Abstract
Biofilms are considered to be highly resistant to antimicrobial agents. Several mechanisms have been proposed to explain this high resistance of biofilms, including restricted penetration of antimicrobial agents into biofilms, slow growth owing to nutrient limitation, expression of genes involved in the general stress response, and emergence of a biofilm-specific phenotype. However, since combinations of these factors are involved in most biofilm studies, it is still difficult to fully understand the mechanisms of biofilm resistance to antibiotics. In this study, the antibiotic susceptibility of Escherichia coli cells in biofilms was investigated with exclusion of the effects of the restricted penetration of antimicrobial agents into biofilms and the slow growth owing to nutrient limitation. Three different antibiotics, ampicillin (100 μg/ml), kanamycin (25 μg/ml), and ofloxacin (10 μg/ml), were applied directly to cells in the deeper layers of mature biofilms that developed in flow cells after removal of the surface layers of the biofilms. The results of the antibiotic treatment analyses revealed that ofloxacin and kanamycin were effective against biofilm cells, whereas ampicillin did not kill the cells, resulting in regrowth of the biofilm after the ampicillin treatment was discontinued. LIVE/DEAD staining revealed that a small fraction of resistant cells emerged in the deeper layers of the mature biofilms and that these cells were still alive even after 24 h of ampicillin treatment. Furthermore, to determine which genes in the biofilm cells are induced, allowing increased resistance to ampicillin, global gene expression was analyzed at different stages of biofilm formation, the attachment, colony formation, and maturation stages. The results showed that significant changes in gene expression occurred during biofilm formation, which were partly induced by rpoS expression. Based on the experimental data, it is likely that the observed resistance of biofilms can be attributed to formation of ampicillin-resistant subpopulations in the deeper layers of mature biofilms but not in young colony biofilms and that the production and resistance of the subpopulations were aided by biofilm-specific phenotypes, like slow growth and induction of rpoS-mediated stress responses.
Reduced susceptibility of biofilm bacteria to antimicrobial agents is a crucial problem for treatment of chronic infections (11, 29, 48). It has been estimated that 65% of microbial infections are associated with biofilms (11, 29, 37), and biofilm cells are 100 to 1,000 times more resistant to antimicrobial agents than planktonic bacterial cells (11, 29, 32).
The molecular nature of this apparent resistance has not been elucidated well, and a number of mechanisms have been proposed to explain the reduced susceptibility, such as restricted antibiotic penetration (47), decreased growth rates and metabolism (7, 52), quorum sensing and induction of a biofilm-specific phenotype (8, 29, 35, 39, 49), stress response activation (7, 52), and an increase in expression of efflux pumps (14). Biofilm resistance has generally been assumed to be due to the fact that the cells in the deeper layers of thick biofilms, which grow more slowly, have less access to antibiotics and nutrients. However, this is not the only reason in many cases. Familiar mechanisms of antibiotic resistance, such as modifying enzymes and target mutations, do not seem to be responsible for the biofilm resistance. Even sensitive bacteria that do not have a known genetic basis for resistance can exhibit profoundly reduced susceptibility when they form biofilms (48).
It was reported previously that changes in gene expression induced a biofilm-specific phenotype (5, 13, 22, 35, 41, 42). Several genes have been proposed to be particularly important for biofilm formation, and the importance of the rpoS gene in Escherichia coli biofilm formation was suggested recently (1, 10, 22, 42). It has been suggested that induction of an rpoS-mediated stress response results in physiological changes that could contribute to antibiotic resistance (29). Although several mechanisms and genes have been proposed to explain biofilm resistance to antibiotics, this resistance is not still fully understood because these mechanisms seem to work together within a biofilm community. In addition, the physiology of biofilm cells is remarkably heterogeneous and varies according to the location of individual cells within biofilms (33, 34, 46).
In this study, susceptibility of E. coli cells in biofilms to antibiotics was investigated. The E. coli cells in the deeper layers of mature biofilms were directly treated with three antibiotics with different molecular targets, the β-lactam ampicillin, the aminoglycoside kanamycin, and the fluoroquinolone ofloxacin. The biofilm biomass was removed before antibiotic treatment, and only the cells located in the deeper layers of the mature biofilms were directly exposed to antibiotics; thus, the effects of restricted antibiotic and nutrient penetration, as well as heterogeneous physiological states in biofilms, were reduced. Although ofloxacin and kanamycin effectively killed the biofilm cells, ampicillin could not kill the cells, which led to regrowth of biofilms. However, the cells in young colony biofilms were completely killed by ampicillin. Therefore, to determine which genes are induced in the mature biofilm cells, allowing increased resistance to ampicillin, global gene expression was analyzed at different stages of biofilm formation, the attachment, colony formation, and maturation stages. Based on the experimental data obtained, possible mechanisms of the increased biofilm resistance to ampicillin are discussed below.
MATERIALS AND METHODS
Bacterial strains and media.
E. coli MG1655 wild-type and rpoS mutant strains (22) were used in this work. An MG1655 rpoS mutant strain was constructed by replacement of a chromosomal gene with a kanamycin marker generated by PCR using plasmid pKD13 as the template and primers rpoS KO F (5′-AGGCTTTTGCTTGAATGTTCCGTCAAGGGATCACGGGTAGGAGCCACCTTGTGTAGGCTGGAGCTGCTTC-3′) and rpoS KO R (5′-GGTCGACGGATCCCCGGAATGTAAGCATCTGTCAGAAAGGCCAGTCTCAAGCGAGGCTGGCCTTTTCTGT-3′), using the method described by Datsenko and Wanner (12). This method is based on the high efficiency of the phage λ Red recombinase (53). Briefly, an FLT-flanked kanamycin marker was amplified and transformed into a strain expressing λ Red recombinase with pKD46. A kanamycin-resistant transformant was selected, and the kanamycin cassette was eliminated using FLP expression plasmid pCP20. Deletion of rpoS was confirmed by PCR performed with primers rpoS F (5′-AAGCCTGCACAAAATTCCAC-3′) and rpoS R (5′-TATCTGGGGTTGTCGGTAGC-3′). The region from the start codon to the stop codon of the rpoS gene was deleted. Planktonic cultures were grown in 3-morpholinopropanesulfonic acid (MOPS) minimal medium (31) supplemented with 2 mg/ml of glucose at 37°C.
Flow cell experiments.
