Abstract
To develop an expression system for the magnetotactic bacterium Magnetospirillum gryphiswaldense, we compared gene expression from the widely used Escherichia coli Plac promoter with that from known and predicted genuine M. gryphiswaldense promoters. With the use of green fluorescent protein as a reporter, the highest expression level was observed with the magnetosomal PmamDC promoter. We demonstrate that this promoter can be used for the expression of modified magnetosome proteins to generate “antibody-binding” magnetosomes.
The formation of magnetosomes, which are subcellular compartments consisting of membrane-bounded magnetite (Fe3O4) nanocrystals in magnetotactic bacteria, is of interdisciplinary interest in fields of microbial cell biology, biotechnology, and nanotechnology. Despite considerable efforts by many researchers, the genetic analysis of magnetotactic bacteria is still cumbersome and many genetic tools are lacking (7). In this study we developed an expression system for Magnetospirillum gryphiswaldense, which is one of the most widely studied magnetotactic organisms (5, 20).
To identify genuine promoters for gene expression in M. gryphiswaldense, we investigated the expression of green fluorescent protein (GFP) from the Escherichia coli Plac promoter and from putative and previously identified M. gryphiswaldense promoters such as PmamDC and PmamAB, which are highly transcribed under magnetite-inducing conditions, i.e., at microaerobiosis and in the presence of micromolar amounts of iron (19). In addition, genomic regions upstream of large ribosomal gene clusters, which potentially encode the strong PrpsJ (rpsJ, MGR3815 ribosomal protein S10) and PrplK (rplK, MGR3801 ribosomal protein L11) promoters, and sequences that putatively encode the PapdA (Pmms16 in “Magnetospirillum magneticum”) and the Pmsp3 promoters, which were described previously in M. magneticum (26), were analyzed.
For the construction of GFP reporter vectors, the egfp gene encoding the GFPmut1 variant (2, 10) was PCR amplified (primers are shown in Table 1), cloned into pGEM-T Easy (Promega), sequenced, and subcloned into pBBR1MCS2 (8) downstream of the Plac promoter to generate pBBRegfp (Table 2). The vector was digested with NsiI and ApaI, blunted with mung bean nuclease (New England Biolabs), and religated to remove the Plac promoter and yield the promoterless GFP reporter plasmid pBBRpl (Table 2). Putative promoter regions that included the intergenic region upstream from the start codon to the next open reading frame were PCR amplified from genomic DNA of M. gryphiswaldense R3/S1 (21). The PCR products were cloned into pGEM-T Easy, sequenced, and subcloned into pBBRpl, resulting in the plasmids pBBRPmamDC, pBBRPmamAB, pBBRPmsp3, pBBRPapdA, pBBRPure, pBBRPrplK, and pBBRPrpsJ (Table 2). The plasmids were transferred into M. gryphiswaldense R3/S1 by conjugation from E. coli BW29427 (K. Datsenko and B. L. Wanner, unpublished data) as described previously (18, 22).
TABLE 1.
Primers used in this studya
| Primer name | Sequencea | Restriction site |
|---|---|---|
| egfpfw | CATATGGTGAGCAAGGGCGAGGAG | NdeI |
| CL2 | GTGGATCCTTACTTGTACAGCTCGTC | BamHI |
| papdAfw | CTCGAGGAGCCTCTCCATTAAACAATG | XhoI |
| papdArev | CATATGCTTGAATTCCTCCAACCGGGGGTATG | NdeI |
| prplKfw | AAGCTTGGCATCAAGGTTTCGGAAG | HindIII |
| prplKrev | CATATGTTTACCCTACCTCTGGTCG | NdeI |
| prpsJfw | CTCGAGGTCCTTCG GGATCGCTTG AC | XhoI |
| prpsJrev | CATATGATTCACGTCATCCGTTAAATC | NdeI |
| purerev | CATATGGTGGTTATGCGCTGCTCAAAATC | NdeI |
| purefw | CTCGAGCTTTTCTCGATCCGGGAAAAATAC | XhoI |
| PmamDCrev | CATATGCTGATCTCCGGCAAGTGTATG | NdeI |
| PmamDCfw | CTCGAGCAATGACCACCACCACCTTA AAC | XhoI |
| PmamABfw | CTCGAGATGGCGCAAAGATGTGACGT C | XhoI |
| PmamABrev | CATATGTCCCGTCACAATTCACCTCC | NdeI |
| pmsp3fw | CTCGAGGAACTCCAAAAGCAAGGCTATTTAC | XhoI |
| pmsp3rev | ATTAATCCGAAAGCTCCTTGAATCAAAAG | VspI |
| PmamDCxlrev | CTCGAGGATCTCCGGCAAGTGTATGCAC | XhoI |
| zzfw | GCTGCACATATGGCGCAACACGATGAAGCC | NdeI |
| zzrev | CCATCTAGAAATATTACCGCCAGCCATTG | XbaI |
Restriction sites that were incorporated in the primer are indicated in bold.
