Abstract
Coaggregation is hypothesized to enhance freshwater biofilm development. To investigate this hypothesis, the ability of the coaggregating bacterium Sphingomonas natatoria to form single- and dual-species biofilms was studied and compared to that of a naturally occurring spontaneous coaggregation-deficient variant. Attachment assays using metabolically inactive cells were performed using epifluorescence and confocal laser scanning microscopy. Under static and flowing conditions, coaggregating S. natatoria 2.1gfp cells adhered to glass surfaces to form diaphanous single-species biofilms. When glass surfaces were precoated with coaggregation partner Micrococcus luteus 2.13 cells, S. natatoria 2.1gfp cells formed densely packed dual-species biofilms. The addition of 80 mM galactosamine, which reverses coaggregation, mildly reduced adhesion to glass but inhibited the interaction and attachment to glass-surface-attached M. luteus 2.13 cells. As opposed to wild-type coaggregating cells, coaggregation-deficient S. natatoria 2.1COGgfp variant cells were retarded in colonizing glass and did not interact with glass-surface-attached M. luteus 2.13 cells. To determine if coaggregation enhances biofilm growth and expansion, viable coaggregating S. natatoria 2.1gfp cells or the coaggregation-deficient variant S. natatoria 2.1COGgfp cells were coinoculated in flow cells with viable M. luteus 2.13 cells and allowed to grow together for 96 h. Coaggregating S. natatoria 2.1gfp cells outcompeted M. luteus 2.13 cells, and 96-h biofilms were composed predominantly of S. natatoria 2.1gfp cells. Conversely, when coaggregation-deficient S. natatoria 2.1COGgfp cells were coinoculated with M. luteus 2.13 cells, the 96-h biofilm contained few coaggregation-deficient S. natatoria 2.1 cells. Thus, coaggregation promotes biofilm integration by facilitating attachment to partner species and likely contributes to the expansion of coaggregating S. natatoria 2.1 populations in dual-species biofilms through competitive interactions.
In nature, most biofilms are not composed of one bacterial species but instead contain multiple species (24). These multispecies communities can be responsible for the fouling of ships (9, 44), the corrosion of liquid-carrying vessels (3, 14), and chronic infections in higher organisms (41, 42, 57). Recent research has demonstrated that in order for multispecies biofilm communities to develop, interbacterial communication is often essential (62) and facilitates the coordination of bacterial activities to promote the formation and to maintain the integrity of multispecies biofilm communities (28, 32, 60). Interspecies communication can be mediated by chemical or physical means. Mechanisms for chemical communication between different species include the secretion and uptake of metabolic by-products (11, 19), the exchange of genetic material (40), and the production and recognition of interspecies signal molecules such as short peptides (36) and autoinducer-2 (10). Mechanisms for interspecies physical communication can involve cell surface structures such as flagella or fimbriae (31, 48) and also include nonspecific adhesion between bacterial species (5) as well as highly specific coaggregations mediated by lectin-saccharide interactions (48).
Coaggregation, the highly specific recognition and adhesion of different bacterial species to one another, was first discovered to occur between human oral bacteria in 1970 (23). Since then, research has shown that coaggregation occurs between specific bacterial species in environments other than the human oral cavity (48). Coaggregation interactions have been detected between bacteria isolated from canine dental plaque (21), the crop of chickens (61), the human female urogenital tract (30), the human intestine (34), and wastewater and freshwater biofilms (27, 37, 53). In particular, Buswell et al. (8) first demonstrated that coaggregation occurred between 19 freshwater strains that were isolated from a drinking water biofilm. Further studies by Rickard et al. demonstrated that coaggregation between these 19 strains was mediated by growth-phase-dependent lectin-saccharide interactions (49, 50) and occurred at the interspecies and intraspecies levels for nine different genera (50). From this aquatic biofilm consortium, coaggregation between the gram-negative bacterium Sphingomonas (Blastomonas) natatoria 2.1 and the gram-positive bacterium Micrococcus luteus 2.13 have been studied further. Coaggregation between this pair is mediated by the growth-phase-dependent expression of a lectin-like adhesin(s) on S. natatoria 2.1 and a complementary polysaccharide-containing receptor(s) on the cell surface of M. luteus 2.13 (47, 49). The addition of millimolar concentrations of galactosamine resulted in the dispersion of the coaggregates (47, 49). Coaggregation between this pair also occurs after growth in artificial biofilm constructs composed of poloxamer (47). These findings suggested that coaggregation may contribute to the integration of S. natatoria 2.1 into freshwater biofilms through specific adhesive interactions with M. luteus 2.13. Indeed, while coaggregation is hypothesized to contribute to the integration of species into freshwater biofilms (31, 32, 48), no direct evidence has yet been presented. If coaggregation promotes the integration of species into a freshwater biofilm, it may contribute to the retention of pathogens in drinking water pipelines (7) as well as the maintenance of the species diversity of aquatic biofilms that are exposed to shear stress (52, 53).
S. natatoria and M. luteus are commonly isolated from moist environments. M. luteus is environmentally ubiquitous and is found in biofilms of aquatic ecosystems (8, 35), in soil (54), and on human and animal skin (17, 29). Cells of M. luteus are gram positive, coccus shaped, arranged in clusters of tetrads, and nonmotile. S. natatoria is indigenous to freshwater environments (55) and has been isolated from swimming pools, deep-ice boreholes, and drinking water systems (1, 50, 56). Cells are gram negative, are rod shaped, and have the propensity to form rosettes containing 4 to 14 cells (55). Each rosette-forming cell has a polar tuft of fimbriae at its nonreproductive pole by which it attaches to other S. natatoria cells and, possibly, solid surfaces (46, 55). Reproduction occurs by asymmetric division (budding) to produce an ovoid daughter cell, which is highly motile, with a single polar flagellum. These ovoid daughter cells do not coaggregate, and only mature cells within rosettes can attach to other species of bacteria. Previous studies indicated that while coaggregation between S. natatoria 2.1 and M. luteus 2.13 is inhibited by the addition of galactosamine, the propensity of S. natatoria 2.1 to form rosettes was unaffected (46, 49).
The aim of this work was to determine if coaggregation enhances the attachment of planktonic S. natatoria 2.1 cells to clean glass surfaces as well as glass surfaces precoated with M. luteus 2.13 cells under static and flowing conditions. This study also aimed to provide insight into whether coaggregation contributes to the expansion of S. natatoria 2.1 populations within dual-species biofilms containing M. luteus 2.13. Epifluorescence microscopy and confocal laser scanning microscopy (CLSM) coupled with three different computer-based analysis programs were used throughout this study. Attachment assays were performed using metabolically inactive planktonic coaggregating or coaggregation-deficient variants of S. natatoria 2.1 that were suspended over or that were flowed across metabolically inactive glass-surface-attached M. luteus 2.13 cells. The potential role of coaggregation in promoting the expansion of S. natatoria 2.1 populations within biofilms containing M. luteus 2.13 was investigated by inoculating flow cells with viable cells and monitoring spatiotemporal development. By achieving these two aims, this work demonstrates that coaggregation contributes to biofilm integration and indicates that there is a possible role for coaggregation interactions in the establishment and expansion of S. natatoria populations in freshwater biofilms.
MATERIALS AND METHODS
Bacterial strains and batch growth conditions.
All strains and plasmids used in this study are shown in Table 1. Micrococcus luteus 2.13 and Sphingomonas (Blastomonas) natatoria 2.1 were grown aerobically at 28°C in liquid R2A medium (45) with shaking at 200 rpm. S. natatoria 2.1gfp and 2.1COGgfp cells were maintained in R2A medium supplemented with 100 μg ml−1 kanamycin and rifampin. Escherichia coli S17-1 λpir and E. coli K-12 HB101 were grown aerobically at 37°C in LB broth or on LB agar containing 100 μg ml−1 kanamycin. Frozen stock cultures of all strains were maintained at −80°C.
TABLE 1.
Strains and plasmids used in this studya
| Strain or plasmid | Genotype or characteristic(s) | Reference or source |
|---|---|---|
| Strains | ||
| Sphingomonas natatoria | ||
| 2.1 | Wild type | 49 |
| 2.1Rf | Rifr | This study |
| 2.1COG | Coaggregation-deficient variant; Rifr | This study |
| 2.1gfp | GFP expressing; Kmr Rifr | This study |
| 2.1COGgfp | GFP expressing, coaggregation deficient; Kmr Rifr | This study |
| Micrococcus luteus | ||
| 2.13 | Wild type | 49 |
| Escherichia coli | ||
| S17-1 λpir | Host for pFAJ1819 | 64 |
| K-12 HB101 | Host for pRK2013 | 4 |
| Plasmids | ||
| pFAJ1819 | Kmr mini-Tn5 gfp gusA nptII | 64 |
| pRK2013 | Kmr Ori (ColE1) OriT (Mob+) Tra+ | 16 |
Mob+, mobilization site (ColE1); Tra+, conjugative phenotype; OriT, origin of transfer (RK2); Rifr, rifampin resistance; Kmr, kanamycin resistance.
Enrichment of coaggregation-deficient variants.
