Abstract
We constructed a novel cell surface display system to control the ratio of target proteins on the Saccharomyces cerevisiae cell surface, using two pairs of protein-protein interactions. One protein pair is the Z domain of protein A derived from Staphylococcus aureus and the Fc domain of human immunoglobulin G. The other is the cohesin (Coh) and dockerin (Dock) from the cellulosome of Clostridium cellulovorans. In this proposed displaying system, the scaffolding proteins (fusion proteins of Z and Coh) were displayed on the cell surface by fusing with the 3′ half of α-agglutinin, and the target proteins fused with Fc or Dock were secreted. As a target protein, a recombinant Trichoderma reesei endoglucanase II (EGII) was secreted into the medium and immediately displayed on the yeast cell surface via the Z and Fc domains. Display of EGII on the cell surface was confirmed by hydrolysis of β-glucan as a substrate, and EGII activity was detected in the cell pellet fraction. Finally, two enzymes, EGII and Aspergillus aculeatus β-glucosidase 1, were codisplayed on the cell surface via Z-Fc and Dock-Coh interactions, respectively. As a result, the yeast displaying two enzymes hydrolyzed β-glucan to glucose very well. These results strongly indicated that the proposed strategy, the simultaneous display of two enzymes on the yeast cell surface, was accomplished by quantitatively controlling the display system using affinity binding.
Microorganisms have been used to enable cell surface display of heterogonous peptides or proteins for various applications (13, 19, 20). For practical purposes, the use of the cell surface display system of the yeast Saccharomyces cerevisiae, which has “generally regarded as safe” status, is suitable and can be employed in many processes, including food and pharmaceutical production. Yeast strains displaying functional peptides or proteins are expected to be used, for instance, as live vaccines (10), matrices for screening of novel proteins from combinatorial libraries (3, 12, 27), whole-cell bioadsorbents (14, 18), reporter substances (23), and whole-cell biocatalysts (8, 9, 11, 16). In yeast-based cell surface display systems, the C-terminal half of α-agglutinin, containing the putative glycosylphosphatidylinositol anchor attachment signal sequence (15), has been used as an anchor protein to successfully display many kinds of proteins on the cell surface. In addition, other surface proteins, such as α-agglutinin (AGA1 and AGA2), Flo1, Sed1, Cwp1, Cwp2, Tip1, and Tir1, were also used for enzyme display (24). To immobilize a target protein at the N terminus, another surface display system has been developed using the flocculation functional domain of Flo1p (16), which is a lectin-like cell wall protein that plays a major role in flocculation (25). These displaying systems could successfully display three kinds of enzymes on the surfaces of yeast cells (8, 9). However, in current yeast-based cell surface display systems, the ratio of the proteins to be displayed on the cell surface cannot be controlled, since the produced target protein attached the protein or cell wall component on the cell surface by only molecular interaction.
A large extracellular polysaccharolytic multicomponent complex called the cellulosome provides a model for the concept of a cell surface displaying system. Cellulosomes from anaerobic and cellulolytic bacteria such as Clostridium, Acetivibrio, Bacteroides, and Ruminococcus have been studied in detail (1). The cellulosomes of the thermophilic organism Clostridium thermocellum and of the mesophilic organisms Clostridium cellulovorans and Clostridium cellulolyticum have been extensively discussed (1, 6). The structure of cellulosomes consists of a nonenzymatic scaffolding protein called CbpA (1, 6), CipA (1, 4), or CipC (1) complexed with a number of cellulosomal enzymes. The scaffolding proteins usually contain a number of cohesin (Coh) domains and cellulose binding domains. On the other hand, cellulosomal enzymes contain a Coh binding site called dockerin (Dock), and the Coh-Dock interaction is an important factor in cellulosome assembly. Some cellulosomes are able to anchor to the cell surface through interaction of other scaffolding proteins containing surface layer homology domains. The cellulosome models that include a cell surface anchoring system are similar to our concept of a cell surface display system.
Based on this concept, we propose a novel method to control the displaying ratio of enzymes for attaining a complicated reaction such as cellulose degradation. Recently, the display of one kind of protein via Z-Fc interaction on the surfaces of yeast cells was successfully achieved (18). According to this proposed method, we used the following two pairs of proteins for assembling specifically with each other: (i) Z domain and the Fc part of immunoglobulin G (IgG) and (ii) Coh and Dock from cellulosomes. The Z domain, which was derived from the B domain of protein A from Staphylococcus aureus, interacts with IgG from various species (21). Coh is a domain of the scaffolding protein from the cellulase complex (cellulosome) from C. cellulovorans (5). Dock is a domain of the cellulosomal enzymes which binds to Coh of the scaffolding protein on the surface of C. cellulovorans. The secreted enzymes fused with Fc or Dock could be displayed on the cell surface through a ZZ-Coh-Coh scaffolding protein (Fig. 1B).
FIG. 1.
