Abstract
The effects of genetically modified (GM), zeaxanthin-accumulating potato plants on microbial communities in the rhizosphere were compared to the effects of different potato cultivars. Two GM lines and their parental cultivar, as well as four other potato cultivars, were grown in randomized field plots at two sites and in different years. Rhizosphere samples were taken at three developmental stages during plant growth and analyzed using denaturing gradient gel electrophoresis (DGGE) fingerprints of Bacteria, Actinobacteria, Alpha- and Betaproteobacteria, Bacillus, Streptomycetaceae, Pseudomonas, gacA, Fungi, and Ascomycetes. In the bacterial DGGE gels analyzed, significant differences between the parental cultivar and the two GM lines were detected mainly for Actinobacteria but also for Betaproteobacteria and Streptomycetaceae, yet these differences occurred only at one site and in one year. Significant differences occurred more frequently for Fungi, especially Ascomycetes, than for bacteria. When all seven plant genotypes were compared, DGGE analysis revealed that different cultivars had a greater effect on both bacterial and fungal communities than genetic modification. The effects of genetic modification were detected mostly at the senescence developmental stage of the plants. The site was the overriding factor affecting microbial community structure compared to the plant genotype. In general, the fingerprints of the two GM lines were more similar to that of the parental cultivar, and the differences observed did not exceed natural cultivar-dependent variability.
Microorganisms play a key role in agriculture because they are important for plant growth and health, turnover of organic material, and maintenance of ecosystem functions. In the rhizosphere, defined as the soil influenced by the plant roots (37), microorganisms benefit from nutrients provided by root exudates and form close relationships with the plants. The plant species and also the plant genotypes have been reported to influence microbial communities in the rhizosphere (15, 17, 22, 28, 29, 36). Despite the importance of soil microbes for soil and plant health, the response of these microbes to large-scale cultivation of genetically modified (GM) crops is still poorly understood. Gene technology offers the possibility of more targeted modification of a plant compared to classical breeding approaches, which might limit effects on associated microbes. Therefore, whether a single genetic modification correlates with a less pronounced effect on microbial communities in the rhizosphere needs to be assessed.
Potatoes with increased zeaxanthin levels in their tubers were designed as a functional food to counteract age-related macular degeneration, which is a major cause of visual impairment in elderly people. It has been shown that dietary intake of a high level of zeaxanthin significantly reduces the risk of suffering from this disease (10, 35). Zeaxanthin is naturally produced in potato plants but is further modified to violaxanthin via the enzyme zeaxanthin epoxidase. Downregulation of the zeaxanthin epoxidase gene resulted in accumulation of zeaxanthin in tubers of GM potato plants (34). However, the possibility that additional plant metabolic processes, as well as root exudation patterns, are affected by the genetic modifications cannot be excluded.
While many studies have aimed at investigating potential impacts of GM plants on their associated microbial communities (for reviews, see references 3 and 26), the majority of studies conducted so far only compared a GM line to a non-GM line (4, 9, 20, 21). However, potential effects of GM plants on microbial communities need to be evaluated in light of natural variation among cultivars of the same plant species. Recently, a study of the rhizosphere communities of fructan-producing GM potatoes compared to those of isogenic controls and conventional cultivars failed to show plant genotype effects (2). However, this result was based only on analysis of Bacteria and did not consider potential effects on different microbial groups.
The objectives of this study were to assess the effects of the growth of zeaxanthin-accumulating potatoes on microbial communities in the rhizosphere and to relate putative effects to natural variation among potato cultivars. Effects were ascertained at two different sites and in several years. Compared to previous studies, this study provides a comprehensive in-depth analysis of the response of various bacterial and fungal groups to potential effects of two GM lines. We investigated the hypothesis that the effects of the genetic modification on rhizosphere communities were less pronounced than the effects of genotype differences among cultivars resulting from conventional breeding.
MATERIALS AND METHODS
Potato cultivars and GM lines.
Two GM potato lines (Solanum tuberosum L.) with altered zeaxanthin levels and their parental cultivar, ‘Baltica’, as well as four additional commercial potato cultivars, ‘Selma’, ‘Désirée’, ‘Ditta’, and ‘Sibu’, were planted. GM lines SR47 (‘Baltica’ cosuppression) and SR48 (‘Baltica’ antisense) accumulate the carotenoid zeaxanthin in their tubers. The tubers of GM lines SR47 and SR48 (referred to by Römer et al. [34] as SR47-18 and SR48-17, respectively) contained up to 40 μg/g (dry weight [dw]) and 17 μg/g (dw) of zeaxanthin, compared to 0.2 μg/g (dw) for ‘Baltica’.
Field design.
The potato cultivars and GM lines were grown at two different field sites in southern Germany, Roggenstein (in 2005 and 2007) and Oberviehhausen (in 2006). The soil characteristics at the two sites differed considerably (Table 1). The potato plants were grown in a randomized block consisting of six replicate plots per cultivar or GM line with 40 plants each (see Fig. S1 in the supplemental material). In addition, only the commercial cultivars ‘Baltica’, ‘Désirée’, and ‘Sibu’ were grown at Roggenstein in 2006 because the GM lines were not permitted at this site in 2006.
TABLE 1.
Characteristics of soils used in this study
| Site | % Clay | % Sand | % Silt | % Organic C | % Total N | pH |
|---|---|---|---|---|---|---|
| Roggenstein | 28.1 | 26.1 | 44.0 | 1.1 | 0.1 | 6.6 |
| Oberviehhausen | 14.1 | 54.6 | 31.3 | 1.9 | 0.2 | 6.5 |
Sampling and sample processing.