Rifampin-resistant S. natatoria 2.1 (2.1Rf) was isolated by successive passages on R2A agar containing 1, 5, 10, 25, 50, and 100 μg ml−1 rifampin. Spontaneous coaggregation-deficient (COG) variants of S. natatoria 2.1Rf were isolated using a modified method described previously by Clemans and Kolenbrander (13). Briefly, after 72 h of growth in batch culture, 2 ml of S. natatoria 2.1Rf cells was mixed with 2 ml of M. luteus 2.13 cells (at an optical density [OD] at 570 nm of 1.0), and coaggregates settled at the bottom of a sterile test tube. The top 10 μl of the clear supernatant was inoculated into liquid R2A medium containing 100 μg ml−1 rifampin (Sigma-Aldrich, St. Louis, MO). M. luteus 2.13 cells were unable to grow in 100 μg ml−1 rifampin, and only S. natatoria 2.1Rf cells grew and were reharvested, after 72 h of growth in batch culture, to test for coaggregation ability. These steps were repeated 10 times, during which S. natatoria 2.1Rf lost the ability to coaggregate with M. luteus 2.13. The resulting coaggregation-deficient variant was named S. natatoria 2.1COG.
Labeling with GFP.
Plasmid pFAJ1819 (64), which carries a mini-Tn5 transposon containing a bifunctional gfp-gusA cassette (mTn5gusA-pgfp21), was used to transform S. natatoria 2.1Rf and S. natatoria 2.1COG. Donor strain E. coli S17-1 λpir carrying pFAJ1819, helper strain E. coli K-12 HB101 carrying pRK2013, and recipient strains S. natatoria 2.1Rf and 2.1COG were grown to late exponential phase. Cells were harvested, washed three times in sterile distilled water, and suspended to an OD at 570 nm of 1.0. Cell suspensions (100 μl) were added to the center of sterile PES Supor-200 0.2-μm filter membranes (Pall Corporation, Ann Arbor, MI) on R2A agar. Mixtures were incubated at 28°C for 48 h. Cells on the filter membrane were resuspended in 10% R2A medium, and 100 μl was spread onto R2A agar supplemented with 100 μg ml−1 kanamycin and 100 μg ml−1 rifampin. Green fluorescent protein (GFP)-expressing colonies were isolated following exposure to a Mineralight 366-nm handheld light source (UVP, Upland, CA).
Negative staining.
To visualize and characterize any possible cell surface appendages on coaggregating S. natatoria 2.1 cells, coaggregation-deficient S. natatoria 2.1 cells, and M. luteus 2.13 cells, 72-h batch cultures of each strain were negatively stained and studied by transmission electron microscopy (TEM). Cells were prepared for negative staining using a modified method described previously by Handley et al. (25), with an additional step of pretreating Formvar-coated TEM grids with poly-l-lysine to increase the attachment of coaggregation-deficient cells. Specifically, this step included the coating of the grids with 10 μl of 1% poly-l-lysine (Ted Pella, Redding, CA) for 5 min, after which the excess liquid was drawn off with filter paper, and 10 μl of cells (OD at 570 nm of 0.5) was then applied and subsequently stained with 1.5% methylamine tungstate (Ted Pella, Redding, CA).
Visual aggregation assay.
A modified visual aggregation assay (51) was used to assess coaggregation. Briefly, suspensions of S. natatoria 2.1gfp, S. natatoria 2.1COGgfp, and M. luteus 2.13 cells were washed in autoclaved distilled water three times. The suspensions were adjusted to a cell density of 5 × 109 cells ml−1, and pairs were combined in equal volumes (200 μl each). The mixtures were mixed by vortexing for 30 s, followed by gentle agitation for 10 s. After leaving mixtures to stand for 30 s, the degree of coaggregation was scored using a semiquantitative assay originally described by Cisar et al. (12).
Adhesion of S. natatoria cells to glass or glass-attached M. luteus 2.13 cells under static conditions.
In order to discriminate coaggregation-mediated adherence from cellular division, stationary-phase (72-h) S. natatoria 2.1gfp, S. natatoria 2.1COGgfp, and M. luteus 2.13 cells were washed and suspended in 0.04% sodium azide (Sigma-Aldrich, St. Louis, MO) at 4°C for 5 days to render them metabolically inactive. Treated cells were unable to grow on R2A agar and did not have altered gross cellular or coaggregation properties. Preserved coaggregating S. natatoria 2.1gfp cells maintained a visual coaggregation score of 4, while coaggregation-deficient S. natatoria 2.1COGgfp cells did not coaggregate with M. luteus 2.13 cells. Prior to attachment studies using glass-bottomed 96-well plates (Greiner Bio-One, Monroe, NC), cell suspensions were washed three times and suspended in 10% R2A medium to an OD at 570 nm of 1.0.
For single-species adhesion assays, 150-μl cell suspensions (OD at 570 nm of 1.0) of S. natatoria 2.1gfp or S. natatoria 2.1COGgfp were added to each well and incubated for 20 min. For sugar inhibition studies, cell suspensions of S. natatoria 2.1gfp or S. natatoria 2.1COGgfp were supplemented with 80 mM galactosamine (80 mM, pH 7.5) prior to addition to the 96-well plates. Plates containing the cell suspensions were incubated for 20 min, and unattached cells were removed by inverting the plate for 20 min at 25°C. For dual-species assays, 150 μl of M. luteus 2.13 cells (OD at 570 nm of 1.0) was added to each well and incubated at room temperature for 20 min. Cell suspensions were removed by inverting the plate for 20 min. Subsequently, 150-μl cell suspensions (OD at 570 nm of 1.0) of S. natatoria 2.1gfp or S. natatoria 2.1COGgfp were added. For sugar inhibition studies, cell suspensions of S. natatoria 2.1gfp or S. natatoria 2.1COGgfp were supplemented with 80 mM galactosamine (80 mM, pH 7.5) prior to addition to the 96-well plates. Plates containing the cell suspensions were incubated for 20 min, and unattached cells were removed by inverting the plate for 20 min at 25°C.
Biofilms were visualized by epifluorescence microscopy, and images were acquired with a 3.3-megapixel cooled charge-coupled-device camera (Tucsen; Fuzhou, Fujian, China). S. natatoria 2.1gfp or S. natatoria 2.1COGgfp biofilm biomasses were determined by measuring the amount of GFP fluorescence, which is proportional to the amount of S. natatoria cells in the biofilms. Quantification of GFP fluorescence was performed using a Victor 3 96-well plate reader (Perkin-Elmer, Waltham, MA). Assays were performed in three biological and four technical replicates.
Adhesion of S. natatoria cells to glass or glass-attached M. luteus 2.13 cells under flowing conditions.
Metabolically inactive S. natatoria 2.1gfp, S. natatoria 2.1COGgfp, and M. luteus 2.13 cell suspensions were prepared as described above. To compare the abilities of coaggregating and coaggregation-deficient S. natatoria 2.1 cells to attach directly to glass under flowing conditions, cell suspensions of S. natatoria 2.1gfp or S. natatoria 2.1COGgfp were adjusted to an OD at 570 nm of 0.1 and flowed (100 μl min−1) over clean glass surfaces within flow cells. Flow cells were made of Teflon and consisted of two flow channels with a chamber volume of 240 μl each (the dimensions of the flow channel are 40 mm long, 3 mm wide, and 2 mm deep). To compare the abilities of coaggregating and coaggregation-deficient S. natatoria 2.1 cells to attach to glass-surface-attached M. luteus 2.13 cells, M. luteus 2.13 cells were suspended in 10% R2A medium to an OD at 570 nm of 0.2 and were first passed through flow cells at a flow rate of 100 μl min−1 for 120 min. During this period, M. luteus 2.13 cells attached to the glass surfaces. Cell suspensions of S. natatoria 2.1gfp or S. natatoria 2.1COGgfp (OD at 570 nm of 0.1) were then flowed (100 μl min−1) over the M. luteus 2.13 cells. For sugar inhibition studies, cell suspensions of S. natatoria 2.1gfp or S. natatoria 2.1COGgfp were supplemented with 80 mM galactosamine (80 mM, pH 7.5) prior to flowing through the flow cell. Surface-attached biofilms composed of M. luteus 2.13 cells were stained with 5 μM Syto 61 (Invitrogen, Carlsbad, CA) for 30 min and washed by the flowing (100 μl min−1) of 10% R2A medium over the surface of the cells for 20 min. Cell suspensions of S. natatoria 2.1gfp or S. natatoria 2.1COGgfp were adjusted to an OD at 570 nm of 0.1 and continually flowed (100 μl min−1) over the stained M. luteus 2.13 biofilms. Flow cells were collected after 20, 60, and 180 min for study by CLSM.
Development of viable biofilms.
S. natatoria 2.1gfp, S. natatoria 2.1COGgfp, and M. luteus 2.13 cells were grown to early stationary phase (66 h). Cells were washed three times in sterile distilled water and suspended in 10% R2A medium to an OD at 570 nm of 0.1. M. luteus 2.13 cells were inoculated first and were incubated at room temperature for 120 min. S. natatoria 2.1gfp and S. natatoria 2.1COGgfp cell suspensions were inoculated and incubated similarly for 5 min. Biofilms, which contained viable cells, were developed by feeding 10% R2A medium through the flow cell. Flow cells were collected after 0 h, 24 h, 42 h, and 96 h; counterstained with Syto 61 (Invitrogen, Carlsbad, CA); and analyzed using CLSM.
CLSM and image analysis.