Flow cell experiments were performed at 37°C with a three-channel flow cell (individual channel dimensions, 1 by 4 by 40 mm; Stovall Life Science, Greensboro, NC). Flow cells were inoculated with overnight cultures grown in MOPS minimal medium supplemented with glucose and diluted to obtain a standard optical density at 600 nm of 0.1. The flow cells were left for 2 h to allow attachment of the cells to glass surfaces. After the initial attachment of the cells, the medium was pumped through the flow cells at a rate of 0.25 ml/min in every chamber using a peristaltic pump.
A confocal laser scanning microscope (CLSM) (LSM510; Zeiss, Thornwood, NY) was used to obtain images of biofilms grown in flow cells. SYTO 60 red fluorescent nucleic acid stain (Invitrogen, Carlsbad, CA) was used to stain the biofilms. For image acquisition with SYTO 60, the excitation wavelength used was 633 nm and the emission wavelength was 650 nm. Three-dimensional images were obtained using IMARIS (Bitplane, Zurich, Switzerland).
Antibiotic susceptibility assay with E. coli cells.
The antibiotic susceptibility was assayed using planktonic E. coli cells and E. coli cells in biofilms. For the planktonic cell culture, cells were obtained at the exponential phase (6 h), early stationary phase (12 h), and late stationary phase (24 h and 72 h). The cells were washed with phosphate-buffered saline and diluted to obtain 107 CFU/ml with MOPS minimal medium with glucose. The cells were treated with different concentrations of antibiotics, including 50 to 200 μg/ml ampicillin, 25 to 150 μg/ml kanamycin, and 1 to 50 μg/ml ofloxacin, in MOPS minimal medium containing or not containing glucose at 37°C for 24 h with agitation. After antibiotic treatment, CFU were counted. The MICs of these antibiotics are 2 to 4 μg/ml for ampicillin, 1 μg/ml for kanamycin, and 0.03 to 0.06 μg/ml for ofloxacin (2). For the biofilm cells, after cultivation of the biofilms (2 h for attachment, 24 h for colony formation, and 72 h for maturation) in the flow cells, the medium was changed to MOPS minimal medium containing an antibiotic (100 μg/ml ampicillin, 25 μg/ml kanamycin, or 10 μg/ml ofloxacin) and pumped through the flow cells at the same flow rate for 24 h. For mature biofilms, the biofilms were flushed away by using the highest flow rate of the peristaltic pump (ca. 4.27 ml/min) and introducing air (air flushing) before incubation with medium containing an antibiotic to remove much of the biofilm biomass. The air was introduced through 0.22-μm sterilized filters into a bubble trap that was placed between the peristaltic pump and the flow cells. After treatment with an antibiotic for 24 h, the medium containing the antibiotic was changed to medium without antibiotics and the preparation was incubated for 72 h to evaluate its ability to form biofilms. The ability to form biofilms was examined with the CLSM. Quantification of biofilms was performed with the image quantification software COMSTAT (20). The average thickness and biovolume were determined. Nine images were used to determine the averages for these parameters for each growth phase of biofilms. Three-dimensional images were obtained using IMARIS (Bitplane).
LIVE/DEAD staining (Invitrogen, Carlsbad, CA) was performed with cells after antibiotic treatment to determine the efficacy of antibiotics. Images were obtained with the CLSM, and the ratio of living cells to total cells was determined with LSM Image Examiner (Zeiss). Nine images were used for this analysis.
DNA microarray analysis.
Planktonic cells were obtained at the exponential phase (6 h) and stationary phase (24 h), and biofilm cells were obtained from glass surfaces of the flow cells. The cells obtained were resuspended immediately in 2 volumes of RNA Protect bacterial reagent (Qiagen, Valencia, CA), centrifuged at 4°C at 5,000 × g for 10 min, and kept at −80°C until they were used according to the manufacturer's protocol. Total RNA was extracted using an RNeasy mini kit (Qiagen). The mRNA was obtained from the total RNA using MICROBExpress (Ambion, Austin, TX) according to the manufacturer's protocol. Double-stranded cDNA was synthesized from the purified mRNA using MessageAmp II-Bacteria (Ambion) according to the manufacturer's protocol. Then GeneChip expression 3′-amplification reagents for IVT labeling (Affymetrix, Santa Clara, CA) was used for amplification and biotinylation of cRNA from the double-stranded cDNA containing the T7 promoter sequence. The biotin-labeled cRNA was fragmented, and 10 μg of the fragmented cRNA was hybridized to an E. coli antisense genome array (Affymetrix) for 16 h at 45°C as described in the Affymetrix manual. Probe arrays were washed and stained with a GeneChip 450 fluidics station (Affymetrix) and were scanned with a GeneChip 3000 scanner (Affymetrix). Three independent biological replicates were used for gene expression analysis with DNA-Chip Analyzer (dChip) (http://www.biostat.harvard.edu/complab/dchip/). Genes whose expression was statistically significantly different in the experimental sample and the control sample (baseline) fulfilled following criteria: (i) a difference in expression of >2.0-fold; (ii) a P value for the t test for equal experimental sample and baseline sample signals of <0.05; and (iii) a P value for a paired t test of <0.05. The comparisons were performed for three stages of biofilm formation, the attachment, colony formation, and maturation stages, for planktonic exponential and stationary phases, and for biofilms and planktonic cells. The relative expression value was calculated with the dChip software. Genes were categorized into functional groups (43).
Microarray data accession number.
The microarray data have been deposited in the NCBI Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/) (16) under accession number GSE13418.
RESULTS AND DISCUSSION
Antibiotic susceptibility of planktonic E. coli cells.
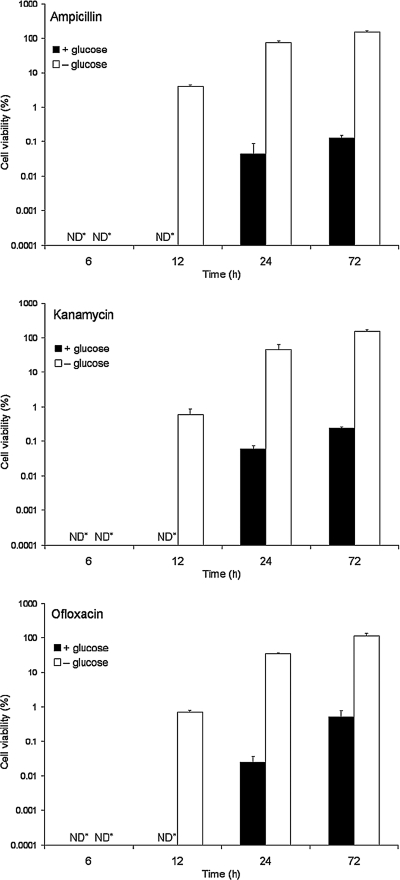
To determine appropriate antibiotic concentrations for the following biofilm experiments, antibiotic susceptibility was examined with planktonic cells. The cell viability decreased as the concentrations of antibiotics increased when cells were incubated with glucose. When cells were incubated without glucose, different trends were observed with the ampicillin treatment and the kanamycin and ofloxacin treatments. Although ampicillin was completely ineffective, kanamycin and ofloxacin killed the majority of the cells at high concentrations. Based on these experimental results, the following antibiotic concentrations were used for the biofilm experiments: 100 μg/ml ampicillin, 25 μg/ml kanamycin, and 10 μg/ml ofloxacin, which killed more than 99.9% of the cells in the presence of glucose.
