Abstract
Pseudomonas mandelii liquid cultures were studied to determine the effect of pH and temperature on denitrification gene expression, which was quantified by quantitative reverse transcription-PCR. Denitrification was measured by the accumulation of nitrous oxide (N2O) in the headspace in the presence of acetylene. Levels of gene expression of nirS and cnorB at pH 5 were 539-fold and 6,190-fold lower, respectively, than the levels of gene expression for cells grown at pH 6, 7, and 8 between 4 h and 8 h. Cumulative denitrification levels were 28 μmol, 63 μmol, and 22 μmol at pH 6, 7, and 8, respectively, at 8 h, whereas negligible denitrification was measured at pH 5. P. mandelii cells grown at 20°C and 30°C exhibited 9-fold and 94-fold increases in levels of cnorB expression between 0 h and 2 h, respectively, and an average 17-fold increase in levels of nirS gene expression. In contrast, induction of cnorB and nirS gene expression for P. mandelii cells grown at 10°C did not occur in the first 4 h. Levels of cumulative denitrification at 10 h were 6.6 μmol for P. mandelii cells grown at 10°C and 20°C and 30 μmol for cells grown at 30°C. Overall, levels of cnorB and nirS expression were relatively insensitive to pH values over the range of pH 6 to 8 but were substantially reduced at pH 5, whereas gene expression was sensitive to temperature, with induction and time to achieve maximum gene expression delayed as the temperature decreased from 30°C. Low pH and temperature negatively affected denitrification activity.
Denitrification is a respiratory microbiological process in which nitrate (NO3−) or nitrite (NO2−) is reduced to gaseous nitric oxide (NO), nitrous oxide (N2O), or molecular nitrogen (N2) under oxygen-limited conditions (33). Denitrification can result in substantial gaseous losses of N, an important plant nutrient, from agricultural fields (7, 14). N2O depletes stratospheric ozone and contributes to global warming (28). An understanding of the environmental controls on denitrifier activity is essential for comprehending the spatial and temporal regulation of denitrification within agricultural production systems.
Denitrification is carried out by various microorganisms belonging to several genera and species of bacteria (3, 4, 27, 33). The strain of Pseudomonas mandelii used in this study was a dominant culturable denitrifier isolated from an agricultural field in a potato production system in New Brunswick, Canada (4).
Several environmental factors control the process of denitrification. These include oxygen availability, substrate availability (i.e., NO2− and NO3−), pH, temperature, and the abundance and species of denitrifiers. The availability of a substrate, the absence of oxygen, and the presence of active denitrifiers are the main controlling factors (24). However, pH and temperature also play a role in influencing denitrifier growth, metabolism, denitrification gene expression, and, subsequently, denitrification rate. A recent review of environmental controls on denitrifying communities and denitrification rates identified the need to link pH and temperature with denitrification gene expression as a step toward an understanding of the relationship between denitrifier community composition and function (29). Most research related to the effect of temperature and pH on denitrification has focused on denitrification rates in soils (9, 15, 16, 19, 22). Several studies have established that denitrification rates tend to decrease at low soil pH values (15, 16, 20). Parkin et al. (16) previously demonstrated a twofold decrease in denitrification rate and a threefold decrease in denitrification enzyme activity when soil pH decreased from pH 6.02 to 4.08, suggesting that a prolonged exposure to low soil pH selected a denitrifier population that was more adapted to the low-pH environment, and subsequently, N2O reduction by the acid-tolerant population was insignificant compared to N2O production rates. Numerous studies have demonstrated that soil pH changed the concentrations of denitrification intermediates and products (8, 12, 19). There are several studies that have investigated the effect of temperature on denitrification rates in soil. Stanford et al. (22) previously established that within a limited temperature range of 15°C to 35°C, the temperature coefficient of denitrification (Q10), was about 2. This value translates to a twofold increase in denitrification for every 10°C increase in temperature in soil. Fischer and Whalen (9) also evaluated the capability of a soil microbial community to denitrify in response to temperature and calculated Q10 values of 1.6 and 2.8 in the temperature intervals of 7°C to 20°C and 20°C to 30°C, respectively. Holtan-Hartwig et al. (10) previously suggested that low temperature (0°C) exerts a challenge to denitrifying communities and affects N2O breakdown more than the reduction of other N-oxides of the denitrification processes.
There is limited research on the effect of pH and temperature on denitrification gene expression in pure cultures. Most studies have focused on identifying the optimal pH for denitrification in terms of enzyme activity, denitrification intermediates and products (11, 26), and induction of mRNA (1). In pure cultures of Pseudomonas species, the optimum pH for denitrification, based on denitrification activity, was found to be in the range of pH 7.0 to 7.5 (11, 25). The accumulations of denitrification intermediates in pure cultures of Paracoccus denitrificans differed at acidic and alkaline pH values (26). The suboptimal pH of 6.8 inhibited denitrification activity in P. denitrificans but did not affect mRNA induction (1). There is no research that has studied the effect of temperature on denitrification gene expression.
This study determined the effect of pH and temperature on the expression of the nirS and cnorB genes and denitrification activity (i.e., N2O emissions in the presence of acetylene) in a pure culture of P. mandelii. It was hypothesized that pH and temperature would differentially affect nirS and cnorB gene expression and decrease denitrification activity under suboptimal conditions.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
P. mandelii strain PD30 was cultured in tryptic soy broth (TSB) medium (Difco, Becton, Dickinson, and Company, Sparks, MD) at 30°C and maintained on tryptic soy agar plates at 4°C for short-term use. For long term use, P. mandelii cultures were stored with 15% (wt/vol) sterile glycerol at −80°C.
Experimental setup to test for pH effects.
