Abstract
Tetracycline-resistant Streptococcus thermophilus isolates from soft cheeses harbored the genes tet(S), tet(M), and tet(L). Molecular analysis of these genes revealed their expression, localization on plasmids or Tn916-Tn1545 family transposons, and their similarity with published sequences. The study highlights the importance of an accurate safety assessment of using S. thermophilus as a starter culture.
Streptococcus thermophilus is a thermophilic lactic acid bacterium of major importance for the dairy industry where strains of this species are commonly used in the manufacture of yoghurt and many varieties of cheeses (3). Albeit phylogenetically close to pathogenic streptococci, this dairy species possesses a long history of safe use, and its innocuous nature has indeed been assessed by comparative genomics analysis (4).
In recent years, an increasing amount of focus has been given to the possibility that lactic acid bacteria could serve as reservoirs of antibiotic resistance determinants with the risk of transferring the genes to many food-borne commensal bacteria and other pathogenic bacteria (15). However, no systematic studies have been carried out to investigate the prevalence of antibiotic-resistant S. thermophilus, despite the abundance of this bacterium in several dairy products. Background information on the level of intrinsic resistance of S. thermophilus to the main classes of antibiotics is available (1, 15), and Tosi et al. (21) have recently developed a new S. thermophilus susceptibility test medium that allows the easy identification of strains with atypical antibiotic resistance. These studies have shown that a few erythromycin-, streptomycin-, and tetracycline-resistant strains can be detected in raw milk or in dairy products. The genetic basis of antibiotic resistance in S. thermophilus is poorly documented, too. Two previous investigations carried out on a limited number of strains have reported the presence of the resistance genes erm(B), tet(S)/(M) (23), or tet(S) (8) in antibiotic-resistant S. thermophilus from retail foods.
In this context, the present study reports the prevalence and magnitude of antibiotic-resistant S. thermophilus populations in a number of largely consumed soft cheeses, a cheese category characterized by its high water content and level of viable bacteria. Thereafter, we focused on the tetracycline-resistant S. thermophilus isolates, assessing their genetic uniqueness and antibiotic resistance gene pattern. In addition, we provided molecular information about the tetracycline resistance genes relative to their sequence diversity, localization on plasmids and Tn916-Tn1545 family transposons, expression, and ability to be transferred by conjugation.
Thirteen samples of Italian soft cheeses (Table 1) were purchased from local grocery chain stores at different periods of time. Samples (25 g) were homogenized in sterile 0.2 g liter−1 sodium citrate solution with a stomacher Lab-Blender 400 (Seward; PBI International, Italy) and serially diluted in physiological solution (0.9 g liter−1 NaCl). For enumeration of S. thermophilus, samples were spread in duplicate on M17 agar plates (Fluka, Italy) containing 100 μg ml−1 cycloheximide (Fluka). For the count and isolation of antibiotic-resistant S. thermophilus, the medium described above was supplemented with one of the following antimicrobials at the breakpoint values (7): 8 μg ml−1 chloramphenicol, 4 μg ml−1 erythromycin, or 4 μg ml−1 tetracycline. Plates were incubated aerobically at 37°C for up to 48 h. Representative colonies of S. thermophilus were isolated from the selective plates and routinely grown in M17 broth with the proper antibiotics. For molecular characterization, extraction of total DNA from the isolates was carried out by using the procedure of Marmur (13). The isolates were maintained as culture stocks in 25% (vol/vol) glycerol at −80°C until further analyses.
TABLE 1.
