp)/repeat size (bp). Flanking regions were deduced from the sequenced PCR products of several strains. The allelic profiles for 12 loci were defined as the number of repeats at each VNTR locus, in the following order: VPTR7, VPTR5, VP1-11, VPTR2, VP1-17, VPTR1, VPTR8, VP1-10, VP2-07, VPTR4, VPTR3, and VPTR6. The designation “not amplified” was given when no PCR amplification was observed at a given locus. The null allele (represented by “0”) designates a locus that contains both flanking sequences but no repeat unit. The similarity of allele numbers across all strains was calculated by the Euclidean distance algorithm on nonnormalized data, and using hierarchical agglomerative clustering, a dendrogram was constructed by the complete linkage method (PRIMER6) (8). The Hunter-Gaston diversity index was calculated as previously described (20), using the VNTR diversity and confidence extractor software (V-DICE) available at the Health Protection Agency bioinformatics tools website (http://www.hpa-bioinfotools.org.uk/cgi-bin/DICI/DICI.pl).
RESULTS
Identification of VNTR loci in V. parahaemolyticus.
Twenty-four likely VNTR loci were identified in the available genome sequence of V. parahaemolyticus RIMD2210633 using Tandem Repeat Finder (4) and the microorganism tandem repeat database (23). These loci were tested for typing on a set of 18 genetically diverse V. parahaemolyticus strains (Table 2). The strains had been characterized previously by DGREA (12) and belonged to different genotype groups. Present approaches rely upon systematic testing, especially for species in which only one genome has been sequenced (24), as is the case for V. parahaemolyticus. Some repeat regions varied in composition between the strains, and others could not be amplified from a number of strains tested (Table 2). Other loci were not tested because the sizes of the products with the predicted primers were too close in size to those of other VNTR products (Table 3).
TABLE 2.
VNTR loci found with Tandem Repeat Finder and microorganism tandem repeat database
| Locus | Physical position (kb) | Chromosome | Unit length (bp) | Copy no. | Total length (bp) | % Matches | % C | % G | % T | % A | GC bias | Commentsa |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| VP1-01 | 384 | 1 | 101 | 4.3 | 433 | 81 | 15 | 26 | 30 | 26 | 0.26 | Not amplified in all strains |
| VP1-02 | 447 | 1 | 6 | 9.5 | 55 | 94 | 16 | 33 | 50 | 0 | 0.34 | Not tested |
| VP1-03 | 452 | 1 | 24 | 3 | 72 | 91 | 22 | 33 | 26 | 18 | 0.19 | Not tested |
| VP1-04 | 772 | 1 | 480 | 3.2 | 1540 | 83 | 24 | 22 | 24 | 27 | 0.04 | Not tested* |
| VP1-05 | 978 | 1 | 166 | 5.8 | 967 | 98 | 23 | 25 | 25 | 25 | 0.04 | Not tested* |
| VP1-06 | 1478 | 1 | 576 | 6.5 | 3743 | 90 | 25 | 28 | 17 | 27 | 0.05 | Not tested* |
| VP1-07 | 1648 | 1 | 33 | 2 | 64 | 81 | 28 | 25 | 14 | 31 | 0.03 | Not tested |
| VP1-08 | 1662 | 1 | 21 | 2.7 | 55 | 80 | 35 | 17 | 8 | 37 | 0.34 | Not amplified in all strains |
| VP1-09 | 1730 | 1 | 366 | 12 | 4395 | 90 | 26 | 25 | 25 | 22 | 0.01 | Not tested* |
| VP1-10 | 2213 | 1 | 114 | 6.8 | 772 | 95 | 26 | 24 | 15 | 33 | 0.03 | Variable in size |
| VP1-11 | 2301 | 1 | 129 | 5.8 | 751 | 85 | 28 | 23 | 30 | 17 | 0.09 | Variable in size |
| VP1-12 | 2339 | 1 | 6 | 21.5 | 131 | 89 | 31 | 31 | 34 | 3 | 0 | Variable in size |
| VP1-13 | 2669 | 1 | 7 | 11.1 | 77 | 100 | 14 | 28 | 42 | 15 | 0.33 | Not amplified in all strains |
| VP1-14 | 2989 | 1 | 238 | 2.1 | 506 | 95 | 25 | 24 | 24 | 26 | 0.02 | Not amplified in all strains |
| VP1-15 | 3065 | 1 | 90 | 1.9 | 173 | 80 | 30 | 28 | 19 | 22 | 0.03 | Not tested |
| VP1-16 | 3077 | 1 | 6 | 28.2 | 168 | 100 | 0 | 33 | 16 | 50 | 0.99 | Variable in size |
| VP1-17 | 3249 | 1 | 30 | 1.9 | 57 | 82 | 22 | 17 | 12 | 47 | 0.12 | Invariable in size |
| VP2-01 | 477 | 2 | 561 | 2 | 1125 | 99 | 19 | 22 | 34 | 23 | 0.07 | Not tested* |
| VP2-02 | 990 | 2 | 306 | 14.1 | 4325 | 96 | 29 | 23 | 26 | 19 | 0.11 | Not tested* |
| VP2-03 | 1292 | 2 | 26 | 2.2 | 56 | 80 | 12 | 14 | 39 | 33 | 0.07 | Invariable in size |
| VP2-04 | 1341 | 2 | 8 | 22.8 | 181 | 100 | 25 | 12 | 12 | 49 | 0.35 | Not amplified in all strains |
| VP2-05 | 1417 | 2 | 27 | 2 | 55 | 85 | 25 | 5 | 34 | 34 | 0.06 | Not amplified in all strains |
| VP2-06 | 1428 | 2 | 960 | 2 | 1901 | 96 | 32 | 26 | 26 | 14 | 0.1 | Not tested* |
| VP2-07 | 1548 | 2 | 6 | 35.2 | 207 | 94 | 32 | 17 | 0 | 50 | 0.3 | Variable in size |
Results of the 18 V. parahaemolyticus strains tested are summarized under Comments. *, not tested further because the target for the PCR was too long and no acceptable primers were found.