P. mandelii PD30 cultures were grown aerobically by inoculating a single colony into 200 ml of TSB medium in a 500-ml Erlenmeyer flask and shaking overnight at 150 rpm and 30°C (New Brunswick Co., Inc., NJ). The culture was centrifuged at 3,600 × g for 10 min at 25°C, resuspended in 200 ml of sterile phosphate-buffered saline (PBS), and shaken aerobically for 1 h in PBS to metabolize carbon sources and traces of metabolites that accumulated in the TSB medium which may influence gene expression. The culture was centrifuged for 10 min and resuspended in 4 ml of PBS. TSB medium was prepared, and the pHs were adjusted to pH 5.00, 6.00, 7.00, and 8.00, respectively. The media were prepared and autoclaved, and the pHs were verified to be 5.06, 6.04, 6.97 and 7.71, respectively, before inoculation.
A randomized complete block design with four treatments and three replicates, and with repeated sampling over time, was used to test for the effect of pH on denitrification gene expression and activity in P. mandelii. Treatments were four levels of pH (pH 5, 6, 7, and 8), and sampling was done every hour up to 8 h, followed by an additional sampling after 24 h. The experiment was conducted twice.
Flask setup and induction of denitrification conditions were performed as previously described (18). A 0-h sample for all treatments and replicates was obtained from P. mandelii cultures by removing a 3-ml culture aliquot from each flask to analyze gene expression and concentrations of NO3− and NO2− and by removing a 12-ml headspace gas aliquot for the N2O concentration prior to the addition of substrate. After sampling, a sufficient volume of gas mixture (10% acetylene-90% helium) was added to maintain the flask at atmospheric pressure. Potassium nitrate was added at a final concentration of 0.1% (wt/vol) for all treatments. The flasks were incubated for 24 h in a 30°C incubator with shaking at 85 rpm.
At each sampling, optical density (OD) measurements were obtained, NO3− and NO2− analyses were performed, and samples were prepared for RNA extraction as previously described (18). The 12-ml headspace gas sampled at each time point was injected into evacuated Exetainers (Labco Limited, United Kingdom) for subsequent N2O and carbon dioxide (CO2) analyses. Cumulative CO2 accumulation was used as a measure of respiration activity.
Experimental setup to test for temperature effects.
A randomized complete block design with three treatments and three replicates, and with repeated sampling over time, was used to test for the effect of temperature on denitrification gene expression and activity in P. mandelii. Treatments consisted of three temperatures (10°C, 20°C, and 30°C), and sampling was conducted hourly to 10 h, followed by an additional sampling at 24 h. The experiment was conducted twice. Three P. mandelii PD30 cultures were established as described above and incubated overnight at 10°C, 20°C, or 30°C (Innova 4240 illuminated refrigerated incubator shaker; New Brunswick Scientific). The P. mandelii cultures were shaken aerobically for 1 h in PBS at 10°C, 20°C, or 30°C, as described above. The cells were inoculated in triplicate into TSB (pH 7.0) to an OD at 600 nm of 0.1. Flasks were sealed and evacuated, and anaerobic conditions were established as described previously (18). Potassium nitrate was added at a final concentration of 0.1% (wt/vol) for all treatments. The flasks were incubated at 10°C, 20°C, or 30°C with shaking at 85 rpm for 24 h. At each sampling, a 3-ml culture aliquot and a 12-ml headspace gas sample were withdrawn using sterile syringes. OD measurements and NO3−, NO2−, and N2O analyses were performed as described below.
Design of P. mandelii nirS quantitative PCR primers.
The sequence of the taxonomically well-defined P. mandelii nirS gene (GenBank accession number DQ518190) was aligned with similar nirS genes available for several Pseudomonas spp. including P. lini (accession number DQ518197), P. migulae (accession number DQ518195), and Pseudomonas spp. isolated from soil (accession numbers DQ518187, DQ518196, DQ518186, and DQ518185) and from uncultured clones (accession numbers AJ811516 and AJ811504) to find unique, conserved regions using MegAlign (Lasergene 7). P. lini and P. migulae have been isolated from soil (bulk and rhizosphere) and have been shown to be taxonomically close to P. mandelii (6). Primers were selected based on standard conditions for real-time quantitative PCR using PrimerSelect (Lasergene 7). The specificities of the primers were tested and verified with P. mandelii genomic DNA in a PCR. The product was sequenced and submitted to BLAST (NCBI). The PCR product had a 100% sequence identity to P. mandelii nirS (accession number DQ518190). The product was cloned the using TOPO kit (Invitrogen, Burlington, Ontario, Canada), sequenced, and verified using BLAST analysis (S. Henderson, unpublished data).
Gene expression quantification.
The RNeasy minikit (Qiagen Inc., Mississauga, Ontario, Canada) was used for total RNA isolation. Modifications to the extraction protocol were described previously (18). RNA was quantified using Ribogreen RNA quantitation reagent (Molecular Probes, Eugene, OR). Gene expression quantification was performed using the Bio-Rad iCycler iQ detection system (Bio-Rad Laboratories, Mississauga, Ontario, Canada). Previously designed primers from our research group targeting the cnorB region were used (5). Conditions for one-step quantitative PCR targeting the P. mandelii cnorB gene were the same as those described previously (5).
The level of P. mandelii nirS gene expression was quantified using one-step quantitative PCR with 50 ng of total RNA template, 12.5 μl of 2× master mix from the Qiagen QuantiTect SYBR green reverse transcription-PCR kit, 300 nM of forward primer (5′-ACCGCGGCCAACAACTCCAACA-3′), 500 nM of reverse primer (5′-CCGCCCTGGCCCTTGAGC-3′), and 0.25 μl of reverse transcriptase enzyme in a final volume of 25 μl. Thermal cycling conditions were as follows: 30 min at 50°C and 15 min at 95°C followed by 40 repeats of 15 s at 95°C, 30 s at 68.4°C, 30 s at 72°C, and 15 s at 83°C. Data collection was performed during the last step of each cycle at a temperature of 83°C. In a 40-cycle PCR, a no-template control was undetected.