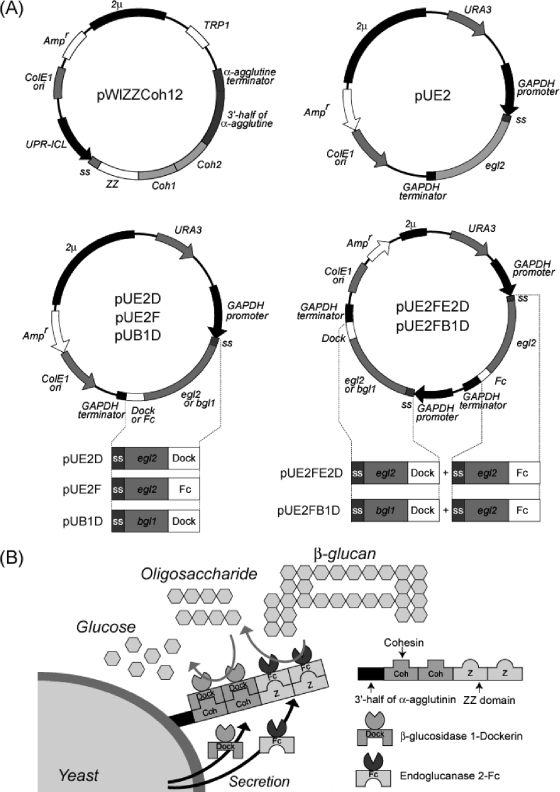
Plasmids and proposed protein-protein interaction cell surface display systems with two pairs of proteins. (A) Maps of plasmids used in this study. (B) Sequential degradation of β-glucan by displayed enzymes. The protein pairs used in this study were (i) the Z domain from Staphylococcus aureus protein A and Fc domain from IgG and (ii) the Coh and Dock domains from the C. cellulovorans cellulosome.
To carry out the proposed strategy, we constructed the cell surface displaying system with Trichoderma reesei endoglucanase II (EGII) and Aspergillus aculeatus β-glucosidase 1 (BGL1). Furthermore, we demonstrated synergistic saccharification of soluble cellulose by integrating two kinds of cellulase enzymes on the cell surface by using the ZZ-Coh-Coh scaffolding protein as a model for a complex reaction.
MATERIALS AND METHODS
Strains and media.
Escherichia coli NovaBlue {endA1 hsdR17(rK-12− mK-12+) supE44 thi-1 gyrA96 relA1 lac recA1 F′ [proAB+ lacIqZΔM15::Tn10(tet)]} (Novagen, Inc., Madison, WI) was used as the host strain for the recombinant DNA manipulations. The yeast S. cerevisiae MT8-1 (MATa ade his3 leu2 trp1 ura3) was used for cell surface display (17). E. coli was grown in LB medium (10 g/liter tryptone, 5 g/liter yeast extract, and 5 g/liter sodium chloride) containing 100 μg/ml ampicillin. Following precultivation, yeast cells were aerobically cultivated at 30°C in a synthetic medium (6.7 g/liter yeast nitrogen base without amino acids [Difco Laboratories, Detroit, MI] and with appropriate supplements, 5 g/liter glucose, and 20 g/liter Casamino Acids [Difco] was added [SDCA medium]).
Construction of plasmids and transformation of yeast cells.
The plasmid pWIZZCoh12, used for display of two Z (ZZ) domains and two Coh domains (Coh1-Coh2), was constructed as follows. The gene encoding Coh1-Coh2 was amplified by PCR with one pair of primers, CbpA-SalI (forward [F]) and CbpA-XhoI (reverse [R]) (Table 1), and pET29-CbpA as a template. The amplified PCR product was digested with SalI and XhoI and subcloned into the XhoI site of the plasmid pMWIZ1 (18), and the resulting plasmid was named pWIZZCoh12 (Fig. 1A). The plasmid pUE2, used for secretory production of EGII, was constructed as follows. A gene encoding EGII was amplified by PCR with one pair of primers, egl2-BglII (F) and egl2-stop-KpnI (R), and pEG23u31H6 as a template. The amplified PCR product was digested with BglII and KpnI and subcloned into the BglII and KpnI sites of pUGP3 (pUE2) (Fig. 1A). Plasmids pUE2F, pUE2D, and pUB1D, used for secretory production of EGII-Fc, EGII-Dock, and BGL1-Dock, respectively, were constructed as follows. A gene encoding EGII or BGL1 was amplified by PCR with two pairs of primers, egl2-BglII (F) and egl2-KpnI (R) or bgl1-XhoI (F) and bgl1-SphI-XhoI (R), and pEG23u31H6 and pBG211 (9) as templates, respectively. The amplified EGII gene was digested with BglII and KpnI and inserted into the BglII and KpnI sites of pUGP3, and the resulting plasmid was designated pUGP3-egl2. The amplified BGL1 gene was digested with XhoI and inserted into the XhoI site of pUGP3, and the resulting plasmids was named pUGP3-bgl1. Each gene encoding the Dock and IgG Fc domains was amplified by PCR with three pairs of primers, Fc-KpnI (F) and Fc-XhoI (R), DE-KpnI (F) and DE-XhoI (R), or DE-SphI (F) and DE-SphI (R), with C. cellulovorans chromosomal DNA or pUC19Fc (22), respectively. Amplified genes corresponding to Fc or Dock were digested with KpnI and XhoI and inserted into the KpnI and XhoI sites of pUGP3-egl2, and the resulting plasmids were named pUE2F and pUE2D, respectively (Fig. 1A). The amplified Dock gene was digested with SphI and inserted into the SphI site of pUGP3-BGL1 (pUB1D) (Fig. 1A).
