Abstract
The wood decay basidiomycete Phanerochaete chrysosporium was grown under standard ligninolytic or cellulolytic conditions and subjected to whole-genome expression microarray analysis and liquid chromatography-tandem mass spectrometry of extracellular proteins. A total of 545 genes were flagged on the basis of significant changes in transcript accumulation and/or peptide sequences of the secreted proteins. Under nitrogen or carbon limitation, lignin and manganese peroxidase expression increased relative to nutrient replete medium. Various extracellular oxidases were also secreted in these media, supporting a physiological connection based on peroxide generation. Numerous genes presumed to be involved in mobilizing and recycling nitrogen were expressed under nitrogen limitation, and among these were several secreted glutamic acid proteases not previously observed. In medium containing microcrystalline cellulose as the sole carbon source, numerous genes encoding carbohydrate-active enzymes were upregulated. Among these were six members of the glycoside hydrolase family 61, as well as several polysaccharide lyases and carbohydrate esterases. Presenting a daunting challenge for future research, more than 190 upregulated genes are predicted to encode proteins of unknown function. Of these hypothetical proteins, approximately one-third featured predicted secretion signals, and 54 encoded proteins detected in extracellular filtrates. Our results affirm the importance of certain oxidative enzymes and, underscoring the complexity of lignocellulose degradation, also support an important role for many new proteins of unknown function.
Global conversion of organic carbon to CO2 with concomitant reduction of molecular oxygen involves the combined metabolic activity of numerous organisms. The most abundant source of carbon is plant biomass, composed primarily of cellulose, hemicellulose, and lignin. Many microorganisms are capable of utilizing cellulose and hemicellulose as carbon and energy sources, but a much smaller group of filamentous fungi has evolved with the ability to depolymerize lignin, the most recalcitrant component of plant cell walls. Collectively known as white rot fungi, they possess the unique ability to efficiently depolymerize lignin in order to gain access to cell wall carbohydrates for carbon and energy sources. These wood decay fungi are common inhabitants of forest litter and fallen trees. As such, white rot fungi play an important, if not pivotal, role in the carbon cycle (for a review, see reference 15).
The ∼30-Mb haploid genome of the most intensively studied white rot fungus, Phanerochaete chrysosporium, has been sequenced by the U.S. Department of Energy's Joint Genome Institute. Analysis of the draft genome (38) and substantially improved assembly and gene models (http://genome.jgi-psf.org/Phchr1/Phchr1.home.html [53]) revealed many genes potentially involved in lignocellulose degradation. These genome resources have facilitated mass spectrometric identification of extracellular proteins in defined (43, 55), semidefined (54), and complex (1, 39, 41) media. In some instances, the investigations affirm the importance of well-characterized oxidative and hydrolytic enzymes, but considerable uncertainty persists with respect to the roles and interactions of hundreds of genes. To provide a more complete view of gene expression, we report here transcriptome and secretome analyses of P. chrysosporium grown under defined ligninolytic and cellulolytic conditions.
MATERIALS AND METHODS
Culture conditions and characterization.
RNA for microarrays was obtained from P. chrysosporium strain RP78 (Forest Mycology Center, Forest Products Laboratory) grown in the following media: (i) RB (replete B3 medium [30, 31]) with sufficient carbon and nitrogen harvested during log-phase growth; (ii) CLB (carbon-limited B3 medium) harvested during stationary phase; (iii) NLB (nitrogen-limited B3 medium) harvested during stationary phase; (iv) HBG (Highley's basal salts medium [20] supplemented with glucose); (v) HBA (Highley's basal salts medium supplemented with cellulose). RB medium contained (per liter): 1.73 g of trans-aconitic acid, 2 g of KH2PO4, 0.5 g of MgSO4·7H2O, 0.1 g of CaCl2·2H2O, 1.6 g of ammonium tartrate, 10 g of glucose, and 1 mg of thiamine. The medium was supplemented with 10 ml of mineral solution containing (per liter): 1.5 g of nitrilotriacetic acid, 3 g of MgSO4·7H2O, 0.5 g of MnSO4·H2O, 1 g of NaCl, 0.1 g of FeSO4·H2O, 0.1 g of CoSO4, 0.1 g of CaCl2, 0.1 g of ZnSO4·7H2O, 0.01 g of CuSO4, 0.01 g of AlK(SO4) 2·12H2O, 0.01 g of H3BO3, and 0.01 g of NaMoO4·2H2O. A total of 50 ml of RB was inoculated with ∼106 spores in 300-ml Erlenmeyer flasks, and the stationary-phase cultures were harvested after 2 days at 37°C. CLB and NLB composition were identical except that glucose and ammonia tartrate were reduced to 2 g and 0.2 g liter−1, respectively. After incubation for 2 days, these nutrient-deprived cultures were flushed with oxygen and tightly stoppered. CLB and NLB cultures were harvested after 4 and 5 days, respectively. Transcript patterns and enzyme systems have been extensively studied using B3 media (reviewed in references 16, 28, and 45). Highley's basal medium contained (per liter): 2 g of NH4NO3, 2 g of KH2PO4, 0.5 g of MgSO4·7H2O, 0.1 g of CaCl2·2H2O, 1 mg of thiamine, and 10 ml of the above-mentioned mineral solution. This basal medium was supplemented with 0.5% (wt/vol) glucose (HBG) or with 0.5% microcrystalline cellulose (Avicel, PH-101; Fluka Chemie, Switzerland) (HBA). Aliquots (250 ml) of medium were inoculated with ∼107 spores in 2-liter Erlenmeyer flasks, and the cultures were harvested after 5 days on a rotary shaker (150 rpm) at 37°C.
Liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis of extracellular proteins included RB, HBA, and WBC or Wood's medium (59). WBC is known to support high yields of cellobiose dehydrogenase (CDH). Comparative analysis also considered protein patterns previously determined in NLB, CLB (53), and NMA (modified Norkran's medium with Avicel [13, 54]) and, characterized by others, in medium containing complex lignocellulose substrates (1, 41).
Culture supernatants with all media were tested for lignin peroxidase (LiP), manganese peroxidase (MnP), glyoxal oxidase (GLX) and CDH enzyme activities. LiP activity, measured by veratryl alcohol oxidation (50), was detectable only in CLB and NLB at 65 and 8.8 nmol min−1 ml−1, respectively. Oxidation of 2,6-dimethoxyphenol was used to measure MnP (57) which, like LiP, was restricted to CLB and NLB at 2.6 and 11 nmol min−1 ml−1. GLX was measured using methylglyoxal as the oxidase substrate and phenol red as the substrate for horseradish peroxidase in a coupled assay (29, 30). CLB and NLB supernatants showed GLX activities of 2.6 and 3.4 nmol min−1 ml−1, respectively. Detection of CDH was based on reduction of 100 μM dichlorophenol-indophenol using cellobiose (500 μM) as the electron donor in 20 mM (pH 4.5) succinate buffer (2). CDH activity was detectable only in WBC (130 nmol min−1 ml−1) and NMA (63 nmol min−1 ml−1) cultures.
Expression microarrays.
From a data set of 10,004 unique alleles, each Roche NimbleGen (Madison, WI) array featured 12 unique 60mer probes per gene, all in triplicate. (Seven gene models composed mostly of repetitive DNA were represented by only 2 to 11 60mers.) Complete design details are available under platform GPL8022 within the Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/index.cgi).