TABLE 2.
Plasmids used in this study
| Plasmid name | Description | Source or reference |
|---|---|---|
| pEGFPN1 | GFP expression vector; Ap | BD Biotech |
| pGEM-T Easy | Cloning vector; Ap | Promega |
| pBBR1MCS-2 | Mobilizable broad-host-range vector; Km | 8 |
| pGEMegfp | pGEM-T Easy + egfp PCR product | This study |
| pBBRegfp | pBBR1MCS-2 + egfp from pGEMegfp | This study |
| pBBRpl | Promoterless GFP reporter vector based on pBBRegfp | This study |
| pGEMPmamDC | pGEM-T Easy + PmamDC-PCR product | This study |
| pGEMPmamAB | pGEM-T Easy + PmamAB-PCR product | This study |
| pGEMPapdA | pGEM-T Easy + PapdA-PCR product | This study |
| pGEMPmsp3 | pGEM-T Easy + Pmsp3-PCR product | This study |
| pGEMPrplK | pGEM-T Easy + PrplK-PCR product | This study |
| pGEMPrpsJ | pGEM-T Easy + PrpsJ-PCR product | This study |
| pBBRPmamDC | pBBRpl + PmamDC from pGEMPmamDC cloned in XhoI and NdeI sites | This study |
| pBBRPmamAB | pBBRpl + PmamAB from pGEMPmamAB cloned in XhoI and NdeI sites | This study |
| pBBRPapdA | pBBRpl + PapdA from pGEMPapdA cloned in XhoI and NdeI sites | This study |
| pBBRPmsp3 | pBBRpl + Pmsp3 from pGEMPmsp3 cloned in XhoI and VspI sites | This study |
| pBBRPrplK | pBBRpl + PrplK from pGEMPrplK cloned in HindIII and NdeI sites | This study |
| pBBRPrpsJ | pBBRpl + PrpsJ from pGEMPrpsJ cloned in XhoI and NdeI sites | This study |
| pEZZ18 | Protein A gene fusion vector; Ap | GE Healthcare |
| pGEMZZ | pGEM-T Easy + ZZ protein domain PCR product | This study |
| pCL6 | pBBR1MCS-2 + MamC-GFP | 9 |
| pBBRCZZ | The ZZ protein domain from pGEMZZ was cloned into the NdeI and BamHI sites of pCL6 to replace GFP with the ZZ domain | This study |
| pJETPdcx1 | pJET1.2 + PmamDC-PCR product | This study |
| pBBRPdcCZZ | pBBRCZZ + PmamDC from pGEMPdcx1 | This study |
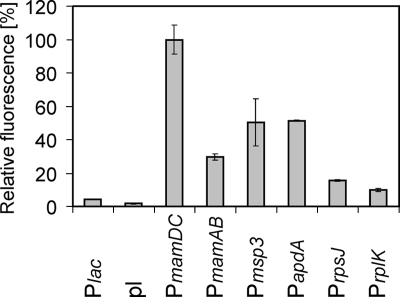
For promoter activity assays, M. gryphiswaldense strains expressing GFP from different promoters were cultivated in triplicate microaerobically in 3-ml culture volumes in six-well culture plates under a microoxic atmosphere (1% oxygen, 99% nitrogen) for 20 to 22 h in FSM medium (6). Cells were washed and resuspended in phosphate-buffered saline to an optical density at 565 nm of 0.5. The expression of GFP was quantified from 100-μl aliquots of the cell suspension with an Infinite 500 plate reader (Tecan). The native promoters were considerably more active than the E. coli Plac promoter that has been used in previous studies with this organism (9, 16-18). Fluorescence quantification and immunoblot analysis (see Fig. S1 in the supplemental material) have shown that the strongest promoter in M. gryphiswaldense was PmamDC, followed by Pmsp3, PapdA, PmamAB, PrpsJ, PrplK, and Plac (Fig. 1). Similarly to other alphaproteobacterial promoters (12, 13, 24), the tested M. gryphiswaldense promoters were inactive in E. coli (see Fig. S2 in the supplemental material).
