Abstract
The rhizobacterium Pseudomonas fluorescens CHA0 promotes the growth of various crop plants and protects them against root diseases caused by pathogenic fungi. The main mechanism of disease suppression by this strain is the production of the antifungal compounds 2,4-diacetylphloroglucinol (DAPG) and pyoluteorin (PLT). Direct plant growth promotion can be achieved through solubilization of inorganic phosphates by the production of organic acids, mainly gluconic acid, which is one of the principal acids produced by Pseudomonas spp. The aim of this study was to elucidate the role of gluconic acid production in CHA0. Therefore, mutants were created with deletions in the genes encoding glucose dehydrogenase (gcd) and gluconate dehydrogenase (gad), required for the conversion of glucose to gluconic acid and gluconic acid to 2-ketogluconate, respectively. These enzymes should be of predominant importance for rhizosphere-colonizing biocontrol bacteria, as major carbon sources provided by plant root exudates are made up of glucose. Our results show that the ability of strain CHA0 to acidify its environment and to solubilize mineral phosphate is strongly dependent on its ability to produce gluconic acid. Moreover, we provide evidence that the formation of gluconic acid by CHA0 completely inhibits the production of PLT and partially inhibits that of DAPG. In the Δgcd mutant, which does not produce gluconic acid, the enhanced production of antifungal compounds was associated with improved biocontrol activity against take-all disease of wheat, caused by Gaeumannomyces graminis var. tritici. This study provides new evidence for a close association of gluconic acid metabolism with antifungal compound production and biocontrol activity in P. fluorescens CHA0.
Plant growth-promoting rhizobacteria (PGPR) (36) are root-colonizing bacteria that enhance the performance of crop plants by several mechanisms. First, they antagonize plant-pathogenic fungi, mainly by the production of antimicrobial metabolites, but also by competition for iron or rhizosphere niches (9, 23, 24, 59). The biocontrol activity of many disease-suppressive microorganisms is also attributed to stimulation of host defense (induced systemic resistance). Other mechanisms by which these rhizobacteria directly promote plant growth are the production of phytohormones and the increase of nutrient, in particular phosphate, availability to plants (18, 37). Certain rhizobacteria are able to solubilize insoluble or poorly soluble mineral phosphates by producing acid phosphatases and organic acids, mainly gluconic acid (2, 34, 60). Some PGPR combine these different plant-beneficial activities and are able to suppress soilborne plant diseases, as well as to increase phosphate availability for plants (72).
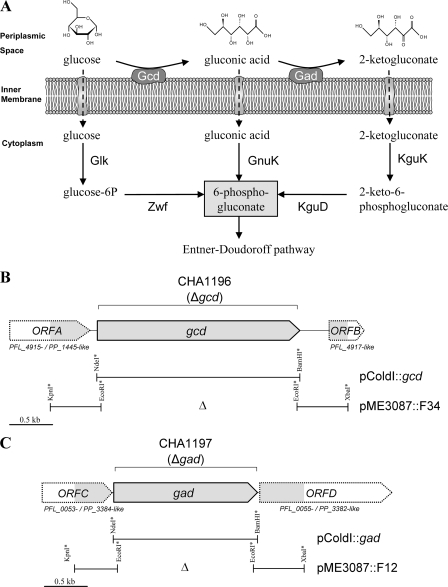
In fluorescent pseudomonads, gluconic acid production is catalyzed by periplasmic oxidation of glucose by membrane-bound glucose dehydrogenase (Gcd) (Fig. 1A) (16, 43). In many gram-negative bacteria, the synthesis of gluconic acid has been shown to be dependent on pyrroloquinoline quinone (PQQ) as an enzymatic cofactor of the Gcd (1, 14). A consecutive oxidation reaction is mediated by gluconate dehydrogenase (Gad), which converts gluconic acid to 2-ketogluconate (Fig. 1A) (11, 12, 44, 50). These enzymes should be of predominant importance for biocontrol soil pseudomonads, as major carbon sources provided by plant root exudates in the rhizosphere are made up of glucose (29, 69, 70). The two enzymes involved in glucose metabolism may have a substantial influence on general nutrient availability in the rhizosphere. First, Gcd and Gad affect glucose levels, and second, they may modulate the availability of soluble phosphates by controlling the amount of gluconic acid released into the rhizosphere. Furthermore, the production of gluconic acid might substantially change the rhizosphere pH. Therefore, Gcd and Gad enzymes produced by fluorescent pseudomonads are likely to be important for soil fertility and to impact the activities of other organisms living in the rhizosphere, e.g., fungal pathogens attacking the roots. Indeed, gluconic acid metabolism has already been linked to antifungal activity. Recently, Kaur et al. (30) proposed that gluconic acid produced by a nonfluorescent Pseudomonas isolate may be important for the biological control of take-all disease.
FIG. 1.
(A) Periplasmic and intracellular glucose catabolism in pseudomonads based on studies with P. aeruginosa (10), P. putida (11, 12), and P. fluorescens (17, 28). Shown are membrane-bound enzymes involved in periplasmic glucose metabolism, Gcd (glucose dehydrogenase) and Gad (gluconate dehydrogenase), and enzymes involved in cytoplasmic glucose metabolism, Glk (glucokinase), Zwf (glucose-6-phosphate 1-dehydrogenase), GnuK (gluconokinase), KguK (2-ketogluconate kinase), and KguD (2-ketogluconate 6-phosphate reductase) (the names of the enzymes are derived from the nomenclature for P. putida KT2440 [12, 54]). (B and C) Physical locations of the gcd (B) and gad (C) genes in the genome of P. fluorescens strain CHA0. The shaded arrows show the sequenced or partly sequenced genes. The representation is based on the sequence data for strain CHA0 obtained by sequencing the chromosomal fragments inserted in the indicated vectors. The designations of the ORFs flanking the gcd and gad genes are based on the corresponding locus tags in the complete annotated sequence of the closely related P. fluorescens strain Pf-5 (56). Δ, region deleted in strains CHA1196 and CHA1197 and in plasmids pME3087::F34 and pME3087::F12. The bars designate the fragments cloned into the vector pME3087 to give pME3087::F34 and pME3087::F12 and into pColdI to give pColdI::gcd and pColdI::gad. Artificial restriction sites on the cloned fragments are marked with asterisks.
Pseudomonas fluorescens CHA0 is a bacterial strain known to be able to suppress various soilborne plant diseases (24). Its biocontrol ability has been linked to the production of the antifungal compounds 2,4-diacetylphloroglucinol (DAPG) (31, 33) and pyoluteorin (PLT) (46, 47). The strain is also able to solubilize mineral phosphate and to improve plant growth under phosphate-limiting conditions (A. von Felten, personal communication). Gluconic acid is supposed to play a predominant role in the phosphate solubilization activity of P. fluorescens CHA0, and we hypothesize that the metabolite also has an impact on the biocontrol activity of this PGPR strain.
The aim of this study was to elucidate the role of gluconic acid production in P. fluorescens CHA0 with respect to its phosphate-solubilizing ability, antifungal metabolite production, and ability to suppress fungal root diseases. To this end, mutants of strain CHA0 carrying deletions in the gcd gene, encoding Gcd, and the gad gene, encoding Gad (Fig. 1), were created. The three in-frame deletion mutants, CHA1196 (Δgcd), CHA1197 (Δgad), and CHA1198 (Δgcd Δgad), were compared with their parental strain for the ability to produce organic acids, to solubilize inorganic phosphate, to produce the antifungal metabolites DAPG and PLT, to inhibit the growth of fungal pathogens, and to suppress different soilborne diseases. We provide evidence that in fact, gluconic acid production by P. fluorescens CHA0 is involved not only in the solubilization of phosphate, but also in the regulation of antifungal compound production and, as a consequence, can influence the level of plant protection provided by the strain.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains and plasmids used in this study are described in Table 1. P. fluorescens strains were cultivated on King's medium B agar (KMB) (35) at 27°C and in Luria broth (LB) (62) at 27°C, unless otherwise specified. Escherichia coli strains were grown on nutrient agar and in LB at 37°C. When required, antibiotics were added to the growth medium at the following concentrations: ampicillin sodium salt, 100 μg/ml; chloramphenicol, 20 μg/ml; kanamycin sulfate, 50 μg/ml; and tetracycline hydrochloride, 25 μg/ml for E. coli and 125 μg/ml for P. fluorescens strains. When appropriate, 5-bromo-4-chloro-3-indolyl-β-d-galactoside (X-Gal) was incorporated into solid media to monitor β-galactosidase expression (62). For monitoring antifungal gene expression, derivatives of P. fluorescens strain CHA0 carrying rhizosphere-stable plasmids with transcriptional fusions of a stable variant of the gfp gene to the DAPG biosynthetic gene phlA (pME7100) or the PLT biosynthetic gene pltA (pME7109) (3) were used.
TABLE 1.
Bacterial strains, plasmids, and oligonucleotides used in this study
| Strain, plasmid, or oligonucleotide | Characteristicsa or sequence (5′→3′)b | Reference(s) and/or source |
|---|---|---|
| P. fluorescens | ||
| CHA0 | Wild type; Cmr | 23, 68 |
| CHA1196 | Δgcd in-frame deletion mutant of CHA0 | This study |
| CHA1197 | Δgad in-frame deletion mutant of CHA0 | This study |
| CHA1198 | Δgcd Δgad in-frame deletion mutant of CHA0 | This study |
| E. coli | ||
| DH5α, HB101, BL21 | Laboratory strains | 62 |
| Plasmids | ||
| pColdI | Cold shock-based expression vector; Apr | 58; Takara Bio |
| pColdI::gcd | pColdI with the gcd gene flanked by an N-terminal His6 tag under the control of the cspA promoter; Apr | This study |
| pColdI::gad | pColdI with the gad gene flanked by an N-terminal His6 tag under the control of the cspA promoter; Apr | This study |
| pME3087 | Suicide vector; ColE1 replicon; RK2-Mob; Tcr | 65 |
| pME3087::F12 | pME3087 carrying the 1,127-bp KpnI-XbaI fragment from pUKF12; Tcr | This study |
| pME3087::F34 | pME3087 carrying the 1,247-bp KpnI-XbaI fragment from pUKF34; Tcr | This study |
| pME7100 | phlA-gfp transcriptional fusion; Tcr | 3 |
| pME7109 | pltA-gfp transcriptional fusion; Tcr | 3 |
| pME497 | Mobilizing plasmid; IncP-1 Tra RepA(Ts) Apr | 66 |
| pUK21 | Cloning vector; lacZα Kmr | 73 |
| pUKF12 | pUK21 carrying a 1,127-bp KpnI-XbaI insert with a deletion in gad; Kmr | This study |
| pUKF34 | pUK21 carrying a 1,247-bp KpnI-XbaI insert with a deletion in gcd; Kmr | This study |
| Oligonucleotides | ||
| Gad1 | GGGGTACCGAATACATCGACCGGCAGATGA; KpnI | This study |
| Gad2 | GGAATTCTGCGTCGACCTTCTTCATCACTG; EcoRI | This study |
| Gad3 | GGAATTCGTTCAGGCATAAGGAGCGATGAC; EcoRI | This study |
| Gad4 | GCTCTAGACTTCTCCTGCATGGTCAGGGCC; XbaI | This study |
| Gcd1 | GGGGTACCGACGTCTACAAGGATGGCAATG; KpnI | This study |
| Gcd2 | GGAATTCGCTCGGACTTGAAGCACCCTCAG; EcoRI | This study |
| Gcd3 | GGAATTCCTGGGCACCAAGATGGGCGACTA; EcoRI | This study |
| Gcd4 | GCTCTAGAGATGTTCTCCAGCAACTGGGTG; XbaI | This study |
| PColdgad-1 | GGAATTCCATATGGCAACAGTGATGAAGAAGGT; NdeI | This study |
| PColdgad-2 | CGGGATCCTTATGCCTGAACCAGCGG; BamHI | This study |
| PColdgcd-1 | GGAATTCCATATGAGCACTGAGGGTGCTTC; NdeI | This study |
| PColdgcd-2 | CGGGATCCTTATTCCGACAGTTTGTAGGCA; BamHI | This study |
Apr, Cmr, Kmr, and Tcr, resistance to ampicillin, chloramphenicol, kanamycin, and tetracycline, respectively.