Total RNA was purified from the five above-mentioned media. In short, cultures were harvested by filtering through Miracloth (Calbiochem/EMD Biosciences, Gibbstown, NJ), squeeze dried, and snap-frozen in liquid nitrogen. Pellets were stored at −80°C until use. Extraction buffer was prepared by combining 10 ml of 690 mM para-aminosalicylic acid (sodium salt) (Sigma-Aldrich, St. Louis, MO) with 10 ml of 56 mM tri-isopropylnapthalene sulfonic acid (sodium salt; Sigma-Aldrich) and placed on ice. To this was added 5 ml of 5× RNB (1.0 M Tris, 1.25 M NaCl, 0.25 M EGTA). The pH of the 5× RNB was adjusted to 8.5 with NaOH. The mixture was kept on ice and shaken just before use.
Frozen fungal pellets were ground to a fine powder with liquid nitrogen in an acid-washed, prechilled mortar and pestle. The ground mycelia were transferred to Falcon 2059 tubes (VWR International, West Chester, PA), and extraction buffer was added to make a thick slurry. The samples were vortex mixed vigorously and placed on ice until all samples were processed. A one-half volume of Tris-EDTA-saturated phenol (Sigma-Aldrich) and a one-quarter volume chloroform (Sigma-Aldrich) were added to each sample, followed by vigorous vortex mixing. Samples were spun at 2,940 × g in a fixed-angle rotor for 5 min. The aqueous layer was moved to a new tube, and phenol-chloroform extractions were repeated until the interface between the aqueous and organic layers was clear. The final aqueous extractions were placed in clean 2059 tubes, to which was added 0.1 volumes of 3 M sodium acetate (pH 5.2; diethyl pyrocarbonate treated) and 2 volumes of absolute ethanol. The tubes were shaken vigorously and stored overnight at −20°C.
The tubes were spun 1 h at 2,940 × g, the supernatants were decanted, and the pellets were resuspended in 4 ml of RNase-free H2O. Total RNA was purified by using an RNeasy Maxi kit (Qiagen, Valencia, CA) according to the manufacturer's protocol for RNA cleanup. RNAs were eluted from the RNeasy spin columns using two spins, for a final volume of 2 ml. The eluted RNAs were ethanol precipitated and stored overnight at −20°C. The RNAs were spun 1 h at 2,940 × g, washed once with 70% ethanol, and resuspended in 50 to 100 μl of RNase-free H2O. Three biological replicates per medium were used (15 separate arrays).
RNA was converted to double-stranded cDNA and labeled with a Cy3 fluorophore for hybridization to the array by Roche NimbleGen (Iceland). In brief, 10 μg of total RNA was incubated with 1× first-strand buffer, 10 mM dithiothreitol, 0.5 mM deoxynucleoside triphosphates, 100 pM oligonucleotide T7 d(T)24 primer, and 400 U of SuperScript II (Invitrogen) for 60 min at 42°C. Second-strand cDNA was synthesized by incubation with 1× second-strand buffer, 0.2 mM deoxynucleoside triphosphates, 0.07 U of DNA ligase/μl, 0.27 U of DNA polymerase I/μl, and 0.013 U of RNase H/μl at 16°C for 2 h. (DNA ligase, DNA polymerase I, and RNase H were obtained from Invitrogen.) Immediately afterward, 10 U of T4 DNA polymerase (Invitrogen) was added for an additional 5 min of incubation at 16°C. Double-stranded cDNA was treated with 27 ng of RNase A (EpiCenter Technologies)/μl for 10 min at 37°C. Treated cDNA was purified by using an equal volume of phenol-chloroform-isoamyl alcohol (Ambion), ethanol precipitated, washed with 80% ethanol, and resuspended in 20 μl of water. Then, 1 μg of each cDNA sample was amplified and labeled with 1 U of Klenow fragment (New England Biolabs)/μl and 1 optical density unit of Cy3 fluorophore (TriLink Biotechnologies, Inc.) for 2 h at 37°C. Array hybridization was carried out with 6 μg of labeled cDNA suspended in NimbleGen hybridization solution for 17 h at 42°C. Arrays were scanned on an Axon 4000B scanner (Molecular Dynamics), and data were extracted from the scanned image by using NimbleScan v2.4. ArrayStar v2.1 (DNAStar, Madison, WI) software was used to quantify and visualize data. Analyses were based on three biological replicates per culture medium. Quantile normalization and robust multiarray averaging (21) were applied to the entire data set. Scatterplots of results are shown in Fig. S1 in the supplemental material. Unless otherwise specified, expression levels are based on log2 values. All MIAME compliant (4) microarray expression data have been deposited in National Center for Biotechnology Information (NCBI)'s GEO and are accessible through GEO series accession number GSE14736.
Competitive reverse transcription-PCR (RT-PCR) was used to quantify transcripts of the copper radical oxidase genes glx and cro2, as previously described (55). For the transcripts of CDH (cdh1), the upstream and downstream primers were 5′-GGCTCGCCCCAACTTCACTT-3′ and 5′-GTTGTACGGCAGCGACGAG-3′, respectively. These primers yielded cDNA and competitive template amplicons of 556 and 709 nucleotides, respectively.
MS.
Soluble extracellular protein was concentrated from culture filtrates as previously reported (53, 54). After sodium dodecyl sulfate-polyacrylamide gel electrophoresis fractionation, “in gel” digestion and MS analysis were performed as described elsewhere (www.biotech.wisc.edu/ServicesResearch/MassSpec/ingel.htm). An in-house licensed Mascot search engine (Matrix Science, London, United Kingdom) identified peptides using the 10,048 gene models in the v2.1 data set (53). To facilitate comparison, Mascot analysis was also extended to data previously analyzed by Spectrum Mill MS (Agilent). Mascot scores of ≥40 were considered highly significant. Throughout, protein similarity scores are based on the Smith-Waterman algorithm (46) using the BLOSUM62 matrix. P. chrysosporium protein model identification numbers are preceded by “Pchr.” Detailed information for each protein can be directly accessed by appending the model number to the following string: http://genome.jgi-psf.org/cgi-bin/dispGeneModel?db=Phchr1&id=X, where X is the one- to five-digit model number. We assigned function or “putative” function only when supported by direct experimental evidence or when comparisons to known proteins revealed conserved catalytic features and/or significant alignment scores (bit scores > 150) to known proteins within the Swiss-Prot database. All other proteins were designated “hypothetical,” and those with significant homology (bit scores > 150) to other conceptual translations within GenBank were considered “conserved hypothetical” proteins.
RESULTS
Overview of gene expression.
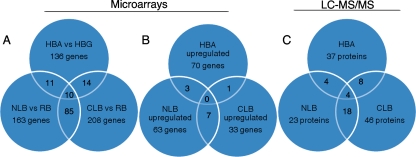
Our analysis focused on transcript profiles in widely used defined ligninolytic (nutrient-limited B3 media) and cellulolytic (cellulose-induced) cultures. Essentially, the microarrays examined three pairwise comparisons: (i) carbon-limited (CLB) versus nutrient-replete B3 media (RB); (ii) nitrogen-limited (NLB) versus RB; and (iii) Highley's basal medium supplemented with avicel (HBA) versus glucose-supplemented basal salts (HBG). Of 10,004 genes represented on arrays, transcripts of 208, 163, and 136 genes exhibited ≥2-fold differences (P < 0.01) in transcript accumulation in the pairwise comparisons of CLB versus RB, NLB versus RB, and HBA versus HBG, respectively (Fig. 1A and see Fig. S1 in the supplemental material). Of these, 33, 63, and 70 genes were upregulated (accumulated ≥2-fold) in CLB, NLB, and HBA, respectively (Fig. 1B). (GEO “superseries” GSE14736 lists log2 and linear response signals for all 10,004 targets in all media, together with expression ratios, genome locations, and InterPro domains.)
FIG. 1.