An external standard curve was constructed using linear regression to obtain a line of best fit for the quantification of nirS gene copy number and transcripts. P. mandelii nirS primers were used to produce nirS PCR products that were then cloned into TOPO according to the manufacturer's instructions (Invitrogen, Burlington, Ontario, Canada). Plasmid DNA was extracted using a plasmid minikit (Qiagen Inc., Mississauga, Ontario, Canada). The plasmid was linearized by digestion with SacI (Roche, Laval, Quebec, Canada) and heat shocking to deactivate the enzymes. The linearized plasmid was quantified using Picogreen (Molecular Probes, Eugene, OR), and the size of the nirS insert was used to calculate the copy number. The curve was linear over a dilution range of 10−3 to 10−9 and sensitive to at least 10 copies of nirS per reaction.
During each run, standard dilutions of digested plasmid carrying a copy of the nirS gene cloned into the PCR2.1-TOPO vector were included to allow for gene quantification. A positive control consisting of P. mandelii genomic DNA isolated as outlined in the DNeasy tissue kit (Qiagen Inc., Mississauga, Ontario, Canada) and quantified by the Quant-iT Picogreen dsDNA assay kit (Molecular Probes, Eugene, OR) was used in each quantitative reverse transcription-PCR run.
NO3−, NO2−, N2O, and CO2 analyses.
Frozen supernatant samples were analyzed for NO3−-N and NO2−-N concentrations at the Soil and Nutrient Laboratory (Laboratory Services Division, University of Guelph, Guelph, Ontario, Canada). N2O and CO2 analyses of headspace gas were performed by gas chromatography as previously described (4). Calculated values of cumulative denitrification (i.e., cumulative N2O emissions in the presence of acetylene) and respiration (i.e., cumulative CO2 emissions) were corrected for dissolved N2O or CO2 in the flask and for changes in pressure and gas concentrations attributed to sampling.
Statistical methods.
All parameters were tested for normality using the univariate function in the SAS System for Windows (version 8; SAS Institute Inc., Cary, NC), and a log transformation was performed when required. Analysis of variance was performed using the mixed procedure of SAS. The statistical model treated duplicate experiments as blocks in order to pool data from the two experiments, and the repeated function was used to account for repeated sampling of flasks over time. Where there were significant differences between pH and temperature treatments over time, treatment means were compared by LSMEANS (P < 0.05) with Tukey's adjustment. Relationships among measured parameters were explored using Pearson correlations on a per-flask basis. Treatment means and standard errors presented in figures were calculated from nontransformed data.
RESULTS
Effects of pH.
P. mandelii cultures grown at pH 5 had no significant change in cell density from 0 to 24 h and showed an average OD at 600 nm of 0.15 (Fig. 1). There was no significant increase in cell density in P. mandelii cultures grown at pH 6, 7, and 8, between 0 and 4 h, with the exception of pH 7, for which the cell density significantly increased between 2 and 4 h. The cell density subsequently increased rapidly after 4 h to reach averages of 0.27 for P. mandelii cultures grown at pH 5 and pH 6 and 0.46 for P. mandelii cultures grown at pH 7 and pH 8, at 6 h. At 8 h, all four treatments were significantly different from one another, with cell densities decreasing in the following order: pH 7 > pH 8 > pH 6 > pH 5. At 24 h, cell density was significantly increased compared to that at 8 h in P. mandelii cultures grown at pH 6 (OD of 0.89) and pH 7 and 8 (average OD of 0.99). In all four treatments, the growth medium became slightly more alkaline at 24 h, with pH values of 5.3, 6.9, 7.3, and 7.8, respectively, than at the beginning of the experiment, most likely due to the accumulation of metabolic by-products.
FIG. 1.

P. mandelii growth in TSB medium at pH 5 (⧫), pH 6 (▪), pH 7(⋄), or pH 8 (□) as measured using OD measurements obtained at 600 nm. Error bars are ±1 standard error of the mean (SEM) (n = 6).
Gene expression levels of nirS and cnorB were differentially affected by pH treatment at different time points in P. mandelii cultures. At pH 5, P. mandelii cultures demonstrated a ninefold increase in the level of nirS expression at 2 h, with an average of 5.9 × 107 transcripts/μg RNA, compared with that at the start of the incubation (Fig. 2A). Similarly, at pH 5, there was a 16-fold increase in the level of cnorB expression at 2 h, where the number of transcripts increased from 3.6 × 104 transcripts/μg RNA to 6.0 × 105 transcripts/μg RNA (Fig. 2B). Gene expression levels for both nirS and cnorB in cultures grown at pH 5 subsequently declined at 4 h and were unchanged to the 24-h time point. In contrast, P. mandelii cultures grown at pH 6, 7, and 8 were not significantly different from one another for both nirS and cnorB genes and exhibited a 171-fold induction of the nirS gene from 0 h to 2 h, with an average of 3.6 × 109 transcripts/μg RNA at 2 h. For the cnorB gene, a 427-fold increase in the level of gene expression at pH 6, 7, and 8 occurred from 0 to 2 h, where levels of cnorB transcripts increased from an average of 2.9 × 105 transcripts/μg RNA to 1.0 × 108 transcripts/μg RNA. nirS and cnorB gene transcripts in cultures grown at pH 6, 7, and 8 remained unchanged from 2 h to 8 h (averages of 2.2 × 109 transcripts/μg RNA and 1.9 × 108 transcripts/μg RNA, respectively) and then decreased to 8.4 × 107 transcripts/μg RNA and 4.9 × 106 transcripts/μg RNA, respectively, at 24 h.
FIG. 2.
nirS (A) and cnorB (B) gene expression in P. mandelii cultures grown in TSB medium at pH 5(⧫), pH 6 (▪), pH 7(⋄), or pH 8 (□), supplemented with 0.1% potassium nitrate. Error bars are ±1 SEM (n = 6). nirS transcript numbers were calculated from the line of best fit described by the linear equations y = −2.65x + 46.2 (r2 = 0.99) for the first experiment and y = −3.01x + 47.6 (r2 = 0.99) for the second experiment. For cnorB, the line of best fit was described by the linear equations y = −3.27x + 35.6 (r2 = 0.98) for the first experiment and y = −2.77x + 34.3 (r2 = 0.99) for the second experiment.