Sampling was carried out at three developmental stages of the plants, young plants (EC30), flowering plants (EC60), and senescent plants (EC90), as described by Hack et al. (16), at the Roggenstein site in 2005 and 2006 and at the Oberviehhausen site in 2006. Because of delayed emergence of the GM lines due to very dry weather conditions, leading to great differences in plant development mainly at EC30, only EC60 and EC90 plants were sampled at Roggenstein in 2007. Five plants per plot were carefully removed from the soil. After the plants were shaken, the roots with adhering soil were combined, cut into pieces, and treated as a composite sample. Four composite samples of each cultivar and GM line were processed further. For isolation of the root-associated microbes, 10 g of root material was transferred into a sterile stomacher bag and homogenized with 30 ml Milli-Q water for 60 s using a stomacher laboratory blender (Seward, West Sussex, United Kingdom) at high speed. This homogenization step was repeated three times, and the combined suspensions were collected in two 50-ml tubes. The first tube was centrifuged for 15 min at 4°C and 10,000 × g, the supernatant was discarded, and the tube was filled with the contents of the second tube prior to another centrifugation. The resulting pellets containing the root-associated bacteria were frozen at −80°C until DNA was extracted.
Extraction of DNA from rhizosphere samples.
For extraction of DNA, 0.5 g of pellets obtained from 10 g root material was used. The bacterial cells were lysed mechanically twice with a FastPrep FP120 bead beating system (Q-Biogene, Carlsbad, CA) for 30 s at high speed. Thereafter, the DNA was extracted with a BIO-101 DNA spin kit for soil (Q-Biogene, Carlsbad, CA) according to the instructions of the manufacturer. The extracted DNA was purified further using a Geneclean spin kit (Q-Biogene, Carlsbad, CA) as described by the manufacturer, except that the DNA was eluted in the same amount of solution that was used at the beginning of the experiment for purification. The yield of genomic DNA was checked by use of a 0.8% agarose gel photographed under UV light after ethidium bromide staining. The DNA yield was estimated using the 1-kb Plus DNA ladder (Invitrogen, Karlsruhe, Germany), and the DNA was diluted 1:5 with the elution buffer from the purification kit.
Development of a Streptomycetaceae-specific primer system.
Primers specific for the family Streptomycetaceae were designed using the PROBE DESIGN and MATCH PROBE subroutines in the ARB software (http://www.arb-home.de). The Probe Match function of Ribosomal Database Project II (http://rdp.cme.msu.edu/) was used for in silico analysis of the primer specificity based on the last 10 nucleotides at the 3′ end. The primer pair was highly specific for the family Streptomycetaceae, showing perfect matches with only five non-Streptomycetaceae sequences related to the genera Microbacterium and Spirillospora.
Amplification of bacterial 16S rRNA and Pseudomonas gacA gene fragments.
Amplification of the bacterial 16S rRNA gene from total community DNA for denaturing gradient gel electrophoresis (DGGE) analysis was carried out using primer pair F984GC/R1378 as described by Heuer et al. (18). For amplification of group-specific 16S rRNA and gacA gene fragments, a nested PCR approach was used. This approach consisted of specific amplification using group-specific primers (Table 2), followed by amplification of the DGGE fragment. Amplification for the Streptomycetaceae family was conducted with a reaction mixture consisting of 1 μl of template DNA (1 to 5 ng), 1× Stoffel buffer II (Applied Biosystems), 0.2 mM deoxynucleoside triphosphates (dNTPs), 2.5 mM MgCl2, 5% (vol/vol) dimethyl sulfoxide (DMSO), 2.5 μg bovine serum albumin, 0.1 μM primer F126, 0.1 μM primer R1423 (Table 2), and 1.25 U AmpliTaq Gold (Applied Biosystems). An initial denaturation at 95°C for 10 min was followed by 30 cycles of 95°C for 1 min, 62°C for 1 min, and 72°C for 1 min and a final extension at 72°C for 10 min. PCR products were diluted 1:10 and used as templates for a PCR with primer pair F984GC/R1378 for 20 cycles.
TABLE 2.
Primers used in this study targeting bacteria and fungi
| Primer pair | Primer | Sequence (5′-3′)a | Taxonomic group | Annealing temp (°C) | Reference |
|---|---|---|---|---|---|
| F984GC/R1378 | F984 | GC clamp-AACGCGAAGAACCTTAC | Bacteria | 53 | 18 |
| R1378 | CGGTGTGTACAAGGCCCGGGAACG | 31 | |||
| F243/R1378 | F243 | GGATGAGCCCGCGGCCTA | Actinobacteria | 63 | 18 |
| F203α/R1492 | F203α | CCGCATACGCCCTACGGGGGAAAGATTTAT | Alphaproteobacteria | 56 | 12 |
| R1492 | TACGG(C/T)TACCTTGTTACGACTT | ||||
| F948β/R1492 | F948β | CGCACAAGCGGTGGATGA | Betaproteobacteria | 64 | 12 |
| F311Ps/R1459Ps | F311Ps | CTGGTCTGAGAGGATGATCAGT | Pseudomonas | 63 | 30 |
| R1459Ps | AATCACTCCGTGGTAACCGT | ||||
| BacF/R1378 | BacF | GGGAAACCGGGGCTAATACCGGAT | Bacillus | 65 | 11 |
| F126/R1423 | F126 | GCCCTGCACTCTGGGACAAGC | Streptomycetaceae | 62 | This study |
| R1423 | GTTAGGCCACCGGCTTCG | ||||
| gacA-1F/gacA2 | gacA-1F | TGATTAGGGTGYTAGTDGTCGA | Pseudomonas gacA gene | 57 | 7 |
| gacA2 | MGYCARYTCVACRTCRCTGSTGAT | 8 | |||
| gacA-1FGC/gacA2R | gacA-1FGC | GC clamp-GATTAGGGTGCTAGTGGTCGA | Pseudomonas gacA gene | 52 | 7 |
| gacA2R | GGTTTTCGGTGACAGGCA | ||||
| ITS1F/ITS4 | ITS1F | CTTGGTCATTTAGAGGAAGTAA | Fungi | 55 | 1 |
| ITS4 | TCCTCCGCTTATTGATATGC | ||||
| ITS1FGC/ITS2 | ITS2 | GCTGCGTTCTTCATCGATGC | Fungi | 55 | 1 |
| ITS1F/ITS4A | ITS4A | CGCCGTTACTGGGGCAATCCCTG | Ascomycetes | 53 | 25 |
The GC clamp sequence was CGCCCGGGGCGCGCCCCGGGCGGGGCGGGGGCACGGGGGG.