Biofilms were visualized using a Carl Zeiss (Thornwood, NY) LSM 510 Meta confocal laser scanning microscope at a total magnification of ×400 with a 40× Neofluar 1.5-numerical-aperture oil immersion lens (Zeiss, Thornwood, NY). Stacks of confocal images were rendered in the third dimension using lMARIS software (Bitplane AG, Zûrich, Switzerland). Biofilm biomasses were computed using COMSTAT (26).
RESULTS
Cellular properties of M. luteus and S. natatoria strains.
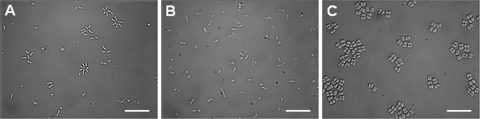
Wild-type S. natatoria 2.1 cells were arranged in rosette structures, and each rosette contained 4 to 14 cells (Fig. 1A). The coaggregation-deficient variant S. natatoria 2.1 did not form rosettes, and the vast majority (>70%) of the cells were arranged in pairs (dimers) or as single cells (Fig. 1B). M. luteus 2.13 cells were coccus-shaped and arranged in tetrads (Fig. 1C). The transformation of wild-type coaggregating and coaggregation-deficient S. natatoria 2.1 cells with the GFP-containing mini-Tn5 transposon (mTn5gusA-pgfp21) to generate S. natatoria 2.1gfp and S. natatoria 2.1COGgfp did not alter growth properties in batch culture, rosette-forming ability, and the ability to coaggregate.
FIG. 1.
Light micrographs showing S. natatoria 2.1gfp cells arranged singularly and within rosettes (A), single rod-shaped cells and cellular dimers of S. natatoria 2.1COGgfp (B), and coccus-shaped M. luteus 2.13 cells arranged in tetrads (C). Bars, 10 μm.
Negative staining revealed that S. natatoria 2.1 cells that were able to coaggregate possessed polar fimbriae, and these structures were typically within or to the side of a mass of aggregated cells (clusters could be >50 cells in size). Cells were often arranged in a polar array and are likely collapsed rosette structures (Fig. 2A). Single cells were seldom observed. Furthermore, the fimbrial structures could reach lengths that were two times the length of the typical cell, exceeding 2.5 μm (data not shown). In contrast, S. natatoria 2.1COG cells lacked polar fimbriae (Fig. 2B). These cells were typically observed in small unarranged clusters (an artifact caused by the drying process during sample preparation on TEM grids), in dimers, or as single cells. The clusters of cells were not arranged in any discernible orientation. M. luteus 2.13 cells were arranged in tetrads and possessed intricate fibrillar material on the cell surfaces (Fig. 2C). This fibrillar material extended away from the cell by approximately 0.4 μm and appeared to be looped on the surface, indicating that the structures had collapsed onto the cell surface during preparation for TEM.
FIG. 2.
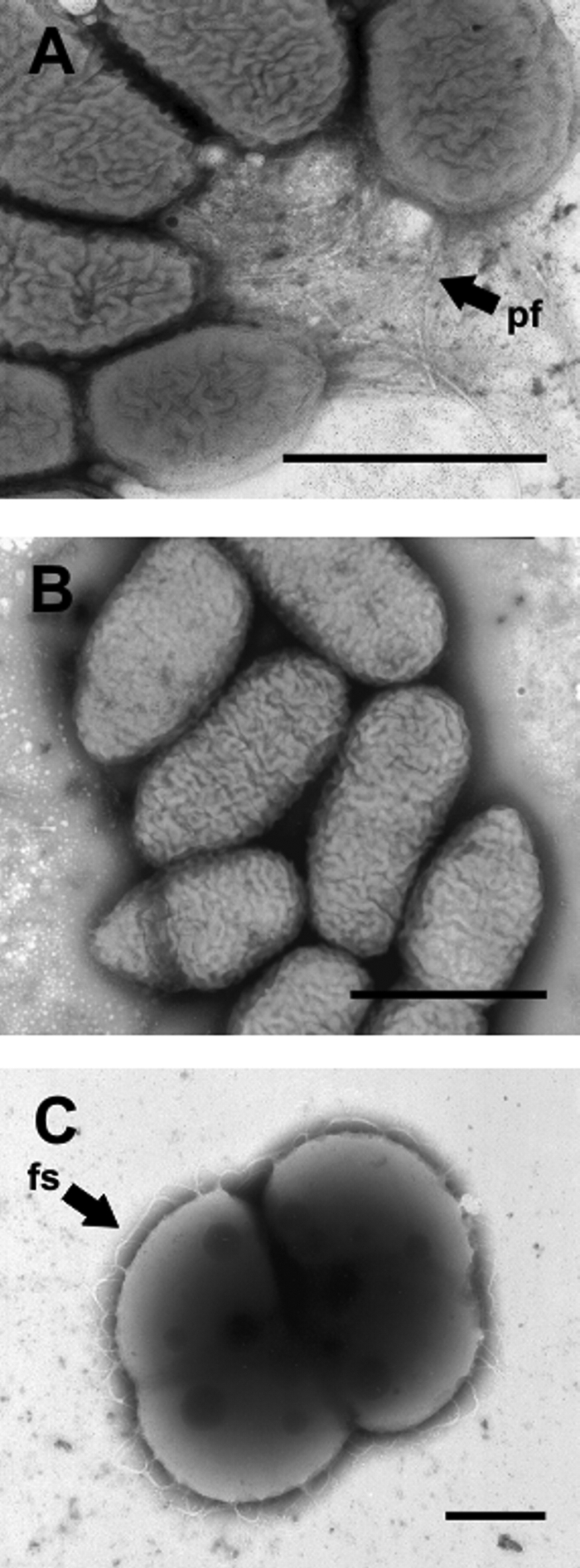
Electron micrographs of negatively stained cells of S. natatoria 2.1 with polar tufts of fimbriae (pf) (A), coaggregation-deficient S. natatoria 2.1 cells which lack fimbriae (B), and M. luteus 2.13 cells (C), which are arranged in tetrads and possess looped fibrillar structures (fs) on the cell surfaces. Cells were harvested after 48 h from batch culture and stained with 1.5% (wt/vol) methylamine tungstate. Bars, 1 μm.
Coaggregation between planktonic cells.
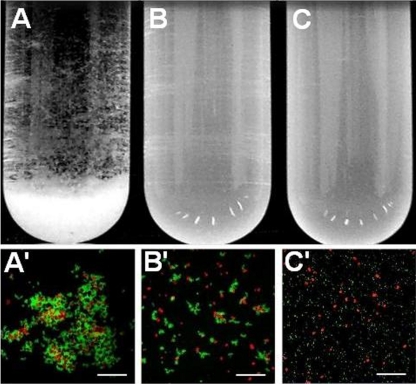
In order to examine coaggregation in planktonic suspensions, S. natatoria 2.1gfp cells were mixed with an equal number of M. luteus 2.13 cells (5 × 109 cells ml−1) in test tubes. Upon mixing, cells of the two species immediately coaggregated and settled to the bottom of the tube within 60 s, leaving a clear supernatant above (Fig. 3A). Fluorescence microscopy of coaggregates containing GFP-expressing S. natatoria 2.1gfp and Syto 61-stained M. luteus 2.13 cells showed that the coaggregates did not possess any defined structure with respect to the spatial arrangement of cells of each species in the coaggregate. The coaggregates varied in diameter from 10 μm to 1 mm (Fig. 3A and A′). The addition of 80 mM galactosamine to coaggregating mixtures and subsequent 30-s vortexing prevented coaggregates from reforming (Fig. 3B). Microscopic examination of the galactosamine-treated mixture revealed that while cells of S. natatoria 2.1 were still arranged as rosettes and cells of M. luteus 2.13 were still arranged as tetrads, very few cells coaggregated (Fig. 3B′). We examined the effect of different concentrations of galactosamine on rosettes and found that concentrations as high as 100 mM did not cause a dissolution of the rosettes (data not shown). The presence of rosettes in concentrations of >80 mM galactosamine but the absence of coaggregates indicated that rosette formation and coaggregation are mediated by different cell surface polymers. S. natatoria 2.1COGgfp was unable to coaggregate with M. luteus 2.13 and was unable to form rosettes (Fig. 3C and C′). The addition of 80 mM galactosamine to S. natatoria 2.1COGgfp and M. luteus 2.13 cells did not restore coaggregation.
FIG. 3.
Visual aggregation assay showing, from left to right, coaggregation between S. natatoria 2.1gfp and M. luteus 2.13 cells (visual coaggregation score of 4) (A), reversal of coaggregation by the addition of 80 mM galactosamine (visual coaggregation score of 0) (B), and the absence of coaggregation between S. natatoria 2.1COGgfp and M. luteus 2.13 cells (visual coaggregation score of 0) (C). Cell suspensions are at a density of 5 × 109 cells/ml. Micrographs from left to right are a coaggregate containing S. natatoria 2.1gfp cells (green) and Syto 61-stained cells of M. luteus 2.13 (red) (A′), the absence of coaggregates after the addition of 80 mM galactosamine to a coaggregating mixture (B′), and the inability of S. natatoria 2.1COGgfp cells to coaggregate with M. luteus 2.13 cells (C′). Bars, 50 μm.
Adhesion of S. natatoria cells to glass or glass-surface-attached M. luteus cells under static conditions.
A microtiter plate assay was developed to determine if coaggregation enhances the adherence of planktonic S. natatoria 2.1gfp cells to glass or glass-surface-attached M. luteus 2.13 cells under static (nonflowing) conditions.