Italian soft cheeses analyzed in this study, their characteristics, and the results of counts of total and antibiotic-resistant S. thermophilus organisms
| Cheese and sample name | Description of the cheese, locationa | Producer |
S. thermophilus counts (log CFU g−1) onb:
|
|
|---|---|---|---|---|
| M17 | M17 plus tetracycline | |||
| Caciotta | Pasteurized cows’ milk cheese, northeast Italy | |||
| 1Ca | A | 8.68 | 4.34 | |
| Caprino | Raw goats’ milk cheese, southern Italy | |||
| 1Cp | B | 4.04 | 3.04 | |
| Mozzarella | Pasteurized cows’ milk cheese, northern Italy | |||
| 1 M | C | 9.96 | 5.96 | |
| Robiola | Pasteurized cows’ milk cheese, northwest Italy | |||
| 1R | D | 2.04 | −c | |
| 2R | D | 2.72 | − | |
| 3R | D | 2.28 | − | |
| Squacquerone | Pasteurized cows’ milk cheese, northern Italy | |||
| 1Sq | E | 9.20 | 3.40 | |
| Stracchino | Pasteurized cows’ milk cheese, northwest Italy | |||
| 1S | C | 9.86 | 2.57 | |
| 2S | C | 8.20 | 2.32 | |
| 3S | C | 8.72 | 2.99 | |
| Taleggio | PDO cheese from raw or pasteurized cows’ milk, northern Italy | |||
| 1T | F | 8.56 | 4.30 | |
| 2T | G | 8.54 | − | |
| 3T | F | 9.28 | 5.66 | |
PDO, protected denomination of origin cheese.
The values are means of two determinations. No chloramphenicol- or erythromycin-resistant S. thermophilus isolates were observed in any sample.
−, no colonies found.
Table 1 summarizes the counts obtained for each cheese sample. The total count of S. thermophilus varied from 102 to 109 CFU g−1, depending on the type of cheese. Except for the low count of Caprino and Robiola cheeses, these values are in accordance with the literature data, which indicate that the streptococcal count of short-ripened cheeses is around 108 to 109 CFU g−1 (3, 9). Regarding antibiotic-resistant S. thermophilus, for all samples no colonies were detected in plates containing chloramphenicol. For the samples 2S and 1T, some colonies appeared on plates containing erythromycin, but the derived isolates did not grow in broth medium in the presence of this antibiotic. Hence, no chloramphenicol- or erythromycin-resistant S. thermophilus isolates were retrieved in the examined cheeses. Conversely, tetracycline-resistant streptococci were found in 9 out of 13 samples, at variable levels, ranging from 102 to 105 CFU g−1, depending on the type of cheese and dairy producer. The size of the resistant population was generally from 4 to 6 logs lower than that of the total S. thermophilus population, with the exception of the Caprino cheese sample. In this sample, the difference was less remarkable (Table 1), and the tetracycline-resistant S. thermophilus was an important component of the streptococcal population. Markedly diverse levels of tetracycline-resistant streptococci were found in the analyzed samples of Taleggio cheese. This finding can be ascribed to the fact that the samples were produced in two dairy plants, and thus, they harbored different indigenous or starter S. thermophilus strains.
A total of 40 colonies were isolated from the M17 plates with tetracycline for phenotypic and molecular characterization. All presumptive S. thermophilus isolates were gram positive and coccoid under a microscope catalase negative and did not grow on kanamycin-esculin-azide agar (Oxoid, Italy), a selective medium for enterococci. This last confirmation was necessary since M17 can be poorly selective for S. thermophilus when a high number of enterococci are expected in the sample (18), as was the case of the analyzed cheeses.
The tetracycline resistance phenotype was checked for all isolates by using the broth dilution method (16) in 90% (vol/vol) Iso-Sensitest broth (Oxoid) plus 10% (vol/vol) M17, 0.5% (wt/vol) lactose (21), and tetracycline. All cultures were able to grow in up to almost 16 μg ml−1 tetracycline (MIC > 16 μg ml−1). The obtained data agree with those reported in recent studies, which indicated tetracycline MICs variable from 16 to >128 μg ml−1 for resistant S. thermophilus isolates (8, 21, 23).
The identities of tetracycline-resistant S. thermophilus isolates were confirmed by species-specific PCR analysis, using the method described by Lick et al. (12) modified with an annealing step of 56°C for 30 s in order to avoid amplification of related lactic acid bacteria, such as lactococci. For some isolates, showing a faint 968-bp band (results not shown) with the procedure described above, identification was further corroborated by sequencing a 16S rRNA gene fragment, as previously reported (19).