TABLE 3.
Molecular characteristics of the tandem repeat loci used in this study
| Locus | Repeat motif | No. of repeatsa | Function | Period size (nt) | Locus position on chromosome | Chromosome |
|---|---|---|---|---|---|---|
| VPTR7 | CTGCTC | 6 | Hypothetical protein | 6 | 2240338-2240373 | 1 |
| VPTR5 | CTCAAA | 7 | Noncoding region | 6 | 3216099-3216282 | 1 |
| VP1-11 | TCGGCCATAGATGCCAATGCATCTTCTTCGTCGAACTCTGGCAGTTCCAGTTCGTCGAAGTTGAA | 5.8 | Hypothetical protein | 129 | 2301238-2301989 | 1 |
| TTCTTCTTCCGCGTCAGCGGTTGGCGCTG CCGCTTCACGTTCAGCTTCTGGTAGCTCAGGCTCA | ||||||
| VPTR1 | ATAGAG | 28.2 | Hypothetical protein | 6 | 3078097-3077865 | 1 |
| VPTR8 | CTTCTG | 7.3 | Cell division protein | 6 | 3160308-3160595 | 1 |
| VP1-10 | GAAGAAACAGCTTCTCAAGAGCCAGCCGAAGATCCGAAGAAAGCCGCAGTCGCTGC | 6.8 | RbfC-related protein | 114 | 2213604-2214373 | 1 |
| TGCTATTGCTCGTGCAAAAGCGCGTAAAGCGCAACAAGAGACTGAATCTCAACCTGTT | ||||||
| VP2-07 | AGCAAC | 35.2 | Hypothetical protein | 6 | 1548365-1548572 | 2 |
| VPTR4 | TGTGTC | 8.8 | Putative hemolysin | 6 | 447433-447638 | 1 |
| VPTR3 | ATCTGT | 7.2 | Putative collagenase | 6 | 743929-744240 | 2 |
| VPTR6 | GCTCTG | 17.8 | Conserved hypothetical protein | 6 | 2339401-2339507 | 1 |
As found in reference strain RIMD2210633.
MLVA of V. parahaemolyticus using capillary electrophoresis.
A total of 113 isolates comprising the genome-sequenced strain (VpKX, RIMD 2210633), the type strain (VpD, ATCC 171802), and clinical and environmental V. parahaemolyticus isolates obtained from Chilean outbreaks were analyzed to generate MLVA typing data. Three multiplex PCRs, comprising 12 loci (Table 1), were performed and subjected to capillary electrophoresis. The obtained electropherograms permitted us to see defined peaks, in which each locus was identified according to its size range and color (Fig. 1). The PCR primers described by Kimura et al. (22) were used to amplify V. parahaemolyticus-specific VNTR loci, named VPTR7, VPTR5, VPTR1, VPTR8, VPTR4, VPTR3, and VPTR6. We designed the following primers to amplify the new loci: VP2-03, VP1-11, VP1-17, VP1-10, and VP2-07 (VP stands for V. parahaemolyticus, and 1 and 2 stand for the chromosome number). For the VPTR7 locus, we redesigned the primers used by Kimura et al. (22) to obtain a 243-bp PCR product instead of a 221-bp product to improve separation by capillary electrophoresis.
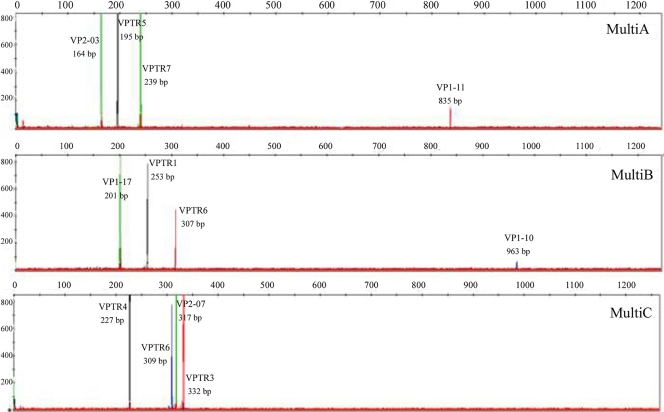
FIG. 1.
Representative electropherograms of the three developed multiplex PCR assays, as separated by capillary electrophoresis and identified according to their sizes and colors. The electropherograms correspond to the PCR products from 12 VNTRs in strain VpKX. (Top electropherogram) MultiA: VP2-03 (VIC [green]), VPTR5 (NED [black]), VPTR7-1 (VIC), VP1-11 (PET [red]). (Middle electropherogram) MultiB: VP1-17 (VIC), VPTR1 (NED), VPTR8 (PET), VP1-10 (6-FAM). (Bottom electropherogram) MultiC: VPTR4 (NED), VPTR6 (6-FAM), VP2-07 (VIC), VPTR3 (PET).