FIG. 1.
Analysis of GFP expression from different promoters in M. gryphiswaldense by fluorometry. The excitation wavelength was 485 nm (20-nm bandwidth), and emission was recorded at 535 nm (25-nm bandwidth). The value for each sample was averaged from 10 reads over an integration period of 20 μs. The error bars reflect the standard deviations calculated from three independent experiments. pl, promoterless control.
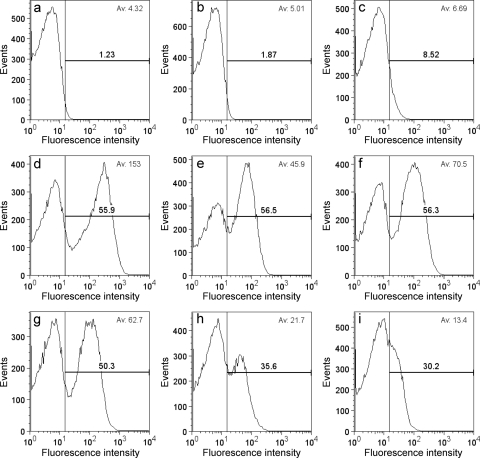
Using GFP as a reporter, we estimated gene expression in individual cells by flow cytometry according to a previously described procedure (9). Comparison of the average fluorescence intensities confirmed the bulk measurements (Fig. 1) and showed that cells containing the PmamDC-GFP construct fluoresced on average twice as much as did cells expressing GFP from Pmsp3 or PapdA (Pmms16) (Fig. 2). In a study of gene expression in M. magneticum, Pmsp3 was identified in a luciferase-based assay as the strongest promoter with an activity more than threefold higher than that of the Pmms16 promoter (26). Even though in our study a different reporter and the homologous promoters from M. gryphiswaldense were used, it was unexpected that the activities of Pmsp3 and PapdA were twofold lower than the activity of PmamDC and that Pmsp3 activity was almost identical to PapdA (Pmms16) activity in M. gryphiswaldense. It is possible that Pmsp3, which is a putative peroxiredoxin promoter, was downregulated due to the microaerobic growth conditions, which are required for the production of magnetosomes.
FIG. 2.
Flow cytometry of M. gryphiswaldense (a) and strains expressing GFP from different plasmids: pBBRpl (promoterless) (b), pBBRegfp (Plac) (c), pBBRPmamDC (d), pBBRPmamH (e), pBBRPmsp3 (f), pBBRPapdA (g), pBBRPrpsJ (h), and pBBRPrplK (i). The proportion of fluorescent cells is shown in bold, and the average fluorescence intensity is displayed in the upper right corner. To estimate the proportion of fluorescent cells, a threshold for fluorescence was set to the fluorescence intensity below which 99% of untransformed M. gryphiswaldense cells, which served as a nonfluorescent standard, were detected. Fifty thousand events were analyzed from each sample.
Flow cytometry also revealed that GFP was not expressed homogeneously in all cells but that a variable fraction of the cells was nonfluorescent (Fig. 2). For the weak and intermediate-strength promoters Plac, PrplK, PrpsJ, and PmamAB, the proportions of fluorescent cells (from 8.52% to 56.5%) correlated well with the increasing average fluorescence intensities, which indicates that intermediate-strength promoters activate GFP expression in a higher proportion of cells than do weak promoters. In contrast, use of the strong Pmsp3, PmamDC, and PapdA promoters did not result in a further increased proportion of fluorescent cells (56.3 to 50.3%). It is unclear why a large proportion of cells was inactive with respect to GFP expression. However, inhomogeneous gene expression in an isogenic population of cells can be frequently observed in bacteria (3, 23) and might be caused by cell cycle-dependent effects, stochasticity of gene expression, or variations of growth rates and protein synthesis between individual cells (4, 15, 25). The heterogeneity of gene expression from strong promoters in M. gryphiswaldense is of relevance for the genetic engineering of magnetosomes for biotechnological applications, as the heterogeneity of gene expression will be reflected by heterogeneously modified magnetosomes. However, a proportion of 50 to 60% of cells that express a gene of interest is sufficient for many practical purposes.