Specified restriction enzyme sites are underlined.
Fungal strains and culture conditions.
Gaeumannomyces graminis var. tritici strain ETH1000, the causal agent of take-all disease of wheat, and Magnaporthe grisea no. 283 (obtained from Syngenta, Stein, Switzerland), the causal agent of rice blast, were grown and maintained on potato dextrose agar (PDA) (24 g/liter of Difco potato dextrose broth and 12 g/liter of Oxoid technical agar, pH 7.0) at 24°C in the dark. Rhizoctonia solani strain 160 (obtained from Ciba-Geigy SA, Basel, Switzerland), the causal agent of root rot of cucumber in this study, was grown and maintained under the same conditions on malt extract agar (15 g/liter of Oxoid malt extract and 12 g/liter of Oxoid technical agar, pH 7.0).
DNA manipulations and sequence analyses.
Chromosomal DNA of P. fluorescens was isolated as described previously (66). Small- and large-scale plasmid preparations were performed by the cetyltrimethylammonium bromide method (62), with the GenElute HP Plasmid Miniprep kit (Sigma-Aldrich Chemie GmbH, Buchs, Switzerland), or with the Jetstar 2.0 kit (Genomed, Basel, Switzerland). Standard techniques were used for restriction, agarose gel electrophoresis, dephosphorylation, generation of blunt ends with T4 DNA polymerase (Boehringer, Ingelheim, Germany), isolation of DNA fragments from low-melting-point agarose gels, and ligation (62, 65, 66). Restriction fragments were purified from agarose gels using the MinElute gel extraction or Qiaquick gel extraction kit (Qiagen, Basel, Switzerland), depending on the size of the fragment. Electroporation of bacterial cells with plasmid DNA was done as described previously (66). Nucleotide primers were designed with the online software Primer3 (v.0.4.0) (61) and purchased from Microsynth (Balgach, Switzerland). PCRs were carried out using GoTaq DNA Polymerase (Promega, WI) and PrimeStar HS DNA Polymerase (Takara Bio Inc., Shiga, Japan), following procedures detailed elsewhere (3, 57). Nucleotide sequences were determined on both strands with the Big Dye terminator cycle-sequencing kit 3.1 (Applied Biosystems, Foster City, CA) and an ABI Prism 3130 Genetic Analyzer (Applied Biosystems). Nucleotide and deduced amino acid sequences were analyzed with the programs Sequencher 4.8 (Gene Codes Corporation, Ann Arbor, MI) and the ExPASy Proteomics Server (http://www.expasy.org). Sequence alignments were performed using Clustal W software (39).
Cloning of the gcd and gad genes from P. fluorescens.
The gcd gene was amplified from chromosomal DNA of P. fluorescens CHA0 using primers PColdgcd-1 and PColdgcd-2 (Table 1). To amplify the gad gene from CHA0, primers PColdgad-1 and PColdgad-2 (Table 1) were used. Primers were designed based on genomic data available for the very closely related P. fluorescens strain Pf-5 (http://www.pseudomonas.com) (56). For gcd, the PCR amplification yielded a 2.4-kb band. The PCR product was digested with NdeI and BamHI and was cloned into the vector pColdI to give pColdI::gcd (Table 1). For gad, the PCR amplification produced a 1.7-kb fragment that was digested with NdeI and BamHI and was cloned into pColdI to give pColdI::gad. For both constructs, the inserts of two of the obtained clones were sequenced to confirm the identities of the cloned fragments.
Construction of Δgcd and Δgad in-frame deletion mutants of P. fluorescens.
To generate the Δgcd mutant CHA1196, a 2,328-bp fragment was deleted in frame in the gcd gene. For this purpose, a 666-bp KpnI-EcoRI fragment including the first nine codons of gcd and the adjacent upstream chromosomal region was amplified by PCR from chromosomal DNA of P. fluorescens CHA0, using primers Gcd1 and Gcd2. A 575-bp EcoRI-XbaI fragment including the last 16 codons of gcd and the adjacent downstream region was amplified by PCR with primers Gcd3 and Gcd4. The resulting upstream and downstream fragments were digested with KpnI and EcoRI and with EcoRI and XbaI, respectively, and were cloned by a triple ligation into pUK21 digested with KpnI and XbaI, giving plasmid pUKF34. The 1.24-kb KpnI-XbaI insert in pMEF34 was checked by sequencing, excised, and cloned into the suicide plasmid pME3087 digested with KpnI-XbaI, producing pME3087::F34. To obtain the Δgcd mutant CHA1196, the suicide plasmid pME3087::F34 was then integrated into the chromosome of strain CHA0 by triparental mating, using E. coli HB101/pME497 as the mobilizing strain, with selection for tetracycline- and chloramphenicol-resistant recombinants (65, 66). Excision of the vector by a second crossing over occurred after enrichment for tetracycline-sensitive cells.
An analogous gene replacement strategy was followed to obtain the 1,731-bp Δgad deletion mutant CHA1197. Using CHA0 DNA as a template, two fragments including the first nine and the last three codons of the gad gene and the respective up- and downstream flanking regions were amplified by PCR with primers Gad1 plus Gad2 and Gad3 plus Gad4, respectively. The 494-bp upstream and 627-bp downstream fragments obtained were digested with KpnI and EcoRI and with EcoRI and XbaI, respectively, and were cloned by a triple ligation into KpnI- and XbaI-digested pUK21. The identity of the insert in the resulting plasmid, pUKF12, was checked by sequencing. The 1.12-kb KpnI-XbaI fragment was then excised from pMEF12 and cloned into pME3087 digested with the same restriction enzymes, giving plasmid pME3087::F12. The suicide plasmid pME3087::F12 then served to delete the gad gene in CHA0, creating CHA1197.
The Δgcd Δgad double mutant CHA1198 was obtained by using the suicide plasmid pME3087::F12 to delete the gad gene in the Δgcd mutant CHA1196. The Δgcd and Δgad mutations were verified by PCR (data not shown).
Assessment of acid production by P. fluorescens and its Δgcd and Δgad mutants.
Strain CHA0 and its mutant derivatives were cultivated in LB overnight. The cells were washed twice with sterile water and adjusted to an optical density at 600 nm (OD600) of 0.1. To visualize the acid production, 5-μl drops of bacterial suspensions were inoculated onto National Botanical Research Institute phosphate growth medium (NBRIP) (52) agar plates containing 0.01% methyl red as a pH indicator. The pH of the medium had been adjusted to 7.0 before it was autoclaved. After 3 days of incubation at 24°C in the dark, the acidification of the agar was visualized by the presence of a blurred red halo around the bacterial colony.
The pH and amount of acid produced by CHA0 and its mutant derivatives were estimated using the titration methods of Schleissner et al. (63) and Kaur et al. (30). For this purpose, 100 ml of fresh LB medium was inoculated with 100 μl from a bacterial overnight LB culture and incubated at 24°C with rotational shaking at 180 rpm for a 24-h period until the cell density had reached 1 × 109 CFU/ml. The bacterial cells were harvested by centrifugation and washed twice with sterile distilled NaCl solution (0.9%). The cells were resuspended in 100 ml of a glucose solution (5.0 g/liter) adjusted to pH 7.0. The bacterial suspensions were incubated at 24°C with shaking at 160 rpm. Aliquots of 5 ml of the suspensions were collected at different time intervals, and the cells were pelleted. The supernatant was used for titration and pH measurement. The titration was done as described by Kaur et al. (30) with 0.001 N sodium hydroxide (NaOH) using phenolphthalein as an indicator. The titration was repeated twice.
Phosphate solubilization assay.
To visualize the phosphate solubilization ability of strain CHA0 and its mutant derivatives, 5-μl drops of washed bacterial suspensions (prepared as for the acid production assay on plates) were inoculated onto NBRIP agar plates containing insoluble tricalcium phosphate. A visible halo/zone appeared on the plate as the phosphate was solubilized by the bacteria. The diameters of the halos were measured after 12 days of incubation at 24°C. The experiment was repeated twice, with eight replicates per treatment.
Assessment of growth characteristics and assay for expression of DAPG and PLT biosynthetic genes of the P. fluorescens Δgcd and Δgad mutants.
Growth rates for P. fluorescens CHA0 and its Δgcd and Δgad mutants, as well as antifungal gene expression in derivatives carrying a phlA-gfp fusion on plasmid pME7100 or a pltA-gfp fusion on pME7109 (3), were monitored in OS minimal medium (3, 4) with 25 mM glucose (OS glucose) (for growth and gene expression assays), 47 mM glycerol (OS glycerol) (for growth assays), or 25 mM gluconate (OS gluconate) (for gene expression assays) as a carbon source (3, 66). P. fluorescens CHA0 and its derivatives were grown in these media without selective antibiotics in 96-well black microtiter plates with flat transparent bottoms. For the assays, 10 ml of the respective medium was inoculated with 20 μl of exponential-growth-phase LB cultures of the bacterial strains diluted to an OD600 of 0.05. For each treatment, six wells of the microtiter plate were then partially filled with aliquots of 200 μl of the respective bacterial culture. The cultures were incubated at 30°C with orbital shaking at 500 rpm in a Thermostar incubator (BMG Labtechnologies, Offenburg, Germany). The OD600 as a parameter of growth and green fluorescence (excitation at 480 nm and emission at 520 nm) as a parameter of antifungal gene expression were measured with a Spectrafluor Plus microplate reader (Tecan Group Ltd., Männedorf, Switzerland) throughout the exponential and stationary growth phases (3). For each individual measurement, the green fluorescence value was divided by the corresponding OD600 value, giving the specific fluorescence of the cells expressed as relative fluorescence units (3). The green fluorescence emitted by cells of wild-type strain CHA0 without the gfp reporter fusion was determined for background correction.
Quantification of antifungal compound production.