TABLE 1.
Oligonucleotides used for plasmid construction
| Primer | Sequence |
|---|---|
| CbpA-SalI (F) | 5′-GCTGTCGACTTAACAAAAGTTCCAGCAGCTGGTTTAGCA-3′ |
| CbpA-XhoI (R) | 5′-GCTCTCGAGACGACCTTCGATTGCAACGTCTGCTGGTGCTTTTTTATCAAAA-3′ |
| egl2-BglII (F) | 5′-CTAAGATCTATGAACAAGTCCGTGGCTCC-3′ |
| egl2-KpnI (R) | 5′-GTCGGTACCCTTTCTTGCGAGACACGAGC-3′ |
| egl2-stop-KpnI (R) | 5′-GTCGGTACCCTACTTTCTTGCGAGACACGAGC-3′ |
| DE-KpnI (F) | 5′-GTCGGTACCAATCTTCTAGGCGATGTAGA-3′ |
| DE-XhoI (R) | 5′-GTCCTCGAGTTATATTGCTTTTTTTAAGAATGC-3′ |
| Fc-KpnI (F) | 5′-CTAGGTACCGGGGGACCGTCAGTCTTCCTCTTCCCCCCA-3′ |
| Fc-XhoI (R) | 5′-GCACTCGAGTCATTTACCCGGAGACAGGGAGAGGCTCTT-3′ |
| bgl1-XhoI (F) | 5′-ATCCTCGAGATGCAACTGTTCAATTTGCCATTGAAAGTT-3′ |
| bgl1-SphI-XhoI (R) | 5′-TACCTCGAGGCATGCTTGCACCTTCGGGAGCGCCGCGTGAAGGGG-3′ |
| DE-SphI (F) | 5′-CTAGCATGCAATCTTCTAGGCGATGTAGACGGTAATGAT-3′ |
| DE-SphI (R) | 5′-GCTGCATGCCTATATTGCTTTTTTTAAGAATGCAAGATCAAT-3′ |
| pro-egl2-BamHI (F) | 5′-GCATGGATCCACCAGTTCTCACACGGAACACCACTAATGG-3′ |
| egl2-term-BamHI (R) | 5′-GCATGGATCCGCGCGCTCAATCAATGAATCGAAAATGTCAT-3′ |
| pro-bgl1-NotI (F) | 5′-GCATGCGGCCGCACCAGTTCTCACACGGAACACCACTAATGG-3′ |
| bgl1-term-NotI (R) | 5′-GCATGCGGCCGCGCGCGCTCAATCAATGAATCGAAAATGTCAT-3′ |
The plasmid pUE2FB1D, used for simultaneous production of EGII and BGL1, was constructed as follows. A gene carrying the EGII-Fc or the BGL1-Dock expression cassette was amplified by PCR with one pair of primers, pro-egl2-BamHI (F) and egl2-term-BamHI (R), or pro-bgl1-NotI (F) and bgl1-term-NotI (R) using pUE2F or pUB1D as a template, respectively. The amplified EGII-Fc expression cassette gene was digested with BamHI and inserted into the BamHI site of the plasmids pRS406-2μm (21), and the resulting plasmid was named pRSE2. Subsequently, the amplified BGL1-Dock expression cassette was digested with NotI and inserted into the NotI site of pRSE2, and finally the resulting plasmid was named pUE2FB1D (Fig. 1A).
Transformation of S. cerevisiae with the constructed plasmids (Fig. 1A) was performed by the lithium acetate method using the Yeastmaker yeast transformation system (Clontech Laboratories, Inc., Palo Alto, CA).
Immunofluorescence analysis of surface-displayed ZZ-Coh-Coh.