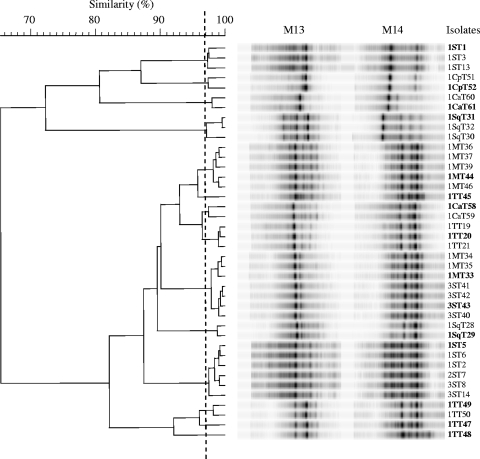
To investigate for the genetic diversity and the presence of clones among the isolates, typing was carried out by randomly amplified polymorphic DNA (RAPD)-PCR with the primers M13 and M14 in separate reactions according to Zapparoli et al. (24). Figure 1 shows S. thermophilus isolates grouped by numerical analysis of combined RAPD-PCR profiles. A high degree of genetic polymorphism among the isolates was found, and two major groups were clearly separated at a similarity level of 65%. Clustering allowed differentiation of 14 additional subgroups of isolates that can be considered different strains. These clonal subgroups are constituted mainly by isolates deriving from the same cheese sample that give almost identical RAPD-PCR profiles. Interestingly, similar profiles were also obtained for some isolates from different cheeses manufactured in the same dairy plant, i.e., mozzarella and Stracchino from producer C, maybe due to the presence of common starters or contaminant strains. Further, a wide genetic heterogeneity among isolates from some cheese samples was observed, thus demonstrating that different strains of tetracycline-resistant S. thermophilus may reside in association in the same product.
FIG. 1.
Genotypic clustering of the 40 isolates of S. thermophilus based on comparison of RAPD-PCR profiles with primers M13 and M14. The vertical dotted line designates the similarity level (97%) chosen as discriminatory at the strain level. The isolates shown in bold were selected for further molecular characterization.
Representative isolates from each clonal subgroup were then investigated by PCR analysis for the presence of six genes most commonly involved in resistance to tetracycline [tet(K), tet(L), tet(M), tet(O), tet(S), and tet(W)]. The isolates 1MT33 and 3ST43 from mozzarella and Stracchino, respectively, included in the same clonal subgroup, were also analyzed to confirm their clonality. The references of the specific primers for tetracycline resistance genes, PCR conditions, and the positive control strains used are reported in Table 2. The occurrence of the Tn916-Tn1545 transposon family, the most frequent family in tet(M)-carrying bacteria, was explored by specific amplification of the integrase gene (int) (6). Both positive and negative controls were included in all experiments. The results of the PCR analyses on the selected tetracycline-resistant isolates are summarized in Table 3. All isolates tested negative for the tet(K), tet(O), and tet(W) genes by PCR analysis, while all except one strain carried the gene tet(S). Ten of the 14 strains possessed two tetracycline resistance genes, tet(S) plus either the gene tet(M) or tet(L). In detail, three strains carried the genes tet(S) and tet(M) that have the same mode of action (ribosomal protection), and seven strains possessed the genes tet(S) and tet(L) with different modes of action (ribosomal protection and efflux, respectively). Genes coding for different resistance mechanisms were found together in a number of gram-positive bacterial strains (5). Until now, the occurrence of one resistance gene, tet(S) or tet(S)/(M), has been described in tetracycline-resistant S. thermophilus (8, 23); the present study provides the first demonstration of the presence of the tet(L) gene in S. thermophilus and broadens the number of tetracycline resistance genes found in this species to three, tet(S), tet(M), and tet(L).
TABLE 2.