Strain CHA0 and its mutants CHA1196 and CHA1197 were grown in 300-ml Erlenmeyer flasks sealed with cotton wool stoppers containing 100 ml of yeast-malt (YM) medium (3 g/liter Oxoid malt extract, 3 g/liter Difco yeast extract, 5 g/liter Difco Bacto peptone amended with 10 g/liter glucose or 10 g/liter gluconate, pH 7.0) (5) or glycerol-Casamino Acids medium (GCM) (48) without antibiotics. The media were inoculated with 100 μl of an overnight LB culture and incubated at 24°C with shaking at 160 rpm. Samples were taken over a period of 3 days. Production of DAPG and PLT was quantified in ethyl acetate extracts of the cultures using high-performance liquid chromatography as previously described in detail (13, 47). Briefly, aliquots of bacterial cultures (30 ml) were acidified with 2 M HCl to pH 2, mixed with 40 ml of ethyl acetate, and shaken vigorously. The organic phase was separated from the aqueous phase by filtering it through silicon-coated filter paper (Macherey-Nagel, Düren, Germany) and brought to dryness in a rotary evaporator (Rotavapor-RE; Büchi, Flawil, Switzerland). The residue was dissolved in 1 ml of methanol and analyzed with a liquid chromatograph (Hewlett-Packard 1090; Hewlett-Packard Co., Palo Alto, CA) equipped with a diode array detector and a column (4 by 100 mm) packed with Nucleosil 120-5-C18 (Macherey-Nagel, Düren, Germany).
Fungal inhibition assays.
The inhibitory effects of CHA0 and its Δgcd and Δgad mutants on the growth of fungal pathogens were monitored on one-fifth-strength PDA. For G. graminis var. tritici, 0.6-cm plugs from a 1-week-old growing agar culture were placed in the centers of one-fifth-strength PDA plates and incubated at 24°C for 2 days in the dark. Suspensions (OD600 = 0.1) of washed bacterial cells grown overnight in LB were then applied to the plate in a square of 5.5-cm side length centered on the fungal plug. After an additional 4 days of incubation at 24°C, the radial growth of the fungi was measured. Bioassays with R. solani and M. grisea were performed in the same manner, except that the fungal agar plug and the bacterial suspensions were placed at the same time. The experiment consisted of six plates per treatment and was repeated twice.
To test the sensitivity of G. graminis var. tritici to gluconic acid, the fungus was grown for 5 days at 24°C on one-fifth-strength PDA with 10-μl drops of gluconic acid solutions of different concentrations (10 to 200 mg/ml distilled water) added. Drops were spotted in the corners of a square of 5.5-cm side length centered on the fungal plug.
Plant disease suppression and root colonization assays.
The effects of gcd and gad gene deletions in P. fluorescens CHA0 on its biocontrol ability were tested in two different plant-pathogen systems: wheat-G. graminis var. tritici and cucumber-R. solani. Seeds of cucumber (Cucumis sativus cv. Chinesische Schlange) and seeds of winter wheat (Triticum aestivum cv. Arina) were surface sterilized for 10 min in 4% NaClO (vol/vol) and subsequently rinsed with autoclaved distilled water. The seeds were germinated on soft agar (Oxoid technical agar at 8.5 g/liter) for 48 h at 24°C in the dark. The gnotobiotic system used for the R. solani plant disease assay has been described in detail elsewhere (31). Briefly, 300 g of artificial soil consisting of quartz sand and vermiculite (32) was added to 1-liter Erlenmeyer flasks and autoclaved. Bacterial cells were harvested from LB overnight cultures by centrifugation, washed twice, and adjusted with sterile 0.9% NaCl solution to a cell density of 2.5 × 108 CFU/ml. Aliquots of 0.2 g of 1-week-old R. solani millet seed inoculum (prepared as described by Maurhofer et al. [45]) and 12 ml of the bacterial suspensions were added to the flasks and mixed into the artificial soil. Control flasks were amended with the same amounts of sterile millet seeds and sterile distilled water. Three sterile-grown, 2-day-old cucumber seedlings were transferred into each flask and supplemented with 15 ml of modified Knop plant nutrition solution (32). The flasks were incubated for 14 days in a growth chamber with 70% relative humidity and 16 h of light (160 microeinsteins/m2/s) at 22°C, followed by an 8-h dark period at 20°C.
For G. graminis var. tritici assays, the artificial soil consisted of 30 g Perlite Horticole (Otto Hauenstein Samen SA, Rafz, Switzerland) added to 1-liter Erlenmeyer flasks. The flasks were sealed with cotton wool stoppers and autoclaved. A G. graminis var. tritici agar plug was inoculated into 30 ml defined liquid minimal medium (8) and grown for 2 to 3 weeks at 24°C without shaking. G. graminis var. tritici mycelium was harvested, rinsed several times with autoclaved distilled H2O, cut into small parts, and deposited in the center of the Perlite bed (adapted from the method of Zogg and Amiet [77]). One flask received the mycelium of a 30-ml culture. The bacterial strains were grown overnight on KMB plates at 27°C. The cells were flooded with autoclaved double-distilled water, scraped off with a spatula, washed twice by centrifugation, and resuspended in sterile 0.9% NaCl solution and were finally adjusted to a density of 3 × 108 CFU/ml. Sterile-grown, 2-day-old wheat seedlings were inoculated in the bacterial suspension for 2 h. Afterwards, three seedlings were transferred into each flask and supplemented with 90 ml of modified Knop plant nutrition solution (32), as well as with 10 ml of the corresponding bacterial suspension. The flasks were incubated for 18 days in a growth chamber with 70% relative humidity and 16 h of light (160 microeinsteins/m2/s) at 22°C, followed by an 8-h dark period at 18°C.
For both take-all and Rhizoctonia root rot, roots were harvested, and the disease incidence was calculated for each plant as the percentage of the root surface infected and was assessed on an eight-class scale: 0%, no disease; 5%, 0% < x ≤ 10% of roots infected; 17.5%, 10% < x ≤ 25% of roots infected; 37.5%, 25% < x ≤ 50% of roots infected; 62.5%, 50% < x ≤ 75% of roots infected; 82.5%, 75% < x ≤ 90% of roots infected; 95%, 90% < x ≤ 100% of roots infected; 100%, plant dead. For the wheat-G. graminis var. tritici assay, only the uppermost 2 cm of the root system was assessed. Thereafter, roots from each flask were pooled, assessed for fresh weight, and transferred into sterile 50-ml Erlenmeyer flasks containing 7 ml of sterile distilled water. The flasks were vigorously shaken at 400 rpm for 20 min. The number of culturable fluorescent pseudomonads was determined by plating serial dilutions of the resulting root washes onto KMB containing cycloheximide (100 μg/ml), ampicillin (40 μg/ml), and chloramphenicol (13 μg/ml) and subsequent CFU counting. Wheat-G. graminis var. tritici assays were conducted three times and the cucumber-R. solani assay one time with eight replicates per treatment and three plants per replicate.
Statistical analysis.
Values describing disease severity (percentages of infected roots) were transformed to arc sines prior to statistical analysis. Statistical analyses were performed using the statistics program Systat, version 10.0 (Systat Inc., Evanston, IL). The results of independent repetitions over time (disease suppression assay and in vitro inhibition assay) were first analyzed by two-way analysis of variance. This analysis revealed significant repetition versus treatment interactions. Therefore, individual repetitions over time were analyzed separately. Each individual repetition was analyzed first by one-way analysis of variance and then by Fisher's protected least significant difference test (P ≤ 0.05).
Accession numbers.
The nucleotide and deduced amino acid sequences for the gcd and gad genes, as well as parts of the respective flanking regions of P. fluorescens CHA0, have been deposited in the GenBank database under accession no. FJ694890 and FJ694891, respectively.
RESULTS
Identification and characterization of the chromosome regions containing gcd and gad in P. fluorescens CHA0 and construction of deletion mutants.
Prior to this study, we had undertaken an approach to identify novel regulators of antibiotic production in P. fluorescens. To this end, a derivative of CHA0 carrying a phlA-gfp fusion on pME7100 (3) had been subjected to random Tn5 insertion mutagenesis. From this collection of Tn5 mutants (4), those with altered antifungal gene expression were characterized and Tn5-flanking regions were amplified by arbitrary PCR, cloned, and sequenced. Several mutants were identified for which the Tn5 insert was located in genes involved in glucose metabolism (L. Rochat and P. de Werra, unpublished data).
Comparison of sequence data with the complete genomic sequence available for the closely related P. fluorescens strain Pf-5 (56) revealed that in one CHA0 mutant the transposon had been inserted into an open reading frame (ORF) that was highly similar to the Pf-5 locus PFL_4916 (http://www.pseudomonas.com), encoding a putative Gcd. The CHA0 locus was termed gcd by analogy with closely related sequences in other pseudomonads (see below). Pf-5 sequence information was used to PCR amplify, clone, and sequence a 3,581-bp chromosomal region in strain CHA0 containing gcd and its surroundings (Fig. 1B). The deduced product of the 2,421-bp gcd of strain CHA0 (806 amino acids; 86.4 kDa) is 99% identical to the putative Gcd of P. fluorescens Pf-5 encoded by PFL_4916 (GenBank accession no. AAY94145.1). Gcd of CHA0 was also found to be 80% identical to the well-characterized Gcd of Pseudomonas putida KT2440 (PP_1444 [12, 15, 54]) and 83% identical to the Gcd of Pseudomonas aeruginosa PAO1 (PA2290 [67]) and to show similarity (46% identity) to the Gcd of E. coli K-12 (GenBank accession no. D12651 [74, 75]). The gcd gene of P. fluorescens CHA0 is located between two ORFs (Fig. 1B) that are also present at the corresponding loci of P. fluorescens Pf-5. The partially sequenced upstream ORF (ORFA) encodes a putative porin B (with a C terminus 100% identical to that of the PFL_4915 product of P. fluorescens Pf-5 and also 85% identical to that of the PP_1445 product of P. putida KT2440). The sequenced part of the downstream ORF (ORFB) is 100% identical to PFL_4917, encoding a hypothetical protein of P. fluorescens Pf-5.
A second CHA0 mutant had the Tn5 transposon inserted in an ORF, which we termed gad by analogy with published sequences (see below), encoding a Gad. We amplified and sequenced a 2,864-bp chromosomal region in strain CHA0 containing gad and its surroundings (Fig. 1C). The deduced product (594 amino acids; 65.2 kDa) of the 1,785-bp gad gene of strain CHA0 is 99% identical to the corresponding ORF encoding the putative Gad in P. fluorescens Pf-5 (locus tag PFL_0054; GenBank accession no. AAY95472.1). Gad of CHA0 was also found to share 89% amino acid similarity with the Gad of P. putida KT2440 encoded by the gene gadB (PP_3383) (11) and 77% with the Gad of P. aeruginosa PAO1 (PA2265 [67]). Moreover, the Gad amino acid sequence of CHA0 is still highly similar (77% identity) to that of the well-described membrane-bound Gad (GADH) of Erwinia cypripedii ATCC 29267 (GenBank accession no. U97665 [76]). The deduced product of the only partially sequenced ORFC (Fig. 1C) upstream of gad shows similarities to the C terminus of a conserved hypothetical protein of P. fluorescens Pf-5 (PFL_0053; 99% identity) and P. putida KT2440 (PP_3384; 76% identity). The adjacent downstream gene (ORFD) was partially sequenced in its 5′ region. Its deduced N terminus amino acid sequence displays similarity to the Gad cytochrome c subunit of P. fluorescens Pf-5 (PFL_0055; 99% identity) and P. putida KT2440 (PP_3382; 74% identity).