Amplification of fungal ITS fragments.
Amplification of the fungal internal transcribed spacer (ITS) fragment prior to DGGE analysis was performed using a nested PCR approach with primer pairs ITS1F/ITS4 and ITS1FGC/ITS2 (1) (Table 2). The reaction mixture for the first PCR (25 μl) was composed of 1 μl of template DNA (1 to 5 ng), 1× Stoffel buffer II (Applied Biosystems), 0.2 mM dNTPs, 3.75 mM MgCl2, 5% (vol/vol) DMSO, 0.1 μM primers, and 2.5 U AmpliTaq Gold (Applied Biosystems). Initial denaturation at 95°C for 5 min was followed by 30 cycles of 95°C for 30 s, 55°C for 30 s, 72°C for 1 min and a final extension at 72°C for 10 min. Samples served as templates for the second PCR. The reaction mixture for the second PCR was the same as that for the first PCR, except that 4% (vol/vol) DMSO and 0.2 μM primers were used. The PCR conditions were the same as those described for the first PCR except for the number of cycles, which was reduced to 25. For Ascomycetes-specific amplification primer pairs ITS1F/ITS4A (25) and ITS1FGC/ITS2 (Table 2) were used. The reaction mixture (25 μl) contained 1 μl of template DNA (1 to 5 ng), 1× Stoffel buffer II (Applied Biosystems), 0.2 mM dNTPs, 3.75 mM MgCl2, 5% (vol/vol) DMSO, 0.1 μM of each primer, and 2.5 U AmpliTaq Gold (Applied Biosystems). The thermal cycle started with denaturation at 95°C for 5 min, which was followed by 30 cycles of 95°C for 30 s, 53°C for 35 s, and 72°C for 2 min and then a final extension at 72°C for 10 min. The samples were diluted 1:20 and served as templates for the second PCR as described above.
DGGE of bacterial and fungal gene fragments.
DGGE analysis was performed using a PhorU2 apparatus (Ingeny, Goes, The Netherlands) with a double gradient for both community and group-specific 16S rRNA gene fragment separation. The gradient was composed of 46.5 to 65% denaturant (100% denaturant was defined as 7 M urea and 40% formamide) and 6.2 to 9% acrylamide (13). In addition, a stacking gel with 15% acrylamide was pipetted on top. Approximately 3-μl aliquots of PCR products were loaded side by side on a gel and run in 1× Tris-acetate-EDTA buffer at a constant voltage of 140 V for 17 h at 58°C, and the gel was silver stained as described by Heuer et al. (19). After electrophoresis the gels were air dried and scanned transmissively (Epson 1680 Pro; Seiko-Epson, Japan). A marker composed of GC-clamped fragments (positions 984 to 1378) of 11 bacterial strains with different electrophoretic mobilities (18) was loaded twice on each gel. For analysis of fungal gene fragments the gradient consisted of 23 to 58% denaturant and 8% acrylamide. Gels were run in 1 × Tris-acetate-EDTA buffer at a constant voltage of 100 V for 18 h at 60°C. Subsequent processing of the gels was performed as described above for bacterial gene fragments.
Analysis of DGGE fingerprints and statistics.
Analysis of DGGE fingerprints was performed using the GelCompar II program, version 4.5 (Applied Maths, Ghent, Belgium), as described by Rademaker et al. (33), to convert and normalize the gel images. Modifications of settings described by Smalla et al. (36) were used. The pairwise similarities of lanes were calculated for each gel by using Pearson correlation. The resulting similarity matrices were used for cluster analysis by the unweighted-pair group method using average linkages and to test for significant treatment effects (24). The similarities of microbial fingerprints of ‘Baltica’ to those of the other potato cultivars were analyzed by using box-whisker plots, using an SAS macro written by Michael Friendly (http://euclid.psych.yorku.ca/ftp/sas/sssg/macros/boxplot.sas) which plots the median, quartiles, 95% confidence limits, and whisker lines that extend to the most extreme similarity values that are no more than 1.5 times the interquartile range beyond the quartiles.
RESULTS
Effect of genetic modification.