Primers and conditions used for gene-specific PCRs
| Targeted gene | Primer sequences (5′ to 3′) | Amplicon size (bp) | Positive control straina | Reference(s) for primers and PCR conditions |
|---|---|---|---|---|
| tet(K) | TCGATAGGAACAGCAGTA | 169 | S. epidermidis DST-SE20 | 17, 19 |
| CAGCAGATCCTACTCCTT | ||||
| tet(L)b | ATAAATTGTTTCGGGTCGGTAAT | 1,077 | E. faecalis DST-ET10 | 22 |
| AACCAGCCAACTAATGACAATGAT | ||||
| tet(M) | GTGGACAAAGGTACAACGAG | 406 | E. faecalis DST-EE12 | 17, 19 |
| CGGTAAAGTTCGTCACACAC | ||||
| tet(O) | AACTTAGGCATTCTGGCTCAC | 515 | E. mundtii DST-ET41 | 17, 19 |
| TCCCACTGCTCCATATCGTCA | ||||
| tet(S)b,c | CATAGACAAGCCGTTGACC | 667 | E. gallinarum DST-ET14 | 17 |
| ATGTTTTTGGAACGCCAGAG | ||||
| tet(W) | GAGAGCCTGCTATATGCCAGC | 168 | B. animalis subsp. lactis DST-Bl | 14 |
| GGGCGTATCCACAATGTTAAC | ||||
| int | GCGTGATTGTATCTCACT | 1,028 | E. faecalis DST-EE12 | 6 |
| GACGCTCCTGTTGCTTCT |
E. gallinarum, Enterococcus gallinarum; B. animalis, Bifidobacterium animalis; E. mundtii, Enterococcus mundtii; S. epidermidis, Staphylococcus epidermidis; DST, Dipartimento Scientifico e Tecnologico (University of Verona, Italy).
PCR analysis was modified using 2 mM MgCl2, 200 μM of each dNTP, and 0.5 μM of each specific primer.
The annealing step of the amplification cycle was modified to 58°C for 30 s.
TABLE 3.
Antibiotic resistance gene pattern of representative isolates of S. thermophilus, presence of the int gene, and genetic location of the genes and their expressione
| Straina | Tetracycline resistance geneb
|
Tn916-1545 int gene | Gene located on plasmidc | Expressed gene(s)d | ||
|---|---|---|---|---|---|---|
| tet(M) | tet(L) | tet(S) | ||||
| 1CaT58 | + | − | + | + | ND | tet(S) |
| 1CaT61 | − | + | + | − | ND | tet(S) |
| 1CpT52 | − | + | + | − | tet(S) | tet(S) |
| 1MT33/3ST43 | − | − | + | − | ND | tet(S) |
| 1MT44 | + | − | + | + | ND | tet(S) |
| 1ST1 | + | − | + | + | tet(S) | tet(S), tet(M) |
| 1ST5 | − | − | + | − | ND | tet(S) |
| 1SqT29 | − | + | + | − | ND | tet(S) |
| 1SqT31 | + | − | − | + | tet(M) | tet(M) |
| 1TT20 | − | − | + | − | tet(S) | tet(S) |
| 1TT45 | − | + | + | − | tet(S) | tet(S) |
| 1TT47 | − | + | + | − | ND | tet(S), tet(L) |
| 1TT48 | − | + | + | − | ND | tet(S), tet(L) |
| 1TT49 | − | + | + | − | ND | tet(S), tet(L) |
Strain name contains the name of the isolation sample followed by the letter T, indicating tetracycline resistance, and a consecutive number.
The genes tet(K), tet(O), and tet(W) were not found in these isolates.
ND, no plasmids were detected in these strains.
Expression analysis was conducted in the presence of 16 μg ml−1 tetracycline.
+, gene present; −, gene not present.
Expression analysis was conducted on the 14 tetracycline-resistant S. thermophilus strains listed in Table 3, grown in the presence of the antibiotic (16 μg ml−1, a concentration greater than the breakpoint defined for this species) to establish the functionality of the tetracycline resistance genes. RNA extraction and reverse transcription-PCR procedures were performed as previously described (20). At the tetracycline concentration tested, tet(S) was always expressed, while expression of the genes tet(M) and tet(L) was found only in a few strains (Table 3). To our knowledge, the expression of tetracycline resistance genes in lactic acid bacteria has not been extensively evaluated. Ammor et al. (2) found in Lactobacillus sakei an increase in the expression level of tet(M) and tet(L) at higher tetracycline concentrations (up to 64 μg ml−1). As the activity of one tetracycline resistance gene, mainly tet(S), can explain the phenotypic resistance observed, we can suppose that in our S. thermophilus strains the expression of the genes tet(M) and tet(L) may be induced as a consequence of a higher concentration of the antibiotic. Further investigations are necessary to corroborate this hypothesis.