In-frame deletion mutations in the gcd and gad genes of P. fluorescens CHA0 (Table 1) were constructed as described in Materials and Methods, and the resulting Δgcd mutant CHA1196, Δgad mutant CHA1197, and Δgcd Δgad double mutant CHA1198 were compared with the parental strain for growth characteristics, acid production, phosphate solubilization, and antifungal and biocontrol activities.
Growth characteristics of Δgcd and Δgad mutants of P. fluorescens CHA0.
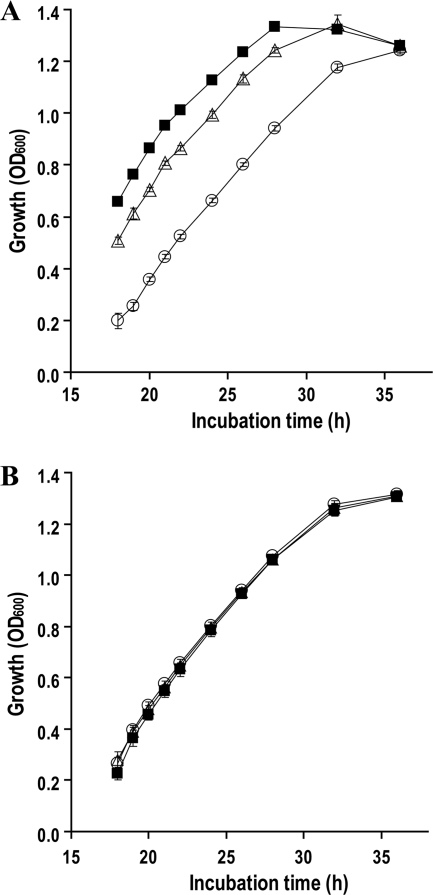
The Δgcd, Δgad, and Δgcd Δgad mutants were indistinguishable from the wild type with respect to their growth characteristics and morphologies in liquid or solid nutrient-rich media, such as KMB and LB. However, in liquid OS minimal medium amended with glucose as the sole carbon source, the loss of Gcd in strain CHA1196 markedly slowed its initial growth phase compared to that of the wild-type strain (Fig. 2A). The lack of Gad in strain CHA1197 also resulted in a slight delay in initial growth. However, the exponential growth curves of both mutants were similar to that of the wild type (Fig. 2A). All strains attained the same final cell density at the stationary growth phase (Fig. 2A). When the strains were grown in OS minimal medium amended with glycerol, no difference in growth was observed between the two mutants and wild-type CHA0 (Fig. 2B).
FIG. 2.
Growth curve of P. fluorescens wild-type CHA0 (▪); its Δgcd mutant, CHA1196 (○); and its Δgad mutant, CHA1197 (▵) in OS glucose (A) and OS glycerol (B) media at 30°C. The data represent the means (± standard errors) of six replicate cultures. The experiment was repeated twice with similar results.
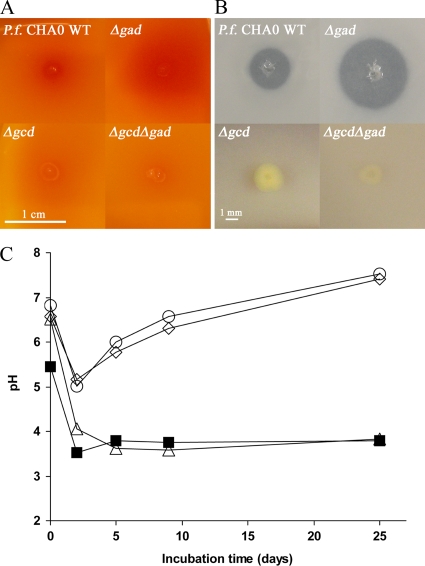
A difference in colony morphology was observed for mutants grown on NBRIP agar plates containing glucose as the sole carbon source (Fig. 3B). Colonies of wild-type CHA0 and the Δgad mutant were less than 1 mm in diameter, whereas the two other mutants, Δgcd and Δgcd Δgad, formed colonies 2 to 2.5 mm in diameter. A second morphological difference was observed on one-fifth-strength PDA. On this medium, the Δgcd mutant had a strong yellow coloration that also diffused into the agar zone surrounding the colony.
FIG. 3.
In vitro acid production (A and C) and phosphate solubilization (B) by P. fluorescens CHA0 and its Δgcd (CHA1196), Δgad (CHA1197), and Δgcd Δgad (CHA1198) mutants. The bacterial strains were grown on NBRIP agar plates with (A) or without (B) a pH indicator. (A) Acidification of the medium resulted in the formation of a blurred red halo. (B) The solubilization of tricalcium phosphate [Ca3(PO4)2] resulted in the formation of cleared zones. (A and B) The experiments were repeated twice with eight replicates per treatment. (C) Acidification of a glucose solution by addition of washed cells of wild-type CHA0 (▪), the Δgad mutant (▵), the Δgcd mutant (○), and the Δgcd Δgad double mutant (⋄). The pHs of the solutions were measured at different time intervals. The experiment was repeated twice with similar results.
Roles of gcd and gad in acid production.
A simple agar plate experiment was performed to visualize the acidification of NBRIP medium amended with a pH indicator by the wild-type CHA0 and its derivatives affected in glucose metabolism. The wild-type strain CHA0 produced a blurred red halo around the colony (Fig. 3A). A stronger and larger red halo was observed for the Δgad mutant, probably due to an accumulation of gluconic acid. In contrast, no red coloration was observed for the Δgcd mutant and the Δgcd Δgad double mutant (Fig. 3A), reflecting the defect in gluconic acid production in these strains.
In order to quantify the amounts of acid produced by the mutants, washed pellets of bacteria were inoculated in a glucose solution as described in Materials and Methods. Wild-type CHA0 and the Δgad mutant converted the glucose into gluconic acid, which strongly acidified the solution. After 5 days, the solution reached a minimal pH of 4.0 and did not change during the following days (Fig. 3C). Using the titration method of Schleissner et al. (63), the wild-type strain and the Δgad mutant were estimated to produce acid at a concentration of about 1.3 mM. As expected, the absence of Gcd in the Δgcd and the Δgcd Δgad mutants resulted in less acidification of the glucose solution, and the pH decreased to only 5.0 after 2 days. This value corresponded to an acid concentration of about 0.6 mM. The glucose solution inoculated with the Δgcd and the Δgcd Δgad mutants recovered a neutral pH during the following days (Fig. 3C). Briefly, the ability of CHA0 to acidify its environment is largely determined by its ability to produce gluconic acid from glucose.
Phosphate solubilization abilities of Δgcd and Δgad mutants of P. fluorescens CHA0.
In order to monitor the influence of Gcd and Gad in P. fluorescens CHA0 on its ability to solubilize mineral phosphate, we conducted an in vitro experiment on NBRIP agar plates with the wild type and its three mutant derivatives. Strain CHA0 produced acid on this medium (Fig. 3A) and thus was able to solubilize phosphate, which resulted in the formation of a clear halo (4.2 ± 0.8 mm in diameter) on the agar plate (Fig. 3B). The Gcd defect in strain CHA1196 and the Δgcd Δgad double mutant, CHA1198, resulted in a loss of phosphate-solubilizing ability on NBRIP agar medium. On the other hand, the absence of Gad in strain CHA1197 impaired the transformation of gluconic acid into 2-ketogluconate. As a consequence, gluconic acid accumulated in larger amounts than in the wild type (Fig. 3A), which resulted in significantly greater phosphate solubilization (halo diameter, 5.6 ± 0.9 mm) on the NBRIP agar plates (Fig. 3B). These results indicate that phosphate solubilization by CHA0 is strongly dependent on its ability to produce gluconic acid.
Effects of gcd and gad deletions on the production of antifungal compounds and on the expression of antifungal genes.
The production of the two antifungal compounds DAPG and PLT by wild-type CHA0 and its Δgcd and Δgad mutants was investigated in YM medium containing glucose or gluconate and GCM containing glycerol as carbon sources. DAPG levels obtained in YM glucose medium for the Δgcd mutant CHA1196 reached 118 μM after 1 day of incubation and were 22-fold higher than those for the wild-type CHA0 (Table 2). After 3 days, the concentration of DAPG in YM glucose medium was still 50% higher in cultures of the Δgcd mutant than in those of the wild type. DAPG production by the Δgad mutant did not differ significantly from that of the wild-type strain. Neither the wild-type CHA0 nor the Δgad mutant CHA1197 produced any PLT, the second antifungal compound studied, in YM glucose medium. The Δgcd mutant, however, was able to form PLT at concentrations of 7 to 12 μM, which did not substantially change over time (Table 2). In YM medium amended with gluconate, the Δgcd mutant produced 10 times larger amounts of DAPG (41 μM) than the Δgad mutant (4 μM) after 1 day of incubation (Table 2). In wild-type extracts, however, DAPG was not detected at this time point. After 2 and 3 days of incubation, there were no differences in the detected amounts of DAPG between the wild type and any of the mutants. Neither the wild-type CHA0 nor the Δgcd or Δgad mutant produced any PLT in YM gluconate medium (Table 2). Extractions performed with GCM cultures revealed no significant difference in DAPG or PLT production between any mutant and the wild type (data not shown). In this medium, production levels for all strains ranged between 70 and 78 μM for DAPG and between 6.8 and 7.2 μM for PLT after 3 days.
TABLE 2.
Production of the antifungal compounds DAPG and PLT by P. fluorescens CHA0 and its Δgcd and Δgad mutants
| Culture medium | Incubation time (days) | DAPG production (μM)a
|
PLT production (μM)a
|
||||
|---|---|---|---|---|---|---|---|
| CHA0 (wild type) | CHA1196 (Δgcd) | CHA1197 (Δgad) | CHA0 (wild type) | CHA1196 (Δgcd) | CHA1197 (Δgad) | ||
| YM glucose | 1 | 5.3 ± 1.2 | 118.1 ± 47.2 | 6.6 ± 1.2 | NDb | 7.5 ± 3.1 | ND |
| 2 | 51.7 ± 2.7 | 94.7 ± 28.2 | 37.7 ± 14.6 | ND | 11.9 ± 3.2 | ND | |
| 3 | 73.7 ± 9.8 | 113.3 ± 35.7 | 72.5 ± 13.8 | ND | 7.2 ± 1.4 | ND | |
| YM gluconate | 1 | ND | 40.9 ± 9.1 | 4.3 ± 5.3 | ND | ND | ND |
| 2 | 66.9 ± 13.5 | 62.1 ± 15.2 | 80.3 ± 4.9 | ND | ND | ND | |
| 3 | 18.2 ± 3.6 | 15.5 ± 1.6 | 12.0 ± 0.2 | ND | ND | ND | |
P. fluorescens wild-type CHA0 and its derivatives, CHA1196 (Δgcd) and CHA1197 (Δgad), were cultivated in 100 ml YM medium amended with 25 mM glucose or 25 mM gluconate and extracted with ethyl acetate. DAPG and PLT contents in extracts were quantified by high-performance liquid chromatography. The data represent the means (± standard errors) of three replicate cultures per treatment.