Figure 1 shows three representative electropherograms of strain VpKX, RIMD2210633, for each multiplex PCR. In each case, we observed four differently colored peaks, each corresponding to a PCR product specific for one locus. The internal size standard displayed uniform band spacing, with sharp and defined peaks from 20 to 1,200 bp, thus allowing a precise determination of the PCR product sizes over the range needed, if the samples were run for 2.5 h. Using normal PCR conditions, MultiA and MultiB showed a series of peaks that were unexpected. We improved amplification of these two multiplexes using a touchdown PCR. The assays and the resulting tandem repeat profiles were highly reproducible, as determined by typing 10 representative strains three times, with the replicates showing identical results (data not shown).
Description of MLVA profiles.
To perform an MLVA, we included in our assay the VNTR loci used by Kimura et al. (22), VPTR1 (the same as VP1-16), VPTR3, VPTR4, VPTR5, VPTR6 (the same as VP1-12), VPTR7, and VPTR8, and the loci that showed the expected polymorphism, VP1-10, VP1-11, and VP2-07 (the same as VPTR2). The molecular characteristics of these loci are summarized in Table 3. The minimum and maximum sizes for all loci were based on the PCR products obtained in some strains, and we used these data for setting the marker sizes in GeneMapper version 3.7. The total flanking regions were calculated according to the obtained sequences. These numbers were used for all strains analyzed to calculate the number of repeats for each locus based on the fragment size (see Table S2 in the supplemental material). This data set was used for dendrogram construction and strain comparison. The same number of repeats in each locus indicates the allele string for each strain in the following order: VPTR7, VPTR5, VP1-11, VPTR1, VPTR8, VP1-10, VP2-07, VPTR4, VPTR3, and VPTR6. For example, for the strain PMA37.5 the allele string is 6-7-5-39-7-7-41-8-7-14, which means that PMA37.5 has 6 repeats in locus VPTR7, 7 in VPTR5, 5 in VP1-11, 39 in VPTR1, 7 in VPTR8, 7 in VP1-10, 41 in VP2-07, 8 in VPTR4, 7 in VPTR3, and 14 in VPTR6. Isolates of the most frequent MLVA type in DGREA group KX were represented by allele numbers 6-7-5-36-7-7-42-8-7-14 (in 10 isolates, PMC68, PMA115.5, PMA15.6, PMA21.6, PMA23.6, PMA24.6, PMA25.6, PMA29.6, and PMA30.6, and PMC18.5) and were often isolated in Quillaipe, Chile, corresponding to different outbreaks during the years 2004 to 2006. In DGREA group 118 the MLVA type 6-3-7-9-8-5-17-5-11-10 (in 4 isolates, PMA33.6, PMA38.7, PMA29.5, and PMA2.5) dominated and also corresponded to isolates from different outbreaks. This indicates the spread of certain clones within the Puerto Montt region over time.
Variability of VNTRs in V. parahaemolyticus isolates and allelic diversity.
To find loci with multiple alleles and variability, we amplified the isolates with 12 sets of VNTR primers (Table 1). Two loci, VP2-03 and VP1-17, did not show polymorphism in the product sizes and were therefore excluded from the assay. No amplification, or null variants, was observed for the locus VP2-07 (Table 1) in all isolates of DGREA group 187 (100%) and for locus VPTR7 in isolates of DGREA group 1.5 (82%). This lack of amplification could be due to some differences in the sequences of the flanking regions. One allele per locus was detected in 72% of the analyzed isolates, and in the remaining 32 isolates (28%), more than one allele was found in some loci, including slightly different-sized alleles, illustrating the greater sensitivity of capillary over conventional gel electrophoresis (see Table S2 in the supplemental material). Coincidentally, the VNTRs (VPTR1, VP2-07, and VPTR6) that had more repeat units in the genome-sequenced strain VpKX (Table 3) showed more allelic diversity in several strains, i.e., higher intragenomic diversity.
Analysis of the genetic diversity of the VNTR loci for V. parahaemolyticus strains varied among the various loci (Table 4). The locus VP2-07 showed the highest and most variable number of alleles between all loci, and also, VPTR1 was polymorphic, as was described previously (21). Among all isolates, VP2-07 had 38 alleles, VPTR1 had 29, VPTR6 had 14, VPTR3 had 16, VPTR8 had 12, VPTR4 had 11, VPTR5 had 10, VPTR6 had 8, VP1-10 had 7, VP1-11 had 5, and VPTR7 had 4. The diversity index for VNTR data measures the variation of the number of each locus and ranges from 0.0 (no diversity) to 1.0 (complete diversity). Hunter and Gaston's index of diversity (V-DICE) was calculated for the combined typing set and for each loci and showed the highest index (D = 0.949) for VP2-07.
TABLE 4.
Allelic diversity of 10 VNTR loci in V. parahaemolyticus isolates
| Locus | HGIa | Confidence intervalb | No. of alleles | Max (PI)c |
|---|---|---|---|---|
| VP2-07 | 0.949 | 0.943-0.955 | 38 | 0.152 |
| VPTR1 | 0.880 | 0.869-0.891 | 29 | 0.219 |
| VPTR8 | 0.834 | 0.821-0.847 | 12 | 0.281 |
| VPTR5 | 0.781 | 0.766-0.797 | 10 | 0.337 |
| VPTR4 | 0.771 | 0.755-0.787 | 11 | 0.320 |
| VPTR3 | 0.749 | 0.722-0.777 | 16 | 0.466 |
| VPTR6 | 0.741 | 0.725-0.758 | 8 | 0.354 |
| VP1-11 | 0.682 | 0.661-0.704 | 5 | 0.472 |
| VP1-10 | 0.646 | 0.628-0.663 | 7 | 0.433 |
| VPTR7 | 0.534 | 0.522-0.546 | 4 | 0.511 |
HGI, Hunter and Gaston index.