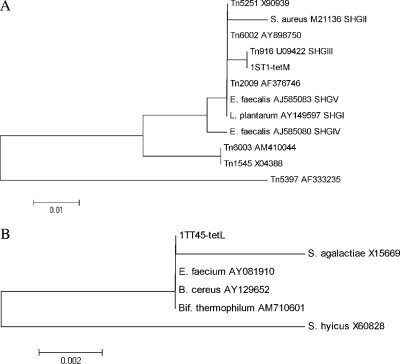
To obtain information about the nucleotide sequences of the tetracycline resistance genes found in our isolates, fragments of tet(S), tet(M), and tet(L) from representative strains were sequenced as previously described (19). The online BLASTN program was used for sequence similarity searches. In general, sequence analysis showed a high level of similarity (99 to 100%) with the sequences corresponding to the genes tet(S), tet(M), and tet(L) available in the NCBI database (http://www.ncbi.nlm.nih.gov/) from various food-borne bacteria belonging mainly to the genera Bacillus, Enterococcus, Lactococcus, Lactobacillus, Listeria, Staphylococcus, and Streptococcus. In more detail, the sequence alignment of the 599-bp tet(S) gene fragment showed a 100% similarity (no nucleotide differences) with seven sequences present in the molecular database (accession no. AB290882, AM039486, AM039488, AM039489, AY534326, DQ377341, and L09756). Phylogenetic trees for the tet(M) and tet(L) genes were constructed using distance analysis as implemented in MEGA4. The trees are shown in Fig. 2. The tet(M) sequences included in the tree (Fig. 2A) comprised those derived from various transposons of the Tn916-Tn1545 family and those corresponding to the five sequence homology groups described by Huys et al. (10). The sequence of the 232-bp tet(M) fragment of S. thermophilus strain 1ST1 was identical to that of the corresponding fragment of the Tn916 transposon, while it differed by 1 to 24 nucleotides from the other sequences included in this analysis. The 738-bp tet(L) sequence of S. thermophilus strain 1TT45 shows 100% similarity to three of the considered sequences (Fig. 2B), and the difference with respect to the others was less than 1.5% (11/738 bp) of the sequence length. The strict similarity observed validates the hypothesis that horizontal gene transfer events occurred from bacteria carrying tetracycline resistance genes to S. thermophilus.
FIG. 2.
Phylogenetic trees based on the sequence analysis of tet(M) (A) and tet(L) (B) gene fragments. Sequence accession numbers are indicated as well as the five sequence homology groups described by Huys et al. (10). Trees were constructed by distance matrix analysis. Bar, number of nucleotide substitutions per site; B. cereus, Bacillus cereus; Bif. thermophilum, Bifidobacterium thermophilum; E. faecalis, Enterococcus faecalis; E. faecium, Enterococcus faecium; L. plantarum, Lactobacillus plantarum; S. aureus, Staphylococcus aureus; S. agalactiae, Staphylococcus agalactiae; S. hyicus, Staphylococcus hyicus.
All four strains that carried the tet(M) gene were positively amplified in the int-specific PCR analysis, demonstrating that tet(M) is located on a Tn916-Tn1545 family transposon that is probably responsible for the acquisition of this gene by the bacterium. This is the first report of the presence of members of this transposon family, which are characterized by a wide host range, in S. thermophilus strains. On the contrary, in our isolates the tet(S) gene was not associated with these transposons. Although a Tn916-like element containing tet(S) has been identified in the related bacterium Streptococcus intermedius (11), our results did not show the presence of the Tn916S transposon, thus suggesting that another mobile element may have directed the transfer of the tet(S) gene.