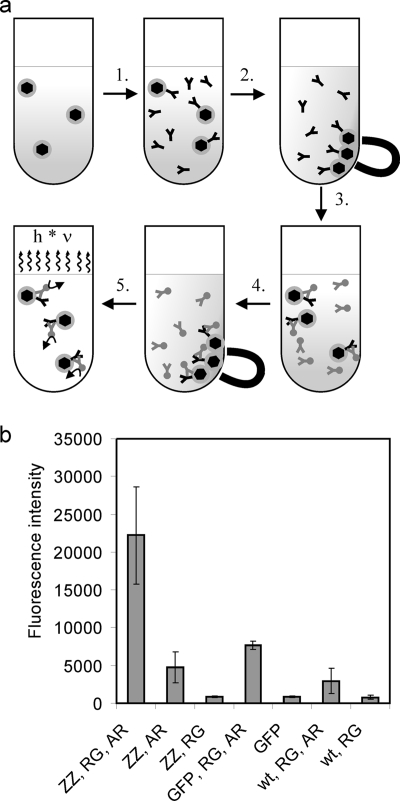
To test the applicability of PmamDC for the expression of fusion proteins in the magnetosome membrane, we expressed a MamC-ZZ fusion protein from PmamDC. The ezz gene, which codes for the antibody-binding ZZ-protein domain (11), was PCR amplified from pEZZ18 (GE Healthcare), cloned into pGEM-T Easy, sequenced, and cloned into the pCL6 vector (9) to create the mamC-ezz fusion construct pBBRCZZ (Table 2). The PmamDC promoter was PCR amplified, cloned into pJET1.2/blunt (Fermentas), sequenced, and cloned into pBBRCZZ to yield pBBRPdcCZZ. This plasmid was transferred to M. gryphiswaldense, and MamC-ZZ magnetosomes were purified as described previously (9) with the only exception that the cells were lysed with a bench top constant cell disruptor (135,000 kPa) (Constant Systems) instead of a French press. After incubation of MamC-ZZ modified, unmodified, and GFP-displaying magnetosomes with rabbit anti-GFP antibody, we detected the highest abundance of the rabbit anti-GFP antibody with a shrimp alkaline phosphatase-labeled goat anti-rabbit antibody on the surface of MamC-ZZ magnetosomes (Fig. 3a and b). The signal was substantially stronger than the signal observed for MamC-GFP modified magnetosomes, which were produced by expression of the MamC-GFP fusion from the Plac promoter (9) (Fig. 3b). These results demonstrate that MamC-ZZ modified magnetosomes efficiently bind rabbit antibodies. In addition, MamC-ZZ magnetosomes that were incubated only with alkaline phosphatase-labeled goat antibody yielded a weak signal (Fig. 3b). This indicates that MamC-ZZ modified antibodies also interact weakly with goat antibody. This was expected, as the staphylococcal protein A, from which the ZZ protein domain was derived, interacts weakly with goat antibodies but strongly with rabbit antibodies (14). While ZZ-modified magnetosomes have been produced in M. magneticum via genetic engineering of the MamC homolog Mms13 previously (27), this is the first time that antibody-binding magnetosomes from M. gryphiswaldense were generated by genetic engineering. One particular advantage of M. gryphiswaldense as a host for production of such genetically functionalized magnetosomes is that several mutant strains with an average magnetosome size of between 24 and 37 nm are available (16), which makes it possible to produce a wide range of magnetosomes with engineered bio- and physicochemical characteristics. The purified ZZ-modified magnetosomes can be used for magnetoimmunoassays (27) or for purification of antibodies, as described with ZZ-modified bacterial polyester granules (1).
FIG. 3.