The PCR-DGGE fingerprints of bacterial rhizosphere communities of the parental cultivar ‘Baltica’ and the two GM lines were compared using both bacterial (Bacteria, Actinobacteria, Alpha- and Betaproteobacteria, Streptomycetaceae, Bacillus, Pseudomonas, and gacA gene) and fungal (Fungi and Ascomycetes) fingerprints (Table 3). For bacteria, the differences between ‘Baltica’ and the GM lines ranged from nondetectable to 4.5%. Significant differences could be detected only for Roggenstein samples collected in 2005. The Actinobacteria fingerprints displayed significant differences at all three growth stages, but the differences were most pronounced at EC90, and the Betaproteobacteria and Streptomycetaceae fingerprints differed only at EC90. None of the other bacterial or group-specific fingerprints could be distinguished for the GM lines and ‘Baltica’. Similarly, comparison of ‘Baltica’ and GM line fingerprints for Oberviehhausen samples in 2006 and Roggenstein samples in 2007 did not reveal significant differences for any of the bacterial groups analyzed at any plant growth stage. For fungi, the differences between the fingerprints of ‘Baltica’ and the GM lines were, in general, more pronounced than the differences for bacteria, with a maximal deviation of 6.6%. In particular, the effects of plant genetic modification were apparent in the Ascomycetes fingerprints. All but two samplings exhibited significant differences. The Fungi fingerprints showed a significant effect of genetic modification for three of eight samplings.
TABLE 3.
Differences between microbial fingerprints of ‘Baltica’ and GM lines and among all potato genotypes
| DGGE gel | Plant growth stage | % Differences for the following comparisonsa:
|
|||||
|---|---|---|---|---|---|---|---|
| Roggenstein, 2005
|
Oberviehhausen, 2006
|
Roggenstein, 2007
|
|||||
| ‘Baltica’ and GM lines | All genotypes | ‘Baltica’ and GM lines | All genotypes | ‘Baltica’ and GM lines | All genotypes | ||
| Bacteria | EC30 | 0.4 | 4.4** | 1.8 | 1.4 | ||
| EC60 | 1.7 | 4.0** | 0.0 | 1.2 | 0.0 | 0.8 | |
| EC90 | 0.6 | 13.1** | 0.5 | 4.5** | 0.0 | 0.9 | |
| Pseudomonas | EC30 | 0.0 | 3.1** | 0.0 | 0.2 | ||
| EC60 | 0.1 | 0.3 | 0.0 | 0.1 | 0.0 | 0.0 | |
| EC90 | 0.2 | 2.4** | 0.0 | 5.7** | 0.0 | 1.4** | |
| gacA | EC30 | 0.0 | 0.0 | 0.0 | 1.0 | ||
| EC60 | 0.2 | 0.0 | 0.1 | 0.3 | 0.0 | 3.8** | |
| EC90 | 0.0 | 5.1** | 0.0 | 3.3* | 0.0 | 0.0 | |
| Actinobacteria | EC30 | 2.6** | 6.4** | 0.0 | 0.6 | ||
| EC60 | 3.0** | 3.7** | 0.0 | 0.0 | 0.0 | 1.4* | |
| EC90 | 4.5** | 9.7** | 0.9 | 0.0 | 0.0 | 0.0 | |
| Streptomycetaceae | EC30 | 0.0 | 0.0 | 0.0 | 0.0 | ||
| EC60 | 0.5 | 0.1 | 0.0 | 1.2 | 0.0 | 0.0 | |
| EC90 | 2.8* | 1.4 | 0.0 | 0.3 | 0.1 | 0.0 | |
| Alphaproteobacteria | EC30 | 0.0 | 1.1* | 1.8 | 0.0 | ||
| EC60 | 0.0 | 0.7 | 1.7 | 0.0 | 0.0 | 0.0 | |
| EC90 | 0.0 | 1.4* | 0.0 | 0.2 | 0.0 | 0.0 | |
| Betaproteobacteria | EC30 | 0.0 | 1.1 | 1.1 | 5.1** | ||
| EC60 | 4.6 | 1.7 | 0.0 | 0.0 | 0.0 | 0.0 | |
| EC90 | 2.8* | 3.1** | 0.0 | 2.7** | 0.0 | 0.0 | |
| Bacillus | EC30 | 3.2 | 0.2 | 0.0 | 0.0 | ||
| EC60 | 0.0 | 1.8* | 0.0 | 0.0 | 0.0 | 0.4 | |
| EC90 | 0.0 | 9.8** | 0.0 | 0.0 | 0.0 | 0.0 | |
| Fungi | EC30 | 5.4* | 14.3** | 0.8 | 3.0** | ||
| EC60 | 1.2 | 3.1** | 2.3** | 3.9** | 1.5 | 3.0** | |
| EC90 | 1.9* | 6.9** | 1.7 | 4.2** | 1.3 | 5.8** | |
| Ascomycetes | EC30 | 4.8** | 13.6** | 0.0 | 3.5** | ||
| EC60 | 3.2 | 6.2** | 1.4** | 2.6** | 2.2** | 5.1** | |
| EC90 | 6.6** | 9.0** | 1.8* | 3.9** | 3.5** | 7.9** | |
*, significant difference (P ≤ 0.05); **, highly significant difference (P ≤ 0.01).
Effect of plant genotype.
The effect of the plant genotype on rhizosphere microbial communities was analyzed by comparing DGGE fingerprints of the five cultivars and the two GM lines (a total of seven plant genotypes). For bacteria, genotype-specific effects were strongly dependent on the site and the year of sampling (Table 3). The most pronounced differences among plant genotypes were found for Roggenstein samples obtained in 2005. Significant differences were observed for Bacteria and Actinobacteria at all plant growth stages. In addition, effects on Pseudomonas, Alphaproteobacteria, and Bacillus were observed at several growth stages. The effect of the plant genotype was especially apparent at plant growth stage EC90, when significant differences were found for all bacterial groups except Streptomycetaceae. The fingerprints for total Bacteria differed by as much as 13% between cultivars and about 10% for the gram-positive groups Actinobacteria and Bacillus.