Venn diagrams illustrating the distribution of regulated genes (A and B) and MS-identified proteins (C). (A) Genes with transcript levels that change ≥2-fold in comparisons of nitrogen-limited and nutrient-replete media (NLB versus RB), carbon-limited and nutrient-replete media (CLB versus RB), and cellulose- and glucose-containing media (HBA versus HBG). (B) Genes with transcripts that are upregulated ≥2-fold in these same comparisons. (C) Genes encoding proteins identified by LC-MS/MS in NLB, CLB, and HBA. “Upregulation” is arbitrarily assigned to genes whose transcripts accumulate in NLB relative to RB, in CLB relative to RB, and in HBA relative to HBG. The corresponding “downregulated” genes are not enumerated in this figure but are listed in Table S3 in the supplemental material. In all cases, values within overlapping regions have not been subtracted from totals. For example, peptides corresponding to a total of 37 protein models were identified in HBA medium. Of the 37, 8 were also present in carbon-limited medium, and 4 of these were common to all three media. An additional 61 proteins were identified in Wood's cellulose-containing medium (WBC), and these are listed in Table S3 in the supplemental material.
In these same media, LC-MS/MS unambiguously identified peptide sequences corresponding to 46, 23, and 37 genes (Fig. 1C). MS data for CLB and NLB was previously acquired and subjected to Spectrum Mill MS (Agilent) analysis (53) but analyzed here by Mascot to simplify comparison. HBA MS data was not previously reported, nor were the 101 and 76 proteins identified in RB and in Wood's cellulose-containing medium (WBC), respectively. Complete listings of peptide sequences, scores, and corresponding gene models are presented in Tables S1 and S2 in the supplemental material. Correlation coefficients between the peptide Mascot score and the microarray signal strength were 0.04, 0.36, 0.77, and 0.1 for RB (n = 101), NLB (n = 23), CLB (n = 46), and HBA (n = 37), respectively.
In aggregate, our microarray and proteome analyses flagged 545 genes based on significant transcript regulation and/or on identification of specific peptide sequences in extracellular fluids. Table S3 in the supplemental material contains detailed information for all of these genes, including putative function, characteristics of the predicted proteins (pI, molecular weight, targeting signals), microarray signals for all replicates, Interpro hits, highest Swiss-Prot hits with scoring, genome localization, and MS scores from all media. For all gene models, supporting evidence, annotations, and analyses are available through the U.S. Department of Energy Joint Genome Institute's genome portal at www.jgi.doe.gov/whiterot.
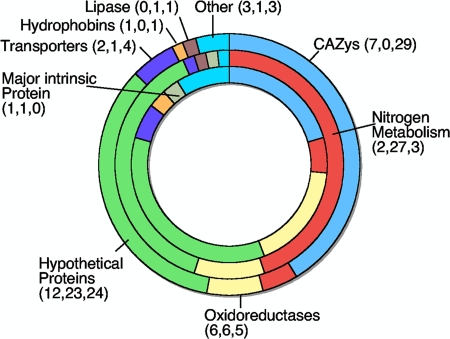
Focusing on genes upregulated in ligninolytic (CLB and NLB) and cellulolytic (HBA) media, substantial differences were observed in the number and distribution of functional categories (Fig. 2). Among the more dramatic differences, the numbers of upregulated genes encoding carbohydrate active enzymes (CAZYs) was significantly higher in HBA, while genes involved in nitrogen metabolism were predominant in NLB.
FIG. 2.
Distribution of P. chrysosporium genes encoding upregulated transcripts in carbon limited B3 medium (CLB, inner ring), nitrogen-limited B3 medium (NLB, center ring), and Highley's basal medium containing avicel (HBA, outer ring). Parenthetical values following each category refer to the number of genes in CLB, NLB, and HBA.
Nitrogen metabolism.
Nitrogen limitation, perhaps analogous to decayed wood (34), was accompanied by the accumulation of an impressive battery of transcripts involved in the mobilization and recycling of nitrogen (Table 1). Among these were various transmembrane proteins involved in the transport of small-molecular-weight nitrogenous compounds and an impressive family of related oligopeptide transporters (OPTs). Recent phylogenetic analysis (37) shows large families of OPT genes in P. chrysosporium, Coprinopsis cinereus, and Laccaria bicolor. Differential regulation of OPT genes has been observed in response to growth conditions and developmental state for the ectomycorrhiza L. bicolor and Hebeloma cylindrosporum (3, 37). In particular, H. cylindrosporum PTR2A is upregulated under nitrogen limitation, and transcripts of the homologous P. chrysosporium gene (protein model Pchr10757) are highly expressed, showing a modest increase in NLB (NLB/RB ratio of 1.89; see Table S3 in the supplemental material). Although coordinately expressed, no physical clustering of OPT genes was observed in the P. chrysosporium v2.0 assembly.
TABLE 1.
Upregulated P. chrysosporium genes involved in nitrogen metabolisma
| Protein ID no. | Putative function, gene | Microarray signal (log2) or ratio
|
LC-MS/MS (medium, score) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CLB | NLB | RB | CLB/RB | NLB/RB | HBA | HBG | HBA/HBG | |||
| 6738b | Acetamidase | 11.9 | 12.7 | 11.6 | 1.18 | 2.15 | 10.9 | 11.0 | 0.93 | |
| 9665 | Acetamidase | 12.2 | 13.3 | 12.1 | 1.04 | 2.31 | 11.6 | 11.7 | 0.95 | |
| 122350 | Amine oxidase | 13.1 | 14.1 | 12.8 | 1.24 | 2.35 | 12.6 | 12.6 | 1.03 | |
| 4526 | Amine oxidase | 11.2 | 12.8 | 11.6 | 0.79 | 2.35 | 10.6 | 10.7 | 0.93 | |
| 135275 | Amino acid permease | 10.7 | 12.3 | 10.6 | 1.09 | 3.47 | 10.5 | 10.5 | 0.98 | |
| 121517 | Ammonia transporter | 13.4 | 14.8 | 13.3 | 1.03 | 2.82 | 13.1 | 13.1 | 1.01 | |
| 124436 | Arginase | 12.6 | 13.5 | 12.2 | 1.31 | 2.48 | 11.6 | 11.4 | 1.14 | |
| 3906 | Aspartokinase | 11.7 | 12.6 | 11.6 | 1.05 | 2.01 | 11.1 | 11.5 | 0.76 | |
| 129869 | Methionine synthase | 12.3 | 11.8 | 11.3 | 2.09 | 1.44 | 10.9 | 10.4 | 1.45 | |
| 135021 | Nitrilase | 12.8 | 13.6 | 12.5 | 1.20 | 2.10 | 12.1 | 12.2 | 0.93 | |
| 37290 | Nucleoside transporter | 11.2 | 13.2 | 11.1 | 1.04 | 4.13 | 11.9 | 11.9 | 0.97 | |
| 10555 | O-Acetylhomoserine sulfhydrylase | 12.8 | 14.0 | 12.6 | 1.16 | 2.77 | 11.4 | 11.3 | 1.06 | |
| 132680 | Oligopeptide transporter | 12.9 | 14.5 | 12.8 | 1.07 | 3.26 | 13.4 | 14.0 | 0.64 | |