Using these antibiotic concentrations, antibiotic susceptibility was examined at different stages of planktonic growth (Fig. 1). Similar patterns of cell viability were observed for all of the antibiotics used. With glucose, all of the cells growing in the exponential and early stationary phases (at 6 h and 12 h) were completely killed, but a small fraction (less than 0.1%) of the cells in the late stationary phase (at 24 h and 72 h) survived, indicating that subpopulations essentially resistant to antibiotics developed. Without glucose, the cell viability increased with culture time, and almost none of the cells were killed in late stationary phase (at 72 h) with all of the antibiotics used. This suggests that the development of resistant subpopulations depends on the growth stage and slow growth or no growth. Therefore, glucose was added during antibiotic treatments in the following biofilm experiments.
FIG. 1.
Antibiotic susceptibility of E. coli cells in the planktonic state. Cells were obtained at exponential phase (6 h), early stationary phase (12 h), and late stationary phase (24 h and 72 h). The cells were washed with PBS and diluted to obtain 107 CFU/ml in MOPS minimal medium. After this the cells were treated with ampicillin (100 μg/ml), kanamycin (25 μg/ml), or ofloxacin (10 μg/ml) in MOPS minimal medium with or without glucose at 37°C for 24 h. After antibiotic treatment, CFU were counted. ND*, not detected. The error bars indicate the standard deviations of triplicate experiments.
Antibiotic susceptibility of E. coli cells in mature biofilms.
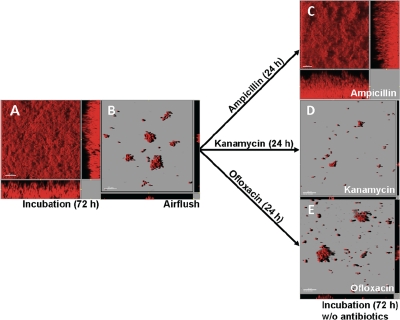
The antibiotic susceptibility of E. coli cells in mature biofilms was determined after removal of the surface layers of the biofilms by air flushing (Fig. 2A and 2B). The air flushing removed much of the biofilms. The results of a COMSTAT analysis showed that the thickness and biovolume of biofilms before the air flushing were 42.59 ± 10.48 μm and 32.91 ± 8.60 μm3/μm2, respectively (Fig. 2A), and decreased to 1.48 ± 1.08 μm and 2.28 ± 1.95 μm3/μm2, respectively, after the air flushing (Fig. 2B). The ampicillin treatment did not inhibit regrowth, and thick biofilms were formed again after 72 h of incubation (Fig. 2C). However, kanamycin and ofloxacin treatments completely inhibited regrowth of the biofilms (Fig. 2D and 2E).
FIG. 2.
Antibiotic susceptibility of cells in the deeper layers of mature biofilms. (A) Mature biofilm after 72 h of incubation. (B) The surface layer of the mature biofilm in panel A was removed by air flushing, resulting in some small cell clusters that remained on the glass surface. (C to D) The remaining cells were treated with 100 μg/ml of ampicillin (C), 25 μg/ml of kanamycin (D), or 10 μg/ml of ofloxacin (E) for 24 h and then incubated with MOPS minimal medium containing no antibiotics for another 72 h. The ampicillin-treated cells grew and formed thick biofilms again (C), whereas the kanamycin- and ofloxacin-treated cells did not grow after 72 h of incubation (D and E).
Effects of ampicillin on E. coli cells during biofilm formation.
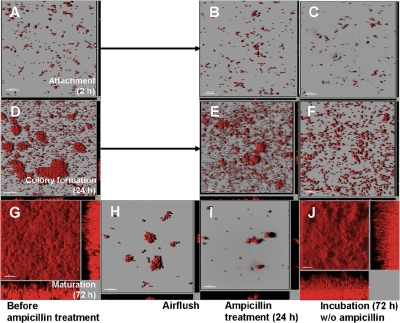
To further examine the increased ampicillin resistance of E. coli cells in the deeper layers of the mature biofilms (Fig. 2C), ampicillin was applied to cells in the different stages of biofilm formation (i.e., the attachment, colony formation, and maturation stages), and then the ability to form biofilms was examined (Fig. 3). For the cells in the maturation stage, the biofilm biomass was removed with strong shear forces and air flushing, which was assumed to eliminate the effect of the restricted penetration of ampicillin and nutrients, as well as the heterogeneous physiological state in the mature biofilms (Fig. 3G and 3H). Therefore, all of the remaining cells were directly exposed to ampicillin, and ampicillin could penetrate throughout the remaining cell clusters. The cells in the attachment (2 h) and colony formation (24 h) stages did not form mature biofilms again after the 24-h ampicillin treatment (Fig. 3A to F). However, the remaining cells in the mature biofilms formed thick biofilms again within 72 h after the ampicillin treatment was discontinued (Fig. 3G to J). The results clearly indicated that some of the cells in the mature biofilms, but not cells in the young colony biofilms, were resistant to ampicillin.
FIG. 3.
Development of ampicillin-resistant cells during biofilm formation. Biofilms in the attachment stage (2 h) (A), the colony formation stage (24 h) (D), and the maturation stage (72 h) (G) were treated with 100 μg/ml of ampicillin for 24 h (B, E, and I) and incubated with MOPS minimal medium containing no antibiotics for another 72 h (C, F, and J, respectively). The surface layers of the mature biofilm in panel G were removed by air flushing (H), and then the preparation was treated with ampicillin. The cells at each biofilm formation stage did not grow during ampicillin treatment (B, E, and I). Only the cells in the deeper layers of the mature biofilms grew again and formed thick biofilms after ampicillin treatment was discontinued (J), whereas newly attached cells and cells in young colony biofilms did not form biofilms again (C and F).
Formation of ampicillin-resistant subpopulations in mature biofilms.