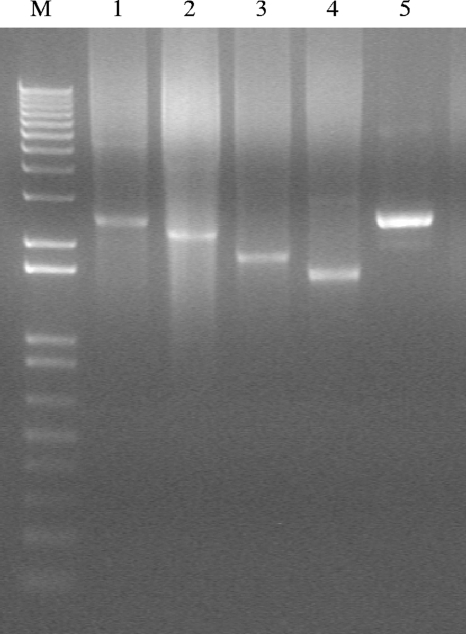
To assess the presence of other mobile genetic elements carrying tetracycline resistance, plasmid DNA was isolated from 4-ml exponential S. thermophilus cultures by using the Wizard Plus SV Minipreps DNA purification system (Promega Corporation, Madison, WI). A preliminary digestion with lysozyme (final concentration, 10 mg ml−1) was conducted at 37°C for 2 hours to allow cell lysis to occur. Five (1CpT52, 1ST1, 1SqT31, 1TT20, and 1TT45) out of the 14 representative strains were found to harbor plasmids of different sizes (Fig. 3). The plasmids were separated on a 1% agarose gel, gel slices were cut, and DNA was purified from the gel with the Wizard SV gel and PCR cleanup system according to the package insert (Promega Corporation). PCRs specific for tet(S), tet(M), and tet(L) conducted on the plasmid DNA extracted from the gel showed tet(S) to be associated with plasmids in four strains and tet(M) in the strain 1SqT31 (Table 3). These data indicate that different plasmids (of different sizes) can serve an important role as a vehicle for resistance gene capture and subsequent dissemination. However, the presence of tetracycline resistance genes in S. thermophilus strains may be due to promiscuous gene transfer systems that include acquisition of plasmids or conjugative transposons or both. These aspects of resistance to antibiotics in lactic acid bacteria need to be deeply explored.
FIG. 3.
Plasmid profiles of tetracycline-resistant Streptococcus thermophilus strains isolated from cheeses. Lane M, 1 kb plus the DNA marker (Invitrogen); lanes 1 through 5, strains 1ST1, 1TT20, 1SqT31, 1TT45, and 1CpT52, respectively.
Finally, conjugation trials were conducted to evaluate if the tetracycline-resistant S. thermophilus strains are able to transfer the resistance genes to other bacteria (both belonging to the same species and to other food-related genera). Filter mating experiments were carried out as described by Huys et al. (10) using the 14 streptococcal strains as donors and Enterococcus faecalis OG1RF (resistant to rifampin and fusidic acid), Listeria innocua LMG 11387T, and S. thermophilus ST72 (resistant to gentamicin), all susceptible to tetracycline and plasmid free, as recipients. No gene transfer events were observed in any experiment with all recipient strains. These findings seem to show that S. thermophilus strains are not able to efficiently disseminate antibiotic resistance under the conditions applied here.
In recent years, the question of antibiotic resistance in nonpathogenic bacteria has been contemplated by the Scientific Panel on Additives and Products or Substances used in Animal Feed (FEEDAP) of the European Commission. The FEEDAP panel considers that strains of bacteria carrying an acquired resistance to antimicrobials (due to the acquisition of added genes) should not be used as a feed additive. In an effort to slow the development of resistance, these criteria should also be applied to bacteria in human food, and this is especially important for cultures such as S. thermophilus which are largely applied in the production of fermented dairy products.