The fingerprints of samples obtained from Oberviehhausen in 2006 and from Roggenstein in 2007 revealed much less evidence of plant genotype-specific effects. Nevertheless, significant differences were detected for Bacteria, Pseudomonas, Actinobacteria, and Betaproteobacteria, mainly at EC90. The maximal difference between cultivars, 5.7%, was observed for Pseudomonas.
When the Fungi and Ascomycetes fingerprints of all potato genotypes were compared, significant differences were found for both sites at all plant growth stages. The differences among the cultivars were as great as 14% at EC30. In general, comparisons of all seven plant genotypes revealed greater differences than comparisons of ‘Baltica’ with the two GM lines for both bacteria and fungi.
Similarity of GM lines and cultivars to ‘Baltica’.
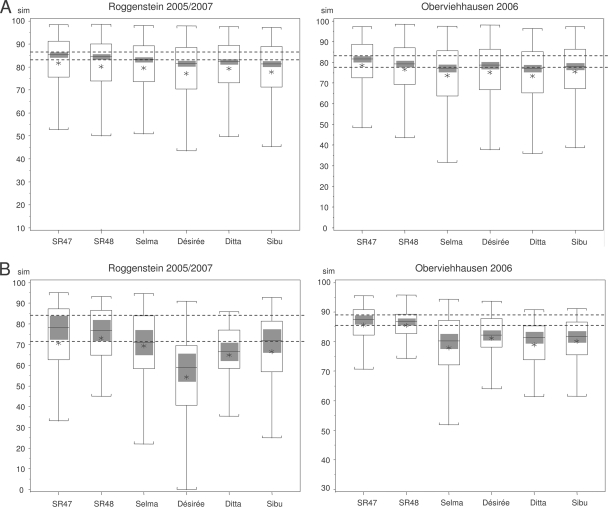
To test the hypothesis that the effect of genetic modification of the GM lines on rhizosphere communities was less pronounced than the effect of genotype differences among cultivars, the similarities of the DGGE fingerprints of all plant genotypes to that of ‘Baltica’ were compared. Analysis of the median similarities of all bacterial fingerprints for samples obtained from both Roggenstein (in 2005 and 2007) and Oberviehhausen (in 2006) revealed that the GM lines were, on average, more similar to the parent ‘Baltica’ than all other cultivars were (Fig. 1A). This suggests that the genetic modifications of the GM lines had a less pronounced effect on the associated bacterial communities than the genetic differences among the potato cultivars had. The GM line SR47 was slightly more similar to ‘Baltica’ than SR48 was, especially at Oberviehhausen in 2006. For the Roggenstein site the confidence intervals for the cultivars ‘Désirée’ and ‘Sibu’ did not overlap with the confidence interval for the two GM lines. This indicated that the difference between these two cultivars and ‘Baltica’ was significantly greater than the differences between the two GM lines and ‘Baltica’. In contrast, for Oberviehhausen samples, the confidence intervals for all cultivars and the two GM lines did overlap. When each bacterial group was analyzed separately, the general trend of higher levels of similarity of the GM lines to ‘Baltica’ was observed for all groups except the Streptomycetaceae (data not shown). In particular, Actinobacteria and Betaproteobacteria contributed to this trend. The analysis of the different plant growth stages revealed more pronounced cultivar-specific differences at EC90 than at EC30 and EC60 (data not shown). At EC90 the commercial cultivars ‘Selma’, ‘Désirée’, and ‘Ditta’ differed from ‘Baltica’ to a significantly higher degree than the GM lines differed from it.
FIG. 1.
Box-whisker plots showing the similarity of the microbial fingerprints for the two GM lines and the commercial cultivars to that for the parental cultivar ‘Baltica’ based on a global comparison of bacterial (Bacteria, Actinobacteria, Alpha- and Betaproteobacteria, Bacillus, Streptomycetaceae, Pseudomonas, and gacA) (A) and fungal (Fungi and Ascomycetes) (B) fingerprints for the Roggenstein site (in 2005 and 2007) and the Oberviehhausen site (in 2006). The SAS macro plots the median, quartiles, 95% confidence intervals (gray bars), and whiskers extending to the most extreme similarity values. The dashed lines indicate the confidence intervals for the two GM lines. Asterisks indicate the means.
For fungi, it was even clearer that genetic modification had a less pronounced effect than genetic differences among the cultivars had. Analysis of all fungal DGGE fingerprints of Roggenstein and Oberviehhausen samples revealed that the median levels of similarity to ‘Baltica’ were higher for the two GM lines than for all other cultivars (Fig. 1B). In Oberviehhausen samples all four cultivars were significantly less similar to ‘Baltica’ than the GM lines were. In Roggenstein samples the same trend was found, with significant differences for the cultivars ‘Désirée’ and ‘Ditta’. Analysis of each plant developmental stage showed that at both EC30 and EC90 all commercial cultivars were significantly more different from ‘Baltica’ than the two GM lines were, while at EC60 the confidence intervals for all cultivars and GM lines overlapped (data not shown).
Effect of environmental factors on bacterial and fungal community fingerprints.
Environmental factors that differed at different field sites and in various years had a much greater impact on microbial rhizosphere communities than plant genotypes had. For example, the DGGE fingerprints of Fungi, Pseudomonas, Streptomycetaceae, and Bacillus for Roggenstein and Oberviehhausen samples obtained in 2006 at plant growth stage EC60 differed by 23%, 40%, 39%, and 15%, respectively. These differences were statistically highly significant (P < 0.0001). Also, comparisons of fingerprints of Streptomycetaceae and Bacillus for Roggenstein samples obtained in 2005 and Oberviehhausen samples obtained in 2006 at EC60 showed similar effects (see Fig. S2b and c in the supplemental material). These fingerprints differed by 31% and 34%, respectively. However, the Pseudomonas-specific fingerprints for this comparison differed only by 6% (see Fig. S2a in the supplemental material).