Precision of the diversity index expressed as 95% upper and lower boundaries.
PI, performance index. Fraction of samples that have the most frequent repeat number in this locus (range 0.0 to 1.0).
Comparison of DGREA and MLVA of V. parahaemolyticus.
On the basis of DGREA, the 113 strains were subdivided into 25 distinct genotypes. Clonal isolates that belong to the same group (by DGREA patterns) are distinguishable by differing VNTR patterns. Also, isolates that differed in their DGREA patterns showed polymorphism in the VNTR profiles. Altogether, 59 different allele combinations (MLVA types) have been found among 81 isolates with unique alleles in each locus that belong to 20 different DGREA groups (see Table S2 in the supplemental material). Thirty isolates that belonged to DGREA group KX were distinguished in 19 different MLVA types, 17 isolates of DGREA group 118 in 9 MLVA types, 2 isolates of DGREA group 128 in 2 MLVA types, 4 isolates of DGREA group 187 in 4 MLVA types, 11 isolates of DGREA group 1.5 in 9 MLVA types, 7 isolates of DGREA group 34.6 in 5 MLVA types, and 11 isolates of different DGREA groups belonged to different MLVA types, except for PMA109.5, which corresponded to an MLVA type found in the KX isolates.
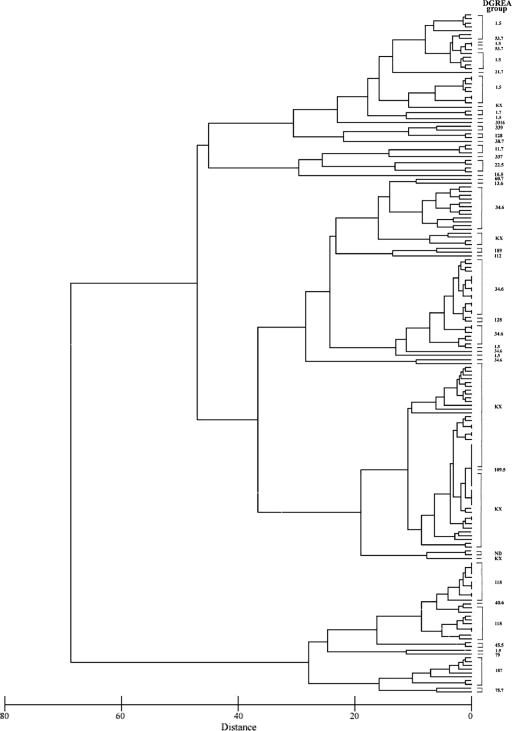
The MLVA types of V. parahaemolyticus were grouped into two main clusters by similarity matrices based on Euclidean distance and a complete linkage clustering (Fig. 2). Generally, isolates in the same DGREA group clustered together, with the exception of PMA109.5, which clustered with the KX group, despite belonging to a different DGREA group. Isolates of the groups KX and 34.6 clustered in one genogroup, and isolates of groups 1.5, 118, 128, and 187 were grouped in a second genogroup (Fig. 2). However, we saw differences between isolates of the same DGREA group. PMC29.7 was the only one that belongs to the KX group but was not clustered with the other isolates of this group. PMA88.5, which belongs to the 1.5 DGREA group, clustered in a different genogroup. Only one isolate of a different DGREA group showed the same MLVA profile as that of isolates from the KX group (PMA109.5). When we constructed a dendrogram with all 113 isolates, including those that had more than one allele in some loci, the genotype diversity increased (Fig. 3). Cluster analysis revealed that each cluster contained subgroups that corresponded with environmental and clinical isolates. Also, clusters showed a diverse spatial distribution of isolates obtained during different outbreaks from 2004 until 2007. Finally, MLVA resulted in a differentiation of the V. parahaemolyticus isolates that belong to the same DGREA group, yielding more genogroups than DGREA.
FIG. 2.
Dendrogram of 81 V. parahaemolyticus isolates, with one allele per tandem repeat, that have been typed previously by DGREA. Each color symbol represents a different DGREA group, which is indicated at the far right column. PMA, Puerto Montt environmental isolate; PMC, Puerto Montt clinical isolate; VpKX, RIMD2210633 strain.
FIG. 3.
Dendrogram of all 113 V. parahaemolyticus isolates studied, i.e., those including the strains with polymorphic alleles. Each bracket at left of the DGREA groups (far right) represents isolates belonging to a DGREA group. PMA, Puerto Montt environmental isolate; PMC, Puerto Montt clinical isolate; VpKX, RIMD2210633 strain.
DISCUSSION
The control of bacterial pathogens requires the development of tools allowing the precise identification of strains at the subspecies level. It is now widely accepted that these tools need to be DNA-based assays (5, 11, 34). Conceivable methods are multilocus sequence typing (up to a few thousand entries for the more than 20 bacterial species) and more recently MLVA (up to a few hundred entries, with assays available for more than 20 pathogens) (27). The genome of V. parahaemolyticus has been shown to contain several polymorphic repetitive DNA elements where DNA motifs exist in multiple copies and can be used to discriminate between isolates. One of these which has been used in molecular typing studies for many bacteria is the analysis of polymorphic minisatellites, now referred to as VNTRs. It is well known that in these repeats, genomic events, such as DNA polymerase slippage and recombination, are often occurring. The result of these events is often a change in the size of the repeated tandem (44). If these events occur with a high enough frequency, they will create variations between strains at the repeat loci.