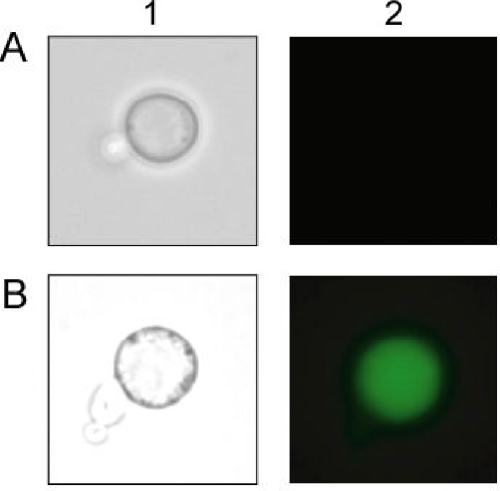
Immunofluorescence labeling of the yeast cells was performed as follows. After cultivation of transformant in SDCA medium at 30°C for 120 h, yeast cells were collected and washed with phosphate-buffered saline (PBS; 50 mM phosphate, 150 mM sodium chloride [pH 7.4]). Then the cells were resuspended in PBS containing 10 g/liter bovine serum albumin at 4°C for 30 min (optical density at 600 nm = 10). The primary antibody (rabbit IgG, dilution rate of 1:100) was added to the suspension, and incubated at 4°C for 1.5 h. Furthermore, the yeast cells were incubated at 4°C for 1 h with the secondary antibody, Alexa Fluor 488 conjugated goat anti-rabbit IgG (H+L) (Molecular Probes, Inc., Eugene, OR). After being washed with PBS, yeast cells labeled with fluorescent antibody were observed by fluorescence microscopy. In addition, the green fluorescence intensity of the cells was analyzed with a flow cytometer (FACSCalibur; Becton Dickinson, Franklin Lakes, NJ) at an even rate of 500 cells/s. The average fluorescence intensity of 10,000 cells was calculated by subtracting the average fluorescence intensity of control cells (MT8-1/pWI3) from that of each sample.
Enzyme assay.
The enzyme activity of each transformant and culture supernatant was measured with barley β-glucan as a substrate. After cultivation in SDCA medium at 30°C for 120 h, the supernatant was separated by centrifugation at 6,000 × g for 10 min at 4°C. Yeast cells were also collected by centrifugation at 6,000 × g for 10 min at 4°C. Yeast cells were washed twice with cold distilled water. Then, 50 μl of supernatants or cell suspension (the optical density at 600 nm was adjusted to 5) was added to 1 ml of reaction mixture (50 mM sodium acetate buffer [pH 5.0], 1 g/liter barley β-glucan [Sigma Chemical Co., St. Louis, MO]) for the appropriate incubation time at 30°C. After the hydrolysis reaction, the supernatants were separated by centrifugation at 20,000 × g for 3 min at 4°C, and the amounts of reducing sugar and total sugar were measured. The amount of reducing sugar released from the substrate was determined as the number of glucose equivalents using the Somogyi-Nelson method (26). Total sugar was measured using the phenol-sulfuric acid method (7).
RESULTS
Expression of ZZ-Coh-Coh gene on the yeast cell surface.
To immobilize target proteins on the yeast cell surface, we constructed a yeast strain (MT8-1/pWIZZCoh12) displaying a scaffolding protein fused with two tandem-aligned ZZ and Coh domains (ZZ-Coh-Coh). The gene encoding the scaffolding protein fused to the gene containing the secretion signal sequence of the Rhizopus oryzae glucoamylase gene and the 3′ end of the α-agglutinin gene was expressed under the control of the 5′ upstream region of the isocitrate lyase of the Candida tropicalis (UPR-ICL) gene (Fig. 1A). To confirm the immobilization of the scaffolding protein on the cell surface, immunofluorescence labeling of yeast cells was performed using the rabbit IgG antibody as the primary antibody and Alexa Fluor 488-conjugated goat anti-rabbit IgG (H+L) as the secondary antibody. The immunostained scaffolding protein was observed on the yeast cells harboring pWIZZCoh12, while there was no green fluorescence in the control strain, MT8-1/pWI3 (Fig. 2). This result indicated that scaffolding protein was successfully anchored on the cell surface. In addition, the production of the scaffolding protein on the yeast cell surface was monitored using flow cytometry. Scaffolding protein-displaying yeast strains (MT8-1/pWIZZCoh12) showed a maximal fluorescence intensity of 35 after 120 h of cultivation, while little fluorescence intensity was detected in the yeast strain not displaying the scaffolding protein (MT8-1/pWI3).
FIG. 2.
Fluorescence microscopy of the transformed yeast cells. The fluorescence antibody used to label yeast cells was observed by phase-contrast microscopy (column 1) and immunofluorescence microscopy (column 2). (A) MT8-1/pWI3 (control); (B) MT8-1/pWIZZCoh12.
Expression of each recombinant egl2 gene.