| 137519 | Oligopeptide transporter | 12.7 | 14.2 | 12.6 | 1.13 | 3.19 | 12.4 | 12.4 | 0.98 | |
| 138073 | Oligopeptide transporter | 12.2 | 13.7 | 11.5 | 1.54 | 4.51 | 10.6 | 10.7 | 0.97 | |
| 131282 | Oligopeptide transporter | 12.3 | 13.5 | 12.1 | 1.14 | 2.66 | 11.7 | 11.7 | 0.98 | |
| 4874 | Oligopeptide transporter | 12.9 | 14.8 | 12.9 | 0.94 | 3.65 | 14.6 | 14.0 | 1.44 | |
| 122372c | Oligopeptide transporter | 12.8 | 14.8 | 12.8 | 0.98 | 3.78 | 12.7 | 12.7 | 0.97 | |
| 10276 | Oligopeptide transporter | 11.6 | 13.5 | 11.7 | 0.92 | 3.50 | 11.6 | 11.4 | 1.16 | |
| 25929 | Oligopeptide transporter | 13.1 | 15.5 | 12.6 | 1.44 | 7.50 | 12.2 | 12.1 | 1.07 | |
| 8470 | Peptidase family A1A, asp5 | 11.8 | 12.7 | 11.6 | 1.15 | 2.14 | 11.2 | 11.0 | 1.16 | NLB, 704; CLB, 415; WBC, 58 |
| 8468 | Peptidase family A1A, asp3 | 10.2 | 13.0 | 9.8 | 1.32 | 10.30 | 9.4 | 9.4 | 1.05 | NLB, 891; CLB, 95 |
| 133689 | Peptidase family M38 | 12.9 | 13.9 | 12.9 | 1.01 | 2.04 | 12.4 | 12.6 | 0.86 | |
| 2647 | Peptidase family S10 | 12.4 | 13.4 | 12.1 | 1.21 | 2.39 | 12.2 | 11.8 | 1.26 | |
| 130748 | Peptidase family S53, prt53B | 12.5 | 14.7 | 12.8 | 0.80 | 3.86 | 11.0 | 10.9 | 1.06 | RB, 821; NLB, 837; CLB, 375 |
| 131262d | Peptidase family G1 | 10.6 | 10.5 | 10.7 | 0.92 | 0.88 | 11.7 | 10.1 | 3.19 | RB, 1445; HBA, 682 |
| 43144d | Peptidase family G1 | 11.9 | 12.0 | 12.1 | 0.91 | 0.93 | 13.0 | 11.3 | 3.20 | HBA, 955 |
| 130601 | Peptidase family T1A | 10.8 | 11.2 | 12.1 | 0.41 | 0.55 | 12.5 | 11.5 | 2.01 | |
| 124439 | Phenylalanine ammonia lyase | 13.5 | 13.5 | 11.8 | 3.37 | 3.30 | 12.3 | 12.8 | 0.71 | |
| 134355 | Urea transporter | 11.2 | 12.9 | 11.2 | 1.03 | 3.34 | 10.6 | 10.6 | 0.99 | |
| 134532 | Urea transporter | 12.3 | 13.6 | 12.5 | 0.85 | 2.19 | 11.9 | 12.0 | 0.94 | |
Protein model numbers (v2.1), putative function, expression levels, expression ratios, and LC-MS/MS Mascot scores for genes with transcripts accumulating >2-fold in CLB, NLB, or HBA. Peptidase family assignment based on MEROPS (http://merops.sanger.ac.uk/). Protein models indicated in boldface denote incomplete and/or inaccurate gene models. The media are defined in Materials and Methods.
Acetamidase model 6738 may be a partial duplication of model 9665 or an assembly error.
Transporter model 122372 is truncated at the 3′ terminus by an assembly gap. The corresponding gene may be a partial duplication of the gene encoding protein 137519 or an assembly error.
Clustering and expression of family G1 protease shown in Fig. 3.
Transcripts of five proteases were upregulated in nitrogen starved B3 cultures, and three of these (asp5, asp3, and prt53B) correspond to previously identified proteins (41, 53). No peptides were detected for the M38 (Pchr133689) and S10 (Pchr2647) families, although an earlier study had identified peptides from other genes representing the S10 peptidase family (53). Within the S53 family, only prt53B transcripts accumulated >2-fold (Table 1), but peptides corresponding to prt53D (Pchr129261), prt53E (Pchr26825), prt53A (Pchr133020), and a previously unidentified gene (Pchr125335) were also detected (see Table S3 in the supplemental material). As recently reported (36), the related basidiomycete C. cinereus contains an expanded family of extracellular fungalysins (MEROPS family M36), which are represented by a single sequence in P. chrysosporium, Pchr37396. The transcript accumulation of the P. chrysosporium gene was modest (NLB/RB ratio of 1.51), and no peptides were detected in any media (see the supplemental table in GEO GSE14736).
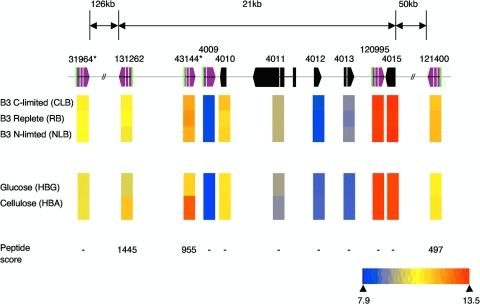
Unexpectedly, we observed transcript accumulation and peptide sequences of two glutamic acid proteases (Pchr131262 and Pchr43144), representatives of which had not been previously observed (53, 54) (Table 1). Peptides corresponding to a third family G1 representative, Pchr121400, were also detected (see Table S3 in the supplemental material), although significant transcript increases were not observed. In their analysis of the v1.0 assembly, Sims et al. (44) noted a large number of glutamic acid proteases, in contrast to other basidiomycetes. These authors also reported clustering of four G1-encoding genes, and the latest assembly extends the linkage to six genes (Fig. 3). Although the three genes encoding detectable protein lie within a 21-kb region, transcriptional regulation among the six linked genes follows no easily discernible pattern. For example, genes corresponding to Pchr120995 and Pchr4009 were expressed under all conditions examined, whereas those encoding Pchr131262 and Pchr43144 were significantly upregulated in cellulose-containing medium.
FIG. 3.
Organization and expression of scaffold 5 cluster of family G1 proteases. Purple filled arrows denote G1 family peptidases. Protein models Pchr31964 and Pchr43144 (indicated by asterisks) are truncated at the 5′ termini, but alternative ab initio models Pchr3963 and Pchr4008, respectively, reveal complete sequence with predicted secretion signals (green fill). Vertical colored bars indicate microarray-derived expression results from nutrient-limited cultures versus nutrient-replete medium and from cellulose-containing medium versus glucose-containing medium. The lower right scale indicates the log2 signal strength. Peptides corresponding to models Pchr131262, Pchr43144, and Pchr121400 were identified by LC-MS/MS, and the corresponding Mascot scores are listed.
Carbohydrate active enzymes.
Genes encoding CAZYs (5) also revealed dramatic differential regulation in response to culture conditions (Table 2). Table 2 lists 70 genes exhibiting significant transcript accumulation and/or peptide scores. Expression of many of these genes was previously recognized in proteome studies, and detailed biochemical analyses have been performed on several endoglucanases (18, 51), cellobiohydrolases (52), endoxylanases (9), endo-1,3(4)-β-glucanase (26), α-galactosidases (17), β-1,3-glucanase (25), and xyloglucanase (22). In other instances, we have inferred putative function by sequence similarity to accessions within the Swiss-Prot database (see Table S3 in the supplemental material).
TABLE 2.