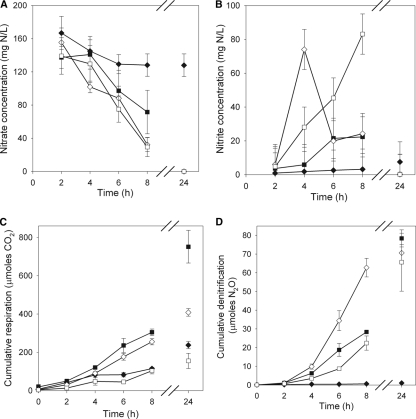
NO3− concentrations were not significantly different among pH treatments at 2 h (Fig. 3A). Although a small amount of nitrate seemed to be reduced at pH 5, there was no significant decrease in the NO3− concentration between 2 and 24 h, with an average value of 139 mg NO3−-N/liter. At 8 h, NO3− concentrations declined to 72 mg NO3−-N/liter for P. mandelii cultures grown at pH 6 and 31 mg NO3−-N/liter for cells grown at pH 7 and 8. By 24 h, NO3− had been reduced to negligible concentrations in P. mandelii cultures grown at pH 6, 7, and 8.
FIG. 3.
NO3− (A) and NO2− (B) concentrations in liquid culture, cumulative respiration (i.e., CO2 accumulation) (C), and cumulative denitrification (i.e., N2O accumulation in the presence of acetylene) (D) in the headspace in TSB medium supporting the growth of denitrifying P. mandelii cells grown at pH 5(⧫), pH 6 (▪), pH 7(⋄), or pH 8 (□) in the presence of NO3−. Error bars are ±1 SEM (n = 6).
There was no significant difference in NO2− concentrations among time points for the pH 5 and 6 treatments, with average NO2− concentrations of 7.7 mg NO2−-N/liter at pH 5 and pH 6 from 2 to 8 h (Fig. 3B). Maximum values of NO2− concentrations were measured at different time points for the treatments at pH 7 and 8. At 4 h, the P. mandelii culture grown at pH 7 had significantly higher NO2− concentrations (74 mg NO2−-N/liter) than did cultures grown at pH 5, 6, and 8 (average, 12 mg NO2−-N/liter). When cultures were grown at pH 8, NO2− concentrations significantly increased at 6 h, and cultures had a higher NO2− concentration at 8 h than with all other treatments, at 83 mg NO2−-N/liter. Cultures grown at pH 5, 6, and 7 were not significantly different at 8 h (average, 16.6 mg NO2−-N/liter), and by 24 h, small concentrations of NO2− (average, 2.0 mg NO2−-N/liter) were measured for all four treatments.
Cumulative respiration did not increase significantly over the first 4 h of the incubation for any treatment (Fig. 3C). Significant increases in the level of cumulative respiration were measured at 6 h for pH 6 and 7 and 24 h for pH 5 and 8. At 8 h, cumulative respiration did not differ between the pH 5 and pH 8 treatments, with an average of 109 μmol, which was significantly lower than that for the pH 6 and 7 treatments, with an average of 280 μmol. The level of cumulative respiration at 24 h in P. mandelii cultures grown at pH 6 (751 μmol) was much greater than that at pH 7 (408 μmol). In comparison, the level of cumulative respiration at 24 h for pH 5 and 8 averaged 196 μmol.
Cumulative denitrification was differentially expressed in various pH treatments in P. mandelii cultures, and the response varied over time. At 8 h, the level of cumulative denitrification did not differ between the pH 6 and pH 8 treatments, with an average of 25 μmol, which was significantly lower than that of the pH 7 treatment, with an average of 63 μmol (Fig. 3D). The level of cumulative denitrification subsequently increased over time for P. mandelii cultures grown at pH 6, 7, and 8, reaching an average value of 72 μmol at 24 h. In contrast, the pH 5 treatment demonstrated negligible cumulative denitrification over time, with an average of 0.52 μmol over the 24-h incubation (Fig. 3D).
The increase in cumulative denitrification over time (Fig. 3D) coincided with a decrease in the NO3− concentration (Fig. 3A), except for the pH 5 treatment, where negligible denitrification was measured. The magnitude of the NO3− loss was proportional to the increase in NO2− at 8 h, which was the time at which significant differences in both NO3− and NO2− concentrations among treatments were observed. Gene expression at 2 h, the time of maximum expression, and denitrification at 8 h, the time of significant differences in cumulative denitrification, demonstrated correlation constants of 0.47 (P = 0.0234) for the nirS gene and 0.53 (P = 0.0092) for the cnorB gene.
Effects of temperature.
The density of P. mandelii cells grown at 10°C did not significantly increase from 0 to 10 h (average OD of 0.20) and then increased significantly at 24 h (OD of 0.77) (Fig. 4). Cell densities of P. mandelii cells grown at 20°C and 30°C demonstrated identical growth patterns; there was no significant change in P. mandelii growth from 0 h to 4 h, after which cell growth increased significantly from an average OD of 0.24 at 4 h to an OD of 0.63 at 10 h and to a final OD of 1.2 at 24 h.
FIG. 4.

P. mandelii growth in TSB medium at 10°C (○), 20°C (▪), or 30°C (▵) as measured using OD measurements obtained at 600 nm. Error bars are ±1 SEM (n = 6).
A P. mandelii culture grown at 10°C demonstrated a small (1.5-fold) increase in the level of nirS expression between 0 h and 2 h, and thereafter, the level of nirS gene expression increased more rapidly, with 2.5 × 107 transcripts/μg RNA at 4 h and an average of 2.0 × 108 transcripts/μg RNA between 6 and 10 h (Fig. 5A). P. mandelii cells grown at 20°C and 30°C demonstrated a 17-fold increase in nirS gene expression between 0 and 2 h, reaching an average of 9.4 × 107 transcripts/μg RNA at 2 h, and maintained an average of 3.6 × 108 transcripts/μg RNA between 2 h and 10 h. The induction of nirS gene expression was more rapid in P. mandelii cells grown at 20°C and 30°C than in cells grown at 10°C; however, by 6 h, there was no significant difference among treatments (Fig. 5A). At 24 h, the 10°C treatment showed significantly higher levels of nirS gene expression (5.9 × 108 transcripts/μg RNA) than did the 20°C and 30°C treatments (average of 6.0 × 106 transcripts/μg RNA).