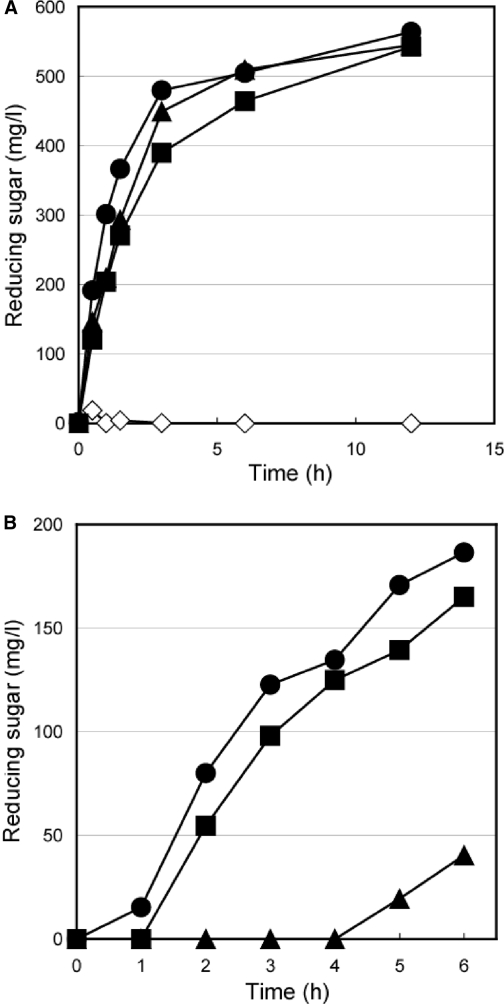
To display EGII on the yeast cell surface via scaffolding protein, we constructed three recombinant yeast strains, which secreted wild-type EGII (MT8-1/pUE2) and a chimeric EGII protein fused with a tandem-aligned Fc domain (EGII-Fc; MT8-1/pUE2F) or Dock domain (EGII-Dock; MT8-1/pUE2D) (Fig. 1B). The genes encoding recombinant EGII were fused to the gene coding for the secretion signal sequence of the R. oryzae glucoamylase and were expressed under the constitutive control of the glyceraldehyde 3-phosphate dehydrogenase (GAPDH) promoter (Fig. 1A). To confirm whether each transformant had endoglucanase production potential, a plate assay was carried out using 0.1% carboxymethyl cellulose as a substrate according to the Congo red staining method (2). The yeast cells harboring the plasmids encoding EGII (MT8-1/pUE2, MT8-1/pUE2F, and MT8-1/pUE2D) hydrolyzed the substrate, and a clear halo around the colony was formed; however, no halo appeared around the colony of the control strain harboring the control plasmid pUGP3 (data not shown). This result indicated that each recombinant EGII was expressed in the active form. The EGII activity of each transformant in the culture supernatant was examined with 0.1% β-glucan as a substrate at 30°C (Fig. 3A). Two of the fused proteins produced, EGII-Fc and EGII-Dock, exhibited high EGII activity toward β-glucan similar to that of wild-type EGII. These results indicated that the fusion domain of Fc and Dock at the C terminus of EGII did not interfere with EGII activity itself.
FIG. 3.
Hydrolysis of soluble cellulose (β-glucan) with EGII by S. cerevisiae. (A) Hydrolysis of β-glucan by secreted EGII. Symbols for transformants: open diamond, MT8-1/pWI3 (control); closed triangle, MT8-1/pUE2; closed circle, MT8-1/pUE2D; closed square, MT8-1/pUE2F. (B) Hydrolysis of β-glucan by anchored EGII on the surfaces of yeast cells. Symbols for transformants: closed triangle, MT8-1/pUE2+pWIZZCoh12; closed circle, MT8-1/pUE2D+pWIZZCoh12; closed square, MT8-1/pUE2F+pWIZZCoh12.
Assembly of recombinant EGII on the yeast cell surface.
For assembling the recombinant secreted EGII on the yeast cell surface, we constructed three yeast strains which simultaneously secreted EGII (wild-type EGII, EGII-Fc, and EGII-Dock) and the scaffolding proteins: MT8-1/pWIZZCoh12+pUE2, MT8-1/pWIZZCoh12+pUE2F, and MT8-1/pWIZZCoh12+pUE2D. To assess whether the recombinant secreted EGII (EGII-Fc or EGII-Dock) was assembled on the cell surface, the endoglucanase activities of MT8-1/pWIZZCoh12+pUE2, MT8-1/pWIZZCoh12+pUE2F, and MT8-1/pWIZZCoh12+pUE2D were examined in their cell pellet fractions (Fig. 3B). As a result, two transformants, MT8-1/pWIZZCoh12+pUE2F and MT8-1/pWIZZCoh12+pUE2D, showed higher activity, whereas little activity was detected with MT8-1/pWIZZCoh12+pUE2. These results strongly indicated that the secreted EGII-Fc and EGII-Dock were assembled with the scaffolding protein (ZZ-Coh-Coh) on the cell surface via Z-Fc and Coh-Dock interaction, respectively.
Assembly of recombinant EGII and BGL1 on the yeast cell surface.