Kimura et al. (22) have shown that MLVA is a valuable typing technique for characterizing recently emerged and highly homogeneous pandemic strains of V. parahaemolyticus. The data demonstrate the utility of this approach for individual strain identification and for discriminating very closely related strains, such as V. parahaemolyticus serotype O3:K6 strains. To this end we have developed the first MLVA multicolor capillary electrophoresis method for high-resolution discrimination of V. parahaemolyticus isolates, which was validated using strains from Chilean outbreaks. Using multicolored dyes, it is possible to give MLVA amplicons different colors so that they can be run together and still be typed individually. Coupled with capillary electrophoresis, this is a very fast way of separating and assigning sizes to various VNTR loci in a single run. By applying this rapid and efficient approach, 32 isolates can be analyzed within 8 h during a run of a 96-well microtiter plate on a sequencer equipped with 16 capillaries.
The MLVA method was applied to 113 V. parahaemolyticus isolates that were genotyped previously by DGREA. The genotypes or profiles determined by MLVA can differentiate between isolates that belong to the same clonal group obtained by DGREA, showing a high discrimination power for the environmental and clinical isolates that were previously classified. All 81 isolates that showed one allele per locus and that belonged to 20 different DGREA groups could be distinguished in 59 different MLVA profiles. In MLVA, for which the target is well defined, the range and polymorphism index of each locus can be calculated and primers can be designed to be very specific. A high diversity index with a narrow confidence interval indicates accurate determination of the highly variable locus. The loci that we identified and used in this analysis were sufficiently variable to be used as molecular indicators to discriminate between samples.
The sizes of the PCR products determined by capillary electrophoresis in our assay deviated slightly in several cases from the reported sizes for genome-sequenced strain RIMD2210633 (Table 1). This inconsistency might be due to the contribution of the different fluorescent dyes to the running distance, especially when we used VIC dye. However, the differences between sizes were reproducible and did not interfere with the genotyping.
The use of capillary electrophoresis in MLVA has many advantages. Slab gel preparation and manual sample loading will be replaced by automated analysis and data acquisition. Slab gels use an external size standard, and a mobility shift might cause data interpretation problems, whereas highly reproducible sizing is achieved by capillary electrophoresis using an internal size marker. The use of multiple dyes allows an easy interpretation of the electropherogram, especially in the case of overlapping amplicon sizes. Different alleles at each locus can be assigned allele numbers, like repeat numbers, which removes the uncertainties introduced when analyzing band patterns from gel images and can be used for strain comparison.
Schouls et al. (42) reported that in some cases the PCR yielded multiple bands, either due to the fact that some of the VNTR regions were present in several copies in the genome or due to a possible nonspecific PCR. These multiple bands could exist because some genomic duplications occurred during the evolution in some strains, most of them from environmental samples. Some of the tandem repeats could be direct repeats. Direct repeats are present in multiple copies in the genome and can be interspersed DNA repeats or tandem repeats. We observed that some tandem repeat loci had more than one allele. This phenomenon cannot be predicted if only one genome sequence of V. parahaemolyticus can be used to find tandem repeats. If more genome sequences are available, the polymorphic tandem repeats can be directly identified.
The high diversity index of locus VP2-07 suggests the highly polymorphic nature of this locus, which grants greater discriminatory power between similar strains. The two loci, VP2-07 and VPTR1, with the highest Hunter and Gaston index have higher copies of repeat numbers between the loci analyzed, and this may confer higher allelic variability, as we observed. Some selected loci appeared to show less diversity than others, and this could be due to the fact that the isolates were taken from a limited geographical area such as the Puerto Montt region. The selective use of some less-variable VNTR loci, such as locus VPTR7, may be especially informative regarding higher levels of evolutionary divergence. Locus VPTR7 is among the least diverse loci, with four alleles. This locus is polymorphic in some isolates of the KX DGREA group. The lack of amplification of locus VP2-07 in some isolates of DGREA group 187 and of VPTR7 in isolates of DGREA group 1.5 may be due to sequence diversity in the flanking regions up- or downstream from the repeat regions or the lack of the VNTR loci altogether. Despite the fact that there were no amplifications or null alleles of these loci, we could distinguish four different MLVA types in 100% of the isolates in the first case and nine MLVA types in 82% of the isolates in the second group using other loci.
In DGREA group KX at least 13 isolates were indistinguishable in their MLVA profiles, in two different subgroups (one with 10 and the other with 3), and these included isolates obtained from clinical and environmental samples and in different years. Some environmental isolates were genetically closer to clinical isolates than to other environmental isolates. This is very important because if nonpandemic strains belong to the pandemic group of strains, the pathogenic potential of the environmental isolates could increase due to gene exchange in the environment, specially if the genes transferred are important for virulence, like tdh and orf8, which are present in these environmental strains that clustered together with pandemic strains. Also, these isolates with the same MLVA types that were obtained in different outbreaks and occurred in different years probably have a common origin, which indicates widespread dissemination and persistence of that genotype over time in this area. Further investigations could add more detail to the depth of knowledge about the V. parahaemolyticus population and to understanding of the molecular epidemiology of this bacterium.