DISCUSSION
Effect of genetic modification.
The present study provided evidence that GM potato plants with increased zeaxanthin contents have an effect on both bacterial and fungal rhizosphere communities (Table 3). Effects were especially apparent for Ascomycetes at both sites and in all years. Actinobacteria were affected at all three sampling times at Roggenstein in 2005. Similarly, GM potatoes with an altered starch composition were shown previously to affect rhizosphere communities at the site in Oberviehhausen (30). At this site responses to genetic modification were found for Pseudomonas but not for Fungi. Effects on Fungi might not have been detected due to the lower resolution of fungal 18S rRNA gene profiles used in the previous study compared to the ITS profiles used in the present study. In accordance with our study, effects on Actinobacteria were not observed at the Oberviehhausen site. The use of multiple analyses targeting different microbial groups at different field sites and in different years allowed detection of small differences between GM plants and their parental cultivar, while other studies failed to detect minor effects (20, 27, 38).
Changes in the soil microbial community composition might occur directly through transgene products or indirectly, e.g., via altered composition of root exudates (for a review, see reference 23). Due to the use of a tuber-specific promoter, a direct effect of zeaxanthin was likely to occur when the tubers were fully developed. Indeed, the impact of GM potatoes on both bacterial and fungal communities became most apparent at growth stage EC90 (Table 3).
Effect of genetic modification in relation to the effect of the plant genotype.
In this study the rhizosphere microbial communities of the cultivars investigated were more dissimilar than the rhizosphere microbial communities of GM lines and the parental cultivar. This confirmed the hypothesis that the effects of genetic modification on rhizosphere microbial communities were less pronounced than the effects of genotype differences among cultivars resulting from conventional breeding. In another study great variation in the tuber metabolome among cultivars was observed, while the metabolomes of GM lines and the parental cultivar were substantially identical (5).
Becker et al. (2) compared the effect of fructan-producing GM potatoes with the effect of isogenic controls and conventional cultivars grown at one field site in three consecutive years by using terminal restriction fragment length polymorphism of bacterial 16S rRNA genes. They detected neither GM effects nor cultivar effects on rhizosphere bacterial communities, probably due to insufficient resolution of bacterial fingerprints. In accordance with our results, several studies have revealed cultivar effects on rhizosphere communities using taxon-specific DGGE fingerprinting (30, 32, 38).
More drastic effects on rhizosphere microbial communities resulting from genetic modification of an existing pathway, like zeaxanthin transformation, are expected from transgene expression of antimicrobial compounds. For two of three GM potato lines expressing the antimicrobial peptide magainin II, the differences in bacterial rhizosphere communities compared to the parental line community were more significant than the differences among cultivars at the first sampling time (32). However, at senescence of the plants, the communities for all plant lines were similar. This is in contrast to the present study, where for Bacteria the differences were most pronounced for the comparison of all seven plant genotypes at EC90. Similarly, Heuer et al. (20) and Lottmann et al. (27) detected differentiating ribotypes for the parental and transgenic lines only at EC90.
In the present study, the effect of the plant genotype on Fungi, especially on Ascomycetes, was more pronounced (Table 3), and the higher level of similarity of the two GM lines to ‘Baltica’ was more obvious than that observed for bacteria (Fig. 1B). Only a few studies have evaluated the effects of GM plants on fungal rhizosphere communities (14, 30). Götz et al. (14) reported differences between T4 lysozyme-producing potato plants and the parental cultivar based on differentiating bands. However, their study focused exclusively on endophytic fungi. In contrast to the present study, DGGE fingerprinting based on 18S rRNA fragments could detect neither effects of potatoes with a modified starch content nor cultivar effects on fungal communities in the rhizosphere (30). In the present study significant differences in the fungal community fingerprints among the plant genotypes were detected at all stages of plant development. This is even more surprising considering the findings of Costa et al. (6), who reported that the effect of the plant species (strawberry and oilseed rape) was less pronounced for fungi than for bacteria. The contrasting findings might result either from the different plant species analyzed or from the lower resolution of the 18S rRNA-based fingerprints.
In conclusion, effects of the growth of zeaxanthin-accumulating potatoes on rhizosphere microbial communities can be detected by using highly sensitive fingerprinting techniques, multiple analyses targeting different microbial groups, and extensive sampling. Evaluating these effects in the context of natural cultivar variability showed that changes in the composition of bacterial and fungal communities associated with GM plants were not greater than differences among cultivars. Moreover, the GM effects were negligible compared to the effects of environmental factors, such as field site or year, which were far greater than the effects of genetic modification or plant genotype (2, 20, 30, 38).
Supplementary Material
Acknowledgments
We thank J. Dennert and F. X. Maidl (Technical University of Munich) for the perfect management of the experimental plots in Roggenstein and Oberviehhausen. We are highly thankful to G. Wenzel (Technical University of Munich) for providing the plant material of the transgenic lines. We also thank I.-M. Jungkurth (Julius Kühn-Institut) for critically reading the manuscript.
This work was funded by grant 0313277B from the Bundesministerium für Bildung und Forschung.