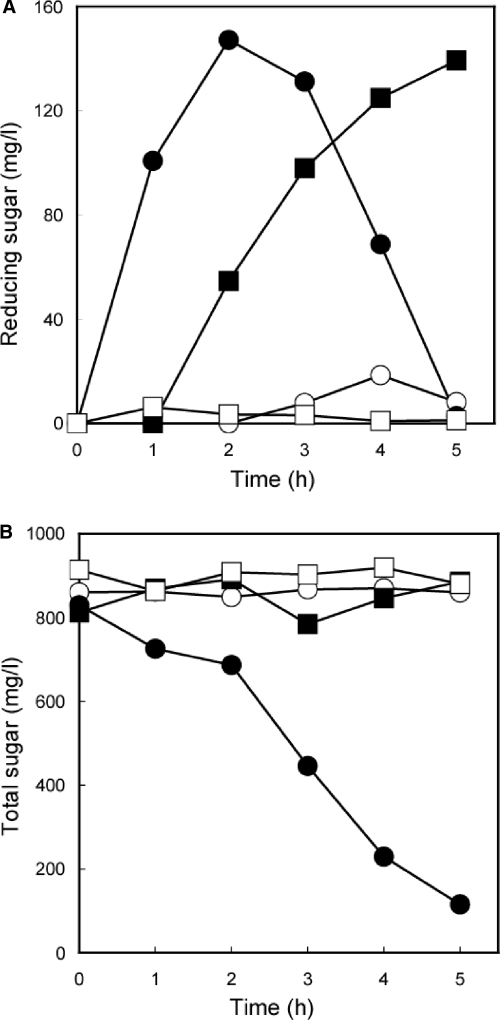
Next, to test for simultaneous assembly of two enzymes (EGII-Fc and BGL1-Dock) on the yeast cell surface (Fig. 1B), a transformant (MT8-1/pWIZZCoh12+pUE2FB1D) which simultaneously produced EGII-Fc, BGL1-Dock, and scaffolding protein (ZZ-Coh-Coh) was constructed. First, by using pNPG as a substrate, the β-glucosidase activity of MT8-1/pWIZZCoh12+pUE2FB1D was estimated to be 4.72 U/g (dry weight). This result indicated that the displayed BGL1 was as active as the EGII discussed above. Next, a hydrolysis experiment was performed with 0.1% β-glucan as a substrate at 30°C (Fig. 4). In the case of the EGII-Fc- and BGL1-Dock-displaying strain (MT8-1/pWIZZCoh12+pUE2FB1D), the amount of reducing sugar in the reaction mixture increased until 2 h, and after that, the concentration of reducing sugar decreased at almost the same rate (Fig. 4A). The yeast displaying EGII-Fc alone (MT8-1/pWIZZCoh12+pUE2F) showed a slower increase in the amount of reducing sugar than the yeast (MT8-1/pWIZZCoh12+pUE2FB1D) displaying two kinds of enzymes. In the yeast strains displaying BGL1-Dock (MT8-1/pWIZZCoh12+pUB1D) and secreting EGII-Fc and BGL1-Dock (MT8-1/pWI3+pUE2FB1D), small amounts of reducing sugars were detected by the Somogyi-Nelson method. In comparing the presence of scaffolding protein on the surfaces of yeast (MT8-1/pWIZZCoh12+pUE2FB1D and MT8-1/pWI3+pUE2FB1D) cells, it was assumed that the scaffolding protein could function and that Fc- and Dock-fused enzymes on those scaffolds resulted in degradation of β-glucan (Fig. 1B).
FIG. 4.
Hydrolysis of soluble cellulose (β-glucan) by the combination of EGII and BGL1 displayed on S. cerevisiae cells (A) and total sugar monitoring by displayed EGII and BGL1 (B). Symbols for transformants: open circle, MT8-1/pUE2FB1D+pWI3; open square, MT8-1/pUB1D+pWIZZCoh12; closed circle, MT8-1/pUE2FB1D+pWIZZCoh12; closed square, MT8-1/pUE2F+pWIZZCoh12.
The two-enzyme-displaying transformant (MT8-1/pWIZZCoh12+pUE2FB1D) exhibited only a decrease in the total sugar in the reaction mixture (Fig. 4B). In addition, only a little decrease in total sugars was detected by the phenol-sulfuric acid method in the reaction mixtures of MT8-1/pWI3+pUE2FB1D, MT8-1/pWIZZCoh12+pUB1D and MT8-1/pWIZZCoh12+pUE2F. These results indicated that MT8-1/pWIZZCoh12+pUE2FB1D, displaying EGII-Fc and BGL1-Dock, only hydrolyzed soluble cellulose to glucose, and the produced glucose was assimilated by the yeast.
DISCUSSION
To reduce the cost of immobilized enzymes, multiple-enzyme-displaying microorganisms have been developed by many researchers (3, 10, 12, 13, 19, 20, 24, 27). However, for performing the sequential reactions by several enzymes in a current cell surface display system, it is necessary to control the ratio of different proteins to be displayed on the cell surface. To overcome this problem, we constructed a novel yeast-based cell surface display system by assembling two pairs of proteins: (i) Z domain and Fc and (ii) Coh and Dock. To display the target proteins, we constructed a yeast strain that simultaneously displayed the scaffolding protein (ZZ-Coh-Coh) and secreted one of the target proteins (EGII and BGL1) fused with Fc or Dock (Fig. 1B). First, we constructed the yeast strain displaying scaffolding protein on the cell surface by a cell surface engineering system using α-agglutinin (13). The gene encoding scaffolding protein was highly expressed after 120 h of cultivation. This result agreed with other results obtained with cell surface expression using the UPR-ICL promoter (8). Next, we constructed a yeast strain secreting EGII fused with Fc (EGII-Fc) or Dock (EGII-Dock) as the target protein (Fig. 3A). The recombinant EGII activity was not suppressed by fusion with Fc or Dock, since it was well known that yeast could produce active EGII fused with any protein at the C terminus of EGII (21).