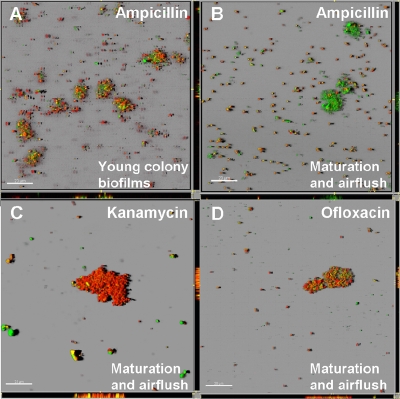
To examine the efficacy of antibiotic treatments for the remaining cells in the mature biofilms, LIVE/DEAD staining was performed after antibiotic treatment for 24 h (Fig. 4). The LIVE/DEAD staining revealed that although 74.4% ± 7.8% of the cells in the young colony biofilms were killed by ampicillin, 44.3% ± 8.2% of the cells in the mature biofilms after air flushing were killed and the majority of the aggregated cells in the mature biofilms after air flushing were alive (Fig. 4A and 4B). However, with the kanamycin and ofloxacin treatments, 93.7% ± 4.7% and 91.5% ± 3.3% of the cells in the mature biofilms after air flushing were dead, respectively (Fig. 4C and 4D). The average thickness and biovolume of biofilms at the colony formation stage were 1.37 ± 0.29 μm and 2.11 ± 0.45 μm3/μm2, respectively, and the average thickness and biovolume at the maturation stage after air flushing were 1.48 ± 1.08 μm and 2.28 ± 1.95 μm3/μm2, respectively. These results showed that the thickness and biovolume of biofilms at the colony formation stage and the thickness and biovolume of mature biofilms after air flushing were not very different. Therefore, ampicillin could penetrate throughout both the remaining cell clusters and the young colony biofilms. These results suggest that the restricted penetration of antibiotics and nutrients into a biofilm cannot always explain the antibiotic resistance. This suggests that there were subpopulations that were resistant to ampicillin in the mature biofilms but not in the young colony biofilms and that these cells probably grew and formed biofilms again after the ampicillin treatment was discontinued.
FIG. 4.
Efficacy of antibiotic treatment of biofilm cells. Young colony biofilms were treated with 100 μg/ml of ampicillin for 24 h (A). The cells remaining in mature biofilms after air flushing were treated with 100 μg/ml of ampicillin (B), 25 μg/ml of kanamycin (C), or 10 μg/ml of ofloxacin (D) for 24 h. After the antibiotic treatments, the cells were stained with LIVE/DEAD stain. Although most of the cells in young colony biofilms were killed by ampicillin (red cells), the majority of the remaining cells, especially the aggregated cells, in the mature biofilms were still alive after ampicillin treatment (green cells).
One of the mechanisms of antibiotic resistance is the formation by subpopulations of microorganisms in biofilms of cells with a unique and highly protective phenotype similar to spore formation. Although most bacteria in biofilms are rapidly killed by antibiotics (18), subpopulations, which might consist of 1% or less of the original population, neither grow nor die in the presence of antibiotics and persist despite continued exposure to antibiotics (48). The existence of resistant subpopulations in the planktonic stationary phase (Fig. 1) has been reported previously (6, 21, 23, 45). A small fraction (ca. 1%) of E. coli bacteria which are a subset of the nonmultiplying bacteria (26) in stationary-phase cultures has been isolated (44). These bacteria had a gene expression profile different from those of exponential-phase and other nonmultiplying bacteria (24, 27). These bacteria expressed some specific genes (30, 40), which implies that they were actively transcribing and metabolically active (9). Therefore, an aminoglycoside, such as kanamycin, must be effective against these cells. A fluoroquinolone, such as ofloxacin, induces a breakdown in iron regulatory dynamics, which promotes formation of reactive oxygen species, leading to cell death (15). Ofloxacin is also reported to be able to kill slowly growing or even nongrowing cells (6, 23), which is consistent with the results showing that ofloxacin was effective in preventing regrowth of biofilms. On the other hand, a β-lactam, such as ampicillin, which interacts with penicillin-binding proteins and glycopeptides that interact with peptidoglycan building blocks, interferes with normal cell wall synthesis and induces cell lysis and death. Because resistant subpopulations do not have cell division and cell wall synthesis activities, ampicillin was not effective against these subpopulations.
Differences in gene expression patterns among the attachment, colony formation, and maturation stages during biofilm formation.
A previous study showed that clumps detached from Staphylococcus aureus biofilms were highly tolerant to oxacillin compared with exponential-phase planktonic cultures, and the antibiotic resistance of the detached biofilm could be attributed to stationary-phase physiology in the clumps (17). It should be noted that only the cells in the deeper layers of mature biofilms, not the cells in the young colony biofilms, were resistant to ampicillin. Therefore, it was speculated that the cell physiology of mature biofilms differs from that of young colony biofilms, which might contribute to the formation of the resistant subpopulations. To verify this hypothesis, global gene expression was analyzed at different stages of biofilm formation, including the attachment stage (2 h), the colony formation stage (24 h), and the maturation stage (72 h).
The gene expression levels for the attachment (2 h), colony formation (24 h), and maturation (72 h) stages were compared. In the attachment stage, there are 99 and 62 genes that show lower and higher levels of expression, respectively, compared to the levels of expression in both the colony formation and maturation stages. In the maturation stage, there are 181 and 184 genes that show lower and higher levels of expression, respectively, compared to the levels of expression in both the attachment and colony formation stages.