It is important to mention that none of the MLVA profiles of the Chilean isolates was identical to that of the pandemic strain VpKX. This result substantiates the hypothesis that during the Chilean outbreaks, a gradual evolution of the original strain occurred with respect to the VNTR loci, thereby causing an increase in diversity of V. parahaemolyticus strains in the region of Puerto Montt in Chile. Therefore, a future task is to obtain more isolates from this region and use the developed technique to understand better the emergence of pathogenic variants of this food-borne pathogen in its natural marine habitat.
In conclusion, MLVA of V. parahaemolyticus was advanced to multiplex PCR using 10 VNTR markers with fluorescent primers and automated capillary electrophoresis. This technology represents a powerful tool for the high-resolution genotyping of V. parahaemolyticus isolates. Its implementation demonstrates the genetic heterogeneity of the VNTR loci among isolates of the same DGREA group and provides the automation necessary for rapid and reliable production and management of the genotyping data. Clonal typing of V. parahaemolyticus might be useful for tracking the strains involved in recent outbreaks as well as for gaining insight into the ecology of the organisms in natural waters. We believe that the application of this automated typing system will enable large-scale genotyping of V. parahaemolyticus and provide a new tool for global epidemiological surveillance, leading to novel comprehensive insights into the population genetics and evolution of this human pathogen.
Supplementary Material
Acknowledgments
We thank Melissa Wos for her support in the statistical analysis and Josefin Draheim, Leila Kahlisch, and Karsten Henne for their technical assistance.
This work was partly supported by funds from the European Commission for the Healthy Water Project (FOOD-CT-2006-036306) and by a grant from FONDECYT (no. 1070658).
The authors are solely responsible for the content of this report. It does not represent the opinion of the European Commission. The European Commission is not responsible for any use that might be made of data appearing herein.
Footnotes
Published ahead of print on 17 April 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Andersen, G. L., J. M. Simchock, and K. H. Wilson. 1996. Identification of a region of genetic variability among Bacillus anthracis strains and related species. J. Bacteriol. 178:377-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ansaruzzaman, M., M. Lucas, J. L. Deen, N. A. Bhuiyan, X. Y. Wang, A. Safa, M. Sultana, A. Chowdhury, G. B. Nair, D. A. Sack, L. von Seidlein, M. K. Puri, M. Ali, C. L. Chaignat, J. D. Clemens, and A. Barreto. 2005. Pandemic serovars (O3:K6 and O4:K68) of Vibrio parahaemolyticus associated with diarrhea in Mozambique: spread of the pandemic into the African continent. J. Clin. Microbiol. 43:2559-2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bag, P. K., S. Nandi, R. K. Bhadra, T. Ramamurthy, S. K. Bhattacharya, M. Nishibuchi, T. Hamabata, S. Yamasaki, Y. Takeda, and G. B. Nair. 1999. Clonal diversity among recently emerged strains of Vibrio parahaemolyticus O3:K6 associated with pandemic spread. J. Clin. Microbiol. 37:2354-2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benson, G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acid Res. 27:573-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Broza, Y. Y., Y. Danin-Poleg, L. Lerner, M. Broza, and Y. Kashi. 2007. Vibrio vulnificus typing based on simple sequence repeats: insights into the biotype 3 group. J. Clin. Microbiol. 45:2951-2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chowdhury, N. R., S. Chakraborty, T. Ramamurthy, M. Nishibuchi, S. Yamasaki, Y. Takeda, and G. B. Nair. 2000. Molecular evidence of clonal Vibrio parahaemolyticus pandemic strains. Emerg. Infect. Dis. 6:631-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chowdhury, N. R., O. C. Stine, J. G. Morris, and G. B. Nair. 2004. Assessment of evolution of pandemic Vibrio parahaemolyticus by multilocus sequence typing. J. Clin. Microbiol. 42:1280-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clarke, K. R., and R. N. Gorley. 2006. PRIMER v6: user manual/tutorial. Primer-E, Plymouth. England.