In conclusion, the results emerged from this study underline that tetracycline-resistant strains belonging to the species S. thermophilus are widespread in the types of cheese here analyzed and the contamination levels of these products, which are readily consumed, are not irrelevant. The high sequence similarity of the tetracycline resistance genes with those reported in the database and the localization of some of them on mobile genetic elements suggest that transfers of antibiotic resistance determinants from species present in the same dairy environment to S. thermophilus can occur. Although further studies are necessary to investigate the transferability of resistance traits from this species in cheese, the results obtained until now suggested that S. thermophilus does not represent an important source for the spread of genes encoding resistance to antimicrobial agents. However, considering that this species is widely used in dairy products and that acquired resistance mediated by added genes may present a risk for public health, an accurate assessment of the safety-related features of S. thermophilus strains used as natural or selected starter cultures is necessary.
Nucleotide sequence accession numbers.
The tet(S), tet(M), and tet(L) fragment sequences have been deposited in the EMBL/GenBank/DDBJ databases under accession no. FM165482 (S. thermophilus 1TT45), FM165483 (S. thermophilus 1ST1), and FM165484 (S. thermophilus 1TT45), respectively.
Acknowledgments
This research was partly supported by a grant from the Italian Ministry of University and Research within the Program PRIN 2006 no. 2006072328_004.
We acknowledge Stefano Carteri and Desj Simeoni for technical assistance.
Footnotes
Published ahead of print on 24 April 2009.
REFERENCES
- 1.Ammor, M. S., A. B. Flórez, and B. Mayo. 2007. Antibiotic resistance in non-enterococcal lactic acid bacteria and bifidobacteria. Food Microbiol. 24:559-570. [DOI] [PubMed] [Google Scholar]
- 2.Ammor, M. S., M. Gueimonde, M. Danielsen, M. Zagorec, A. H. van Hoek, C. G. de Los Reyes-Gavilán, B. Mayo, and A. Margolles. 2008. Two different tetracycline resistance mechanisms, plasmid-carried tet(L) and chromosomally located transposon-associated tet(M), coexist in Lactobacillus sakei Rits 9. Appl. Environ. Microbiol. 74:1394-1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beresford, T. P., N. A. Fitzsimons, N. L. Brennan, and T. M. Cogan. 2001. Recent advances in cheese microbiology. Int. Dairy J. 11:259-274. [Google Scholar]
- 4.Bolotin, A., B. Quinquis, P. Renault, A. Sorokin, S. D. Ehrlich, S. Kulakauskas, A. Lapidus, E. Goltsman, M. Mazur, G. D. Pusch, M. Fonstein, R. Overbeek, N. Kyprides, B. Purnelle, D. Prozzi, K. Ngui, D. Masuy, F. Hancy, S. Burteau, M. Boutry, J. Delcour, A. Goffeau, and P. Hols. 2004. Complete sequence and comparative genome analysis of the dairy bacterium Streptococcus thermophilus. Nat. Biotechnol. 22:1554-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chopra, I., and M. Roberts. 2001. Tetracycline antibiotics: mode of action, applications, molecular biology and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 65:232-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doherty, N., K. Trzcinski, P. Pickerill, P. Zawadzki, and C. G. Dowson. 2000. Genetic diversity of the tet(M) gene in tetracycline-resistant clonal lineages of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 44:2979-2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.European Food Safety Authority. 2005. Opinion of the Scientific Panel on Additives and Products or Substances used in Animal Feed on the updating of the criteria used in the assessment of bacteria for resistance to antibiotics of human or veterinary importance. EFSA J. 223:1-12. [Google Scholar]
- 8.Ge, B., P. Jiang, F. Han, N. K. Saleh, N. Dhiman, D. P. Fedorko, N. A. Nelson, and J. Meng. 2007. Identification and antimicrobial susceptibility of lactic acid bacteria from retail fermented foods. J. Food Prot. 70:2606-2612. [DOI] [PubMed] [Google Scholar]