FIG. 5.
nirS (A) and cnorB (B) gene expression in P. mandelii cultures grown in TSB medium at 10°C (○), 20°C (▪), or 30°C (▵), supplemented with 0.1% potassium nitrate. Error bars are ±1 SEM (n = 6). nirS transcript numbers were calculated from the line of best fit described by the linear equations y = −2.90x + 44.1 (r2 = 0.99) for the first experiment and y = −3.39x + 46.0 (r2 = 0.99) for the second experiment. For cnorB, the line of best fit was described by the linear equations y = −4.20x + 44.4 (r2 = 0.99) for the first experiment and y = −3.27x + 38.8 (r2 = 0.99) for the second experiment.
At 10°C, there was a delay in the induction of cnorB gene for the first 4 h (Fig. 5B). The level of cnorB gene expression then increased to 1.1 × 109 transcripts/μg RNA at 8 h and remained significantly unchanged from 8 h to 24 h, with an average of 1.3 × 109 transcripts/μg RNA. At 20°C, P. mandelii cells demonstrated a ninefold increase in the level of cnorB gene expression between 0 and 2 h, where the cnorB transcript number reached 4.6 × 107 transcripts/μg RNA at 2 h, increased to 2.6 × 109 transcripts/μg RNA at 4 h, and stayed constant until 10 h. The level of gene expression of cnorB subsequently declined to 3.2 × 107 transcripts/μg RNA at 24 h. P. mandelii cells grown at 30°C demonstrated a 94-fold increase in the level of cnorB gene expression between 0 and 2 h, reaching 4.6 × 108 transcripts/μg RNA at 2 h (Fig. 5B). Subsequently, there was no significant change in cnorB gene expression levels for the 30°C treatment between 2 h and 10 h, with an average of 9.5 × 108 transcripts/μg RNA. The level of cnorB expression at 2 h for the 30°C treatment was 10-fold higher than that for the 20°C treatment and 73-fold higher than that for the 10°C treatment. Although the induction of cnorB gene expression was delayed in the 10°C treatment, by 8 h, there was no significant difference among all three temperature treatments, with an average of 1.5 × 109 transcripts/μg RNA. At 24 h, the level of cnorB expression significantly decreased compared with that at 10 h in P. mandelii cells grown at 20°C and 30°C, with an average of 4.0 × 107 transcripts/μg RNA, while the level of cnorB expression in the 10°C treatment remained similar to that of gene expression observed at 8 h, with 1.0 × 109 transcripts/μg RNA.
NO3− concentrations were not significantly different among treatments between 2 and 4 h (Fig. 6A). Nonlimiting NO3− concentrations were measured in P. mandelii cultures grown at 10°C for 10 h, with an average of 133 mg NO3−-N/liter (Fig. 6A), which subsequently declined to 4.2 mg NO3−-N/liter at 24 h. A significant decline in NO3− concentrations was measured at 6 and 8 h in P. mandelii cultures grown at 20°C and 30°C, respectively. At 10 h, NO3− concentrations declined to an average of 40.9 mg NO3−-N/liter and finally to a negligible concentration of 0.1 mg NO3−-N/liter at 24 h in P. mandelii cultures grown at 20°C and 30°C.
FIG. 6.
NO3− (A) and NO2− (B) concentrations in liquid culture, cumulative respiration (i.e., CO2 accumulation) (C), and cumulative denitrification (i.e., N2O accumulation in the presence of acetylene) (D) in the headspace in TSB supporting growth of denitrifying P. mandelii cultures grown at 10°C (○), 20°C (▪), or 30°C (▵) in the presence of NO3−. Error bars are ±1 SEM (n = 6).
NO2− concentrations were not affected by temperature between 2 and 4 h for any treatment (Fig. 6B). Significant increases in NO2− concentrations were measured at 10, 6, and 8 h in P. mandelii cultures grown at 10°C, 20°C, and 30°C, respectively. At 10 h, NO2− concentrations increased to averages of 35.2 mg NO2−-N/liter for P. mandelii cultures grown at 10°C and 30°C and 114 mg NO2−-N/liter for P. mandelii cultures grown at 20°C. P. mandelii cultures grown at 20°C and 30°C showed essentially complete reduction of NO2− by 24 h (average of 0.12 mg NO2−-N/liter), unlike P. mandelii culture grown at 10°C (96 mg NO2−-N/liter). Maximum values of NO2− concentrations were measured at different time points among temperature treatments. The loss of NO3− was proportional to the increase in the NO2− concentration at 10 h, which was the time at which significant differences in both NO3− and NO2− concentrations among treatments were observed.
Temperature did not affect respiration between 0 and 6 h, with average emissions of 6.7 μmol, among treatments (Fig. 6C). The level of cumulative respiration was significantly higher in P. mandelii cells grown at 20°C and 30°C (average, 52.9 μmol) than in cells grown at 10°C (average, 20.4 μmol). At 24 h, a significant increase in the level of cumulative respiration was measured for P. mandelii cultures grown at 30°C only, with 383 μmol CO2. Cultures grown at 10°C and 20°C maintained an average of 75 μmol CO2 at 24 h. Temperature coefficients (Q10 values) for CO2 of 2.2 between 10°C and 20°C and 0.9 between 20°C and 30°C at 10 h were obtained.