ND, not detected.
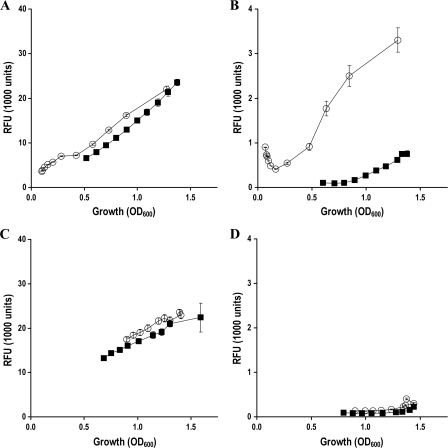
Since the Δgcd mutant, CHA1196, showed a remarkably increased production of antifungal compounds in a medium amended with glucose (Table 2), we investigated the effect of a gcd deletion on the expression of the DAPG and PLT biosynthetic genes phlA and pltA in OS minimal medium amended with either glucose or gluconate as a carbon source (Fig. 4). In OS glucose medium, the loss of Gcd in strain CHA1196 resulted in enhanced (by 20%) expression of the phlA-gfp fusion carried by pME7100 throughout the exponential growth phase in comparison to the wild-type CHA0 (Fig. 4A). The expression of the PLT biosynthetic gene pltA was markedly more affected by a lack of Gcd. The expression of the pltA-gfp fusion carried by pME7109 was strongly increased throughout growth. Expression levels of pltA in the Δgcd mutant CHA1196 were up to 10-fold higher during exponential growth and about fourfold higher at the early stationary growth phase compared to levels of the wild type (Fig. 4B). When grown in OS gluconate medium, slightly higher DAPG biosynthetic gene expression was measured in the Δgcd mutant than in the wild-type strain, CHA0 (Fig. 4C). The expression of the PLT biosynthetic gene pltA did not differ between the Δgcd mutant and the wild type and was very poor in both strains (Fig. 4D).
FIG. 4.
Kinetics of the expression of DAPG (A and C) and PLT (B and D) biosynthetic genes in growing cultures of P. fluorescens CHA0 (▪) and its Δgcd mutant, CHA1196 (○), both carrying either a phlA-gfp reporter fusion on pME7100 (A and C) or a pltA-gfp fusion on pME7109 (B and D). Gene expression is shown as relative fluorescence units (RFU), reflecting green fluorescence per bacterial density. The strains were grown in OS glucose (A and B) or OS gluconate (C and D) medium at 30°C. The data represent the means (± standard errors) of six replicate cultures. The experiment was repeated twice with similar results.
Together, these results indicate that the presence of gluconic acid, either produced by CHA0 or added to the medium, completely prevents the biosynthesis of PLT and partially prevents that of DAPG.
Biocontrol activities of P. fluorescens CHA0 and its Δgcd and Δgad mutants.
In order to determine the relative contributions of Gcd and Gad to the biocontrol ability of P. fluorescens CHA0, we investigated the abilities of the Δgcd and Δgad mutants to inhibit the growth of fungal pathogens in vitro (Table 3) and to suppress take-all of wheat (Table 4), as well as Rhizoctonia root rot of cucumber, under gnotobiotic conditions. The radial growth of G. graminis var. tritici, the causal agent of take-all, on one-fifth-strength PDA was strongly inhibited (by 72%) by strain CHA0 after 5 days (Table 3). The Δgcd mutant reduced the growth of G. graminis var. tritici significantly more strongly (by 83%) than wild-type CHA0. No difference in fungal growth reduction was observed between the Δgad mutant and the parental strain (Table 3). To test the inhibitory activity of gluconic acid on the growth of G. graminis var. tritici, we confronted the fungus growing on one-fifth-strength PDA with 10-μl drops of gluconic acid solution at different concentrations. Only the highest tested concentrations of 100 and 200 mg/ml inhibited fungal growth (data not shown).
TABLE 3.
Growth inhibition of different plant-pathogenic fungi by P. fluorescens CHA0 and its Δgcd and Δgad mutants
| Fungusa | Radial growth of fungal colony (radius in mm)b
|
|||
|---|---|---|---|---|
| No bacteria added | CHA0 (wild type) | CHA1196 (Δgcd) | CHA1197 (Δgad) | |
| G. graminis var. tritici | 29a | 8b | 5c | 7b |
| M. grisea | 17a | 12b | 9c | 13b |
| R. solani | 43a | 26b | 24b | 25b |
G. graminis var. tritici, M. grisea, and R. solani were inoculated in the center of a one-fifth-strength PDA plate. P. fluorescens wild-type CHA0 and its derivatives, CHA1196 (Δgcd) and CHA1197 (Δgad), were streaked out in a square of 5.5-cm lateral length around the fungal plugs and grown for 5 days at 24°C.
The data are means of eight replicates. Means for the same fungus within the same row followed by different letters are significantly different at a P value of ≤0.05 according to Fisher's least significant difference test. The experiment was repeated twice with similar results.
TABLE 4.
Suppression of take-all of wheat by P. fluorescens CHA0 and its Δgcd and Δgad mutants
| Treatmenta | Expt 1
|
Expt 2
|
Expt 3
|
Avg of expt 1-3 (ratio to wild type) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Disease severity
|
Root fresh wt (g) | CFU/g of root (log10) | Disease severity
|
Root fresh wt (g) | CFU/g of root (log10) | Disease severity
|
Root fresh wt (g) | CFU/g of root (log10) | |||||
| %b | Ratio to wild typec | % | Ratio to wild type | % | Ratio to wild type | ||||||||
| Control | 0ad | NA | 0.72a | ND | 0a | NA | 0.73a | ND | 0a | NA | 0.77a | ND | NA |
| G. graminis var. tritici | 81.3b | 6.5 | 0.43b | ND | 93.1b | 1.2 | 0.41b | ND | 89.4b | 2.2 | 0.46b | ND | 3.3 |
| G. graminis var. tritici + CHA0 | 12.5c | 1.0 | 0.61a | 7.84a | 79.0c | 1.0 | 0.52b | 8.58a | 40.6cd | 1.0 | 0.59bc | 8.31a | 1.0 |
| G. graminis var. tritici + Δgcd | 9.1c | 0.73 | 0.64a | 7.48a | 47.9d | 0.61 | 0.74a | 8.34b | 30.0d | 0.74 | 0.63c | 8.36a | 0.69e |
| G. graminis var. tritici + Δgad | 47.5d | 3.80 | 0.58ab | 7.77a | 70.8c | 0.90 | 0.54b | 8.47ab | 50.1c | 1.23 | 0.51bc | 8.31a | 1.98 |
Plants were grown under gnotobiotic conditions for 18 days in Perlite: without addition of G. graminis var. tritici (control), with addition of G. graminis var. tritici, and with addition of G. graminis var. tritici and the wild-type CHA0, its Δgcd mutant CHA1196, or its Δgad mutant CHA1197.
Plant disease severity is expressed as a percentage of infected roots as detailed in Materials and Methods.
Values are the ratio of the disease severity of the respective treatment to the disease severity of the treatment with wild-type CHA0.
Means for the same experiment within the same column followed by a different letter are significantly different (P ≤ 0.05) according to Fisher's least significant difference test. NA, calculation of the ratio was not applicable; ND, no bacteria were detected.
Significantly different from the treatments with wild-type strain CHA0 (Fisher's least significant difference test; P ≤ 0.05).
A similar effect of the gcd deletion was observed on the inhibition of M. grisea. The Δgcd mutant, but not the Δgad mutant, reduced the growth of M. grisea significantly more (47% growth reduction) than the wild type (29% growth reduction) (Table 3). Interestingly, no such difference in fungal growth inhibition between the wild type and the Δgcd mutant was observed in the assay with R. solani (Table 3).
In summary, the loss of Gcd activity in CHA0 resulted in increased inhibition of two out of three tested fungal plant pathogens on agar plates.
The results for the three repetitions of the take-all biocontrol assay are presented individually in Table 4. Due to differences in general take-all severity levels among the three experiments, the protection provided by CHA0 was also subject to high variation and ranged from 15% (experiment 2, with the highest take-all incidence in the G. graminis var. tritici control) to 85% (experiment 1, with the lowest take-all incidence in the G. graminis var. tritici control) reduction of disease severity (Table 4). Root weights were increased by 27% to 41% when plants were protected against G. graminis var. tritici by the wild-type strain CHA0. When wheat plants were protected by the Δgcd mutant, the disease severity index was even lower compared to wild-type treatments in all three independent experiments (the Δgcd/CHA0 ratio ranged from 0.61 to 0.74). This effect was statistically significant in only one (experiment 2) out of three experiments. However, when the ratios of disease severities between Δgcd and CHA0 treatments in all three experiments were analyzed together (average Δgcd/CHA0 ratio, 0.69), the decrease in biocontrol ability due to lack of Gcd was significant (Table 4). Since after 2 weeks G. graminis var. tritici infected only the uppermost 2 cm of the root system, better disease protection did not necessarily result in much higher root weights. Therefore, roots protected by the Δgcd mutant had only slightly higher fresh weights than roots protected by CHA0 (Table 4). In all three experiments, take-all disease severity was significantly higher in the Δgad mutant treatments than in the Δgcd treatments. In the absence of G. graminis var. tritici, plant growth was not significantly affected by the addition of any of the P. fluorescens strains (data not shown). Root colonization by the tested bacterial strains ranged between 7.48 and 7.84 log10 CFU/g of root in experiment 1 and between 8.31 and 8.58 log10 CFU/g of root in experiments 2 and 3. No significant difference between the individual strains was observed, with the exception of experiment 2, where root colonization by the Δgcd mutant was about two times lower than that of the wild-type strain (Table 4). Likewise, there was also no difference in root colonization levels between plants with or without pathogen infection (data not shown).
In the cucumber-R. solani disease suppression assay, both the Δgcd and the Δgad mutants provided plant protection similar to that of wild-type CHA0. The average root fresh weight of healthy control plants was 3.1 ± 0.2 g, and that of plants attacked with R. solani was 0.6 ± 0.4 g. When protected by either the wild type or one of the two mutants, root fresh weights of cucumber plants ranged between 2.9 and 3.1 g (data not shown).
In summary, the deletion of the gcd gene in CHA0 resulted in improved protection of wheat against take-all, but not of cucumber against R. solani.
DISCUSSION
In the present study, we constructed in-frame deletions in the gcd (CHA1196) and gad (CHA1197) genes in P. fluorescens CHA0 in order to study the influence of gluconic acid production on different characteristics of the strain. Our results show that alteration of gluconic acid production in the two mutants has an impact not only on the metabolism of the bacterium, but also on its interaction with fungal pathogens.
First, the loss of Gcd activity strongly impaired strain CHA0 in its ability to produce acids. Mutants impaired in Gcd activity (CHA1196 and CHA1198) were unable to acidify the surrounding solid medium (Fig. 3A) or showed a strongly reduced ability to produce acids in glucose solution (Fig. 3C). We suggest that these effects were due to the loss of Gcd activity, which resulted in an inability to transform glucose into gluconic acid. Interestingly, the Δgcd mutant and the Δgcd Δgad double mutant, both impeded in acid production, grew faster and formed bigger colonies on NBRIP agar plates than the wild-type strain CHA0 and the Δgad mutant. We suppose that the difference in colony development between the strains is due to the lack of gluconic acid production. On a solid medium containing only glucose as the carbon source (NBRIP medium) (Fig. 3A and B), gluconic acid formation by the wild-type strain and the Δgad mutant resulted in strong acidification of the medium, which may have hampered colony development.