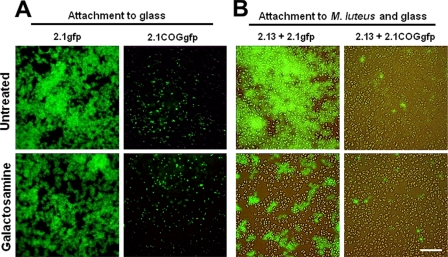
To determine if coaggregation contributes to the adhesion of S. natatoria 2.1gfp or S. natatoria 2.1COGgfp cells to glass surfaces, attachment assays were performed in the absence or presence of 80 mM galactosamine (Fig. 4A). Based upon microscopic analysis, the ability of S. natatoria 2.1gfp cells to adhere to glass was greater than the ability of S. natatoria 2.1COGgfp cells to do so. Coaggregating S. natatoria 2.1gfp cells formed loosely packed (diaphanous) single-species biofilms that covered the majority of the glass surface, while S. natatoria 2.1COGgfp cells were unable to generate confluent biofilms and formed sparse microcolonies of approximately 10 to 20 cells (Fig. 4A). This observation was confirmed by quantifying the amount of GFP fluorescence from the adherent coaggregating and coaggregation-deficient S. natatoria populations (Table 2). The emission of GFP fluorescence from coaggregating S. natatoria 2.1gfp cells was 13-fold greater than the fluorescence from coaggregation-deficient S. natatoria 2.1COGgfp cells. In the presence of 80 mM galactosamine, the ability of S. natatoria 2.1gfp cells to adhere to glass was only mildly reduced, by <17% (Fig. 4A and Table 2). The addition of 80 mM galactosamine to suspensions of S. natatoria 2.1COGgfp cells further reduced attachment to glass and was below detectable levels in our fluorometric attachment assay (Table 2). Thus, the ability to coaggregate does not substantially contribute to adhesion to glass, and a non-galactosamine-inhibitable mechanism predominates.
FIG. 4.
Epifluorescence micrographs showing the ability of coaggregating S. natatoria 2.1gfp cells or coaggregation-deficient variant S. natatoria 2.1COGgfp cells to attach directly to glass (A) or to adhere to glass-surface-attached M. luteus 2.13 cells under static conditions (B). Attachment assays were performed using 10% R2A medium (untreated) or 80 mM galactosamine-treated S. natatoria 2.1gfp cells (fluorescent green). Bar, 20 μm.
TABLE 2.
GFP fluorescence of S. natatoria 2.1gfp or S. natatoria 2.1COGgfp cells that adhered to glass or to glass covered with M. luteus 2.13 cellsa
| Biofilm and strain | Mean fluorescence (arbitrary units) ± SE
|
|
|---|---|---|
| Without galactosamine | With galactosamine | |
| Attachment to glass | ||
| 2.1gfp | 4,117 ± 151 | 3,470 ± 68 |
| 2.1COGgfp | 315 ± 21 | 0b |
| Attachment to glass covered with M. luteus 2.13 | ||
| 2.1gfp + 2.13 | 5,393 ± 84 | 2,881 ± 22 |
| 2.1COGgfp + 2.13 | 0b | 16 ± 0.20 |
The adhesion assay was performed under static conditions. Values are expressed as arbitrary units of GFP fluorescence during excitation at 488 nm and detection of emission at 540 nm. Shown are the averages of data from four replicates and the standard errors.
Fluorescence intensity was below the level of detection.
The contribution of coaggregation to the adherence of S. natatoria 2.1gfp and S. natatoria 2.1COGgfp cells to glass surfaces coated with M. luteus 2.13 was evaluated. This evaluation was performed in the absence or in the presence of 80 mM galactosamine (Fig. 4B). Visual inspection showed that the ability of S. natatoria 2.1gfp cells to adhere to surface-attached M. luteus 2.13 cells was much greater than the ability of S. natatoria 2.1COGgfp cells to adhere to surface-attached M. luteus 2.13 cells (Fig. 4B). S. natatoria 2.1gfp cells formed a thick covering over the surface of M. luteus 2.13 cells, while S. natatoria 2.1COGgfp cells sparsely covered the surfaces of M. luteus 2.13 cells and were attached mostly to exposed glass surfaces. Fluorometric analyses showed that the fluorescence from S. natatoria 2.1gfp cells was much greater than the fluorescence from S. natatoria 2.1COGgfp cells, which was below the threshold of detection using our fluorometric detection system (Table 2). The addition of 80 mM galactosamine reduced the amount of S. natatoria 2.1gfp cells that adhered to M. luteus 2.13 cells by 47% (Table 2). Galactosamine (80 mM) did not quantifiably reduce the poor adherence of S. natatoria 2.1COGgfp cells to M. luteus 2.13 cells or exposed glass surfaces (Fig. 4B and Table 2). Thus, under static conditions, galactosamine-inhibitable coaggregation substantially enhanced adhesion to glass-surface-attached M. luteus 2.13 cells and contributed to a lesser extent to the ability to adhere to glass.
Adhesion of S. natatoria cells to glass or glass-surface-attached M. luteus cells under flowing conditions.
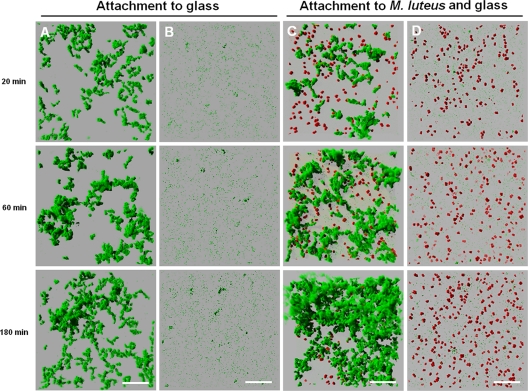
Most freshwater environments are flowing. In order to examine the propensity of coaggregating or coaggregation-deficient S. natatoria 2.1 cells to attach to glass under flowing conditions, metabolically inactive S. natatoria 2.1gfp or S. natatoria 2.1COGgfp cells were flowed over a clean glass surface (Fig. 5A and B), and the development of biofilms was assessed using CLSM. Wild-type S. natatoria 2.1gfp cells rapidly adhered to glass, and clusters of cells were seen after 20 min (Fig. 5A). By 60 min, additional S. natatoria 2.1gfp cells were recruited, and most of the clusters of S. natatoria 2.1gfp cells had enlarged. After 180 min, some of these clusters had increased slightly in size, but mostly small clusters remained. During the 180-min exposure, the biomass occupied by S. natatoria 2.1gfp cells increased from 0 μm3/μm2 to 1.99 ± 0.09 μm3/μm2. In comparison, S. natatoria 2.1COGgfp cells did not readily adhere to glass and did not develop in biomass during 180 min of constant flow over glass surfaces. After flowing coaggregation-deficient S. natatoria 2.1COGgfp cells over a clean glass surface for 20 min, S. natatoria 2.1COGgfp cells were seen to be attached to the glass only as single cells or as dimers and very occasionally formed small (<20 μm) cellular clusters (Fig. 5B). Similarly, after 60 min, sparse patches of singularly attached cells dominated. Only after 180 min were the occasional larger glass-attached clusters observed. S. natatoria 2.1COGgfp cells generated adherent cell populations with a biomass that was 10-fold less than that of S. natatoria 2.1gfp cells (0.17 ± 0.02 μm3/μm2 versus 1.99 ± 0.09 μm3/μm2, respectively).
FIG. 5.
Confocal micrographs showing the time-resolved changes in single- and dual-species biofilm formation through the continued supply and adhesion of metabolically inactive S. natatoria cells to metabolically inactive M. luteus 2.13 cells. Images were taken after 20 min, 60 min, and 180 min of continued flow. (A) Attachment of S. natatoria 2.1gfp cells to glass; (B) attachment of S. natatoria 2.1COGgfp cells to glass; (C) attachment of S. natatoria 2.1gfp cells to glass and adhesion to glass-surface-attached M. luteus 2.13 cells; (D) attachment of S. natatoria 2.1COGgfp cells to glass but no adhesion to glass-surface-attached M. luteus 2.13 cells. S. natatoria cells are green, and M. luteus 2.13 cells are red. Dimensions of the regions shown are 230 μm by 230 μm. Bars, 50 μm.