P. chrysosporium CAZY genes exhibiting >2-fold upregulation and/or encoding peptides detected by LC-MS/MSa
| Protein ID no. | CAZY family, putative function, gene | Reference(s) | Comment(s) | Array ratio
|
LC-MS/MS score
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CLB/RB | NLB/ RB | HBA/ HBG | RB | NLB | CLB | NMA | HBA | WBC | ||||
| 127920 | GH1, β-glucosidase | 1.16 | 1.08 | 3.57 | ||||||||
| 135385 | GH2, β-mannosidase | 0.88 | 0.70 | 1.04 | 1573 | |||||||
| 129849 | GH3, β-glycosidase, gly3B | 1, 39, 41, 54 | 1.13 | 1.07 | 1.52 | 120 | ||||||
| 122884 | GH5, glucan 1,3-β-glucosidase | 0.80 | 0.84 | 0.85 | 110 | |||||||
| 4361 | GH5, endoglucanase, cel5B | 41, 51, 54 | CBM1; EG44 | 1.22 | 1.04 | 5.21 | 283 | 1638 | ||||
| 5115 | GH5, β-mannanase, man5C | 39, 41, 54 | CBM1 | 1.19 | 1.12 | 1.35 | 1011 | |||||
| 6458 | GH5, endoglucanase, cel5A | 51, 54 | CBM1; EG38/36 | 1.06 | 0.99 | 5.82 | 128 | 724 | 1534 | |||
| 135724 | GH5, glucan 1,3-β-glucosidase | 1.13 | 0.90 | 3.34 | 563 | |||||||
| 140501 | GH5, β-mannanase, man5D | 54 | CBM1 | 1.19 | 1.08 | 5.01 | 96 | 1322 | ||||
| 121774 | GH5, β-glycosidase | 1.24 | 1.06 | 2.14 | ||||||||
| 133052 | GH6, cellobiohydrolase II, cel6A | 41, 52 | CBM1; CBH50 | 1.60 | 0.61 | 7.44 | 5444 | 214 | 4480 | |||
| 129072 | GH7, cellobiohydrolase I, celF/celG | 53, 54 | CBM1 | 1.35 | 1.24 | 2.83 | 55 | |||||
| 137372 | GH7, cellobiohydrolase I, cel7D | 39, 41, 52 | CBM1; CBH58 | 1.43 | 1.15 | 4.39 | 346 | 2954 | 37 | 6435 | ||
| 137216 | GH7, cellobiohydrolase I, cel7E | CBM1 | 1.21 | 1.06 | 9.22 | 114 | ||||||
| 127029 | GH7, cellobiohydrolase I, cel7C | 39, 41, 52 | CBM1; CBH62 | 1.26 | 1.08 | 5.71 | 670 | 683 | 90 | 2497 | ||
| 139732 | GH10, xylanase, xyn10E | 53 | CBM1 | 1.19 | 1.20 | 1.13 | 66 | 62 | ||||
| 138345 | GH10, xylanase A, xyn10A | 9, 41, 54 | CBM1 | 1.20 | 1.16 | 2.04 | 47 | 509 | 96 | |||
| 138715 | GH10, xylanase, xyn10B | 9, 41, 54 | CBM1 | 1.26 | 1.16 | 3.95 | 45 | |||||
| 125669 | GH10, xylanase, xyn10D | 1, 54 | 1.66 | 1.03 | 1.03 | 163 | 2034 | 142 | ||||
| 7045** | GH10, xylanase, xyn10C | 54 | CBM1 | 3.53 | 1.04 | 1.08 | 80 | 270 | 1830 | |||
| 133788 | GH11, xylanase B, xyn11A | 9, 41, 54 | CBM1 | 1.62 | 1.28 | 13.35 | 170 | 179 | 1459 | |||
| 7048** | GH12, endo-/xylo-glucanase, cel12B | 54 | 1.27 | 1.10 | 2.75 | 48 | 935 | 263 | 1068 | |||
| 8466 | GH12, endoglucanase, cel12A | 18, 54 | EG28 | 1.17 | 1.09 | 2.44 | 55 | 340 | ||||
| 38357 | GH13, α-amylase | 0.20 | 0.36 | 0.69 | 603 | |||||||
| 7087 | GH13, α-amylase | CBM20 | 1.31 | 1.20 | 1.28 | 229 | ||||||
| 138813 | GH15, glucoamylase, gla15A | 54 | CBM20 | 1.09 | 0.89 | 1.60 | 2265 | 2469 | 625 | |||
| 2630 | GH16, β-glycosidase | 0.54 | 0.86 | 0.81 | 81 | |||||||
| 123909 | GH16, β-glycosidase, gly16A | 41, 54 | 2.04 | 0.98 | 1.45 | 1690 | 168 | 793 | 1092 | |||
| 138982 | GH16, β-glycosidase | 0.76 | 0.82 | 0.91 | 187 | |||||||
| 7122 | GH16, β-glycosidase | 0.75 | 0.86 | 0.95 | 377 | |||||||
| 10833 | GH16, β-1,3-glucanase, lam16A | 26 | 2.42 | 0.92 | 1.78 | 837 | 39 | |||||
| 4127 | GH17, β-glycosidase | 1.95 | 0.80 | 0.98 | 608 | 40 | ||||||
| 40899 | GH18, chitinase | 0.63 | 0.38 | 2.06 | 338 | 219 | ||||||
| 2991 | GH18, chitinase, chi18B | 54 | 1.28 | 1.65 | 2.45 | 156 | 150 | |||||
| 39872 | GH18, chitinase | 0.53 | 1.14 | 1.01 | 1542 | |||||||
| 6412 | GH18, chitinase, chi18A | 54 | 1.44 | 1.32 | 2.07 | 184 | ||||||
| 134311 | GH18, chitinase | 1.51 | 1.46 | 1.44 | 126 | 1085 | ||||||
| 136630 | GH25, lysozyme-like | 1.20 | 1.86 | 2.24 | 89 | 129 | 92 | |||||
| 4422 | GH27, α-galactosidase | 1.07 | 0.88 | 0.99 | 101 | |||||||
| 125033 | GH27, α-galactosidase, aga27A | 17 | 1.04 | 0.88 | 1.01 | 44 | 73 | |||||
| 4449 | GH28, polygalacturonase, epg28B | 53 | 1.00 | 0.84 | 1.13 | 138 | 69 | 72 | ||||
| 29397 | GH28, rhamnogalacturonidase, rhg28C | 41, 54 | CBM1 [7840] | 1.30 | 1.07 | 1.97 | 41 | 311 | ||||
| 3805 | GH28, polygalacturonase, epg28A | 41, 54 | 1.39 | 0.90 | 2.66 | 588 | 196 | 125 | 1822 | 514 | ||
| 9011 | GH30, β-1,6-glucanase | 41 | 2.49 | 1.22 | 0.50 | 592 | ||||||
| 968 | GH31, α-glucosidase | 1.13 | 0.45 | 1.87 | 42 | |||||||
| 125462 | GH31, α-glucosidase | 41 | 0.80 | 0.67 | 1.24 | 1488 | 76 | |||||
| 9466 | GH35, β-galactosidase, lac35A | 41, 53 | 1.18 | 1.11 | 1.05 | 1822 | 437 | 61 | ||||
| 140627 | GH37, trehalase | 39, 41 | GPI anchor? | 1.16 | 0.51 | 0.61 | 208 | |||||
| 297 | GH43, β-glycosidase, gly43A | 54 | 1.39 | 1.34 | 1.06 | 38 | 113 | |||||
| 133070 | GH43, glycosidase | 1.00 | 0.96 | 1.32 | 399 | 68 | ||||||
| 4822 | GH43, endo-1,5-α-l-arabinanase | 1.90 | 0.95 | 1.95 | 91 | |||||||
| 4550 | GH47, α-mannosidase, msd47A | 53 | 1.64 | 0.73 | 1.56 | 3547 | 43 | |||||
| 3651 | GH51, α-l-arabinofuranosidase, arb51A | 54 | 1.17 | 1.12 | 1.10 | 125 | ||||||
| 8072 | GH55, β-1,3-glucanase, exg55A, lam55A | 25, 41, 53, 54 | 13.16 | 1.55 | 1.72 | 5112 | 118 | 3172 | 858 | 141 | 581 | |
| 41123 | GH61, endoglucanase, cel61C | 53 | 1.80 | 0.97 | 1.07 | 323 | 254 | 57 | 1301 | |||
| 129325 | GH61, endoglucanase | CBM1 | 1.30 | 1.23 | 3.50 | |||||||
| 121193 | GH61, endoglucanase, cel61B | 54 | CBM1 | 1.19 | 1.14 | 20.09 | 97 | 39 | 781 | |||
| 31049 | GH61, endoglucanase | CBM1 | 1.20 | 1.10 | 5.78 | 65 | 287 | |||||
| 41563* | GH61, endoglucanase | CBM1 | 1.22 | 1.05 | 1.03 | 46 | ||||||
| 41650* | GH61, endoglucanase | CBM1 | 1.13 | 1.04 | 2.60 | |||||||
| 6893 | GH71, α-1,3-glucanase | 0.79 | 1.38 | 2.11 | ||||||||
| 138266 | GH74, xyloglucanase, gly74A | 54 | 1.18 | 1.02 | 3.27 | 1112 | ||||||
| 134556 | GH74, xyloglucanase, gly74B, xgh74B | 22, 39, 41, 54 | CBM1 | 1.21 | 1.15 | 2.31 | 802 | |||||
| 840 | GH88, glucuronyl hydrolase, gly88A | 39, 41, 54 | 1.66 | 1.01 | 1.51 | 480 | 384 | 225 | ||||
| 3431 | GH92, α-1,2-mannosidase | 39, 41 | 0.91 | 0.81 | 1.08 | 3250 | ||||||
| 133585 | GH92, α-1,2-mannosidase | 1.06 | 1.05 | 1.60 | 122 | |||||||
| 130517 | CE?, glucuronyl esterase | CBM1 | 1.77 | 0.97 | 1.89 | 244 | 51 | 873 | ||||
| 126075 | CE1, feruloyl esterase, axe1 | 41, 54 | CBM1 [7224] | 1.92 | 1.09 | 5.11 | 1052 | 56 | 291 | 666 | ||
| 964 | PL14, alginate lyase-like | 53 | 2.80 | 0.98 | 1.27 | 382 | 550 | |||||
| 6736 | PL8, chondroitin lyase-like | 2.49 | 1.23 | 1.03 | ||||||||