It has been established that in pseudomonads glucose can enter the cell either by a direct oxidative or by a phosphorylative pathway (Fig. 1A) (10, 12, 40). Along the direct oxidative route, glucose is transformed to gluconic acid and 2-ketogluconate in the periplasmic space by Gcd and Gad, which are located in the inner membrane. Both products can be transported through the inner membrane and are then transformed to 6-phosphogluconate (6-PGA) in the cytoplasm. Along the phosphorylative route, glucose is directly transported into the cell and converted to 6-PGA via glucokinase (Glk) and glucose-6-phosphate dehydrogenase (Zwf) in the cytoplasm (Fig. 1A). Berka et al. (6) showed for Burkholderia cepacia that Gcd-deficient strains were blocked in the direct oxidative pathway and glucose was forced to be catabolized entirely by the phosphorylative route. In the same manner, a gcd isogenic mutant of P. putida KT2440 did not produce any gluconate (12), and this indicated that 6-PGA was produced only from glucose-6-phosphate. The phosphorylative pathway is also the only route available in cells during growth under anaerobic (denitrifying) conditions because Gcd activity is not expressed (25, 26, 51).
Interestingly, Fuhrer et al. (17) showed that for P. fluorescens 52-1C, extracellular glucose was mainly converted to gluconate and 2-ketogluconate, suggesting that the oxidative pathway is preferred to the phosphorylative pathway for this strain. Since we have identified gcd and gad genes in P. fluorescens CHA0, it is likely that in our strain also, glucose can pass through the inner membrane in three different forms: as glucose itself or as its oxidized forms gluconic acid and 2-ketogluconate (Fig. 1A). It is evident that the proportion of each form varies between the wild type and the different mutants investigated in this study. In a medium with glucose as the sole carbon source, the Δgcd mutant can take up only the glucose form because the oxidative pathway, and thus production of gluconic acid and 2-ketogluconate, is blocked. This may slow down growth and can explain the delayed initial growth observed for the Δgcd mutant in OS glucose medium (Fig. 2A). Such a reduced growth rate of a gcd mutant was also observed for P. putida KT2440 by del Castillo and coworkers (12). Even a partial inhibition of the oxidative pathway slightly delayed growth, as observed for the Δgad mutant, which is blocked in 2-ketogluconate formation. When mutants were grown in OS glycerol medium (Fig. 2B), no difference in growth was observed between any of the mutants and the wild type. As in pseudomonads glycerol enters the cell by a different route (10, 40), mutations in the gcd and gad genes did not affect bacterial growth in this medium.
Several bacterial genera are known to hydrolyze mineral phosphates to organic phosphates and thus increase phosphate availability for plants (60). The bacterial production of organic acids as the main mechanism of phosphate solubilization has been well documented. Goldstein (19, 20) has proposed that direct periplasmic oxidation of glucose to gluconic acid forms the metabolic basis of the mineral phosphate solubilization in some gram-negative bacteria. Gluconic acid seems to be the most frequent organic acid involved in mineral phosphate solubilization by microorganisms such as Pseudomonas spp. and Penicillium spp. (27, 71), Pantoea agglomerans (41), B. cepacia (21), and P. aeruginosa (7). In this study, we provide evidence that gluconic acid is responsible for phosphate solubilization by P. fluorescens wild-type CHA0, as well as by the Δgad mutant (Fig. 3). The degree of phosphate solubilization is correlated with the degree of acid production. The Δgad mutant, which produced larger amounts of acid (Fig. 3A) than the wild type, also showed better ability to solubilize phosphate (Fig. 3B). In contrast, the Δgcd mutant blocked in gluconic acid production was impaired in phosphate solubilization on NBRIP agar plates (Fig. 3B).
Our results show that the absence of Gcd in the Δgcd mutant enhances the production of both DAPG and PLT in a glucose-based medium (Table 2 and Fig. 4). Similar observations were made previously with a PQQ mutant of P. fluorescens CHA0. PQQ is a cofactor of Gcd (14), and a lack of PQQ in CHA0 leads to inactivation of Gcd and to overproduction of PLT (64). Because the Δgcd mutant is not able to produce gluconic acid, we attributed the change in the strain's antifungal compound production to the absence of this metabolite. So far, little is known about the specific role of gluconic acid in the regulation of antifungal compounds. Several groups have shown that glucose metabolism and carbon sources in general play a role in the regulation of antifungal metabolites. James and Gutterson (28), for example, have postulated that glucose and gluconate might have important roles in the regulation of antibiotic biosynthesis in P. fluorescens strain HV37a, which shows biocontrol activity against Pythium ultimum. Gutterson (22) proposed that glucose may block antibiotic production through repression of dehydrogenases that catalyze glucose oxidation, a reaction that transfers electrons from the enzyme cofactor PQQ to electron transport. Duffy and Défago (13) and other authors showed that carbon sources found in root exudates have different influences on the production of various antifungal compounds in P. fluorescens. In strain CHA0, production of PLT is stimulated by glycerol and the production of DAPG by glucose (13, 46, 66). Glucose, however, represses PLT production (13, 66). The stimulation of PLT production by glycerol and its inhibition by glucose were also described for P. fluorescens Pf-5 by Kraus and Loper and Nowak-Thompson et al. (38, 55). All these studies suggest that glucose interferes with PLT production in CHA0 and the closely related strain Pf-5. However, we postulate here that it is the presence of gluconic acid rather than that of glucose that inhibits PLT production in CHA0 for several reasons. First, the lack of Gcd activity and subsequently the lack of gluconic acid production enable a Δgcd mutant to synthesize PLT in a medium containing glucose (Table 2), in which the wild type cannot produce the antifungal compound. Moreover, in a medium with glucose as the sole carbon source, a PLT biosynthetic gene was expressed at a much higher level in the Δgcd mutant than in the wild type (Fig. 4B). Finally, in OS gluconate medium, the expression of PLT is not only completely repressed in the wild type, but also in the Δgcd mutant (Fig. 4D). How exactly gluconic acid interacts with the PLT biosynthetic pathway still remains unclear; however, the interaction seems to occur at the transcriptional level. It is also unclear why the absence of Gcd results in enhanced DAPG production and DAPG biosynthetic gene expression (Table 2 and Fig. 4A). The lack of gluconic acid formation cannot explain this observation because gluconic acid inhibits neither DAPG production (Table 2) nor the expression of the DAPG biosynthetic gene phlA (Fig. 4C). The only slight difference we have observed in the gluconate-based medium in comparison to the glucose-based medium is a delay of DAPG production in the wild-type strain. Further studies are needed to assess the exact role of Gcd for the regulation of antifungal metabolite production in P. fluorescens CHA0. In the Δgcd mutant, the oxidative pathway is thought to be completely inhibited, and we suppose that all the glucose is forced to enter the cell via the phosphorylative pathway. It is likely that this leads to many modifications within the cell and to the accumulation of different compounds, which might impact the regulation of secondary-metabolite production.
The lack of Gcd in strain CHA1196 resulted in 30% better protection (in terms of disease severity) of wheat against take-all disease than the wild type (Table 4). We suggest that the improved ability to suppress the disease is due to enhanced production of antifungal compounds by the Δgcd mutant, which results in enhanced antibiosis. This suggestion is supported by the fact that the Δgcd mutant produces larger amounts of DAPG and PLT in vitro and indeed shows an enhanced ability to inhibit the growth of G. graminis var. tritici on agar plates (Tables 2 and 3). As a major part of root exudates are composed of glucose (29, 42, 69, 70), it is tempting to assume that more DAPG and PLT is produced in the rhizospheres of plants inoculated with the Δgcd mutant. Since no significant difference in root colonization was observed for the tested bacterial strains, the improved plant protection could not be attributed to higher rhizosphere colonization by the Δgcd mutant. However, the increased ability of the Δgcd mutant to suppress disease seems to be pathogen specific. In the cucumber-R. solani system, the lack of Gcd did not result in improved plant protection. Also, on agar plates, increased inhibition by the Δgcd mutant was observed only for G. graminis var. tritici and M. grisea and not for R. solani. The reasons for such specificity remain unclear; however, different sensitivities of individual plant-pathogenic fungi to DAPG and PLT have been observed. Maurhofer et al. (45) showed that G. graminis var. tritici is more sensitive to DAPG and PLT than R. solani. Even different isolates of G. graminis vary in sensitivity to DAPG (49). Moreover, biocontrol activity is not only defined by antibiosis (24), and for the protection of cucumber against R. solani, other mechanisms might also be important. The direct role of gluconic acid production in the biocontrol activity of CHA0 remains unclear at present. Gluconic acid itself has been proposed to be an antifungal agent and to be involved in the biocontrol ability of an Australian nonfluorescent, non-DAPG-producing pseudomonad (30). A transposon insertion mutant of strain AN5 which had lost its ability to produce gluconic acid was partially impaired in its ability to inhibit G. graminis var. tritici in vitro (30, 53), yet the genetic basis of the mutation was not described. The authors suggested that gluconic acid production is involved in the suppression of take-all disease by the bacterium, probably by lowering the pH in the wheat rhizosphere. In our study, we could not demonstrate that gluconic acid produced by CHA0 is involved in inhibition of G. graminis var. tritici or suppression of take-all. First, the lack of gluconic acid production in the Δgcd mutant did not result in reduced inhibition of G. graminis var. tritici (Table 3). Still, it is possible that the effect of loss of gluconic acid production was masked by the effect of increased antifungal compound production in the Δgcd mutant. Second, the Δgad mutant, which produces the same DAPG and PLT levels as the wild type (Table 2) but increased amounts of gluconic acid (Fig. 3A), did not either better inhibit G. graminis var. tritici in vitro (Table 3) or better suppress take-all of wheat (Table 4). However, we do not know whether the Δgad mutant in fact produces higher levels of gluconic acid in the rhizosphere. Moreover, the G. graminis var. tritici isolate used in this study appears not to be as sensitive to gluconic acid as the isolate used by Kaur et al. (30). The growth of the isolate used by Kaur et al. was already inhibited at a concentration of 6 mg/ml gluconic acid, whereas growth of our G. graminis var. tritici isolate was inhibited only at concentrations of 100 mg/ml and higher. Therefore, we suggest that the direct role of gluconic acid produced by a biocontrol bacterium in the suppression of take-all may also be dependent on the sensitivity of the fungus to the acid.
Taken together, our results demonstrate that gluconic acid production is involved in the regulation of antimicrobial compound production in a rhizosphere-associated pseudomonad and as a consequence indirectly modulates the bacterium's biocontrol activity.
Acknowledgments
We thank Geneviève Défago, Plant Pathology, Institute of Integrative Biology, Swiss Federal Institute of Technology, for advice on the take-all disease infection protocol.
We gratefully acknowledge financial support from the Swiss National Science Foundation (project no. 3100A0-105881).