- 9.DePaola, A., C. A. Kaysner, J. Bowers, and D. W. Cook. 2000. Environmental investigations of Vibrio parahaemolyticus in oysters after outbreaks in Washington, Texas, and New York (1997 and 1998). Appl. Environ. Microbiol. 66:4649-4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DePaola, A., J. Ulaszek, C. A. Kaysner, B. J. Tenge, J. L. Nordstrom, J. Wells, N. Puhr, and S. M. Gendel. 2003. Molecular, serological, and virulence characteristics of Vibrio parahaemolyticus isolated from environmental, food, and clinical sources in North America and Asia. Appl. Environ. Microbiol. 69:3999-4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Francois, P., A. Hochmann, A. Huyghe, E.-J. Bonetti, G. Renzi, S. Harbarth, C. Klingenberg, D. Pittet, and J. Schrenzel. 2008. Rapid and high-throughput genotyping of Staphylococcus epidermidis isolates by automated multilocus variable-number of tandem repeats: a tool for real-time epidemiology. J. Microbiol. Methods 72:296-305. [DOI] [PubMed] [Google Scholar]
- 12.Fuenzalida, L., C. Hernandez, J. Toro, M. L. Rioseco, J. Romero, and R. T. Espejo. 2006. Vibrio parahaemolyticus in shellfish and clinical samples during two large epidemics of diarrhoea in southern Chile. Environ. Microbiol. 8:675-683. [DOI] [PubMed] [Google Scholar]
- 13.Fuenzalida, L., L. Armijo, B. Zabala, C. Hernandez, M. L. Rioseco, C. Riquelme, and R. T. Espejo. 2007. Vibrio parahaemolyticus strains isolated during investigation of the summer 2006 seafood related diarrhea outbreaks in two regions of Chile. Int. J. Food Microbiol. 117:270-275. [DOI] [PubMed] [Google Scholar]
- 14.Gendel, S. M., J. Ulaszek, M. Nishibuchi, and A. DePaola. 2001. Automated ribotyping differentiates Vibrio parahaemolyticus O3:K6 strains associated with a Texas outbreak from other clinical strains. J. Food Prot. 64:1617-1620. [DOI] [PubMed] [Google Scholar]
- 15.González-Escalona, N., V. Cachicas, C. Acevedo, M. L. Rioseco, J. A. Vergara, F. Cabello, J. Romero, and R. T. Espejo. 2005. Vibrio parahaemolyticus diarrhea, Chile, 1998 and 2004. Emerg. Infect. Dis. 11:129-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.González-Escalona, N., J. Martinez-Urtaza, J. Romero, R. T. Espejo, L. A. Jaykus, and A. DePaola. 2008. Determination of molecular phylogenetics of Vibrio parahaemolyticus strains by multilocus sequence typing. J. Bacteriol. 190:2831-2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 18.Hara-Kudo, Y., K. Sugiyama, M. Nishibuchi, A. Chowdhury, J. Yatsuyanagi, Y. Ohtomo, A. Saito, H. Nagano, T. Nishina, H. Nakagawa, H. Konuma, M. Miyahara, and S. Kumagai. 2003. Prevalence of pandemic thermostable direct hemolysin-producing Vibrio parahaemolyticus O3:K6 in seafood and the coastal environment in Japan. Appl. Environ. Microbiol. 69:3883-3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harth, E., L. Matsuda, C. Hernández, M. L. Rioseco, J. Romero, N. González-Escalona, J. Martínez-Urtaza, and R. T. Espejo. 2009. Epidemiology of Vibrio parahaemolyticus outbreaks, southern Chile. Emerg. Infect. Dis. 15:163-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hunter, P. R., and M. A. Gaston. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jackson, P. J., E. A. Walthers, A. S. Kalif, K. L. Richmond, D. M. Adair, K. K. Hill, C. R. Kuske, G. L. Andersen, K. H. Wilson, M. Hugh-Jones, and P. Keim. 1997. Characterization of the variable-number tandem repeats in vrrA from different Bacillus anthracis isolates. Appl. Environ. Microbiol. 63:1400-1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kimura, B., Y. Sekine, H. Takahashi, Y. Tanaka, H. Obata, A. Kai, S. Morozumi, and T. Fujii. 2008. Multiple-locus variable-number of tandem-repeats analysis distinguishes Vibrio parahaemolyticus pandemic O3:K6 strains. J. Microbiol. Methods 72:313-320. [DOI] [PubMed] [Google Scholar]
- 23.Le Flèche, P., Y. Hauck, L. Onteniente, A. Prieur, F. Denoeud, V. Ramisse, P. Sylvestre, G. Benson, F. Ramisse, and G. Vergnaud. 2001. A tandem repeats database for bacterial genomes: application to the genotyping of Yersinia pestis and Bacillus anthracis. BMC Microbiol. 1:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Le Flèche, P., M. Fabre, F. Denoeud, J. L. Koeck, and G. Vergnaud. 2002. High resolution, on-line identification of strains from the Mycobacterium tuberculosis complex based on tandem repeat typing. BMC Microbiol. 2:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lindstedt, B. A., T. Vardund, and G. Kapperud. 2004. Multiple-locus variable number tandem-repeat analysis of Escherichia coli O157 using PCR multiplexing and multi-colored capillary electrophoresis. J. Microbiol. Methods 58:213-222. [DOI] [PubMed] [Google Scholar]
- 26.Lindstedt, B. A., T. Vardund, L. Aas, and G. Kapperud. 2004. Multiple-locus variable-number tandem-repeats analysis of Salmonella enterica subsp. enterica serovar Typhimurium using PCR multiplexing and multicolour capillary electrophoresis. J. Microbiol. Methods 59:163-172. [DOI] [PubMed] [Google Scholar]
- 27.Lindstedt, B. A. 2005. Multiple-locus variable number tandem repeats analysis for genetic fingerprinting of pathogenic bacteria. Electrophoresis 26:2567-2582. [DOI] [PubMed] [Google Scholar]
- 28.Lindstedt, B. A., L. T. Brandal, L. Aas, T. Vardund, and G. Kapperud. 2007. Study of polymorphic variable-number of tandem repeats loci in the ECOR collection and in a set of pathogenic Escherichia coli and Shigella isolates for use in a genotyping assay. J. Microbiol. Methods 69:197-205. [DOI] [PubMed] [Google Scholar]