Temperature did not affect denitrification between 0 and 8 h, with average emissions of about 1.9 μmol among treatments (Fig. 6D). P. mandelii cultures grown at 10°C demonstrated no significant increase in levels of cumulative denitrification between 0 and 24 h, with average emissions of 3.3 μmol (Fig. 6D). The levels of denitrification increased significantly at 24 and 10 h in P. mandelii cultures grown at 20°C and 30°C, respectively. At 24 h, the level of denitrification increased to an average of 69 μmol in cells grown at 20°C and 30°C. There was no correlation between cnorB gene expression at 8 h, the time that it took to reach maximal denitrification gene expression for all three temperature treatments, and denitrification at 10 h, which coincided with high denitrification activity (R = 0.11; P = 0.6529), or between nirS gene expression at 8 h and denitrification at 10 h (R = 0.35; P = 0.1612). Temperature coefficients (Q10 values) for N2O of 3.2 between 10°C and 20°C and 3.0 between 20°C and 30°C at 10 h were obtained.
DISCUSSION
In our study, levels of nirS and cnorB gene expression were not affected by pH treatment in P. mandelii cultures grown at pH 6, 7, and 8. A significant increase in levels of nirS and cnorB gene transcripts was observed between 0 and 2 h and was comparable to the increase in cnorB expression levels previously observed for P. mandelii cultures when NO3− was present in the medium (18). Very limited information is available on the effect of variable pH on denitrification gene expression in pure culture. pH 5 was the least favorable for denitrification gene expression. Although a 16-fold induction of cnorB and a 9-fold induction of nirS were observed at 2 h, negligible denitrification occurred. This observation is in agreement with data from research where an inhibition of denitrification activity was observed in P. denitrificans at a suboptimal pH of 6.8, which, however, conflicts with the conclusion from that same study, where no effect on mRNA induction was observed at pH 6.8 (1). The difference in the level of induction of denitrification genes observed in this study also suggests that pH had a stronger effect in negatively regulating nirS expression, resulting in a lower level of induction than that for cnorB expression.
Differences in NO3− utilization and NO2− accumulation were measured between 2 and 8 h in P. mandelii cultures grown at pH 6, 7, and 8. These differences may be attributed to the effect of pH on denitrification enzymes, particularly NO3− reductase and NO2− reductase (NIR). The magnitude of the NO3− loss was proportional to the increase in NO2− concentrations observed at 8 h among treatments. This observation implies that most of the NO3− added to the system was channeled through the denitrification process. The rate of NO3− utilization also varied among treatments, with a sharper decrease in NO3− concentrations observed at 8 h for P. mandelii cultures grown at pH 7 and 8 than for cells grown at pH 6. The timing of maximum NO2− accumulations also differed among treatments, with a higher level of NO2− accumulation at 8 h in cultures grown at pH 8 than at 4 h in cultures grown at pH 7. Similar to this study, NO2− accumulations in P. denitrificans cultures grown under denitrification conditions in a continuous-culture bioreactor were measured (1). Subsequently, NO2− accumulation was proposed to be the cause of the inactivation of the NIR enzyme in P. denitrificans through the formation of toxic nitrous acid (HNO2), since the expression of the NO2− reductase gene was not affected by suboptimal pH. A separate study investigating the accumulation of denitrification intermediates in Pseudomonas alcaligenes and Pseudomonas fluorescens concluded that nitrite accumulation was dependent on the relative rates of nitrate and nitrite reduction (2). Nitrite accumulation may also be attributed to a lag in the synthesis of nitrite reductase (23, 30) or nitrate inhibition of NO reductase (17). In our study, it was hypothesized that enzyme activity was affected over a pH range of 6 to 8, thus causing differences in NO3− utilization and NO2− accumulation among pH treatments over time.
The rate of N2O emissions varied during the incubation, as indicated by higher denitrification activity at 6 h and 8 h for P. mandelii cultures grown at pH 7 than for other pH treatments. Since gene expression was not affected at pH 6, 7, and 8, these differences are likely due to an effect of pH on nitrate reductase, NIR, or N2O reductase activity, resulting in variable denitrification. P. mandelii cultures grown at pH 5 produced negligible denitrification activity compared with that for all other pH treatments. Although there have been no previous studies that investigated the effect of pH on denitrification activity in pure culture, as measured by N2O accumulations using an acetylene block, there is evidence that the overall denitrification rate and denitrification enzyme activity were reduced by an acidic soil pH (16, 19, 21). Previous studies established that neutral-to-alkaline conditions were optimal for denitrification in cultures of Pseudomonas species (20, 25).
It is interesting that although negligible denitrification was observed in P. mandelii cultures grown at pH 5, 237 μmol CO2 was produced at 24 h. Our NO3 data reveal a decrease in the NO3 concentration in cultures grown at pH 5, even though the numbers from 2 to 24 h were not statistically different. This leads to the possibility that a small amount of nitrate is consumed, and in the process, energy and CO2 are produced. Moreover, due to the fact that CO2 solubility increases with increasing pH, one would expect more CO2 in the headspace of P. mandelii cultures grown at pH 5.
The induction of gene expression and the time to reach maximum expression were delayed as the temperature decreased from 30°C. P. mandelii cultures grown at 10°C demonstrated a long lag phase in growth and a subsequent delay in nirS and cnorB induction of gene expression compared with those of P. mandelii cells grown at 20°C and 30°C. P. mandelii cells grown at 30°C demonstrated a 10-fold-greater induction of cnorB expression at 2 h than did cells grown at 20°C; however, once maximum cnorB gene expression was reached at 4 h, there was no effect of temperature on cnorB gene expression between both treatments. The induction of nirS gene expression also did not differ between 20°C and 30°C treatments. Although the induction of nirS and cnorB gene expression was delayed in the 10°C treatment, by 8 h, all three temperature treatments led to the expression of maximum nirS and cnorB transcripts, thus indicating that temperature did not inhibit the transcription of denitrification genes. Temperature did, however, have a differential effect on denitrification gene expression, as is evident by differences in levels and timings of nirS and cnorB gene induction among temperature treatments. This observation suggests an independent regulation of the nirS and cnorB genes in P. mandelii. There have been no studies to date that quantify the effect of temperature on denitrification gene expression.