To confirm the displaying of EGII-Fc and EGII-Dock on the cell surface via the scaffolding protein, we performed the hydrolysis experiment using a yeast strain displaying ZZ-Coh-Coh and secreting EGII-Fc or EGII-Dock (MT8-1/pWIZZCoh12+pUE2Fc and MT8-1/pWIZZCoh12+pUE2D) (Fig. 3B). Two transformants (MT8-1/pWIZZCoh12+pUE2F and MT8-1/pWIZZCoh12+pUE2D) showed higher EGII activity toward β-glucan than MT8-1/pWIZZCoh12+pUE2. We suggest that the accessibility of the substrate for EGII or a slight difference in the affinities of EGII-Fc and EGII-Dock for ZZ-Coh-Coh resulted in the difference.
The hydrolysis experiment showed that the amount of reducing sugar produced by the yeast strain codisplaying EGII-Fc and BGL1-Dock increased until 2 h (Fig. 4A) and that this increase was followed by a decrease in the amount of total sugar (Fig. 4B). This degradation potential was comparable to our previous finding (9), and it was assumed that the regulation of the display ratio of enzymes will permit us to construct bioconversion systems that are more efficient than those constructed with nonregulated displaying systems.
In conclusion, the scaffold ZZ-Coh-Coh was successfully displayed on the yeast cell surface, and simultaneously produced Fc or Dock fusion proteins were assembled via ZZ-Coh-Coh. This is the first report of the integrated protein-displaying cell surface system. These results demonstrate an effective codisplay of target proteins by the novel system in a ratio-controlling manner and will permit us to construct an integrated protein-displaying system.
Acknowledgments
This work was partially supported by the Research and Development Program for New Bio-industry Initiatives and Special Coordination Funds for Promoting Science and Technology, Creation of Innovation Centers for Advanced Interdisciplinary Research Areas (Innovative Bioproduction Kobe), MEXT, Japan.
Footnotes
Published ahead of print on 1 May 2009.
REFERENCES
- 1.Bayer, E. A., J. P. Belaich, Y. Shoham, and R. Lamed. 2004. The cellulosomes: multienzyme machines for degradation of plant cell wall polysaccharides. Annu. Rev. Microbiol. 58:521-554. [DOI] [PubMed] [Google Scholar]
- 2.Béguin, P. 1983. Detection of cellulase activity in polyacrylamide gels using Congo red-stained agar replicas. Anal. Biochem. 131:333-336. [DOI] [PubMed] [Google Scholar]
- 3.Boder, E. T., and K. D. Wittrup. 1997. Yeast surface display for screening combinatorial polypeptide libraries. Nat. Biotechnol. 15:553-557. [DOI] [PubMed] [Google Scholar]
- 4.Demain, A. L., M. Newcomb, and J. H. Wu. 2005. Cellulase, clostridia, and ethanol. Microbiol. Mol. Biol. Rev. 69:124-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doi, R. H., J. S. Park, C. C. Liu, L. M. Malburg, Y. Tamaru, A. Ichiishi, and A. Ibrahim. 1998. Cellulosome and noncellulosomal cellulases of Clostridium cellulovorans. Extremophiles 2:53-60. [DOI] [PubMed] [Google Scholar]
- 6.Doi, R. H., and A. Kosugi. 2004. Cellulosomes: plant-cell-wall-degrading enzyme complexes. Nat. Rev. Microbiol. 2:541-551. [DOI] [PubMed] [Google Scholar]
- 7.Dubois, M., K. A. Gilles, J. K. Hamilton, P. A. Reberse, and F. Smith. 1956. Colorimetric method for determination of sugars and related substances. Anal. Chem. 28:350-356. [Google Scholar]
- 8.Fujita, Y., S. Takahashi, M. Ueda, A. Tanaka, H. Okada, Y. Morikawa, T. Kawaguchi, M. Arai, H. Fukuda, and A. Kondo. 2002. Direct and efficient production of ethanol from cellulosic material with a yeast strain displaying cellulolytic enzymes. Appl. Environ. Microbiol. 68:5136-5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fujita, Y., J. Ito, M. Ueda, H. Fukuda, and A. Kondo. 2004. Synergistic saccharification, and direct fermentation to ethanol, of amorphous cellulose by use of an engineered yeast strain codisplaying three types of cellulolytic enzyme. Appl. Environ. Microbiol. 70:1207-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Georgiou, G., C. Stathopoulos, P. S. Daugherty, A. R. Nayak, B. L. Iverson, and R. Curtiss III. 1997. Display of heterologous proteins on the surface of microorganisms: from the screening of combinatorial libraries to live recombinant vaccines. Nat. Biotechnol. 15:29-34. [DOI] [PubMed] [Google Scholar]