There were two patterns of gene expression (Table 1). One of these patterns included 129 genes whose expression gradually decreased during biofilm formation. The other pattern included 199 genes whose expression gradually increased during biofilm formation. In the group of genes whose expression gradually decreased (attachment > maturation), many genes were classified as genes for “biosynthesis of cofactors, prosthetic groups and carriers,” “cell structure,” “central intermediary metabolism,” “energy metabolism,” and “nucleotide biosynthesis and metabolism,” indicating that metabolic activities and cell growth decreased during biofilm maturation. This group included 33 genes negatively regulated by rpoS (22, 28) (Table 2). These genes are involved in energy metabolism (aceE, acnB, cyoABC, cyoE, mdh, nuoG, and sucAB) and flagellum synthesis (flgC, flgN, fliA, and fliM). The levels of expression of these genes decreased as the level of rpoS expression increased during biofilm formation (22). In the group of genes whose expression gradually increased (attachment < maturation), many genes were classified as genes for “carbon compound catabolism,” “energy metabolism,” “putative regulatory proteins,” and “putative transport proteins,” indicating that the cells in the mature biofilms were still metabolically active but not active in cell division. This group included 39 genes positively regulated by rpoS (22, 25, 28, 36, 50, 51) (Table 3). These genes include a heat shock protein gene (hslJ), genes involved in energy metabolism with anaerobic respiration (hyaABCDE, hycF, hycI, and narY) and multidrug resistance (yhiU and yhiV), and a gene previously reported to be upregulated in persister cells (wrbA) (44). The levels of expression of these genes increased as the level of rpoS expression increased during biofilm formation (22). The group of genes whose expression gradually increased included several genes involved in efflux pumps, such as ABC superfamily genes (ccmB, ccmE, nikBCD, oppD, oppF, pstA, yaeE, yfhH, and yrbD), a major facilitator super family gene (ycdD), and a small multidrug resistance family gene (emrE), which may decrease the concentrations of antibiotics in the periplasmic spaces (see Table S1 in the supplemental material). It also included genes encoding other transport proteins (b1601, feoB, ybgH, ydhE, yeaS, yebM, and yhiM), heat shock proteins (dnaJ and hslU), a growth inhibitor (chpB), and proteins involved in metal resistance (cutF and ylcAB) and genes previously reported to be upregulated in persister cells (glgC, umuD, and ygiU) (24, 44). Induction of heat shock proteins suppressed autolysis of E. coli by a number of β-lactams that inhibit the synthesis of peptidoglycan (26, 38). The increased expression of the heat shock proteins in mature biofilms may also have contributed to the resistance to ampicillin in this study. It has been reported that metal and antibiotic resistance genes are linked and share structural and functional characteristics of prokaryotic metal and antibiotic resistance systems (3). This implies that E. coli acquires metal and antibiotic resistance during the maturation of biofilms, which might contribute to the increased resistance of biofilms to ampicillin.
TABLE 1.
Numbers of genes induced significantly at early stages of biofilm formation (attachment > maturation) and at late stages of biofilm formation (attachment < maturation)
| Function(s) | No. of genes induced
|
|
|---|---|---|
| Attachment > maturation | Attachment < maturation | |
| Amino acid biosynthesis and metabolism | 4 | 4 |
| Biosynthesis of cofactors, prosthetic groups, and carriers | 7 | 3 |
| Carbon compound catabolism | 0 | 9 |
| Cell processes (including adaptation and protection) | 8 | 8 |
| Cell structure | 8 | 4 |
| Central intermediary metabolism | 7 | 2 |
| DNA replication, recombination, modification, and repair | 1 | 5 |
| Energy metabolism | 17 | 38 |
| Fatty acid and phospholipid metabolism | 1 | 2 |
| Hypothetical, unclassified, unknown | 31 | 71 |
| Membrane proteins | 0 | 1 |
| Nucleotide biosynthesis and metabolism | 9 | 1 |
| Other known genes | 2 | 1 |
| Phage, transposon, or plasmid | 1 | 2 |
| Putative enzymes | 9 | 8 |
| Putative regulatory proteins | 2 | 11 |
| Putative transport proteins | 3 | 6 |
| Regulatory function | 0 | 2 |
| rRNA | 0 | 2 |
| Structural proteins | 1 | 1 |
| Transcription, RNA processing, and degradation | 0 | 1 |
| Translation and posttranslational modification | 1 | 3 |
| Transport and binding proteins | 17 | 14 |
| Total | 129 | 199 |
TABLE 2.
Genes induced significantly at early stages of biofilm formation (attachment > maturation)a
| Gene | Gene product | Relative expressionb
|
||
|---|---|---|---|---|
| Attachment | Colony formation | Maturation | ||
| aceAc | Isocitrate lyase | 0.61 | −0.06 | −1.10 |
| aceEc | Pyruvate dehydrogenase E1 component | 0.87 | 0.08 | −0.78 |
| acnBc | Aconitate hydrase B | 0.83 | −0.18 | −0.97 |
| aroPc | Aromatic amino acid transport protein | 0.53 | 1.10 | −0.54 |
| bioDc | Dethiobiotin synthetase | 1.11 | 0.38 | −0.64 |
| codAc | Cytosine deaminase | 1.27 | 0.05 | −0.64 |
| cyoAc | Cytochrome o ubiquinol oxidase subunit II | 1.16 | −0.19 | −0.96 |
| cyoBc | Ubiquinol oxidase polypeptide I | 0.96 | −0.10 | −0.96 |
| cyoCc | Cytochrome o ubiquinol oxidase subunit III | 0.90 | 0.01 | −0.91 |
| cyoEc | Protoheme IX farnesyltransferase (heme O biosynthesis) | 1.00 | 0.29 | −1.00 |
| flgCc,d | Flagellar basal body rod protein | 1.73 | −0.07 | −0.90 |
| flgNc | Flagellum synthesis protein | 1.54 | 0.54 | −0.51 |
| fliAc,d | RNA polymerase sigma factor for flagellar operon | 1.80 | −0.18 | −0.80 |
| fliMd | Flagellar motor switch protein | 1.47 | 0.28 | −0.64 |
| guaCc | GMP reductase | 0.90 | 0.20 | −0.60 |
| icdAc | e14 prophage, isocitrate dehydrogenase | 0.67 | 0.19 | −0.92 |
| ilvBc | Acetolactate synthase isozyme I large subunit | 0.98 | −0.15 | −0.94 |
| ilvNc | Acetolactate synthase I small subunit | 1.22 | −0.26 | −1.03 |
| lysPc | Lysine-specific permease | 1.38 | 0.63 | −0.79 |
| mdhc | Malate dehydrogenase | 0.57 | −0.27 | −0.94 |
| nuoGc | NADH dehydrogenase I chain G | 0.52 | 0.29 | −0.77 |
| ppac | Inorganic pyrophosphatase | 0.61 | 1.03 | −0.65 |
| pyrd | Aspartate carbamoyltransferase catalytic chain | 0.84 | 0.71 | −0.56 |
| pyrCd | Dihydroorotase | 1.40 | 0.20 | −0.77 |
| pyrLc | PyrBI operon leader peptide | 0.80 | −0.17 | −0.87 |
| sucAc | 2-Oxoglutarate dehydrogenase E1 component | 0.56 | −0.36 | −0.92 |
| sucBc | 2-Oxoglutarate dehydrogenase | 0.72 | −0.29 | −0.88 |
| tigc | Molecular chaperone involved in cell division | 1.19 | 0.24 | −0.52 |
| tpxc | Thiol peroxidase | 0.81 | 0.24 | −1.06 |
| ybhCc | Putative pectinesterase | 0.57 | 0.57 | −0.59 |
| yccAc | Putative carrier/transport protein | 0.62 | 1.13 | −0.61 |
| yceDc | Hypothetical protein | 0.67 | 0.77 | −0.67 |
| yjbCc | Pseudouridine synthase | 0.57 | 1.32 | −0.55 |
Genes whose expression was statistically significantly different in the attachment, colony formation, and maturation stages fulfilled the following criteria: (i) the ratio of the signal of the experimental sample to the signal of the baseline sample was >2.0 or the ratio of the signal of the baseline sample to the signal of the experimental sample was >2.0, which means that there was a >2.0-fold change; (ii) the P value for a t test for equal experimental sample and baseline sample signals was <0.05; and (iii) the P value for a paired t test was <0.05.