To determine if the ability of S. natatoria 2.1 cells to adhere to a glass-surface-attached population of M. luteus 2.13 cells under flowing conditions is enhanced by coaggregation, metabolically inactive S. natatoria 2.1gfp or S. natatoria 2.1COGgfp cells were flowed over glass surfaces that were covered with metabolically inactive M. luteus 2.13 cells (Fig. 5C and D). Within 20 min, glass-surface-attached dual-species aggregates predominated (Fig. 5C). Upon closer inspection and analysis by rotating three-dimensional reconstructions of CLSM image stacks of the dual-species population (Fig. 6A), it was clear that the vast majority of S. natatoria 2.1gfp cells were arranged in rosettes, and most cells were in intimate contact with M. luteus 2.13 cells. After 60 min of flowing S. natatoria 2.1gfp cells over M. luteus 2.13 cells, more colocalized dual-species clusters were observed, and many of the M. luteus 2.13 cells were completely covered by S. natatoria 2.1gfp cells. By 180 min, the dual-species clusters had developed further as a consequence of continued integrations of S. natatoria 2.1gfp cells. By this time, some of the M. luteus 2.13 cells had detached from the glass surface and had become part of the stacks of S. natatoria 2.1gfp cells that radiated into the lumen of the flow cell. These observations were quantified using COMSTAT (Table 3), and by flowing S. natatoria 2.1gfp cells over M. luteus 2.13 cells, an adherent population of S. natatoria 2.1gfp cells generated a biomass of 5.51 ± 0.10 μm3/μm2. This is 2.77-fold greater than the biomass that developed by flowing S. natatoria 2.1gfp cells over a glass surface for the same amount of time (Table 3). As opposed to the ability of S. natatoria 2.1gfp cells to adhere to M. luteus 2.13 cells and form dense dual-species assemblages, S. natatoria 2.1COGgfp cells did not attach to M. luteus 2.13 cells, and only a few cells were attached singularly to the glass substratum (Fig. 5D). Even after 180 min of continually flowing across the glass surface coated with M. luteus 2.13 cells, very few adherent S. natatoria 2.1COGgfp cells were visible. Quantification of the biomass showed that S. natatoria 2.1COGgfp cells occupied a volume that was 68.9-fold less than the biomass of S. natatoria 2.1gfp cells that developed in the presence of glass-attached M. luteus 2.13 cells (0.08 ± 0.01 μm3/μm2 versus 5.51 ± 0.01 μm3/μm2, respectively) after 180 min.
FIG. 6.
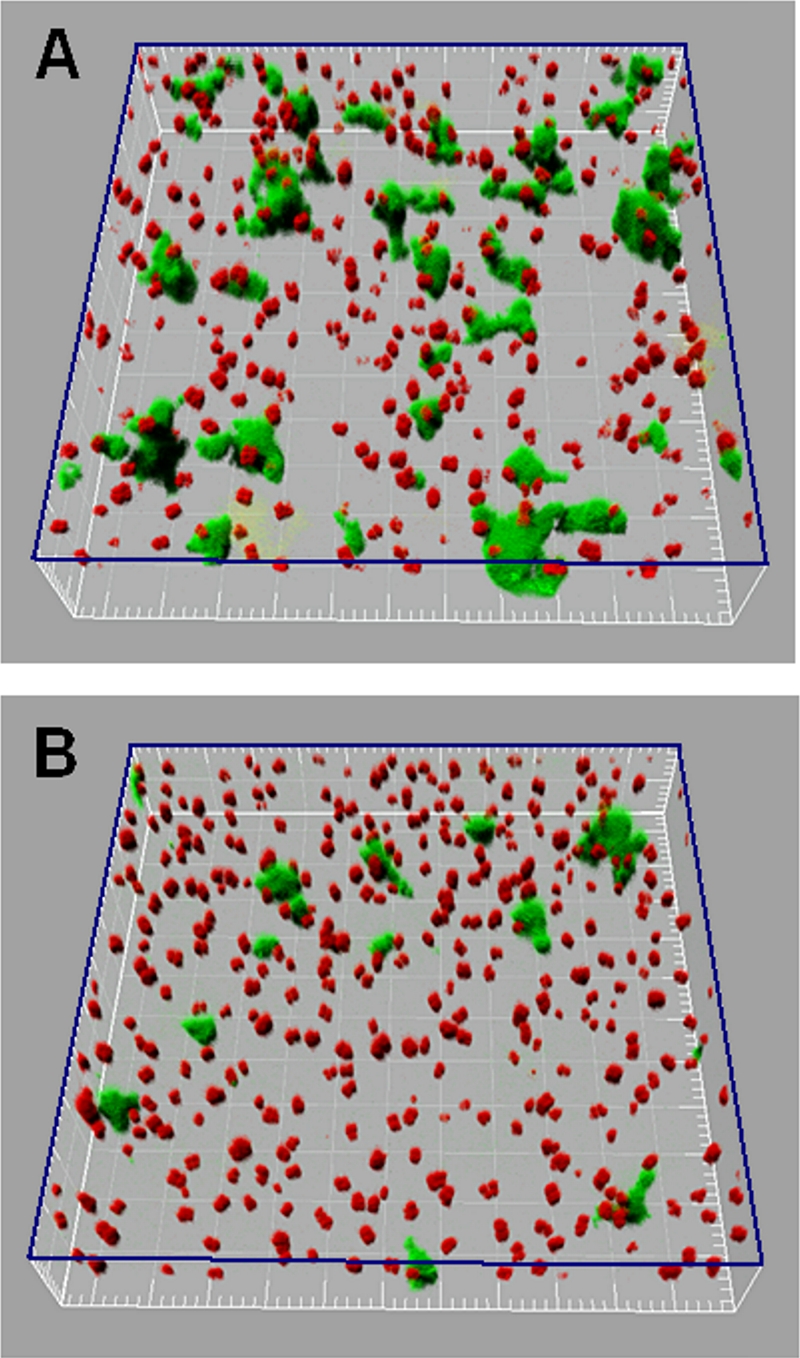
Three-dimensional CLSM reconstructions of dual-species biofilms formed after 20 min by continually flowing S. natatoria 2.1gfp cells over glass-surface-attached M. luteus 2.13 cells. (A) Spatial positions of coaggregating S. natatoria 2.1gfp cells that adhered to glass and glass-surface-attached M. luteus 2.13 cells in 10% R2A medium. (B) Spatial positions of the S. natatoria 2.1gfp cells that adhered to glass and often non-glass-surface-attached M. luteus 2.13 cells in 10% R2A medium supplemented with 80 mM galactosamine. The three-dimensional plane indicated in blue represents the glass surface. Green cells are S. natatoria 2.1gfp cells, and red cells are M. luteus 2.13 cells. Dimensions of the boxed regions are 230 μm by 230 μm.
TABLE 3.
Biomass of S. natatoria 2.1gfp or S. natatoria 2.1COGgfp cells that attached to glass or to glass-surface-attached M. luteus 2.13 cells under flowing conditionsa
| Biofilm and strain | Mean biomass size (μm3/μm2) ± SE
|
||
|---|---|---|---|
| 20 min | 60 min | 180 min | |
| Attached to glass | |||
| 2.1gfp | 1.49 ± 0.09 | 1.85 ± 0.17 | 1.99 ± 0.09 |
| 2.1COGgfp | 0.08 ± 0.01 | 0.09 ± 0.01 | 0.17 ± 0.02 |
| Attached to M. luteus 2.13 and glass | |||
| 2.1gfp + 2.13 | 1.34 ± 0.09 | 3.19 ± 0.31 | 5.51 ± 0.10 |
| 2.1COGgfp + 2.13 | 0.10 ± 0.01 | 0.07 ± 0.002 | 0.08 ± 0.01 |
Shown are the averages of data from four replicates and the standard errors.
In order to confirm that coaggregation contributes to the development of dual-species biofilms under flowing conditions, S. natatoria 2.1gfp cells were flowed over glass-attached M. luteus 2.13 cells in the presence or absence of 80 mM galactosamine. These experiments were done in parallel and only for 20 min due to the limited availability of galactosamine. In the presence of galactosamine, the amount of S. natatoria 2.1gfp cells that adhered to M. luteus 2.13 cells was significantly reduced (Fig. 6A versus B). A closer inspection of the treated biofilms indicates that the majority of the S. natatoria 2.1gfp cells adhered to the glass surface and not to the M. luteus 2.13 cells. Image analysis showed that in the absence of this sugar, S. natatoria 2.1gfp cells occupied a biomass of 0.93 ± 0.08 μm3/μm2. In the presence of galactosamine, S. natatoria 2.1gfp cells occupied a biomass of 0.30 ± 0.03 μm3/μm2. Therefore, the addition of galactosamine reduced the amount of S. natatoria 2.1gfp cells that adhered, via coaggregation interactions, to glass-surface-attached M. luteus 2.13 cells by greater than threefold within 20 min under flowing conditions.
Development of viable single- and dual-species biofilms.
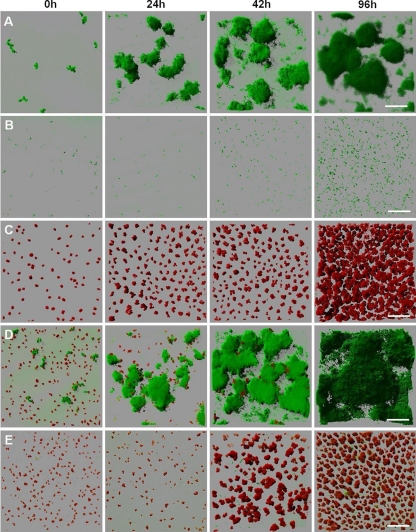
To evaluate whether the ability to coaggregate contributes to the successful colonization and expansion of S. natatoria 2.1 cells in single- and dual-species biofilms containing M. luteus 2.13 cells, viable S. natatoria 2.1gfp or S. natatoria 2.1COGgfp cells were inoculated or coinoculated with M. luteus 2.13 cells into flow cells and grown in 10% R2A medium over 4 days. CLSM images were collected, and the spatiotemporal developments of the growing single- and dual-species biofilms were compared.