NO3− utilization, NO2− accumulation, and denitrification activity differed among temperature treatments. The decrease in NO3− concentrations was proportional to the increase in NO2− concentrations in P. mandelii cultures grown at 10°C, 20°C, and 30°C, suggesting N flux mainly through the denitrification pathway. The timing of maximum NO2− accumulations also differed among treatments and was observed at 24 h in P. mandelii cultures grown at 10°C and at 10 h in cells grown at 20°C and 30°C. The level of denitrification was higher in cells grown at 30°C at 10 h than in P. mandelii cells grown at 10°C and 20°C. These observations suggest that temperature had a significant effect on enzyme activity. Low temperature resulted in slower enzyme rates of reaction and subsequently influenced the rate of growth and metabolism. Holtan-Hartwig et al. (10) also concluded that lower temperatures may exert a challenge to the denitrifying community, thus resulting in lower denitrification rates, as measured by N2O production and reduction, at cooler temperatures (0°C). Alternatively, NirS synthesis may be negatively affected in P. mandelii at temperatures below 30°C, thus causing nitrite accumulations in the growth medium.
The pattern of CO2 production in P. mandelii cultures grown at different pH values and temperatures was investigated. Previous studies have well established the concept of Q10, defined as the change in denitrification, or respiration with a 10°C change in temperature (9, 13, 31, 32). When CO2 accumulations were analyzed in P. mandelii cultures grown at 10°C, 20°C, and 30°C, Q10 values of 2.2 at temperatures between 10°C and 20°C and of 0.9 at temperatures between 20°C and 30°C were obtained. These values are comparable to those reported in previous studies, where Q10 values between 1.6 and 2.6 for CO2 production were observed in soils treated with animal slurry (13). The patterns of CO2 accumulation in P. mandelii cultures grown at pH 6 and pH 7 were significantly different from those of CO2 accumulations in cultures grown at pH 5 and pH 8 (Fig. 3C). This suggests that both pH and temperature had an effect on the respiration rate in P. mandelii.
In our study, P. mandelii cultures did not exhibit a twofold increase in denitrification, as measured by N2O emissions, with temperature increases of 10°C, averaged over 10 h. This observation is contrary to data from previous studies that established a temperature coefficient of denitrification of approximately 2 for every 10°C temperature increase in soil (9, 22). This disparity could be due to the fact that our study was conducted with pure cultures of P. mandelii under completely anaerobic conditions, which differ from conditions of a soil community of denitrifiers composed of members that are denitrifying at variable rates and that exist in aerobic and anaerobic microcosms.
In conclusion, a pH value of 5 negatively impacted denitrification gene expression in the P. mandelii strain used; however, levels of cnorB and nirS gene expression were not affected by pH values over the range of pH 6 and 8. Gene expression was sensitive to temperature, since induction and the time to reach maximum expression were delayed as the temperature decreased from 30°C. Temperature and pH influenced NO3− utilization, NO2− accumulation, as well as cumulative denitrification, leading us to conclude that both pH and temperature had a significant effect on denitrification enzyme activity.
Acknowledgments
We are grateful to Drucie Janes and Jan Zeng for providing technical support. We are also grateful to Stephen Bowley for providing guidance in statistical analysis of data.
Funding for this project was supplied by the GAPS program of Agriculture and Agri-Food Canada and an NSERC strategic team grant. Saleema Saleh-Lakha was the recipient of an NSERC doctoral scholarship. Kelly E. Shannon was the recipient of an Ontario graduate scholarship. Infrastructure and equipment grants from the Canadian Foundation Innovation and the Ontario Innovation Trust are sincerely acknowledged by Jack T. Trevors.
Footnotes
Published ahead of print on 17 April 2009.
REFERENCES
- 1.Baumann, B., J. R. van der Meer, M. Snozzi, and A. J. B. Zehnder. 1997. Inhibition of denitrification activity but not of mRNA induction in Paracoccus denitrificans by nitrite at a suboptimal pH. Antonie van Leeuwenhoek 72:183-189. [DOI] [PubMed] [Google Scholar]
- 2.Betlach, M. R., and J. M. Tiedje. 1981. Kinetic explanation for accumulation of nitrite, nitric oxide, and nitrous oxide during bacterial denitrification. Appl. Environ. Microbiol. 42:1074-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chèneby, D., L. Philippot, A. Hartmann, G. Hénault, and J. Germon. 2000. 16S rDNA analysis for characterization of denitrifying bacteria isolated from three agricultural soils. FEMS Microbiol. Ecol. 34:121-128. [DOI] [PubMed] [Google Scholar]
- 4.Dandie, C. E., D. L. Burton, B. J. Zebarth, J. T. Trevors, and C. Goyer. 2007. Analysis of denitrification genes and comparison of nosZ, cnorB, and 16S rDNA from culturable denitrifying bacteria in potato cropping systems. Syst. Appl. Microbiol. 30:128-138. [DOI] [PubMed] [Google Scholar]
- 5.Dandie, C. E., M. N. Miller, D. L. Burton, B. J. Zebarth, J. T. Trevors, and C. Goyer. 2007. Nitric oxide reductase-targeted real-time PCR quantification of denitrifier populations in soil. Appl. Environ. Microbiol. 73:4250-4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delorme, S., P. Lemarceau, R. Christen, T. Corberand, J. M. Meyer, and L. Garden. 2002. Pseudomonas lini sp. nov., a novel species from bulk and rhizospheric soils. Int. J. Syst. Evol. Microbiol. 52:513-523. [DOI] [PubMed] [Google Scholar]
- 7.Firestone, M. K. 1982. Biological denitrification, p. 289-326. In F. J. Stevenson (ed.), Nitrogen in agricultural soils. American Society of Agronomy, Madison, WI.