The relative expression value was calculated with dChip software.
This gene is also negatively regulated by rpoS according to Ito et al. (22).
This gene is also negatively regulated by rpoS according to Loewen et al. (28).
TABLE 3.
Genes induced significantly at late stages of biofilm formation (attachment < maturation)a
| Gene | Gene product | Relative expressionb
|
||
|---|---|---|---|---|
| Attachment | Colony formation | Maturation | ||
| appAe | Phosphoanhydride phosphorylase | −0.65 | 0.24 | 1.64 |
| appBg | Cytochrome bdII oxidase subunit II | −0.62 | 0.03 | 1.81 |
| b1428c | Hypothetical protein YdcK | −1.23 | −0.17 | 1.39 |
| b2880c | Hypothetical protein YgfM | −0.70 | −0.01 | 1.67 |
| bfrc,d,f,h | Bacterioferritin | −1.56 | 0.39 | 0.65 |
| cfae | Cyclopropane-fatty acyl phospholipid synthase | −0.90 | 0.28 | 1.64 |
| deoBc | Phosphopentomutase | −1.00 | −0.44 | 1.48 |
| glgSe | Glycogen synthesis protein GlgS | −0.90 | −0.69 | 1.06 |
| hepAc | RNA polymerase-associated protein | −1.00 | −0.46 | 1.56 |
| hslJc | Heat shock protein HslJ | −0.76 | −0.87 | 0.53 |
| hyaAe | Hydrogenase 1 small chain precursor | −0.68 | 0.09 | 1.74 |
| hyaBe | Hydrogenase 1 large subunit | −0.66 | 0.06 | 1.79 |
| hyaCe | Probable Ni/Fe hydrogenase 1 B-type cytochrome subunit | −0.71 | 0.07 | 1.73 |
| hyaDe | Processing of HyaA and HyaB proteins | −0.69 | 0.01 | 1.77 |
| hyaEe | Processing of HyaA and HyaB proteins | −0.70 | 0.01 | 1.79 |
| hycFh | Formate hydrogen lyase subunit 6 | −0.77 | −0.28 | 1.72 |
| hycIh | Protease involved in processing the C-terminal end of HycE | −0.68 | −0.05 | 1.66 |
| melBc | Melibiose permease II | −1.14 | −0.15 | 0.87 |
| narYg,h | Cryptic nitrate reductase 2 beta subunit | −0.69 | 0.24 | 1.72 |
| pflAc | Pyruvate formate lyase 1 activating enzyme | −0.88 | −0.45 | 1.49 |
| pphAc | Protein phosphatase 1 | −1.07 | −0.10 | 1.16 |
| Rncc | RNase III | −0.76 | 0.35 | 1.62 |
| slpc,f,h | FKBP-type 16-kDa peptidyl-prolyl cis-trans isomerase | −1.06 | 0.20 | 1.65 |
| treFh | Cytoplasmic trehalase | −1.14 | −0.59 | 0.56 |
| wrbAc,d,e,f,h | Flavoprotein WrbA | −0.81 | −0.36 | 1.71 |
| yadQc | Voltage-gated ClC-type chloride channel EriC | −0.94 | 0.90 | 1.29 |
| ybaSf,h | Putative glutaminase | −0.85 | 0.23 | 1.64 |
| ybaTf | Hypothetical transport protein YbaT | −0.89 | 0.31 | 1.61 |
| ybcOc | DLP12 prophage | −0.98 | −0.28 | 1.20 |
| yeaQc | Hypothetical protein YeaQ | −1.36 | −0.13 | 1.29 |
| yffGc | Putative oxidoreductase Fe-S subunit | −0.98 | 1.15 | 0.95 |
| ygaMc,h | Hypothetical protein | −0.97 | 0.18 | 1.66 |
| ygaUc,f,g,h | Unknown protein | −1.45 | −0.61 | 0.76 |
| yhaHc | Hypothetical protein YhaH | −0.93 | −0.66 | 1.56 |
| yheLc | Hypothetical protein | −0.58 | 0.16 | 1.75 |
| yhiUg,h | Multidrug resistance protein (lipoprotein) | −0.81 | 0.81 | 1.37 |
| yhiVg | Multidrug transport protein, RpoS dependent (RND family) | −0.83 | 1.13 | 1.05 |
| yhiXc | Putative AraC-type regulatory protein | −0.91 | 1.03 | 0.80 |
| yjdIc,f,h | Hypothetical protein | −1.29 | −0.67 | 0.70 |
Genes whose expression was statistically significant different in the attachment, colony formation, and maturation stages fulfilled the following criteria: (i) the ratio of the signal of the experimental sample to the signal of the baseline sample was >2.0 or the ratio of the signal of the baseline sample to the signal of the experimental sample was >2.0, which means that there was a >2.0-fold change; (ii) the P value for a t test for equal experimental sample and baseline sample signals was <0.05; and (iii) the P value for a paired t test was <0.05.
The relative expression value was calculated with dChip software.
This gene is also positively regulated by rpoS according to Ito et al. (22).
This gene is also positively regulated by rpoS according to Lacour and Landini (25).
This gene is also positively regulated by rpoS according to Loewen et al. (28).
This gene is also positively regulated by rpoS according to Patten et al. (36).
This gene is also positively regulated by rpoS according to Vijayakumar et al. (50).
This gene is also positively regulated by rpoS according to Weber et al. (51).
RpoS dependence.
The number of resistant cells was very low in the lag or early exponential phase but increased dramatically in mid- to late exponential phases (Fig. 1), which has been reported previously for E. coli, Pseudomonas aeruginosa, and S. aureus (4, 19, 23). A previous study has shown that the level of rpoS expression increased in late exponential to early stationary phase (22), which correlated with the increase in the number of resistant cells. This suggested that formation of resistant subpopulations depends on the growth stage and growth rate (23) and might be regulated by rpoS expression (29). It has also been reported that rpoS is important for E. coli biofilm formation through regulation of global gene expression (22). The rpoS expression in biofilms changes the physiological state of E. coli, including induction of a stress response and suppression of energy metabolism and motility. Involvement of RpoS in multidrug resistance has been suggested previously (40).