Before dual-species biofilm experiments were performed, the growth and development of single-species biofilms were assessed over 96 h. Following inoculation, clusters of single and aggregated cells of S. natatoria 2.1gfp grew on glass surfaces and generated microcolonies within 24 h (Fig. 7A). These single-species microcolonies continued to develop to form large stacks of biofilm cells that extended >100 μm from the glass surface into the flow cell lumen (by 96 h). In comparison, S. natatoria 2.1COGgfp was retarded in its biofilm-forming ability, and very few aggregates were observed regardless of the period of growth in the flow cell (Fig. 7B). At any time point, the vast majority of S. natatoria 2.1COGgfp cells were scattered randomly as single cells attached to the glass surface. This indicates that either S. natatoria 2.1COGgfp cells grow poorly (a reduced mean doubling time compared to that of S. natatoria 2.1gfp cells) or the cells adhered poorly to the glass surface. As both wild-type and coaggregation-deficient strains have identical mean doubling times in batch cultures with 10% R2A medium (210 min per doubling), the failure of S. natatoria 2.1COGgfp cells to form substantial biofilms must be due to an inability to adhere to glass surfaces. Thus, the loss of S. natatoria 2.1COGgfp cells was equal or similar to the rate of growth and division (the net gain of biomass during 96 h). Inoculation of sterile flow cells with M. luteus 2.13 cells revealed that micrococcal tetrads multiplied during 96 h of incubation and grew as a biofilm to cover >70% of the glass surface and extended up to 37 μm into the lumen (Fig. 7C).
FIG. 7.
Development of viable single-species and dual-species biofilms over 96 h. Following inoculation with early-stationary-phase cells, biofilms were visualized by CLSM every 24 h. Flow cells were inoculated with S. natatoria 2.1gfp (A), S. natatoria 2.1COGgfp (B), M. luteus 2.13 (C), S. natatoria 2.1gfp and M. luteus 2.13 (D), and S. natatoria 2.1COGgfp and M. luteus 2.13 (E) cells. S. natatoria cells are green, and M. luteus cells are red. Dimensions of the regions shown are 230 μm by 230 μm. Bars, 50 μm.
The spatiotemporal development of dual-species biofilms containing coaggregating S. natatoria 2.1gfp and M. luteus 2.13 cells was very different from the development of dual-species biofilms containing M. luteus 2.13 and coaggregation-deficient S. natatoria 2.1COGgfp cells (Fig. 7D versus E). Coinoculation of flow cells with coaggregating S. natatoria 2.1gfp and M. luteus 2.13 cells resulted in the rapid development of the S. natatoria 2.1gfp biofilm community at the expense of the M. luteus community (Fig. 7D). In comparison to 96-h single-species biofilms of S. natatoria 2.1gfp, the dual-species biofilm communities contained greater amounts of S. natatoria 2.1gfp cells. Furthermore, the dual-species community contained very few M. luteus 2.13 cells, unlike the closely packed single-species biofilm of M. luteus at 96 h. In these 96-h dual-species biofilms, only very occasional M. luteus 2.13 cells could be visualized inside the biofilm and never in between the stacks of S. natatoria 2.1gfp cells. Coinoculation of flow cells with S. natatoria 2.1COGgfp and M. luteus 2.13 cells resulted in very poor biofilm development. Neither the S. natatoria 2.1COGgfp nor the M. luteus 2.13 cells formed substantial dual-species biofilms by 96 h (Fig. 7E). S. natatoria 2.1COGgfp cells did not adhere in high numbers to the glass surface and did not adhere to M. luteus 2.13 cells (Fig. 7E). Only by 96 h were the occasional microcolonies of S. natatoria 2.1GOGgfp observed. Even though there were much fewer S. natatoria 2.1COGgfp cells in the dual-species biofilm (than in the dual-species biofilm of coaggregating S. natatoria 2.1gfp and M. luteus 2.13 cells), there was a reduction in the numbers of M. luteus 2.13 cells in the biofilm (96 h) (Fig. 7E). Compared to the single-species biofilms formed by M. luteus 2.13 cells, the numbers of M. luteus 2.13 cells in the dual-species biofilms containing S. natatoria 2.1COGgfp cells were much smaller (Fig. 7C versus E). This reduction was not as extreme as was observed in dual-species biofilms containing coaggregating S. natatoria 2.1gfp cells (Fig. 7D versus E).
DISCUSSION
We have demonstrated that coaggregation enhances adhesion between planktonic and biofilm freshwater bacteria and likely contributes to the development of biofilms in static and flowing environments. This was achieved by using a combination of microscopic and quantitative techniques to study S. natatoria 2.1, an aquatic species that is indigenous to many freshwater ecosystems (22, 50, 53), and the environmentally ubiquitous species M. luteus 2.13 (8, 29, 54).
In order to demonstrate that coaggregation promotes the specific adhesion of S. natatoria 2.1gfp cells to glass-attached M. luteus 2.13 cells and subsequent biofilm development, we enriched for a naturally occurring variant of S. natatoria 2.1 that was unable to coaggregate. This approach was used previously to select for coaggregation-deficient variants of oral bacteria (13) but never before in studies of freshwater bacterial coaggregation. An unforeseen outcome was that our freshwater coaggregation-deficient variant not only was unable to coaggregate but was also unable to form rosettes and adhere to glass. A comparison of the contributions of nonspecific adhesion to glass surfaces and coaggregation interactions with M. luteus 2.13 in biofilm development was therefore difficult. It is well known that nonspecific interactions facilitate attachment to glass surfaces and other cells, although these interactions are physicochemically complex and difficult to measure (5, 6, 20). Consequently, we used galactosamine to determine if it prevented S. natatoria 2.1gfp cells from adhering to glass surfaces or prevented adhesion to M. luteus 2.13 cells that covered glass surfaces. In a study of coaggregating oral bacteria reported previously by Kolenbrander et al. (33), galactosamine was shown to inhibit coaggregation between seven strains of Treponema denticola and Fusobacterium nucleatum PK1594 (33). In parallel to their proposal, we also suggest that galactosamine inhibits coaggregation by competing against the lectin binding sites on coaggregation adhesins. Such a suggestion, that galactosamine plays an important role in mediating coaggregation between S. natatoria 2.1 and M. luteus 2.13, is also supported by data described previously by Stewart-Tull (58), who demonstrated that galactosamine is present in the cell wall of M. luteus. In context with previous findings reported by Rickard et al. (49) and findings described here, we propose that S. natatoria 2.1 expresses coaggregation adhesins that enhance biofilm integration and expansion. Indeed, under static or flowing conditions, galactosamine substantially reduced adhesion between coaggregating S. natatoria 2.1gfp and glass-surface-attached M. luteus 2.13 cells but only mildly reduced the adhesion of S. natatoria 2.1gfp cells to glass (Fig. 4 and 6). Galactosamine did not affect rosette formation (Fig. 3). Nonspecific attachment to glass and rosette formation likely require a mechanism that is independent from galactosamine-reversible coaggregation. Negative staining and visualization with TEM showed that S. natatoria 2.1 possesses polar fimbriae (Fig. 2A). We hypothesize that these polar fimbriae promote attachment to glass and to other S. natatoria 2.1 cells to form rosettes but do not promote coaggregation. The polar tufts of fimbriae are morphologically similar to those of type IV fimbriae, which are known to enhance adhesion to surfaces and enhance microcolony formation and may aid in rosette formation (15, 43, 46). S. natatoria 2.1COGgfp cells lacked these fimbriae, and this is likely the reason for the reduced ability of these cells to adhere to glass surfaces and their inability to form rosettes. While not visible by TEM and taking into account basic characterization studies of the S. natatoria 2.1 coaggregation adhesin(s) (46, 49), we hypothesize that membrane-bound galactosamine-reversible lectin-like adhesins, which are not fimbriae, recognize polysaccharides present on the surface of M. luteus 2.13 cells. Membrane-bound coaggregation adhesins in coaggregating oral bacteria have been characterized and identified (18, 32, 38), but no coaggregation adhesins from any bacteria outside of the oral cavity have been identified, and this includes S. natatoria 2.1.
The work presented here indicates that the ability to coaggregate has an impact upon the spatiotemporal development of biofilms as well as the numerical dominance of a species within a dual-species biofilm. This was demonstrated by comparing the single- and dual-species biofilm-forming abilities of the coaggregating and coaggregation-deficient variants of S. natatoria 2.1gfp. While both strains possessed the same batch culture growth characteristics, only S. natatoria 2.1gfp was able to numerically dominate in dual-species biofilms at the expense of M. luteus 2.13 (Fig. 7). Numerical dominance must be due to the coaggregation ability of S. natatoria 2.1gfp cells as well as an ability to adhere to glass and other S. natatoria 2.1gfp cells (rosette formation). An interesting possibility to consider is that S. natatoria 2.1gfp succeeds in dominating because it will juxtapose, through coaggregation, to M. luteus 2.13. Juxtaposition may enhance growth of S. natatoria 2.1gfp as a consequence of cross-species feeding between S. natatoria 2.1gfp and M. luteus 2.13 or because M. luteus 2.13 produces growth-inhibitory substances to which coaggregation-deficient S. natatoria 2.1 cells (and not coaggregating cells) are susceptible. We have not found evidence, through cross-streaking assays on R2A agar, that wild-type or coaggregation-deficient S. natatoria 2.1 or M. luteus 2.13 cells produce inhibitory substances in order to compete against one another (data not shown). With respect to the potential of cross-species feeding to improve the competitiveness of a species, Sphingomonas species are well known to be metabolically versatile and can assimilate many different compounds (2, 59, 63) and even long-chain polysaccharides (39). The ability to coaggregate may facilitate juxtaposition to target species (such as M. luteus 2.13) and allow for cross-feeding through the assimilation of components on cell surfaces or substances secreted by cells. In other ecosystems, coaggregation between Streptococcus gordonii and Veillonella atypica facilitates the improved metabolism of lactate by V. atypica by facilitating the juxtaposition of the two species (19). S. gordonii produces lactate through the fermentation of carbohydrates, and V. atypica uses lactate as a preferred source of carbon and energy. Another study that raises the importance of juxtaposition, nutrition, and competition was presented previously by Christensen et al. (11), who demonstrated that when aquatic dual-species biofilms containing Pseudomonas putida R1 and Acinetobacter sp. strain C6 were fed benzyl alcohol, P. putida R1 cells persisted in the biofilm, and the dual-species population expanded. This was because Acinetobacter sp. strain C6 assimilates benzyl alcohol and produces benzoate, which is used by juxtaposed P. putida R6 cells. If the cells were not juxtaposed, such as in planktonic chemostats, P. putida R6 cannot maintain a niche. We are currently using proteomic and genomic techniques to explore the mechanism behind the interspecies competitive nature of S. natatoria 2.1.