Footnotes
Published ahead of print on 17 April 2009.
REFERENCES
- 1.Ameyama, M., E. Shinagawa, K. Matsushita, and O. Adachi. 1981. d-Glucose dehydrogenase of Gluconobacter suboxydans: solubilization, purification and characterization. Agric. Biol. Chem. 45:851-861. [Google Scholar]
- 2.Babu-Khan, S., T. C. Yeo, W. L. Martin, M. R. Duron, R. D. Rogers, and A. H. Goldstein. 1995. Cloning of a mineral phosphate-solubilizing gene from Pseudomonas cepacia. Appl. Environ. Microbiol. 61:972-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baehler, E., M. Bottiglieri, M. Péchy-Tarr, M. Maurhofer, and C. Keel. 2005. Use of green fluorescent protein-based reporters to monitor balanced production of antifungal compounds in the biocontrol agent Pseudomonas fluorescens CHA0. J. Appl. Microbiol. 99:24-38. [DOI] [PubMed] [Google Scholar]
- 4.Baehler, E., P. de Werra, L. Y. Wick, M. Péchy-Tarr, S. Mathys, M. Maurhofer, and C. Keel. 2006. Two novel MvaT-like global regulators control exoproduct formation and biocontrol activity in root-associated Pseudomonas fluorescens CHA0. Mol. Plant-Microbe Interact. 19:313-329. [DOI] [PubMed] [Google Scholar]
- 5.Bangera, M. G., and L. S. Thomashow. 1996. Characterization of a genomic locus required for synthesis of the antibiotic 2,4-diacetylphloroglucinol by the biological control agent Pseudomonas fluorescens Q2-87. Mol. Plant-Microbe Interact. 9:83-90. [DOI] [PubMed] [Google Scholar]
- 6.Berka, T. R., P. Allenza, and T. G. Lessie. 1984. Hyperinduction of enzymes of the phosphorylative pathway of glucose dissimilation in Pseudomonas cepacia. Curr. Microbiol. 11:143-148. [Google Scholar]
- 7.Buch, A., G. Archana, and G. Naresh Kumar. 2008. Metabolic channeling of glucose towards gluconate in phosphate-solubilizing Pseudomonas aeruginosa P4 under phosphorus deficiency. Res. Microbiol. 159:635-642. [DOI] [PubMed] [Google Scholar]
- 8.Caesar-Tonthat, T. C., F. V. Kloeke, G. G. Geesey, and J. M. Henson. 1995. Melanin production by a filamentous soil fungus in response to copper and localization of copper sulfide by sulfide-silver staining. Appl. Environ. Microbiol. 61:1968-1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cook, R. J. 1993. Making greater use of introduced microorganisms for biological control of plant pathogens. Annu. Rev. Phytopathol. 31:53-80. [DOI] [PubMed] [Google Scholar]
- 10.Cuskey, S. M., and P. V. Phibbs. 1985. Chromosomal mapping of mutations affecting glycerol and glucose catabolism in Pseudomonas aeruginosa PAO. J. Bacteriol. 162:872-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.del Castillo, T., E. Duque, and J. L. Ramos. 2008. A set of activators and repressors control peripheral glucose pathways in Pseudomonas putida to yield a common central intermediate. J. Bacteriol. 190:2331-2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.del Castillo, T., J. L. Ramos, J. J. Rodríguez-Herva, T. Fuhrer, U. Sauer, and E. Duque. 2007. Convergent peripheral pathways catalyze initial glucose catabolism in Pseudomonas putida: genomic and flux analysis. J. Bacteriol. 189:5142-5152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duffy, B. K., and G. Défago. 1999. Environmental factors modulating antibiotic and siderophore biosynthesis by Pseudomonas fluorescens biocontrol strains. Appl. Environ. Microbiol. 65:2429-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duine, J. A. 1991. Quinoproteins: enzymes containing the quinonoid cofactor pyrroloquinoline quinone, topaquinone or tryptophan-tryptophan quinone. Eur. J. Biochem. 200:271-284. [DOI] [PubMed] [Google Scholar]
- 15.Duque, E., A. J. Molina-Henares, J. de la Torre, M. A. Molina-Henares, T. del Castillo, J. Lam, and J. L. Ramos. 2007. Towards a genome-wide mutant library of Pseudomonas putida strain KT2440, p. 227-251. In J. L. Ramos and A. Filloux (ed.), Pseudomonas, vol. V. Springer, Dordrecht, The Netherlands. [Google Scholar]
- 16.Eisenberg, R. C., S. J. Butters, S. C. Quay, and S. B. Friedman. 1974. Glucose uptake and phosphorylation in Pseudomonas fluorescens. J. Bacteriol. 120:147-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fuhrer, T., E. Fischer, and U. Sauer. 2005. Experimental identification and quantification of glucose metabolism in seven bacterial species. J. Bacteriol. 187:1581-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaur, A. C., and K. P. Ostwal. 1972. Influence of phosphate dissolving bacilli on yield and phosphate uptake of wheat crop. Indian J. Exp. Biol. 10:393-394. [Google Scholar]
- 19.Goldstein, A. H. 1994. Involvement of the quinoprotein glucose dehydrogenase in the solubilization of exogenous mineral phosphates by gram-negative bacteria, p. 197-203. In A. Torriani-Gorini, E. Yagil, and S. Silver (ed.), Phosphate in microorganisms: cellular and molecular biology. ASM Press, Washington, DC.
- 20.Goldstein, A. H. 1995. Recent progress in understanding the molecular genetics and biochemistry of calcium phosphate solubilization by gram negative bacteria. Biol. Agric. Horticult. 12:185-193. [Google Scholar]
- 21.Goldstein, A. H., R. D. Rogers, and G. Mead. 1993. Mining by microbe. Bio/Technology 11:1250-1254. [Google Scholar]
- 22.Gutterson, N. 1990. Microbial fungicides: recent approaches to elucidating mechanisms. Crit. Rev. Biotechnol. 10:69-91. [Google Scholar]
- 23.Haas, D., and G. Défago. 2005. Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat. Rev. Microbiol. 3:307-319. [DOI] [PubMed] [Google Scholar]
- 24.Haas, D., and C. Keel. 2003. Regulation of antibiotic production in root-colonizing Pseudomonas spp. and relevance for biological control of plant disease. Annu. Rev. Phytopathol. 41:117-153. [DOI] [PubMed] [Google Scholar]
- 25.Hunt, J. C., and P. V. Phibbs, Jr. 1981. Failure of Pseudomonas aeruginosa to form membrane-associated glucose dehydrogenase activity during anaerobic growth with nitrate. Biochem. Biophys. Res. Commun. 102:1393-1399. [DOI] [PubMed] [Google Scholar]
- 26.Hunt, J. C., and P. V. Phibbs, Jr. 1983. Regulation of alternate peripheral pathways of glucose catabolism during aerobic and anaerobic growth of Pseudomonas aeruginosa. J. Bacteriol. 154:793-802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Illmer, P., and F. Schinner. 1992. Solubilization of inorganic phosphates by microorganisms isolated from forest soils. Soil Biol. Biochem. 24:389-395. [Google Scholar]
- 28.James, D. W., and N. I. Gutterson. 1986. Multiple antibiotics produced by Pseudomonas fluorescens HV37a and their differential regulation by glucose. Appl. Environ. Microbiol. 52:1183-1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kamilova, F., L. V. Kravchenko, A. I. Shaposhnikov, T. Azarova, N. Makarova, and B. Lugtenberg. 2006. Organic acids, sugars, and l-tryptophane in exudates of vegetables growing on stonewool and their effects on activities of rhizosphere bacteria. Mol. Plant-Microbe Interact. 19:250-256. [DOI] [PubMed] [Google Scholar]
- 30.Kaur, R., J. Macleod, W. Foley, and M. Nayudu. 2006. Gluconic acid: an antifungal agent produced by Pseudomonas species in biological control of take-all. Phytochemistry 67:595-604. [DOI] [PubMed] [Google Scholar]
- 31.Keel, C., U. Schnider, M. Maurhofer, C. Voisard, J. Laville, U. Burger, P. Wirthner, D. Haas, and G. Défago. 1992. Suppression of root diseases by Pseudomonas fluorescens CHA0: importance of the bacterial secondary metabolite 2,4-diacetylphloroglucinol. Mol. Plant-Microbe Interact. 5:4-13. [Google Scholar]
- 32.Keel, C., C. Voisard, C. H. Berling, G. Kahr, and G. Défago. 1989. Iron sufficiency, a prerequisite for the suppression of tobacco black root-rot by Pseudomonas fluorescens strain CHA0 under gnotobiotic conditions. Phytopathology 79:584-589. [Google Scholar]
- 33.Keel, C., P. Wirthner, T. Oberhänsli, C. Voisard, U. Burger, D. Haas, and G. Défago. 1990. Pseudomonads as antagonists of plant pathogens in the rhizosphere: role of the antibiotic 2,4-diacetylphloroglucinol in the suppression of black root-rot of tobacco. Symbiosis 9:327-341. [Google Scholar]
- 34.Kim, C. H., S. H. Han, K. Y. Kim, B. H. Cho, Y. H. Kim, B. S. Koo, and Y. C. Kim. 2003. Cloning and expression of pyrroloquinoline quinone (PQQ) genes from a phosphate-solubilizing bacterium Enterobacter intermedium. Curr. Microbiol. 47:457-461. [DOI] [PubMed] [Google Scholar]
- 35.King, E. O., M. K. Ward, and D. E. Raney. 1954. Two simple media for the demonstration of pyocyanin and fluorescin. J. Lab. Clin. Med. 44:301-307. [PubMed] [Google Scholar]
- 36.Kloepper, J. W., and M. N. Schroth. 1978. Plant growth-promoting rhizobacteria on radishes, p. 879-882. In Proceedings of the 4th International Conference on Plant Pathogenic Bacteria, vol. II. Gilbert-Clay, Tours, France. [Google Scholar]
- 37.Krasilnikov, N. A. 1961. On the role of soil bacteria in plant nutrition. J. Gen. Appl. Microbiol. 7:128-144. [Google Scholar]
- 38.Kraus, J., and J. E. Loper. 1995. Characterization of a genomic region required for production of the antibiotic pyoluteorin by the biological control agent Pseudomonas fluorescens Pf-5. Appl. Environ. Microbiol. 61:849-854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Larkin, M. A., G. Blackshields, N. P. Brown, R. Chenna, P. A. McGettigan, H. McWilliam, F. Valentin, I. M. Wallace, A. Wilm, R. Lopez, J. D. Thompson, T. J. Gibson, and D. G. Higgins. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947-2948. [DOI] [PubMed] [Google Scholar]
- 40.Lessie, T. G., and P. V. Phibbs, Jr. 1984. Alternative pathways of carbohydrate utilization in pseudomonads. Annu. Rev. Microbiol. 38:359-388. [DOI] [PubMed] [Google Scholar]