- 11.Ito, J., Y. Fujita, M. Ueda, H. Fukuda, and A. Kondo. 2004. Improvement of cellulose-degrading ability of a yeast strain displaying Trichoderma reesei endoglucanase II by recombination of cellulose-binding domains. Biotechnol. Prog. 20:688-691. [DOI] [PubMed] [Google Scholar]
- 12.Kieke, M. C., E. V. Shusta, E. T. Boder, L. Teyton, K. D. Wittrup, and D. M. Kranz. 1999. Selection of functional T cell receptor mutants from a yeast surface-display library. Proc. Natl. Acad. Sci. USA 96:5651-5656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kondo, A., and M. Ueda. 2004. Yeast cell-surface display-applications of molecular display. Appl. Microbiol. Biotechnol. 64:28-40. [DOI] [PubMed] [Google Scholar]
- 14.Kuroda, K., S. Shibasaki, M. Ueda, and A. Tanaka. 2001. Cell surface-engineered yeast displaying a histidine oligopeptide (hexa-His) has enhanced adsorption of and tolerance to heavy metal ions. Appl. Microbiol. Biotechnol. 57:697-701. [DOI] [PubMed] [Google Scholar]
- 15.Lipke, P. N., D. Wojciechowicz, and J. Kurjan. 1989. AGα1 is the structural gene for the Saccharomyces cerevisiae α-agglutinin, a cell surface glycoprotein involved in cell-cell interactions during mating. Mol. Cell. Biol. 9:3155-3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsumoto, T., H. Fukuda, M. Ueda, A. Tanaka, and A. Kondo. 2002. Construction of yeast strains with high cell surface lipase activity by using novel display systems based on the Flo1p flocculation functional domain. Appl. Environ. Microbiol. 68:4517-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murai, T., M. Ueda, T. Kawaguchi, M. Arai, and A. Tanaka. 1998. Assimilation of cellooligosaccharides by a cell surface-engineered yeast expressing β-glucosidase and carboxymethylcellulase from Aspergillus aculeatus. Appl. Environ. Microbiol. 64:4857-4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakamura, Y., S. Shibasaki, M. Ueda, and A. Tanaka, H. Fukuda, and A. Kondo. 2001. Development of novel whole-cell immunoadsorbents by yeast surface display of the IgG-binding domain. Appl. Microbiol. Biotechnol. 57:500-505. [DOI] [PubMed] [Google Scholar]
- 19.Samuelson, P., E. Gunneriusson, P. A. Nygren, and S. Ståhl. 2002. Display of proteins on bacteria. J. Biotechnol. 96:129-154. [DOI] [PubMed] [Google Scholar]
- 20.Schreuder, M. P., A. T. Mooren, H. Y. Toschka, C. T. Verrips, and F. M. Klis. 1996. Immobilizing proteins on the surface of yeast cells. Trends Biotechnol. 14:115-120. [DOI] [PubMed] [Google Scholar]
- 21.Shibasaki, S., A. Kawabata, J. Ishii, S. Yaga, T. Kadonosono, M. Kato, N. Fukuda, A. Kondo, and M. Ueda. 2007. Construction of a novel synergistic system for production and recovery of secreted recombinant proteins by the cell surface engineering. Appl. Microbiol. Biotechnol. 75:821-828. [DOI] [PubMed] [Google Scholar]
- 22.Shibasaki, S., K. Kuroda, H. D. Nguyen, T. Mori, W. Zou, and M. Ueda. 2006. Detection of protein-protein interactions by a combination of a novel cytoplasmic membrane targeting system of recombinant proteins and fluorescence resonance energy transfer. Appl. Microbiol. Biotechnol. 70:451-457. [DOI] [PubMed] [Google Scholar]
- 23.Shibasaki, S., Y. Ninomiya, M. Ueda, M. Iwahashi, T. Katsuragi, Y. Tani, S. Harashima, and A. Tanaka. 2001. Intelligent yeast strains with the ability to self-monitor the concentrations of intra- and extracellular phosphate or ammonium ion by emission of fluorescence from the cell surface. Appl. Microbiol. Biotechnol. 57:702-707. [DOI] [PubMed] [Google Scholar]
- 24.Van der Vaart, J. M., R. T. Biesebeke, J. W. Chapman, H. Y. Toschka, F. M. Klis, and C. T. Verrips. 1997. Comparison of cell wall proteins of Saccharomyces cerevisiae as anchors for cell surface expression of heterologous proteins. Appl. Environ. Microbiol. 63:615-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Watari, J., Y. Takata, M. Ogawa, H. Sahara, S. Koshino, M. L. Onnela, U. Airaksinen, R. Jaatinen, M. Penttilä, and S. Keränen. 1994. Molecular cloning and analysis of the yeast flocculation gene FLO1. Yeast 10:211-225. [DOI] [PubMed] [Google Scholar]
- 26.Wood, T. M., and K. M. Bhat. 1988. Methods for measuring cellulase activities. Methods Enzymol. 160:87-112. [Google Scholar]
- 27.Zou, W., M. Ueda, and A. Tanaka. 2002. Screening of a molecule endowing Saccharomyces cerevisiae with n-nonane-tolerance from a combinatorial random protein library. Appl. Microbiol. Biotechnol. 58:806-812. [DOI] [PubMed] [Google Scholar]