Footnotes
Published ahead of print on 17 April 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Anderson, I. C., and J. W. Carney. 2004. Diversity and ecology of soil fungal communities: increased understanding through the application of molecular techniques. Environ. Microbiol. 6:769-779. [DOI] [PubMed] [Google Scholar]
- 2.Becker, R., U. Behrendt, B. Hommel, S. Kropf, and A. Ulrich. 2008. Effects of transgenic fructan-producing potatoes on the community structure of rhizosphere and phyllosphere bacteria. FEMS Microbiol. Ecol. 66:411-425. [DOI] [PubMed] [Google Scholar]
- 3.Bruinsma, M., G. A. Kowalchuk, and J. A. van Veen. 2003. Effects of genetically modified plants on microbial communities and processes in soil. Biol. Fertil. Soils 37:329-337. [Google Scholar]
- 4.Brusetti, L., P. Francia, C. Bertolini, A. Pagliuca, S. Borin, C. Sorlini, A. Abruzzese, G. Sacchi, C. Viti, L. Giovannetti, E. Giuntini, M. Bazzicalupo, and D. Daffonchio. 2004. Bacterial communities associated with the rhizosphere of transgenic Bt 176 maize (Zea mays) and its non transgenic counterpart. Plant Soil 266:11-21. [Google Scholar]
- 5.Catchpole, G. S., M. Beckmann, D. P. Enot, M. Mondhe, B. Zywicki, J. Taylor, N. Hardy, A. Smith, R. D. King, D. B. Kell, O. Fiehn, and J. Draper. 2005. Hierarchical metabolomics demonstrates substantial compositional similarity between genetically modified and conventional potato crops. Proc. Natl. Acad. Sci. USA 102:14458-14462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Costa, R., M. Götz, N. Mrotzek, J. Lottmann, G. Berg, and K. Smalla. 2006. Effects of site and plant species on rhizosphere community structure as revealed by molecular analysis of microbial guilds. FEMS Microbiol. Ecol. 56:236-249. [DOI] [PubMed] [Google Scholar]
- 7.Costa, R., N. C. M. Gomes, E. Krögerrecklenfort, K. Opelt, G. Berg, and K. Smalla. 2007. Pseudomonas community structure and antagonistic potential in the rhizosphere: insights gained by combining phylogenetic and functional gene-based analyses. Environ. Microbiol. 9:2260-2273. [DOI] [PubMed] [Google Scholar]
- 8.De Souza, J. T., M. Mazzola, and J. M. Raaijmakers. 2003. Conservation of the response regulator gene gacA in Pseudomonas species. Environ. Microbiol. 5:1328-1340. [DOI] [PubMed] [Google Scholar]
- 9.Dunfield, K. E., and J. J. Germida. 2003. Seasonal changes in the rhizosphere microbial communities associated with field-grown genetically modified canola (Brassica napus). Appl. Environ. Microbiol. 69:7310-7318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gale, C. R., N. F. Hall, D. I. W. Phillips, and C. N. Martyn. 1994. Lutein and zeaxanthin status and risk of age-related macular degeneration. Investig. Ophthalmol. Vis. Sci. 44:2461-2465. [DOI] [PubMed] [Google Scholar]
- 11.Garbeva, P., J. A. van Veen, and J. D. van Elsas. 2003. Predominant Bacillus spp. in agricultural soil under different management regimes detected via PCR-DGGE. Microb. Ecol. 45:302-316. [DOI] [PubMed] [Google Scholar]
- 12.Gomes, N. C. M., H. Heuer, J. Schönfeld, R. Costa, L. Mendonça-Hagler, and K. Smalla. 2001. Bacterial diversity of the rhizosphere of maize (Zea mays) grown in tropical soil studied by temperature gradient gel electrophoresis. Plant Soil 232:167-180. [Google Scholar]
- 13.Gomes, N. C. M., I. A. Kosheleva, W.-R. Abraham, and K. Smalla. 2005. Effects of the inoculant strain Pseudomonas putida KT2442(pNF142) and of naphthalene contamination on the soil bacterial community. FEMS Microbiol. Ecol. 54:21-33. [DOI] [PubMed] [Google Scholar]
- 14.Götz, M., H. Nirenberg, S. Krause, H. Wolters, S. Draeger, A. Buchner, J. Lottmann, G. Berg, and K. Smalla. 2006. Fungal endophytes in potato roots studied by traditional isolation and cultivation-independent DNA-based methods. FEMS Microbiol. Ecol. 58:404-413. [DOI] [PubMed] [Google Scholar]
- 15.Grayston, S. J., S. Wang, C. D. Campbell, and A. C. Edwards. 1998. Selective influence of plant species on microbial diversity in the rhizosphere. Soil Biol. Biochem. 30:369-378. [Google Scholar]
- 16.Hack, H., H. Gal, T. Klemke, R. Klose, U. Meier, R. Stauß, and A. Witzenberger. 1993. Phänologische Entwicklungsstadien der Kartoffel (Solanum tuberosum L.). Nachrichtenbl. Dtsch. Pflanzenschutzd. 45:11-19. [Google Scholar]
- 17.Haichar, F. E., C. Marol, O. Berge, J. I. Rangel-Castro, J. I. Prosser, J. Balesdent, T. Heulin, and W. Achouak. 2008. Plant host habitat and root exudates shape soil bacterial community structure. ISME J. 2:1221-1230. [DOI] [PubMed] [Google Scholar]
- 18.Heuer, H., M. Krsek, P. Baker, K. Smalla, and E. M. H. Wellington. 1997. Analysis of actinomycete communities by specific amplification of genes encoding 16S rRNA and gel electrophoretic separation in denaturing gradients. Appl. Environ. Microbiol. 63:3233-3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heuer, H., J. Wieland, J. Schönfeld, A. Schönwälder, N. C. M. Gomes, and K. Smalla. 2001. Bacterial community profiling using DGGE or TGGE analysis, p. 177-190. In P. Rouchelle (ed.), Environmental molecular microbiology: protocols and applications. Horizon Scientific Press, Wymondham, United Kingdom.