- 41.Liu, S. T., L. Y. Lee, C. Y. Tai, C. H. Hung, Y. S. Chang, J. H. Wolfram, R. Rogers, and A. H. Goldstein. 1992. Cloning of an Erwinia herbicola gene necessary for gluconic acid production and enhanced mineral phosphate solubilization in Escherichia coli HB101: nucleotide sequence and probable involvement in biosynthesis of the coenzyme pyrroloquinoline quinone. J. Bacteriol. 174:5814-5819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lugtenberg, B. J. J., L. V. Kravchenko, and M. Simons. 1999. Tomato seed and root exudate sugars: composition, utilization by Pseudomonas biocontrol strains and role in rhizosphere colonization. Environ. Microbiol. 1:439-446. [DOI] [PubMed] [Google Scholar]
- 43.Matsushita, K., and M. Ameyama. 1982. d-Glucose dehydrogenase from Pseudomonas fluorescens, membrane-bound. Methods Enzymol. 89:149-154. [DOI] [PubMed] [Google Scholar]
- 44.Matsushita, K., E. Shinagawa, and M. Ameyama. 1982. d-Gluconate dehydrogenase from bacteria, 2-keto-d-gluconate-yielding, membrane-bound. Methods Enzymol. 89:187-193. [DOI] [PubMed] [Google Scholar]
- 45.Maurhofer, M., C. Keel, D. Haas, and G. Défago. 1995. Influence of plant species on disease suppression by Pseudomonas fluorescens strain CHA0 with enhanced antibiotic production. Plant Pathol. 44:40-50. [Google Scholar]
- 46.Maurhofer, M., C. Keel, D. Haas, and G. Défago. 1994. Pyoluteorin production by Pseudomonas fluorescens strain CHA0 is involved in the suppression of Pythium damping-off of cress but not of cucumber. Eur. J. Plant Pathol. 100:221-232. [Google Scholar]
- 47.Maurhofer, M., C. Keel, U. Schnider, C. Voisard, D. Haas, and G. Défago. 1992. Influence of enhanced antibiotic production in Pseudomonas fluorescens strain CHA0 on its disease suppressive capacity. Phytopathology 82:190-195. [Google Scholar]
- 48.Maurhofer, M., C. Reimmann, P. Schmidli-Sacherer, S. Heeb, D. Haas, and G. Défago. 1998. Salicylic acid biosynthetic genes expressed in Pseudomonas fluorescens strain P3 improve the induction of systemic resistance in tobacco against tobacco necrosis virus. Phytopathology 88:678-684. [DOI] [PubMed] [Google Scholar]
- 49.Mazzola, M., D. K. Fujimoto, L. S. Thomashow, and R. J. Cook. 1995. Variation in sensitivity of Gaeumannomyces graminis to antibiotics produced by fluorescent Pseudomonas spp. and effect on biological control of take-all of wheat. Appl. Environ. Microbiol. 61:2554-2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McIntire, W., T. P. Singer, M. Ameyama, O. Adachi, K. Matsushita, and E. Shinagawa. 1985. Identification of the covalently bound flavins of d-gluconate dehydrogenases from Pseudomonas aeruginosa and Pseudomonas fluorescens and of 2-keto-d-gluconate dehydrogenase from Gluconobacter melanogenus. Biochem. J. 231:651-654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mitchell, C. G., and E. A. Dawes. 1982. The role of oxygen in the regulation of glucose metabolism, transport and the tricarboxylic-acid cycle in Pseudomonas aeruginosa. J. Gen. Microbiol. 128:49-59. [DOI] [PubMed] [Google Scholar]
- 52.Nautiyal, C. S. 1999. An efficient microbiological growth medium for screening phosphate solubilizing microorganisms. FEMS Microbiol. Lett. 170:265-270. [DOI] [PubMed] [Google Scholar]
- 53.Nayudu, M., K. A. E. Groom, J. Fernance, P. T. W. Wong, and K. Turnbull. 1994. The genetic nature of biological control of the take-all fungal pathogen by Pseudomonas, p. 122-124. In M. H. Ryder, P. M. Stephen, and G. D. Bowen (ed.), Improving plant productivity with rhizosphere bacteria. CSIRO, Glen Osmond, South Australia, Australia.
- 54.Nelson, K. E., C. Weinel, I. T. Paulsen, R. J. Dodson, H. Hilbert, V. A. P. M. dos Santos, D. E. Fouts, S. R. Gill, M. Pop, M. Holmes, L. Brinkac, M. Beanan, R. T. DeBoy, S. Daugherty, J. Kolonay, R. Madupu, W. Nelson, O. White, J. Peterson, H. Khouri, I. Hance, P. C. Lee, E. Holtzapple, D. Scanlan, K. Tran, A. Moazzez, T. Utterback, M. Rizzo, K. Lee, D. Kosack, D. Moestl, H. Wedler, J. Lauber, D. Stjepandic, J. Hoheisel, M. Straetz, S. Heim, C. Kiewitz, J. Eisen, K. N. Timmis, A. Düsterhöft, B. Tümmler, and C. M. Fraser. 2002. Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environ. Microbiol. 4:799-808. [DOI] [PubMed] [Google Scholar]
- 55.Nowak-Thompson, B., S. J. Gould, J. Kraus, and J. E. Loper. 1994. Production of 2,4-diacetylphloroglucinol by the biocontrol agent Pseudomonas fluorescens Pf-5. Can. J. Microbiol. 40:1064-1066. [Google Scholar]
- 56.Paulsen, I. T., C. M. Press, J. Ravel, D. Y. Kobayashi, G. S. A. Myers, D. V. Mavrodi, R. T. DeBoy, R. Seshadri, Q. H. Ren, R. Madupu, R. J. Dodson, A. S. Durkin, L. M. Brinkac, S. C. Daugherty, S. A. Sullivan, M. J. Rosovitz, M. L. Gwinn, L. W. Zhou, D. J. Schneider, S. W. Cartinhour, W. C. Nelson, J. Weidman, K. Watkins, K. Tran, H. Khouri, E. A. Pierson, L. S. Pierson, L. S. Thomashow, and J. E. Loper. 2005. Complete genome sequence of the plant commensal Pseudomonas fluorescens Pf-5. Nat. Biotechnol. 23:873-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Péchy-Tarr, M., M. Bottiglieri, S. Mathys, K. B. Lejbølle, U. Schnider-Keel, M. Maurhofer, and C. Keel. 2005. RpoN (σ54) controls production of antifungal compounds and biocontrol activity in Pseudomonas fluorescens CHA0. Mol. Plant-Microbe Interact. 18:260-272. [DOI] [PubMed] [Google Scholar]
- 58.Qing, G. L., L. C. Ma, A. Khorchid, G. V. T. Swapna, T. K. Mal, M. M. Takayama, B. Xia, S. Phadtare, H. P. Ke, T. Acton, G. T. Montelione, M. Ikura, and M. Inouye. 2004. Cold-shock induced high-yield protein production in Escherichia coli. Nat. Biotechnol. 22:877-882. [DOI] [PubMed] [Google Scholar]
- 59.Raaijmakers, J. M., M. Vlami, and J. T. de Souza. 2002. Antibiotic production by bacterial biocontrol agents. Antonie van Leeuwenhoek 81:537-547. [DOI] [PubMed] [Google Scholar]
- 60.Rodríguez, H., and R. Fraga. 1999. Phosphate solubilizing bacteria and their role in plant growth promotion. Biotechnol. Adv. 17:319-339. [DOI] [PubMed] [Google Scholar]
- 61.Rozen, S., and H. J. Skaletsky. 2000. Primer3 on the WWW for general users and for biologist programmers, p. 365-386. In S. Krawetz and S. Misener (ed.), Bioinformatics methods and protocols: methods in molecular biology. Humana Press, Totowa, NJ. [DOI] [PubMed]
- 62.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 63.Schleissner, C., A. Reglero, and J. M. Luengo. 1997. Catabolism of d-glucose by Pseudomonas putida U occurs via extracellular transformation into d-gluconic acid and induction of a specific gluconate transport system. Microbiology 143:1595-1603. [DOI] [PubMed] [Google Scholar]
- 64.Schnider, U., C. Keel, C. Voisard, G. Défago, and D. Haas. 1995. Tn5-directed cloning of pqq genes from Pseudomonas fluorescens CHA0: mutational inactivation of the genes results in overproduction of the antibiotic pyoluteorin. Appl. Environ. Microbiol. 61:3856-3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schnider-Keel, U., K. B. Lejbølle, E. Baehler, D. Haas, and C. Keel. 2001. The sigma factor AlgU (AlgT) controls exopolysaccharide production and tolerance towards desiccation and osmotic stress in the biocontrol agent Pseudomonas fluorescens CHA0. Appl. Environ. Microbiol. 67:5683-5693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schnider-Keel, U., A. Seematter, M. Maurhofer, C. Blumer, B. Duffy, C. Gigot-Bonnefoy, C. Reimmann, R. Notz, G. Défago, D. Haas, and C. Keel. 2000. Autoinduction of 2,4-diacetylphloroglucinol biosynthesis in the biocontrol agent Pseudomonas fluorescens CHA0 and repression by the bacterial metabolites salicylate and pyoluteorin. J. Bacteriol. 182:1215-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. L. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. S. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. W. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 68.Stutz, E. W., G. Défago, and H. Kern. 1986. Naturally occurring fluorescent pseudomonads involved in suppression of black root rot of tobacco. Phytopathology 76:181-185. [Google Scholar]
- 69.Vančura, V. 1964. Root exudates of plants I. Analysis of root exudates of barley and wheat in their initial phases of growth. Plant Soil 21:231-248. [Google Scholar]
- 70.Vančura, V., and A. Hovadík. 1965. Root exudates of plants II. Composition of root exudates of some vegetables. Plant Soil 22:21-32. [Google Scholar]
- 71.Vassilev, N., M. Fenice, and F. Federici. 1996. Rock phosphate solubilization with gluconic acid produced by immobilized Penicillium variabile P16. Biotechnol. Tech. 10:585-588. [Google Scholar]
- 72.Vassilev, N., M. Vassileva, and I. Nikolaeva. 2006. Simultaneous P-solubilizing and biocontrol activity of microorganisms: potentials and future trends. Appl. Microbiol. Biotechnol. 71:137-144. [DOI] [PubMed] [Google Scholar]
- 73.Vieira, J., and J. Messing. 1991. New pUC-derived cloning vectors with different selectable markers and DNA replication origins. Gene 100:189-194. [DOI] [PubMed] [Google Scholar]
- 74.Yamada, M., S. Asaoka, M. H. Saier, and Y. Yamada. 1993. Characterization of the gcd gene from Escherichia coli K-12 W3110 and regulation of its expression. J. Bacteriol. 175:568-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yamada, M., M. Elias, K. Matsushita, C. T. Migita, and O. Adachi. 2003. Escherichia coli PQQ-containing quinoprotein glucose dehydrogenase: its structure comparison with other quinoproteins. Biochim. Biophys. Acta 1647:185-192. [DOI] [PubMed] [Google Scholar]
- 76.Yum, D. Y., Y. P. Lee, and J. G. Pan. 1997. Cloning and expression of a gene cluster encoding three subunits of membrane-bound gluconate dehydrogenase from Erwinia cypripedii ATCC 29267 in Escherichia coli. J. Bacteriol. 179:6566-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zogg, H., and J. A. Amiet. 1980. Laboratory studies on decline with different foot and root-rot pathogens of wheat. J. Phytopathol. 97:193-213. [Google Scholar]