To investigate the contribution of rpoS to antibiotic susceptibility, wild-type and rpoS mutant strains were exposed to antibiotics in late stationary phase in the planktonic state, and the cell viability was determined. The results showed that the rpoS mutant strain was about 20 times more susceptible to ampicillin than the wild-type strain, which indicated that rpoS contributed to the ampicillin resistance of E. coli cells. The rpoS mutant strain was also more susceptible to kanamycin and ofloxacin than the wild-type strain. An rpoS mutant of P. aeruginosa was also reported to be much more sensitive to the effects of antibiotics than wild-type bacteria (30).
In summary, E. coli cells in the deeper layers of mature biofilms, but not in young colony biofilms, exhibited resistance to ampicillin. The observed resistance to ampicillin was probably due to the emergence of the resistant subpopulations in the mature biofilms, which were still alive after 24 h of ampicillin treatment as determined by LIVE/DEAD staining. Furthermore, global gene expression analyzed at different stages of biofilm formation suggested that the formation and resistance of the subpopulations were aided by induction of biofilm-specific phenotypes, including slow growth or no growth, and rpoS-mediated stress responses.
Supplementary Material
Acknowledgments
This research was carried out as part of the 21st Century COE Program “Sustainable Metabolic System of Water and Waste for Area-Based Society.” A.I. was financially supported by the 21st Century COE Program.
Footnotes
Published ahead of print on 17 April 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Adams, J. L., and R. J. C. McLean. 1999. Impact of rpoS deletion on Escherichia coli biofilms. Appl. Environ. Microbiol. 65:4285-4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrews, J. M. 2001. Determination of minimum inhibitory concentrations. J. Antimicrob. Chemother. 48:5-16. [DOI] [PubMed] [Google Scholar]
- 3.Baker-Austin, C., M. S. Wright, R. Stepanauskas, and J. V. McArthur. 2006. Co-selection of antibiotic and metal resistance. Trends Microbiol. 14:176-182. [DOI] [PubMed] [Google Scholar]
- 4.Bassler, B. L. 2002. Small talk: cell-to-cell communication in bacteria. Cell 109:421-424. [DOI] [PubMed] [Google Scholar]
- 5.Beloin, C., J. Valle, P. Latour-Lambert, P. Faure, M. Kzreminski, D. Balestrino, J. A. J. Haagensen, S. Molin, G. Prensier, B. Arbeille, and J. M. Ghigo. 2004. Global impact of mature biofilm lifestyle on Escherichia coli K-12 gene expression. Mol. Microbiol. 51:659-674. [DOI] [PubMed] [Google Scholar]
- 6.Brooun, A., S. Liu, and K. Lewis. 2000. A dose-response study of antibiotic resistance in Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 44:640-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown, M. R. W., D. G. Allison, and G. Peter. 1988. Resistance of bacterial biofilms to antibiotics: a growth-rate related effect? J. Antimicrob. Chemother. 22:777-783. [DOI] [PubMed] [Google Scholar]
- 8.Brown, M. R. W., and P. Gilbert. 1993. Sensitivity of biofilms to antimicrobial agents. J. Appl. Bacteriol. Symp. Suppl. 74:87S-97S. [DOI] [PubMed] [Google Scholar]
- 9.Coates, A. R., and Y. Hu. 2008. Targeting non-multiplying organisms as a way to develop novel antimicobials. Trends Pharmacol. Sci. 29:143-150. [DOI] [PubMed] [Google Scholar]
- 10.Corona-Izquierdo, F. P., and J. Membrillo-Hernández. 2002. A mutation in rpoS enhances biofilm formation in Escherichia coli during exponential phase of growth. FEMS Microbiol. Lett. 211:105-110. [DOI] [PubMed] [Google Scholar]
- 11.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 12.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Domka, J., J. Lee, T. Bansal, and T. K. Wood. 2007. Temporal gene-expression in Escherichia coli biofilms. Environ. Microbiol. 9:332-346. [DOI] [PubMed] [Google Scholar]
- 14.Drenkard, E. 2003. Antimicrobial resistance of Pseudomonas aeruginosa biofilms. Microbes Infect. 5:1213-1219. [DOI] [PubMed] [Google Scholar]
- 15.Dwyer, D. J., M. A. Kohanski, B. Hayete, and J. J. Collins. 2007. Gyrase inhibitors induce an oxidative damage cellular death pathway in Escherichia coli. Mol. Syst. Biol. 3:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edgar, R., M. Domracheb, and A. E. Lash. 2002. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30:207-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fux, C. A., S. Wilson, and P. Stoodley. 2004. Detachment characteristics and oxacillin resistance of Staphylococcus aureus biofilm emboli in an in vitro catheter infection model. J. Bacteriol. 186:4486-4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goto, T., Y. Nakame, M. Nishida, and Y. Ohi. 1999. In vitro bactericidal activities of beta-lactamases, amikacin, and fluoroquinolones against Pseudomonas aeruginosa biofilm in artificial urine. Urology 53:1058-1062. [DOI] [PubMed] [Google Scholar]
- 19.Hastings, J. W., and E. P. Greenberg. 1999. Quorum sensing: the explanation of a curious phenomenon reveals a common characteristic of bacteria. J. Bacteriol. 181:2667-2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heydorn, A., A. T. Nielsen, M. Hentzer, C. Sternberg, M. Givskov, B. K. Ersbøll, and S. Molin. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146:2395-2407. [DOI] [PubMed] [Google Scholar]
- 21.Hogan, D., and R. Kolter. 2002. Why are bacteria refractory to antimicrobials? Curr. Opin. Microbiol. 5:472-477. [DOI] [PubMed] [Google Scholar]
- 22.Ito, A., T. May, K. Kawata, and S. Okabe. 2008. Significance of rpoS during maturation of Escherichia coli biofilms. Biotechnol. Bioeng. 99:1462-1471. [DOI] [PubMed] [Google Scholar]
- 23.Keren, I., N. Kaldalu, A. Spoering, Y. Wang, and K. Lewis. 2004. Persister cells and tolerance to antimicrobials. FEMS Microbiol. Lett. 230:13-18. [DOI] [PubMed] [Google Scholar]