Protein model numbers (v2.1), putative function, expression ratios, and LC-MS/MS Mascot scores. Family assignment based on CAZY (http://www.cazy.org/ [5]). Protein models indicated in boldface denote incomplete and/or inaccurate gene models. Under Comments, bracketed numbers refer to the preferred protein model. The media are described in Materials and Methods. Additional abbreviations: NMA, modified Norkran's medium with Avicel (13, 54); GH, glycoside hydrolase; PL, polysaccharide lyase; CE, carbohydrate esterase; CBM, carbohydrate-binding module. EG44, EG38/36, CBH50, CBH58, CBH62, and EG28 are protein designations derived from the preceding references. Protein identification numbers designated with “*” and “**” correspond to closely linked genes; 41563/41650 lie 3.4 kb apart, and 7045/7048 lie 11.9 kb apart. Blank LC-MS/MS entries indicate that no significant peptide matches (Mascot scores > 40) were observed.
Certain glycoside hydrolase families are functionally heterogeneous. The P. chrysosporium v2.1 genome contains at least 13 sequences belonging to family 61, and six of these genes are clearly expressed under the conditions examined here (Table 2). The gene encoding Pchr121193 was dramatically upregulated and highly expressed in media containing microcrystalline cellulose as the sole carbon source (HBA). This observation, together with the clear presence of a highly conserved cellulose-binding domain, certainly support a role in cellulose degradation, possibly as an endoglucanase. Still, a recent investigation (24) points out the need for additional examination of the structure and activities of enzymes within family 61.
Upregulated CAZY-encoding genes also included two polysaccharide lyases (PL families 8 and 14) and a carbohydrate esterase family 1 representative. The most accurate model for the latter, Pchr7224, features separate cellulose binding and catalytic domains. Based on similarity to Penicillium feruloyl esterases (32), the protein was tentatively identified as an acetyl xylanase esterase, possibly involved in hydrolyzing ester bonds in arabinoxylans. Peptides matching another carbohydrate esterase, Pchr130517, were detected in several media, and modest transcript accumulation was observed in CLB (ratio 1.77) and in HBA (ratio 1.89). The corresponding enzyme, a glucuronyl esterase, appears to belong to a new and distinct carbohydrate esterase family (11). The lyase sequences showed relatively little similarity to proteins of known function, although both are closely related (BLASTP bit scores of >350) to conceptual translations from L. bicolor and C. cinereus.
β-1,3-Glucan and mixed-linkage β-1,3-1,4-glucans are common constituents of cell walls of certain cereal grains, grasses, and related plants, and chitin is a major component of the cell walls of yeasts and other fungi. Chitinases (GH18), α-trehalases (GH37), and other glycosidases may be involved in cell wall morphogenesis.
Degradation of lignin and aromatic compounds.
The biochemistry and genetics of LiP and MnP have been intensively studied, and the observed influence of medium composition (Table 3) is consistent with previously reported Northern blot and quantitative RT-PCR (qRT-PCR) results (reviewed in references 16 and 28). Earlier studies (47) had failed to identify any clear relationship between clustering and the transcriptional regulation of 8 LiP genes, and in the present study we extended the analysis to 16 additional genes interspersed within the lip cluster (see Fig. S2 in the supplemental material). In short, no obvious relationship(s) were gleaned from this analysis.
TABLE 3.
Upregulated P. chrysosporium genes potentially involved in degradation of lignin and aromatic compoundsa
| Protein ID no. | Putative function, gene | Reference(s) | Comment(s) | Microarray signal (log2) or ratio
|
LC-MS/MS score
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CLB | NLB | RB | CLB/RB | NLB/RB | HBA | HBG | HBA/HBG | RB | NLB | CLB | NMA | HBA | WBC | ||||
| 11055 | Aryl alcohol dehydrogenase aad | 40 | Q01752 | 12.3 | 13.1 | 11.6 | 1.64 | 2.79 | 11.0 | 11.1 | 0.95 | ||||||
| 11098 | CDH cdh1 | 35, 54 | Q01738 | 13.1 | 13.0 | 12.9 | 1.22 | 1.12 | 15.4 | 12.7 | 6.45 | 92 | 3490 | 2507 | |||
| 147 | Cellulose-binding iron reductase cir1 | 60 | AAU12274; CBM1 | 13.0 | 13.0 | 12.8 | 1.1 | 1.17 | 13.7 | 12.6 | 2.19 | ||||||
| 11068 | GLX glx1 | 27, 53 | AAA33747 | 13.9 | 13.0 | 12.3 | 2.93 | 1.56 | 11.5 | 11.6 | 0.95 | 742 | 400 | 534 | |||
| 2255 | Linoleate diol synthase | 11.5 | 11.5 | 11.2 | 1.2 | 1.22 | 12.9 | 11.1 | 3.43 | ||||||||
| 137435 | P450 | Bifunctional P-450 | 12.4 | 12.3 | 12.5 | 0.9 | 0.87 | 13.6 | 12.4 | 2.86 | |||||||
| 138737 | P450 | CYP67 | 11.8 | 12.8 | 11.6 | 1.12 | 2.37 | 12.3 | 12.2 | 1.10 | |||||||
| 34295 | Peroxidase | Peroxidase 2 superfamily | 11.5 | 11.3 | 11.3 | 1.2 | 1.02 | 12.0 | 10.6 | 2.68 | |||||||
| 3589 | Peroxidase mnp2 | 39, 53 | Mn peroxidase | 12.9 | 14.8 | 12.9 | 0.96 | 3.71 | 12.1 | 12.1 | 1.04 | 244 | 506 | 278 | |||
| 121822 | Peroxidase lipB | Lignin peroxidase | 13.1 | 12.5 | 12.0 | 2.16 | 1.40 | 11.6 | 11.4 | 1.12 | |||||||
| 131738 | Peroxidase lipC | 53 | Lignin peroxidase | 12.1 | 14.1 | 11.9 | 1.16 | 5.04 | 11.5 | 11.5 | 1.03 | 123 | |||||
| 6811 | Peroxidase lipD | 39, 53 | Lignin peroxidase | 14.6 | 13.3 | 12.7 | 3.69 | 1.46 | 12.5 | 12.4 | 1.04 | 84 | 378 | 1145 | 85 | ||
| 11110 | Peroxidase lipE | 53 | Lignin peroxidase | 14.8 | 14.5 | 13.7 | 2.22 | 1.76 | 13.7 | 13.6 | 1.05 | 531 | |||||
| 131709 | Peroxidase lipJ | Lignin peroxidase | 11.3 | 13.5 | 11.0 | 1.26 | 5.90 | 10.7 | 10.6 | 1.03 | 41 | ||||||
| 140708 | Peroxidase mnp1 | Mn peroxidase MnP1 | 12.3 | 12.8 | 10.6 | 3.36 | 4.51 | 9.9 | 9.8 | 1.08 | 35 | 58 | |||||
| 137275 | Pyranose 2-oxidase, pox | 10 | AY522922 | 12.5 | 10.6 | 10.9 | 3.20 | 0.82 | 12.3 | 12.1 | 1.11 | ||||||
Protein model numbers (v2.1), putative function, expression levels (log2 signal strength) and ratios, and LC-MS/MS Mascot scores are presented. The protein model indicated in boldface is inaccurate, and the alternative model Pchr2001 is preferred. The media are defined in Materials and Methods. The listing includes only genes with transcripts accumulating ≥2-fold in CLB versus RB, NLB versus RB, or HBA versus HGA. Additional relevant genes not upregulated ≥2-fold, including some with high signal strength and/or LC-MS/MS evidence, are listed in Table S3 in the supplemental material.