Antibody-binding assay of MamC-ZZ magnetosomes. (a) Schematic illustration of the antibody-binding assay procedure. 1. MamC-ZZ modified magnetosomes and controls were diluted to a concentration of 1 mM Fe in 500 μl blocking solution (TBS [20 mM Tris, 0.5 M NaCl, pH 7.5] plus 0.5% [wt/vol] milk powder). The samples were incubated for 30 min at room temperature before rabbit anti-GFP antibody was added at a 1:2,000 dilution. 2. After incubation for 45 min, magnetosomes were magnetically collected and resuspended in TBS. 3. After a second magnetic separation step, a conjugate of shrimp alkaline phosphatase and goat anti-rabbit antibody was added in a 1:2,000 dilution in TBS. 4. After an incubation period of 45 min, the magnetosomes were magnetically separated and washed with TBS three times. 5. The particles were resuspended in 200 μl TBS, of which 100 μl was incubated with 100 μl Attophos shrimp alkaline phosphatase detection reagent (Roche) for 5 min. Fluorescence was detected with an Infinite 500 plate reader (Tecan). The excitation wavelength was 430 nm (20-nm bandwidth), and emission was recorded at 535 nm (25-nm bandwidth). (b) Antibody-binding assay of MamC-ZZ modified magnetosomes. The assay was performed with magnetosomes purified from M. gryphiswaldense pBBRPdcCZZ (ZZ), with MamC-GFP modified magnetosomes (GFP) (from M. gryphiswaldense pCL6 [9]), or with wild-type magnetosomes (wt). The magnetosomes were treated either with a primary rabbit anti-GFP antibody (RG), with a shrimp alkaline phosphatase conjugate of a goat anti-rabbit antibody (AR), or with both antibodies (RG, AR). Standard deviations calculated from three replicates are indicated.
In conclusion, we demonstrate that PmamDC is a powerful tool for the genetic engineering of magnetosome proteins to generate functionalized magnetic nanoparticles for bio- and nanotechnological applications.
Supplementary Material
Acknowledgments
We are grateful to Markus Kador and Michael Boshart for providing help with and access to the flow cytometer. We also thank Torsten Pirch and Kirsten Jung for access to the fluorescence reader and Ivaylo Kostadinov for help with cloning.
The project was funded by German Research Foundation grant SPP 1104 and the “Nanobiotechnology” program of the German Federal Ministry of Education and Research.
Footnotes
Published ahead of print on 24 April 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Brockelbank, J. A., V. Peters, and B. H. A. Rehm. 2006. Recombinant Escherichia coli strain produces a ZZ domain displaying biopolyester granules suitable for immunoglobulin G purification. Appl. Environ. Microbiol. 72:7394-7397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cormack, B. P., R. H. Valdivia, and S. Falkow. 1996. FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173:33-38. [DOI] [PubMed] [Google Scholar]
- 3.Davidson, C. J., and M. G. Surette. 2008. Individuality in bacteria. Annu. Rev. Genet. 42:253-268. [DOI] [PubMed] [Google Scholar]
- 4.Elowitz, M. B., A. J. Levine, E. D. Siggia, and P. S. Swain. 2002. Stochastic gene expression in a single cell. Science 297:1183-1186. [DOI] [PubMed] [Google Scholar]
- 5.Faivre, D., and D. Schüler. 2008. Magnetotactic bacteria and magnetosomes. Chem. Rev. 108:4875-4898. [DOI] [PubMed] [Google Scholar]
- 6.Heyen, U., and D. Schüler. 2003. Growth and magnetosome formation by microaerophilic Magnetospirillum strains in an oxygen-controlled fermentor. Appl. Microbiol. Biotechnol. 61:536-544. [DOI] [PubMed] [Google Scholar]
- 7.Jogler, C., and D. Schüler. 2006. Genetic analysis of magnetosome biomineralization, p. 133-161. In D. Schüler (ed.), Microbiology monographs—magnetoreception and magnetosomes in bacteria. Springer, Heidelberg, Germany.