- 24.Keren, I., D. Shah, A. Spoering, N. Kaldalu, and K. Lewis. 2004. Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. J. Bacteriol. 186:8172-8180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lacour, S., and P. Landini. 2004. σS-Dependent gene expression at the onset of stationary phase in Escherichia coli: function of σS-dependent genes and identification of their promoter sequences. J. Bacteriol. 186:7186-7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lewis, K. 2000. Programmed death in bacteria. Microbiol. Mol. Biol. Rev. 64:503-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lewis, K. 2007. Persister cells, dormancy and infectious disease. Nat. Rev. Microbiol. 5:48-56. [DOI] [PubMed] [Google Scholar]
- 28.Loewen, P. C., B. Hu, J. Strutinsky, and R. Sparling. 1998. Regulation in the rpoS regulon of Escherichia coli. Can. J. Microbiol. 44:707-717. [DOI] [PubMed] [Google Scholar]
- 29.Mah, T.-F. C., and G. A. O'Toole. 2001. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 9:34-39. [DOI] [PubMed] [Google Scholar]
- 30.Murakami, K., T. Ono, D. Viducic, S. Kayama, M. Mori, K. Hirota, K. Nemoto, and Y. Miyake. 2005. Role for rpoS gene of Pseudomonas aeruginosa in antibiotic tolerance. FEMS Microbiol. Lett. 242:161-167. [DOI] [PubMed] [Google Scholar]
- 31.Neidhardt, F. C., P. L. Bloch, and D. F. Smith. 1974. Culture medium for enterobacteria. J. Bacteriol. 119:736-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nickel, J. C., I. Ruseska, J. B. Wricht, and J. W. Costerton. 1985. Tobramycin resistance of Pseudomonas aeruginosa cells growing as a biofilm on urinary catheter material. Antimicrob. Agents Chemother. 27:619-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okabe, S., H. Satoh, and Y. Watanabe. 1999. In situ analysis of nitrifying biofilms as determined by in situ hybridization and the use of microelectrodes. Appl. Environ. Microbiol. 65:3182-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Okabe, S., T. Itoh, H. Satoh, and Y. Watanabe. 1999. Analyses of spatial distributions of sulfate-reducing bacteria and their activity in aerobic wastewater biofilms. Appl. Environ. Microbiol. 65:5107-5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Olson, M. E., H. Ceri, D. W. Morck, A. G. Buret, and R. R. Read. 2002. Biofilm bacteria: formation and comparative susceptibility to antibiotics. Can. J. Vet. Res. 66:86-92. [PMC free article] [PubMed] [Google Scholar]
- 36.Patten, C. L., M. G. Kirchhof, M. R. Schertzberg, R. A. Morton, and H. E. Schellhorn. 2004. Microarray analysis of RpoS-mediated gene expression in Escherichia coli K-12. Mol. Genet. Genomics 272:580-591. [DOI] [PubMed] [Google Scholar]
- 37.Potera, C. 1999. Forging a link between biofilms and disease. Science 283:1837-1839. [DOI] [PubMed] [Google Scholar]
- 38.Powell, J. K., and K. D. Young. 1991. Lysis of Escherichia coli by β-lactams which bind penicillin-binding proteins 1a and 1b: inhibition by heat shock proteins. J. Bacteriol. 173:4021-4026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pratt, L. A., and R. Kolter. 1999. Genetic analyses of bacterial biofilm formation. Curr. Opin. Microbiol. 2:598-603. [DOI] [PubMed] [Google Scholar]
- 40.Rami, A., C. M. Toutain, and A. Jacq. 2005. An increased level of alternative sigma factor RpoS partially suppresses drug hypersensitivity associated with inactivation of the multidrug resistance pump AcrAB in Escherichia coli. Res. Microbiol. 156:356-360. [DOI] [PubMed] [Google Scholar]
- 41.Ren, D., L. A. Bedzyk, S. M. Thomas, R. W. Ye, and T. K. Wood. 2004. Gene expression in Escherichia coli biofilms. Appl. Microbiol. Biotechnol. 64:515-524. [DOI] [PubMed] [Google Scholar]
- 42.Schembri, M. A., K. Kjærgaard, and P. Klemm. 2003. Global gene expression in Escherichia coli biofilms. Mol. Microbiol. 48:253-267. [DOI] [PubMed] [Google Scholar]
- 43.Serres, M. H., S. Gopal, L. A. Nahum, P. Liang, T. Gaasterland, and M. Riley. 2001. A functional update of the Escherichia coli K-12 genome. Genome Biol. 2:RESEARCH0035.1-RESEARCH0035.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shah, D., Z. Zhang, A. B. Khodursky, N. Kaldalu, K. Kurg, and K. Lewis. 2006. Persisters: a distinct physiological state of E. coli. BMC Microbiol. 6:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spoering, A. L., and K. Lewis. 2001. Biofilms and planktonic cells of Pseudomonas aeruginosa have similar resistance to killing by antimicrobials. J. Bacteriol. 183:6746-6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sternberg, C., B. B. Christensen, T. Johansen, A. T. Nielsen, J. B. Andersen, M. Givskov, and S. Molin. 1999. Distribution of bacterial growth activity in flow-chamber biofilms. Appl. Environ. Microbiol. 65:4108-4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stewart, P. S. 1996. Theoretical aspects of antibiotic diffusion into microbial biofilms. Antimicrob. Agents Chemother. 40:2517-2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stewart, P. S., and J. W. Costerton. 2001. Antibiotic resistance of bacteria in biofilms. Lancet 358:135-138. [DOI] [PubMed] [Google Scholar]
- 49.Tresse, O., T. Jouenne, and G.-A. Junter. 1995. The role of oxygen limitation in the resistance of agar-entrapped, sessile-like Escherichia coli to aminoglycoside and β-lactam antibiotics. J. Antimicrob. Chemother. 36:521-526. [DOI] [PubMed] [Google Scholar]
- 50.Vijayakumar, S. R. V., M. G. Kirchhof, C. L. Patten, and H. E. Schellhorn. 2004. RpoS-regulated genes of Escherichia coli identified by random lacZ fusion mutagenesis. J. Bacteriol. 186:8499-8507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weber, H., T. Polen, J. Heuveling, V. F. Wendisch, and R. Hengge. 2005. Genome-wide analysis of the general stress response network in Escherichia coli: σS-dependent genes, promoters, and sigma factor selectivity. J. Bacteriol. 187:1591-1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu, K. D., P. S. Stewart, F. Xia, C.-T. Huang, and G. A. McFeters. 1998. Spatial physiological heterogeneity in Pseudomonas aeruginosa biofilms is determined by oxygen availability. Appl. Environ. Microbiol. 64:4035-4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang, Y., F. Buchholz, J. P. Muyrers, and A. F. Stewart. 1998. A new logic for DNA engineering using recombination in Escherichia coli. Nature 20:123-128. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.