- 20.Heuer, H., R. M. Kroppenstedt, J. Lottmann, G. Berg, and K. Smalla. 2002. Effects of T4 lysozyme release from transgenic potato roots on bacterial rhizosphere communities are negligible relative to natural factors. Appl. Environ. Microbiol. 68:1325-1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jung, S., S. Park, D. Kim, and S. B. Kim. 2008. Denaturing gradient gel electrophoresis analysis of bacterial community profiles in the rhizosphere of cry1AC-carrying Brassica rapa subsp. pekinensis. J. Microbiol. 46:12-15. [DOI] [PubMed] [Google Scholar]
- 22.Kowalchuk, G. A., D. S. Buma, W. De Boer, P. G. L. Klinkhammer, and H. van Veen. 2002. Effects of above-ground plant species composition and diversity on the diversity of soil-borne microorganisms. Antonie van Leeuwenhoek J. Microbiol. 81:509-520. [DOI] [PubMed] [Google Scholar]
- 23.Kowalchuk, G. A., M. Bruinsma, and J. A. van Veen. 2003. Assessing responses of soil microorganisms to GM plants. Trends Ecol. Evol. 18:403-410. [Google Scholar]
- 24.Kropf, S., H. Heuer, M. Grüning, and K. Smalla. 2004. Significance test for comparing microbial community fingerprints using pairwise similarity measures. J. Microbiol. Methods 57:187-195. [DOI] [PubMed] [Google Scholar]
- 25.Larena, I., O. Salazar, V. González, M. C. Julián, and V. Rubio. 1999. Design of a primer for ribosomal DNA internal transcribed spacer with enhanced specificity for ascomycetes. J. Biotechnol. 75:187-194. [DOI] [PubMed] [Google Scholar]
- 26.Liu, B., Q. Zeng, F. Yan, H. Xu, and C. Xu. 2005. Effects of transgenic plants on soil microorganisms. Plant Soil 271:1-13. [Google Scholar]
- 27.Lottmann, J., H. Heuer, K. Smalla, and G. Berg. 1999. Influence of transgenic T4-lysozyme-producing potato plants on potentially beneficial plant-associated bacteria. FEMS Microbiol. Ecol. 29:365-377. [Google Scholar]
- 28.Marschner, P., C. H. Yang, R. Lieberei, and D. E. Crowley. 2001. Soil and plant specific effects on bacterial community composition in the rhizosphere. Soil Biol. Biochem. 33:1437-1445. [Google Scholar]
- 29.Marschner, P., Z. Solaiman, and Z. Rengel. 2006. Rhizosphere properties of Poaceae genotypes under P-limiting conditions. Plant Soil 283:11-24. [Google Scholar]
- 30.Milling, A., K. Smalla, F. X. Maidl, M. Schloter, and J. C. Munch. 2004. Effects of transgenic potatoes with an altered starch composition on the diversity of soil and rhizosphere bacteria and fungi. Plant Soil 266:23-39. [Google Scholar]
- 31.Nübel, U., B. Engelen, A. Felske, J. Snaidr, A. Wiesenhuber, R. I. Amann, W. Ludwig, and H. Backhaus. 1996. Sequence heterogeneities of genes encoding 16S rRNAs in Paenibacillus polymyxa detected by temperature gradient gel electrophoresis. J. Bacteriol. 178:5636-5643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O'Callaghan, M., E. M. Gerard, N. L. Bell, N. W. Waipara, L. T. Aalders, D. B. Baird, and A. J. Conner. 2008. Microbial and nematode communities associated with potatoes genetically modified to express the antimicrobial peptide magainin and unmodified potato cultivars. Soil Biol. Biochem. 40:1446-1459. [Google Scholar]
- 33.Rademaker, J. L. W., F. J. Louws, U. Rossbach, P. Vinuesa, and F. J. de Bruijn. 1999. Computer-assisted pattern analysis of molecular fingerprints and database construction, p. 33. In A. D. L. Akkermans, J. D. van Elsas, and F. J. de Bruijn (ed.), Molecular microbial ecology manual. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 34.Römer, S., J. Lübeck, F. Kauder, S. Steiger, C. Adomat, and G. Sandmann. 2002. Genetic engineering of zeaxanthin-rich potato by antisense inactivation and cosuppression of carotenoid epoxidation. Metab. Eng. 4:263-272. [DOI] [PubMed] [Google Scholar]
- 35.Seddon, J. M., U. A. Ajani, R. D. Sperduto, R. Hiller, N. Blair, T. C. Burton, M. D. Farber, E. S. Gragoudas, J. Haller, D. T. Miller, L. A. Yannuzzi, and W. Willet. 1994. Dietary carotenoids, vitamin A, C and E, and advanced age-related macular degeneration. JAMA 272:1413-1420. [PubMed] [Google Scholar]
- 36.Smalla, K., G. Wieland, A. Buchner, A. Zock, J. Parzy, S. Kaiser, N. Roskot, H. Heuer, and G. Berg. 2001. Bulk and rhizosphere soil bacterial communities studied by denaturing gradient gel electrophoresis: plant-dependent enrichment and seasonal shifts revealed. Appl. Environ. Microbiol. 67:4742-4751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sørensen, J. 1997. The rhizosphere as a habitat for soil microorganisms, p. 21-45. In J. D. van Elsas, J. T. Trevors, and E. M. H. Wellington (ed.), Modern soil microbiology. Marcel Dekker, New York, NY.
- 38.van Overbeek, L., and J. D. van Elsas. 2008. Effects of plant genotype and growth stage on the structure of bacterial communities associated with potato (Solanum tuberosum L.). FEMS Microbiol. Ecol. 64:283-296. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.