S. natatoria 2.1 was originally proposed to be a bridging organism because of the ability of this strain to coaggregate with partner strains from eight other genera in a freshwater biofilm (50). Here, we have demonstrated that the ability to coaggregate with one of these original partner freshwater biofilm strains, M. luteus 2.13, contributes to the integration of S. natatoria 2.1gfp into a biofilm as well as the expansion of the S. natatoria 2.1 population in growing biofilms. Recently, Simoes et al. (53) indicated that Acinetobacter calcoaceticus is also a bridging bacterium in freshwater biofilms. This was demonstrated by adding this species to a multispecies biofilm and showing that the biofilm biomass increased. However, these results are tempered by the ability of this species to strongly autoaggregate (a visual aggregation score of 2+). Nonetheless, the work by Simoes et al. (53) is intriguing and raises the question of whether freshwater bridging organisms such as A. calcoaceticus and S. natatoria can aid the retention of microbial pathogens or if the ability to coaggregate with many species (i.e., bridge) is a mechanism to outcompete other species in freshwater multispecies biofilms. In the case of S. natatoria 2.1gfp, the data presented here suggest that the ability of S. natatoria 2.1gfp to coaggregate and bridge would enhance overall multispecies biofilm biomass, but unlike A. calcoaceticus, which seemingly favors the expansion of all species of a mixed community, S. natatoria 2.1gfp would numerically dominate at the expense of other species in the biofilm.
Acknowledgments
We acknowledge financial support provided by a new lecturer research grant (to A. H. Rickard) from the Society for Applied Microbiology (United Kingdom).
We acknowledge Pauline Handley (University of Manchester, United Kingdom) for her invaluable research support and advice for the TEM studies.
Footnotes
Published ahead of print on 17 April 2009.
REFERENCES
- 1.Alekhina, I. A., D. Marie, J. R. Petit, V. V. Lukin, V. M. Zubkov, and S. A. Bulat. 2007. Molecular analysis of bacterial diversity in kerosene-based drilling fluid from the deep ice borehole at Vostok, East Antarctica. FEMS Microbiol. Ecol. 59:289-299. [DOI] [PubMed] [Google Scholar]
- 2.Balkwill, D. L., J. K. Fredrickson, and M. Romine. 2006. Sphingomonas and related genera, p. 605-629. In M. Dworkin, S. Falkow, E. Rosenberg, K. Schleifer, and E. Stackebrandt (ed.), The prokaryotes, vol. 7. Proteobacteria: delta, epsilon subclass. Springer, New York, NY. [Google Scholar]
- 3.Beech, I. B., and J. Sunner. 2004. Biocorrosion: towards understanding interactions between biofilms and metals. Curr. Opin. Biotechnol. 15:181-186. [DOI] [PubMed] [Google Scholar]
- 4.Boyer, H. W., and D. Roulland-Dussoix. 1969. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J. Mol. Biol. 41:459-472. [DOI] [PubMed] [Google Scholar]
- 5.Busscher, H. J., M. M. Cowan, and H. C. van der Mei. 1992. On the relative importance of specific and non-specific approaches to oral microbial adhesion. FEMS Microbiol. Rev. 8:199-209. [DOI] [PubMed] [Google Scholar]
- 6.Busscher, H. J., and H. C. van der Mei. 1997. Physico-chemical interactions in initial microbial adhesion and relevance for biofilm formation. Adv. Dent. Res. 11:24-32. [DOI] [PubMed] [Google Scholar]
- 7.Buswell, C. M., Y. M. Herlihy, L. M. Lawrence, J. T. McGuiggan, P. D. Marsh, C. W. Keevil, and S. A. Leach. 1998. Extended survival and persistence of Campylobacter spp. in water and aquatic biofilms and their detection by immunofluorescent-antibody and -rRNA staining. Appl. Environ. Microbiol. 64:733-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buswell, C. M., Y. M. Herlihy, P. D. Marsh, C. W. Keevil, and S. A. Leach. 1997. Coaggregation amongst aquatic biofilm bacteria. J. Appl. Microbiol. 83:477-484. [Google Scholar]
- 9.Callow, M. E., and J. E. Callow. 2002. Marine biofouling: a sticky problem. Biologist (London) 49:10-14. [PubMed] [Google Scholar]
- 10.Camilli, A., and B. L. Bassler. 2006. Bacterial small-molecule signaling pathways. Science 311:1113-1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christensen, B. B., J. A. Haagensen, A. Heydorn, and S. Molin. 2002. Metabolic commensalism and competition in a two-species microbial consortium. Appl. Environ. Microbiol. 68:2495-2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cisar, J. O., P. E. Kolenbrander, and F. C. McIntire. 1979. Specificity of coaggregation reactions between human oral streptococci and strains of Actinomyces viscosus or Actinomyces naeslundii. Infect. Immun. 24:742-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clemans, D. L., and P. E. Kolenbrander. 1995. Isolation and characterization of coaggregation-defective (Cog−) mutants of Streptococcus gordonii DL1 (Challis). J. Ind. Microbiol. 15:193-197. [DOI] [PubMed] [Google Scholar]
- 14.Coetser, S. E., and T. E. Cloete. 2005. Biofouling and biocorrosion in industrial water systems. Crit. Rev. Microbiol. 31:213-232. [DOI] [PubMed] [Google Scholar]
- 15.Craig, L., and J. Li. 2008. Type IV pili: paradoxes in form and function. Curr. Opin. Struct. Biol. 18:267-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ditta, G., T. Schmidhauser, E. Yakobson, P. Lu, X. W. Liang, D. R. Finlay, D. Guiney, and D. R. Helinski. 1985. Plasmids related to the broad host range vector, pRK290, useful for gene cloning and for monitoring gene expression. Plasmid 13:149-153. [DOI] [PubMed] [Google Scholar]
- 17.Eady, E. A., P. Coates, J. I. Ross, A. H. Ratyal, and J. H. Cove. 2000. Antibiotic resistance patterns of aerobic coryneforms and furazolidone-resistant gram-positive cocci from the skin surface of the human axilla and fourth toe cleft. J. Antimicrob. Chemother. 46:205-213. [DOI] [PubMed] [Google Scholar]
- 18.Egland, P. G., L. D. Du, and P. E. Kolenbrander. 2001. Identification of independent Streptococcus gordonii SspA and SspB functions in coaggregation with Actinomyces naeslundii. Infect. Immun. 69:7512-7516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Egland, P. G., R. J. Palmer, Jr., and P. E. Kolenbrander. 2004. Interspecies communication in Streptococcus gordonii-Veillonella atypica biofilms: signaling in flow conditions requires juxtaposition. Proc. Natl. Acad. Sci. USA 101:16917-16922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ellen, R. P., G. Lepine, and P. M. Nghiem. 1997. In vitro models that support adhesion specificity in biofilms of oral bacteria. Adv. Dent. Res. 11:33-42. [DOI] [PubMed] [Google Scholar]
- 21.Elliott, D. R., M. Wilson, C. M. Buckley, and D. A. Spratt. 2006. Aggregative behavior of bacteria isolated from canine dental plaque. Appl. Environ. Microbiol. 72:5211-5217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Furuhata, K., Y. Kato, K. Goto, K. Saitou, J. Sugiyama, M. Hara, and M. Fukuyama. 2007. Identification of yellow-pigmented bacteria isolated from hospital tap water in Japan and their chlorine resistance. Biocontrol Sci. 12:39-46. [DOI] [PubMed] [Google Scholar]
- 23.Gibbons, R. J., and M. Nygaard. 1970. Interbacterial aggregation of plaque bacteria. Arch. Oral Biol. 15:1397-1400. [DOI] [PubMed] [Google Scholar]
- 24.Hall-Stoodley, L., J. W. Costerton, and P. Stoodley. 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2:95-108. [DOI] [PubMed] [Google Scholar]
- 25.Handley, P. S., P. L. Carter, J. E. Wyatt, and L. M. Hesketh. 1985. Surface structures (peritrichous fibrils and tufts of fibrils) found on Streptococcus sanguis strains may be related to their ability to coaggregate with other oral genera. Infect. Immun. 47:217-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heydorn, A., A. T. Nielsen, M. Hentzer, C. Sternberg, M. Givskov, B. K. Ersboll, and S. Molin. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146:2395-2407. [DOI] [PubMed] [Google Scholar]
- 27.Ivanov, V., X. H. Wang, S. T. Tay, and J. H. Tay. 2006. Bioaugmentation and enhanced formation of microbial granules used in aerobic wastewater treatment. Appl. Microbiol. Biotechnol. 70:374-381. [DOI] [PubMed] [Google Scholar]