- 9.Gobbetti, M., S. Lowney, E. Smacchia, B. Battistotti, P. Damiani, and P. F. Fox. 1997. Microbiology and biochemistry of Taleggio cheese during ripening. Int. Dairy J. 7:509-517. [Google Scholar]
- 10.Huys, G. H., K. D'Haene, J. M. Collard, and J. Swings. 2004. Prevalence and molecular characterization of tetracycline resistance in Enterococcus isolates from food. Appl. Environ. Microbiol. 70:1555-1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lancaster, H., A. P. Roberts, R. Bedi, M. Wilson, and P. Mullany. 2004. Characterization of Tn916S, a Tn916-like element containing the tetracycline resistance determinant tet(S). J. Bacteriol. 186:4395-4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lick, S., M. Keller, W. Bockelmann, and K. J. Heller. 1996. Rapid identification of Streptococcus thermophilus by primer-specific PCR amplification based on its lacZ gene. Syst. Appl. Microbiol. 19:74-77. [Google Scholar]
- 13.Marmur, J. 1961. A procedure for the isolation of deoxyribonucleic acid from microorganisms. J. Mol. Biol. 3:208-218. [Google Scholar]
- 14.Masco, L., K. Van Hoorde, E. De Brandt, J. Swings, and G. Huys. 2006. Antimicrobial susceptibility of Bifidobacterium strains from humans, animals and probiotic products. J. Antimicrob. Chemother. 58:85-94. [DOI] [PubMed] [Google Scholar]
- 15.Mathur, S., and R. Singh. 2005. Antibiotic resistance in food lactic acid bacteria—a review. Int. J. Food Microbiol. 105:281-305. [DOI] [PubMed] [Google Scholar]
- 16.National Committee for Clinical Laboratory Standards. 2004. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. NCCLS M7-A6. National Committee for Clinical Laboratory Standards, Villanova, PA.
- 17.Ng, L. K., I. Martin, M. Alfa, and M. Mulvey. 2001. Multiplex PCR for the detection of tetracycline-resistant genes. Mol. Cell Probes 15:209-215. [DOI] [PubMed] [Google Scholar]
- 18.Randazzo, C. L., S. Torriani, A. D. L. Akkermans, W. M. de Vos, and E. E. Vaughan. 2002. Diversity, dynamics, and activity of bacterial communities during production of an artisanal Sicilian cheese as evaluated by 16S rRNA analysis. Appl. Environ. Microbiol. 68:1882-1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rizzotti, L., D. Simeoni, P. Cocconcelli, S. Gazzola, F. Dellaglio, and S. Torriani. 2005. Contribution of enterococci to the spread of antibiotic resistance in the production chain of swine meat commodities. J. Food Prot. 68:955-965. [DOI] [PubMed] [Google Scholar]
- 20.Torriani, S., V. Gatto, S. Sembeni, R. Tofalo, G. Suzzi, N. Belletti, F. Gardini, and S. Bover-Cid. 2008. Rapid detection and quantification of tyrosine decarboxylase gene (tdc) and its expression in gram-positive bacteria associated with fermented foods using PCR-based methods. J. Food Prot. 71:93-101. [DOI] [PubMed] [Google Scholar]
- 21.Tosi, L., G. Berruti, M. Danielsen, A. Wind, G. Huys, and L. Morelli. 2007. Susceptibility of Streptococcus thermophilus to antibiotics. Antonie van Leeuwenhoek 92:21-28. [DOI] [PubMed] [Google Scholar]
- 22.Trzcinski, K., B. S. Cooper, W. Hryniewicz, and C. G. Dowson. 2000. Expression of resistance to tetracyclines in strains of methicillin-resistant Staphylococcus aureus. J. Antimicrob. Chemother. 45:763-770. [DOI] [PubMed] [Google Scholar]
- 23.Wang, H. H., M. Manuzon, M. Lehman, K. Wan, H. Luo, T. E. Wittum, A. Yousef, and L. O. Bakaletz. 2006. Food commensal microbes as a potentially important avenue in transmitting antibiotic resistance genes. FEMS Microbiol. Lett. 254:226-231. [DOI] [PubMed] [Google Scholar]
- 24.Zapparoli, G., C. Reguant, A. Bordons, S. Torriani, and F. Dellaglio. 2000. Genomic DNA fingerprinting of Oenococcus oeni strains by pulsed-field gel electrophoresis and randomly amplified polymorphic DNA-PCR. Curr. Microbiol. 40:351-355. [DOI] [PubMed] [Google Scholar]