GLX, thought to be physiologically coupled to extracellular peroxidases via peroxide generation, was also upregulated under nutrient starvation (Table 3) as previously established by Northern blots (48). Peptides corresponding to a structurally related copper radical oxidase gene, cro2 (Pchr134241), were identified, and while transcript levels were generally high, the gene was slightly downregulated in NLB, CLB, and HBA (see Table S3 in the supplemental material). The GMC oxidoreductases pyranose 2 oxidase (pox, Pchr137275) and aryl alcohol dehydrogenase (aad, Pchr11055) are also potentially involved in peroxide generation, and transcripts accumulated in CLB as previously reported (10, 40). No significant transcript accumulation was noted for several related GMC oxidoreductases, although peptides were detected for three genes distantly related to glucose oxidase (see Table S3 in the supplemental material).
Other upregulated genes possibly related to lignin degradation include a family 67 P450 (Pchr138737) and a family 2 peroxidase (Pchr34295) (Table 3). These enzymes may be involved in the intracellular metabolism of low-molecular-weight lignans and other breakdown products. We also observed upregulation of genes encoding O-methyltransferase (see Table S3 in the supplemental material) and phenylalanine ammonia lyase (Table 1) in both CLB and NLB. These enzymes might be involved in multiple pathways, including the biosynthesis of veratryl alcohol (23). Secreted by P. chrysosporium, veratryl alcohol has been implicated in lignin depolymerization, although the precise mechanism remains uncertain (reviewed in reference 16).
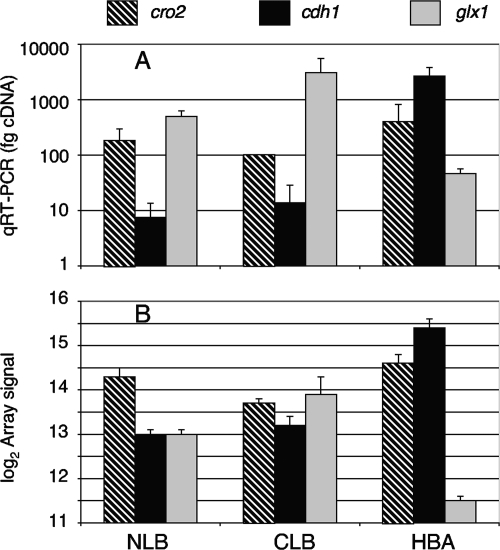
Also implicated in lignin degradation, CDH transcript accumulation was most pronounced in cellulose-containing medium (Table 3). This enzyme contains separate iron reductase and dehydrogenase domains and can generate highly reactive hydroxyl radical capable of depolymerizing cellulose and lignin (for a review, see reference 19). Although high transcript levels were observed in HBA, no protein was detected by LC-MS/MS, and no activity could be measured in that medium. Transcript accumulation was confirmed by qRT-PCR (Fig. 4). In contrast, the protein and activity were easily detected in NMA and WBC cultures (Table 3), and we therefore presume some form of posttranscriptional regulation is operative. In this regard, it is noteworthy that the production of active CDH in cultures is particularly sensitive to growth buffers such as succinate, acetate, and phosphate, a factor that could also be physiologically relevant (2). The CBM1-containing gene cir1, closely related to the reductase domain of cdh1 (60), was also upregulated in HBA, but no peptides were detected.
FIG. 4.
Transcript levels of copper radical oxidase (cro2), CDH (cdh1), and copper radical oxidase (glx) as determined by qRT-PCR (A) and microarray (B).
Hypothetical proteins.
An impressive 193 genes encoding proteins of unknown function were regulated ≥2-fold and/or detected by MS (for full details, see Table S3 in the supplemental material). Of these, 111 were closely related (bit scores > 150) to at least one NCBI conceptual translation, often derived from the basidiomycetes C. cinereus, L. bicolor, or Postia placenta. Transcript accumulation >2-fold was observed for 12, 23, and 24 “hypothetical” genes in CLB versus RB, NLB versus RB, and HBA versus HBG, respectively. Peptide sequences corresponding to 55 genes were detected, and 40 of these were not previously reported. Transmembrane helices (TMHMM v.2.0 [www.cbs.dtu.dk/services/TMHMM/]) were present in ≥18 predicted proteins, and secretion signals (SignalP v.3.0; www.cbs.dtu.dk/services/SignalP/) were observed in 61 models. Also potentially relevant to their function, we identified putative glycosyl phosphatidylinositol (GPI) anchors (http://mendel.imp.ac.at/sat/gpi/fungi_server.html), mitochondrial signal sequences (TargetP v.1.1;/www.cbs.dtu.dk/services/TargetP/), and carbohydrate binding modules (www.cazy.org/) in nine, three, and three sequences, respectively. BlastP and CLUSTAL W (49) analysis of all 193 hypothetical proteins distributed 37 proteins into 16 groups each containing between two and four members each. Fifty genes encoding hypothetical proteins were located within 10 kb of one another (25 pairs). In several instances, the closely linked genes were structurally related and/or similarly regulated (Table 4).
TABLE 4.