- 28.Jayaraman, A., and T. K. Wood. 2008. Bacterial quorum sensing: signals, circuits, and implications for biofilms and disease. Annu. Rev. Biomed. Eng. 10:145-167. [DOI] [PubMed] [Google Scholar]
- 29.Kloos, W. E., R. J. Zimmerman, and R. F. Smith. 1976. Preliminary studies on the characterization and distribution of Staphylococcus and Micrococcus species on animal skin. Appl. Environ. Microbiol. 31:53-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kmet, V., and F. Lucchini. 1997. Aggregation-promoting factor in human vaginal Lactobacillus strains. FEMS Immunol. Med. Microbiol. 19:111-114. [DOI] [PubMed] [Google Scholar]
- 31.Kolenbrander, P. E. 2000. Oral microbial communities: biofilms, interactions, and genetic systems. Annu. Rev. Microbiol. 54:413-437. [DOI] [PubMed] [Google Scholar]
- 32.Kolenbrander, P. E., R. N. Andersen, D. S. Blehert, P. G. Egland, J. S. Foster, and R. J. Palmer, Jr. 2002. Communication among oral bacteria. Microbiol. Mol. Biol. Rev. 66:486-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kolenbrander, P. E., K. D. Parrish, R. N. Andersen, and E. P. Greenberg. 1995. Intergeneric coaggregation of oral Treponema spp. with Fusobacterium spp. and intrageneric coaggregation among Fusobacterium spp. Infect. Immun. 63:4584-4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ledder, R. G., A. S. Timperley, M. K. Friswell, S. Macfarlane, and A. J. McBain. 2008. Coaggregation between and among human intestinal and oral bacteria. FEMS Microbiol. Ecol. 66:630-636. [DOI] [PubMed] [Google Scholar]
- 35.Lopez, L., C. Pozo, B. Rodelas, C. Calvo, B. Juarez, M. V. Martinez-Toledo, and J. Gonzalez-Lopez. 2005. Identification of bacteria isolated from an oligotrophic lake with pesticide removal capacities. Ecotoxicology 14:299-312. [DOI] [PubMed] [Google Scholar]
- 36.Lyon, G. J., and R. P. Novick. 2004. Peptide signaling in Staphylococcus aureus and other gram-positive bacteria. Peptides 25:1389-1403. [DOI] [PubMed] [Google Scholar]
- 37.Malik, A., M. Sakamoto, T. Ono, and K. Kakii. 2003. Coaggregation between Acinetobacter johnsonii S35 and Microbacterium esteraromaticum strains isolated from sewage activated sludge. J. Biosci. Bioeng. 96:10-15. [PubMed] [Google Scholar]
- 38.Manch-Citron, J. N., J. Allen, M. Moos, Jr., and J. London. 1992. The gene encoding a Prevotella loescheii lectin-like adhesin contains an interrupted sequence which causes a frameshift. J. Bacteriol. 174:7328-7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mishima, Y., K. Momma, O. Miyake, W. Hashimoto, B. Mikami, and K. Murata. 2002. A super-channel in bacteria: macromolecule uptake and depolymerization systems of Sphingomonas sp. A1 with a special cell surface structure. Biotechnol. Genet. Eng. Rev. 19:105-119. [DOI] [PubMed] [Google Scholar]
- 40.Molin, S., and T. Tolker-Nielsen. 2003. Gene transfer occurs with enhanced efficiency in biofilms and induces enhanced stabilisation of the biofilm structure. Curr. Opin. Biotechnol. 14:255-261. [DOI] [PubMed] [Google Scholar]
- 41.Murray, T. S., M. Egan, and B. I. Kazmierczak. 2007. Pseudomonas aeruginosa chronic colonization in cystic fibrosis patients. Curr. Opin. Pediatr. 19:83-88. [DOI] [PubMed] [Google Scholar]
- 42.Parsek, M. R., and P. K. Singh. 2003. Bacterial biofilms: an emerging link to disease pathogenesis. Annu. Rev. Microbiol. 57:677-701. [DOI] [PubMed] [Google Scholar]
- 43.Pelicic, V. 2008. Type IV pili: e pluribus unum? Mol. Microbiol. 68:827-837. [DOI] [PubMed] [Google Scholar]
- 44.Qian, P. Y., S. C. Lau, H. U. Dahms, S. Dobretsov, and T. Harder. 2007. Marine biofilms as mediators of colonization by marine macroorganisms: implications for antifouling and aquaculture. Mar. Biotechnol. (New York) 9:399-410. [DOI] [PubMed] [Google Scholar]
- 45.Reasoner, D. J., and E. E. Geldreich. 1985. A new medium for the enumeration and subculture of bacteria from potable water. Appl. Environ. Microbiol. 49:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rickard, A. H. 2001. Coaggregation between freshwater biofilm bacteria. University of Manchester, Manchester, United Kingdom.
- 47.Rickard, A. H., P. Gilbert, and P. S. Handley. 2004. Influence of growth environment on coaggregation between freshwater biofilm bacteria. J. Appl. Microbiol. 96:1367-1373. [DOI] [PubMed] [Google Scholar]
- 48.Rickard, A. H., P. Gilbert, N. J. High, P. E. Kolenbrander, and P. S. Handley. 2003. Bacterial coaggregation: an integral process in the development of multi-species biofilms. Trends Microbiol. 11:94-100. [DOI] [PubMed] [Google Scholar]
- 49.Rickard, A. H., S. A. Leach, C. M. Buswell, N. J. High, and P. S. Handley. 2000. Coaggregation between aquatic bacteria is mediated by specific-growth-phase-dependent lectin-saccharide interactions. Appl. Environ. Microbiol. 66:431-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rickard, A. H., S. A. Leach, L. S. Hall, C. M. Buswell, N. J. High, and P. S. Handley. 2002. Phylogenetic relationships and coaggregation ability of freshwater biofilm bacteria. Appl. Environ. Microbiol. 68:3644-3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rickard, A. H., A. J. McBain, R. G. Ledder, P. S. Handley, and P. Gilbert. 2003. Coaggregation between freshwater bacteria within biofilm and planktonic communities. FEMS Microbiol. Lett. 220:133-140. [DOI] [PubMed] [Google Scholar]
- 52.Rickard, A. H., A. J. McBain, A. T. Stead, and P. Gilbert. 2004. Shear rate moderates community diversity in freshwater biofilms. Appl. Environ. Microbiol. 70:7426-7435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Simoes, L. C., M. Simoes, and M. J. Vieira. 2008. Intergeneric coaggregation among drinking water bacteria: evidence of a role for Acinetobacter calcoaceticus as a bridging bacterium. Appl. Environ. Microbiol. 74:1259-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sims, G. K., L. E. Sommers, and A. Konopka. 1986. Degradation of pyridine by Micrococcus luteus isolated from soil. Appl. Environ. Microbiol. 51:963-968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sly, L. I., and M. M. Cahill. 1997. Transfer of Blastobacter natatorius (Sly 1985) to the genus Blastomonas gen. nov. as Blastomonas natatoria comb. nov. Int. J. Syst. Bacteriol. 47:566-568. [DOI] [PubMed] [Google Scholar]
- 56.Sly, L. I., and M. H. Hargreaves. 1984. Two unusual budding bacteria isolated from a swimming pool. J. Appl. Bacteriol. 56:479-486. [DOI] [PubMed] [Google Scholar]
- 57.Socransky, S. S., and A. D. Haffajee. 2005. Periodontal microbial ecology. Periodontol. 2000 38:135-187. [DOI] [PubMed] [Google Scholar]
- 58.Stewart-Tull, D. E. 1968. Determination of amino sugars in mixtures containing glucosamine, galactosamine and muramic acid. Biochem. J. 109:13-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stolz, A. 2009. Molecular characteristics of xenobiotic-degrading sphingomonads. Appl. Microbiol. Biotechnol. 81:793-811. [DOI] [PubMed] [Google Scholar]
- 60.Stoodley, P., K. Sauer, D. G. Davies, and J. W. Costerton. 2002. Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 56:187-209. [DOI] [PubMed] [Google Scholar]
- 61.Vandevoorde, L., H. Christiaens, and W. Verstraete. 1992. Prevalence of coaggregation reactions among chicken lactobacilli. J. Appl. Bacteriol. 72:214-219. [DOI] [PubMed] [Google Scholar]
- 62.West, S. A., A. S. Griffin, A. Gardner, and S. P. Diggle. 2006. Social evolution theory for microorganisms. Nat. Rev. Microbiol. 4:597-607. [DOI] [PubMed] [Google Scholar]
- 63.White, D. C., S. D. Sutton, and D. B. Ringelberg. 1996. The genus Sphingomonas: physiology and ecology. Curr. Opin. Biotechnol. 7:301-306. [DOI] [PubMed] [Google Scholar]
- 64.Xi, C., M. Lambrecht, J. Vanderleyden, and J. Michiels. 1999. Bi-functional gfp- and gusA-containing mini-Tn5 transposon derivatives for combined gene expression and bacterial localization studies. J. Microbiol. Methods 35:85-92. [DOI] [PubMed] [Google Scholar]