- 8.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop II, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 9.Lang, C., and D. Schüler. 2008. Expression of green fluorescent protein fused to magnetosome proteins in microaerophilic magnetotactic bacteria. Appl. Environ. Microbiol. 74:4944-4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Larrainzar, E., F. O'Gara, and J. P. Morrissey. 2005. Applications of autofluorescent proteins for in situ studies in microbial ecology. Annu. Rev. Microbiol. 59:257-277. [DOI] [PubMed] [Google Scholar]
- 11.Löwenadler, B., B. Jansson, S. Paleus, E. Holmgren, B. Nilsson, T. Moks, G. Palm, S. Josephson, L. Philipson, and M. Uhlén. 1987. A gene fusion system for generating antibodies against short peptides. Gene 58:87-97. [DOI] [PubMed] [Google Scholar]
- 12.MacLellan, S. R., A. M. MacLean, and T. M. Finan. 2006. Promoter prediction in the rhizobia. Microbiology 152:1751-1763. [DOI] [PubMed] [Google Scholar]
- 13.Malakooti, J., S. P. Wang, and B. Ely. 1995. A consensus promoter sequence for Caulobacter crescentus genes involved in biosynthetic and housekeeping functions. J. Bacteriol. 177:4372-4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Richman, D. D., P. H. Cleveland, M. N. Oxman, and K. M. Johnson. 1982. The binding of staphylococcal protein A by the sera of different animal species. J. Immunol. 128:2300-2305. [PubMed] [Google Scholar]
- 15.Roostalu, J., A. Jõers, H. Luidalepp, N. Kaldalu, and T. Tenson. 2008. Cell division in Escherichia coli cultures monitored at single cell resolution. BMC Microbiol. 8:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scheffel, A., A. Gärdes, K. Grünberg, G. Wanner, and D. Schüler. 2008. The major magnetosome proteins MamGFDC are not essential for magnetite biomineralization in Magnetospirillum gryphiswaldense but regulate the size of magnetosome crystals. J. Bacteriol. 190:377-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scheffel, A., M. Gruska, D. Faivre, A. Linaroudis, P. L. Graumann, J. M. Plitzko, and D. Schüler. 2006. An acidic protein aligns magnetosomes along a filamentous structure in magnetotactic bacteria. Nature 440:110-115. [DOI] [PubMed] [Google Scholar]
- 18.Scheffel, A., and D. Schüler. 2007. The acidic repetitive domain of the Magnetospirillum gryphiswaldense MamJ protein displays hypervariability but is not required for magnetosome chain assembly. J. Bacteriol. 189:6437-6446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schübbe, S., C. Würdemann, J. Peplies, U. Heyen, C. Wawer, F. O. Glöckner, and D. Schüler. 2006. Transcriptional organization and regulation of magnetosome operons in Magnetospirillum gryphiswaldense. Appl. Environ. Microbiol. 72:5757-5765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schüler, D. 2008. Genetics and cell biology of magnetosome formation in magnetotactic bacteria. FEMS Microbiol. Rev. 32:654-672. [DOI] [PubMed] [Google Scholar]
- 21.Schultheiss, D., M. Kube, and D. Schüler. 2004. Inactivation of the flagellin gene flaA in Magnetospirillum gryphiswaldense results in nonmagnetotactic mutants lacking flagellar filaments. Appl. Environ. Microbiol. 70:3624-3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schultheiss, D., and D. Schüler. 2003. Development of a genetic system for Magnetospirillum gryphiswaldense. Arch. Microbiol. 179:89-94. [DOI] [PubMed] [Google Scholar]
- 23.Siegele, D. A., and J. C. Hu. 1997. Gene expression from plasmids containing the araBAD promoter at subsaturating inducer concentrations represents mixed populations. Proc. Natl. Acad. Sci. USA 94:8168-8172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smit, J., and N. Agabian. 1984. Cloning of the major protein of the Caulobacter crescentus periodic surface layer: detection and characterization of the cloned peptide by protein expression assays. J. Bacteriol. 160:1137-1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Strovas, T. J., L. M. Sauter, X. F. Guo, and M. E. Lidstrom. 2007. Cell-to-cell heterogeneity in growth rate and gene expression in Methylobacterium extorquens AM1. J. Bacteriol. 189:7127-7133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoshino, T., and T. Matsunaga. 2005. Development of efficient expression system for protein display on bacterial magnetic particles. Biochem. Biophys. Res. Commun. 338:1678-1681. [DOI] [PubMed] [Google Scholar]
- 27.Yoshino, T., and T. Matsunaga. 2006. Efficient and stable display of functional proteins on bacterial magnetic particles using Mms13 as a novel anchor molecule. Appl. Environ. Microbiol. 72:465-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.