- 29.Lista, F., G. Faggioni, S. Valjevac, A. Ciammaruconi, J. Vaissaire, C. le Doujet, O. Gorgé, R. De Santis, A. Carattoli, A. Ciervo, A. Fasanella, F. Orsini, R. D'Amelio, C. Pourcel, A. Cassone, and G. Vergnaud. 2006. Genotyping of Bacillus anthracis strains based on automated capillary 25-loci multiple locus variable-number tandem repeats analysis. BMC Microbiol. 6:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marshall, S., C. G. Clark, G. Wang, M. Mulvey, M. T. Kelly, and W. M. Johnson. 1999. Comparison of molecular methods for typing Vibrio parahaemolyticus. J. Clin. Microbiol. 37:2473-2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martinez-Urtaza, J., L. Simental, D. Velasco, A. DePaola, M. Ishibashi, Y. Nakaguchi, M. Nishibuchi, D. Carrera-Flores, C. Rey-Alvarez, and A. Pousa. 2005. Pandemic Vibrio parahaemolyticus O3:K6, Europe. Emerg. Infect. Dis. 11:1319-1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matsumoto, C., J. Okuda, M. Ishibashi, M. Iwanaga, P. Garg, T. Rammamurthy, H. C. Wong, A. DePaola, Y. B. Kim, M. J. Albert, and M. Nishibuchi. 2000. Pandemic spread of an O3:K6 clone of Vibrio parahaemolyticus and emergence of related strains evidenced by arbitrarily primed PCR and toxRS sequence analyses. J. Clin. Microbiol. 38:578-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Metzgar, D., E. Thomas, C. Davis, D. Field, and C. Wills. 2001. The microsatellites of Escherichia coli: rapidly evolving repetitive DNAs in a non-pathogenic prokaryote. Mol. Microbiol. 39:183-190. [DOI] [PubMed] [Google Scholar]
- 34.Nederbragt, A. J., A. Balasingham, R. Sirevåg, H. Utkilen, K. S. Jakobsen, and M. J. Anderson-Glenna. 2008. Multiple-locus variable-number tandem repeat analysis of Legionella pneumophila using multi-colored capillary electrophoresis. J. Microbiol. Methods 73:111-117. [DOI] [PubMed] [Google Scholar]
- 35.Noller, A. C., M. C. McEllistrem, A. G. F. Pacheco, D. J. Boxrud, and L. H. Harrison. 2003. Multilocus variable-number tandem repeat analysis distinguishes outbreak and sporadic Escherichia coli O157:H7 isolates. J. Clin. Microbiol. 41:5389-5397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Okuda, J., M. Ishibashi, E. Hayakawa, T. Nishino, Y. Takeda, A. K. Mukhopadhyay, S. Garg, S. K. Bhattacharya, G. B. Nair, and M. Nishibuchi. 1997. Emergence of a unique O3:K6 clone of Vibrio parahaemolyticus in Calcutta, India, and isolation of strains from the same clonal group from Southeast Asian travelers arriving in Japan. J. Clin. Microbiol. 35:3150-3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Onteniente, L., S. Brisse, P. T. Tassios, and G. Vergnaud. 2003. Evaluation of polymorphisms associated with tandem repeats for Pseudomonas aeruginosa strain typing. J. Clin. Microbiol. 41:4991-4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Potasman, I., A. Paz, and O. Majed. 2002. Infectious outbreaks associated with bivalve shellfish consumption: a worldwide perspective. Clin. Infect. Dis. 35:921-928. [DOI] [PubMed] [Google Scholar]
- 39.Pourcel, C., P. Visca, B. Afshar, S. D'Arezzo, G. Vergnaud, and N. K. Fry. 2007. Identification of variable-number tandem-repeat (VNTR) sequences in Legionella pneumophila and development of an optimized multiple-locus VNTR analysis typing scheme. J. Clin. Microbiol. 45:1190-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Quilici, M. L., A. Robert-Pillot, J. Picart, and J. M. Fournier. 2005. Pandemic Vibrio parahaemolyticus O3:K6 spread, France. Emerg. Infect. Dis. 11:1148-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sabat, A., J. Krzyszton-Russjan, W. Strzalka, R. Filipek, K. Kosowska, W. Hryniewicz, J. Travis, and J. Potempal. 2003. New method for typing Staphylococcus aureus strains: multiple-locus variable-number tandem repeat analysis of polymorphism and genetic relationships of clinical isolates. J. Clin. Microbiol. 41:1801-1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schouls, L. M., A. van der Ende, M. Damen, and I. van de Pol. 2006. Multiple-locus variable-number tandem repeat analysis of Neisseria meningitidis yields groupings similar to those obtained by multilocus sequence typing. J. Clin. Microbiol. 44:1509-1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Slack, A., M. Symonds, M. Dohnt, and L. Smythe. 2006. An improved multiple-locus variable number of tandem repeats analysis for Leptospira interrogans serovar Australis: a comparison with fluorescent amplified fragment length polymorphism analysis and its use to redefine the molecular epidemiology of this serovar in Queensland, Australia. J. Med. Microbiol. 55:1549-1557. [DOI] [PubMed] [Google Scholar]
- 44.Tuntiwechapikul, W., and M. Salazar. 2002. Mechanism of in vitro expansion of long DNA repeats: effect of temperature, repeat length, repeat sequence, and DNA polymerases. Biochemistry 41:854-860. [DOI] [PubMed] [Google Scholar]
- 45.Valcheva, V., I. Mokrousov, O. Narvskaya, N. Rastogi, and N. Markova. 2008. Utility of new 24-locus variable-number tandem-repeat typing for discriminating Mycobacterium tuberculosis clinical isolates collected in Bulgaria. J. Clin. Microbiol. 46:3005-3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Belkum, A., S. Scherer, L. van Alphen, and H. Verbrugh. 1998. Short-sequence DNA repeats in prokaryotic genomes. Microbiol. Mol. Biol. Rev. 62:275-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.