- 8.Firestone, M. K., R. B. Firestone, and J. M. Tiedje. 1980. Nitrous oxide from soil denitrification: factors controlling its biological production. Science 208:749-751. [DOI] [PubMed] [Google Scholar]
- 9.Fischer, E. N., and S. C. Whalen. 2005. Rates and controls on denitrification in an agricultural soil fertilized with liquid lagoonal swine waste. Nutr. Cycl. Agroecosys. 71:271-287. [Google Scholar]
- 10.Holtan-Hartwig, L., P. Dörsch, and L. R. Bakken. 2002. Low temperature control of soil denitrifying communities: kinetics of N2O production and reduction. Soil Biol. Biochem. 34:1797-1806. [Google Scholar]
- 11.Knowles, R. 1982. Denitrification. Microbiol. Rev. 46:43-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koskinen, W. C., and D. R. Keeney. 1982. Effect of pH on the rate of gaseous products of denitrification in a silt loam soil. Soil Sci. Soc. Am. J. 46:1165-1167. [Google Scholar]
- 13.Maag, M., and F. P. Vinther. 1999. Effect of temperature and water on gaseous emissions from soils treated with animal slurry. Soil Sci. Soc. Am. J. 63:858-865. [Google Scholar]
- 14.Mosier, A., C. Kroeze, C. Nevison, O. Oenema, S. Seitzinger, and O. van Cleemput. 1998. Closing the global N2O budget: nitrous oxide emissions through the agricultural nitrogen cycle. Nutr. Cycl. Agroecosys. 52:225-248. [Google Scholar]
- 15.Müller, M. M., V. Sundman, and J. Skujins. 1980. Denitrification in low pH spodosols and peats determined with the acetylene inhibition method. Appl. Environ. Microbiol. 40:235-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parkin, T. B., A. J. Sexstone, and J. M. Tiedje. 1985. Adaptation of denitrifying population to low soil pH. Appl. Environ. Microbiol. 49:1053-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Payne, W. J., and P. S. Riley. 1969. Suppression by nitrate of enzymatic reduction of nitric oxide. Proc. Soc. Exp. Biol. Med. 132:258-260. [DOI] [PubMed] [Google Scholar]
- 18.Saleh-Lakha, S., K. E. Shannon, C. Goyer, J. T. Trevors, B. J. Zebarth, and D. L. Burton. 2008. Nitric oxide reductase gene expression and nitrous oxide production in nitrate-grown Pseudomonas mandelii. Appl. Environ. Microbiol. 74:6876-6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Šimek, M., and J. E. Cooper. 2002. The influence of soil pH on denitrification: progress towards the understanding of this interaction over the last 50 years. Eur. J. Soil Sci. 53:345-354. [Google Scholar]
- 20.Šimek, M., L. Jíšová, and D. W. Hopkins. 2002. What is the so-called optimum pH for denitrification in soil? Soil Biol. Biochem. 34:1227-1234. [Google Scholar]
- 21.Šimek, M., and D. W. Hopkins. 1999. Regulation of potential denitrification by soil pH in long-term fertilized arable soils. Biol. Fertil. Soil 30:41-47. [Google Scholar]
- 22.Stanford, G., S. Dzienia, and R. A. Vander Pol. 1975. Effect of temperature on denitrification rate in soils. Soil Sci. Soc. Am. Proc. 39:867-870. [Google Scholar]
- 23.Swain, H. M., H. J. Somerville, and J. A. Cole. 1978. Denitrification during growth of Pseuomonas aeruginosa on octane. J. Gen. Microbiol. 107:103-112. [Google Scholar]
- 24.Tate, R. L., III. 2000. Denitrification, p. 404-432. Soil microbiology, 2nd ed. John Wiley & Sons, New York, NY.
- 25.Thomas, K. L., D. Lloyd, and L. Boddy. 1994. Effects of oxygen, pH and nitrate concentration on denitrification by Pseudomonas species. FEMS Microbiol. Lett. 118:181-186. [DOI] [PubMed] [Google Scholar]
- 26.Thomsen, J. K., T. Geest, and R. P. Cox. 1994. Mass spectrometric studies of the effect of pH on the accumulation of intermediates in denitrification by Paracoccus denitrificans. Appl. Environ. Microbiol. 60:536-541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Verhille, S., N. Baida, F. Dabboussi, D. Izard, and H. Leclerc. 1999. Taxonomic study of bacteria isolated from natural mineral waters: Proposal of Pseudomonas jessenii sp. nov. and Pseudomonas mandelii sp. nov. Syst. Appl. Microbiol. 22:45-58. [DOI] [PubMed] [Google Scholar]
- 28.Waibel, A. E., T. H. Peter, K. S. Carslaw, H. Oelhaf, G. Wetzel, P. J. Crutzen, U. Poschl, A. Tsias, V. Reimer, and H. Fischer. 1999. Arctic ozone loss due to denitrification. Science 283:2064-2069. [DOI] [PubMed] [Google Scholar]
- 29.Wallenstein, M. D., D. D. Myrold, M. Firestone, and M. Voytek. 2006. Environmental controls on denitrifying communities and denitrification rates: insights from molecular methods. Ecol. Appl. 16:2143-2152. [DOI] [PubMed] [Google Scholar]
- 30.Williams, D. R., J. J. Rowe, P. Romero, and R. G. Eagon. 1978. Denitrifying Pseudomonas aeruginosa: some parameters of growth and active transport. Appl. Environ. Microbiol. 367:257-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu, M., and Y. Qi. 2001. Spatial and seasonal variations of Q10 determined by soil respiration measurements at a Sierra Nevadan forest. Global Biogeochem. Cycles 15:687-696. [Google Scholar]
- 32.Yuste, J. C., I. A. Janssens, A. Carrara, and R. Ceulemans. 2004. Annual Q10 of soil respiration reflects plant phenological patterns as well as temperature sensitivity. Global Change Biol. 10:161-169. [Google Scholar]
- 33.Zumft, W. G. 1997. Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 61:533-616. [DOI] [PMC free article] [PubMed] [Google Scholar]