Tandemly arranged paralogs encoding hypothetical proteins with secretion signalsa
| Protein ID no. | Location | Group no. | Comment | Microarray signal (log2) or ratio
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CLB | RB | NLB | HBG | HBA | CLB/RB | NLB/RB | HBA/HBG | ||||
| 1449 | 2:1254101-1255446 | 1 | His rich | 12.19 | 11.63 | 13.13 | 12.19 | 12.64 | 1.47 | 2.94 | 1.36 |
| 1450 | 2:1256515-1257396 | 1 | His rich | 11.67 | 11.40 | 12.88 | 13.31 | 13.53 | 1.21 | 2.89 | 1.17 |
| 2035* | 3:80684-81631 | 13 | 10.95 | 10.47 | 11.47 | 12.54 | 10.46 | 1.40 | 2.10 | 0.23 | |
| 2036* | 3:82673-83620 | 13 | 12.19 | 11.89 | 12.04 | 12.90 | 11.66 | 1.23 | 1.11 | 0.42 | |
| 4931* | 8:357446-358764 | 4 | 3′ TMH | 13.52 | 14.48 | 14.26 | 13.21 | 13.07 | 0.51 | 0.86 | 0.91 |
| 4932* | 8:359380-360704 | 4 | 3′ TMH | 13.98 | 14.29 | 13.93 | 14.74 | 14.54 | 0.80 | 0.78 | 0.87 |
| 8377 | 16:485910-487103 | 2 | 11.79 | 11.65 | 12.00 | 11.20 | 12.43 | 1.10 | 1.28 | 2.34 | |
| 8378 | 16:487758-488977 | 2 | 12.56 | 12.34 | 13.55 | 13.01 | 13.74 | 1.16 | 2.44 | 1.67 | |
Protein model numbers (v2.1), location, expression levels and ratios. The media CLB, RB, NLB, HBG, and HBA, are defined in Materials and Methods. TMH, transmembrane helices. Protein identification numbers designated with “*” were identified by LC-MS/MS (see Table S3 in the supplemental material). As described in the text, CLUSTAL W analysis of hypothetical proteins distinguished 16 “groups” of related sequences.
DISCUSSION
A complete characterization of the P. chrysosporium secretome will illuminate the repertoire of enzymes involved in lignocellulose depolymerization. Computational analysis of the v2.1 annotations had previously predicted 769 secreted proteins (53). Although useful, such predictions are likely incomplete since many models feature inaccurate 5′ termini, and some proteins with signal peptides are secreted into the cell wall or into vacuoles or are retained in the endoplasmic reticulum. In addition, certain proteins may be secreted in the absence of a recognizable signal. Clearly, direct experimental support for secretion is preferred.
Substantial progress has been made using MS-based protein identification (1, 39, 41, 43, 54, 55), but these approaches have limitations. Low-abundance, low-molecular-weight, and unstable proteins are less likely to be detected. Highly glycosylated proteins and/or those adhering to cell walls or substrate may also escape detection. Incomplete annotation may be problematic under some circumstances, as previously observed for a GH45-encoding gene (54).
Microarrays offer a complementing perspective on gene expression, although here too, limitations must be considered. Although there is generally a good correlation between expression levels and LC-MS/MS identification of secreted proteins, exceptions such as CDH were noted. Other examples of highly transcribed genes with putative secretion signals, but without LC-MS/MS support, include a putative aldose-1-epimerase. In contrast, high scoring peptides were sometimes associated with relatively low transcript levels. For example, peptides corresponding to class 3 lipase Pchr8996 were easily detected (Mascot score 652) in CLB, but the microarray signal for the gene (log2 11.7) was somewhat lower than the genomewide average signal (log2 12.39 ± 1.18). Similarly, multiple, high-scoring peptides were assigned to hypothetical models Pchr5499 (Mascot score 1128) and Pchr5500 (Mascot score 673) in HBA, but the array values of log2 10.8 and 10.5, respectively, were well below the average signal in this medium (12.14 ± 1.30). Interestingly, these tandemly arranged hypothetical proteins do not feature canonical secretion signals, but SecretomeP (www.cbs.dtu.dk/services/SecretomeP/) analysis predicts potential “nonclassical” signals. Possibly, such proteins are stable and accumulate during growth, although transcript turnover/instability may also be involved.
Irrespective, transcript and LC-MS/MS patterns alone cannot firmly establish the role of specific genes in lignocellulose depolymerization. In most instances, enzyme characterization and/or genetic suppression are necessary. However, such approaches are technically challenging with filamentous basidiomycetes, particularly on a high-throughput basis. Complementary approaches such as microarrays and LC-MS/MS, albeit imperfect, provide a filter for focusing on the more promising genes.
A distinguishing feature of the P. chrysosporium genome is the occurrence of large and complex families of related sequences including genes encoding cytochrome P450s, peroxidases, glycoside hydrolases, proteases, copper radical oxidases, and multicopper oxidases. In some instances, a close linkage has been observed, but there is scant evidence relating transcriptional regulation and such clustering. This was previously demonstrated for the lignin peroxidases (47) and again here for the G1 family proteases (Fig. 3). The role of genetic multiplicity remains unclear. Structurally related genes may encode proteins with subtle differences in function, and such diversity may provide the flexibility needed in changing environmental conditions (pH, temperature, and ionic strength), substrate composition and accessibility, and wood species. Alternatively, some or all of the genetic multiplicity may merely reflect functional redundancy. Functional characterization of individual genes has been limited.
Nutrient-limited B3 medium has been widely used in studies of ligninolysis, and our results provide a broad view of potentially important proteins. Supporting the close physiological connections between GLX and the extracellular peroxidases of P. chrysosporium, coordinate upregulation was observed (Table 3). GLX has a regulatory mechanism responsive to peroxidase, peroxidase substrates, and peroxidase products (e.g., phenolics resulting from ligninolysis) (33). The oxidase becomes inactive in the absence of a coupled peroxidase system but is reactivated by lignin peroxidase and nonphenolic peroxidase substrates. Conversely, phenolics prevent activation by lignin peroxidase. GLX is also activated by lignin in the coupled reaction with LiP (33). The basis for this modulation in GLX activity is probably related, at least in part, to the redox interconversion (E1/2 = 0.64 V versus NHE, pH 7) of active and inactive forms of the enzyme (58).
Two glucose oxidases have been reported in P. chrysosporium cultures: glucose 1-oxidase from P. chrysosporium ME-446 and glucose 2-oxidase or pyranose 2-oxidase from P. chrysosporium K3 (reviewed in reference 28). While predominantly intracellular in liquid cultures of P. chrysosporium, evidence supports an important role for pyranose 2-oxidase in wood decay, with no evidence for glucose 1-oxidase (8). Pyranose 2-oxidase is preferentially localized in the periplasmic space and associated membranous materials and, as expected, peptides were not detected in extracellular fluids. The P. chrysosporium pyranose 2-oxidase transcript patterns are similar to lignin peroxidases and GLX, supporting a role in lignocellulose degradation (10). Peptides corresponding to three glucose-oxidase-like sequences have now been detected (see Table S3 in the supplemental material), but none exhibited substantial upregulation under nutrient limitation, and only transcripts matching the Pchr6270-encoding gene showed an increase in the cellulose-containing medium, HBA. Previously designated gox1, this model is related (Smith-Waterman score = 480) to Aspergillus niger glucose-1-oxidase (38). The function of gox1 and related genes is yet to be determined.
Structurally related to glucose oxidases (56), several putative aryl alcohol oxidase genes have been identified in P. chrysosporium. Specifically, Pchr135972, Pchr37188, and Pchr6199 are 46.3, 43.8, and 39.0% identical, respectively, to the aryl alcohol oxidases from Pleurotus eryngii (gi3851524). However, none of the corresponding transcripts accumulated under the conditions examined. The precise role(s) of these enzymes remain uncertain, but they may support a redox cycle by supplying extracellular peroxide, perhaps coupled to intracellular aryl alcohol dehydrogenase (56). Transcripts matching the latter protein model (Pchr11055) were significantly upregulated under nutrient limitation (Table 3).
Of course genes highly expressed, but not necessarily upregulated, may also play a crucial role in lignin depolymerization. For example, genes encoding a methanol oxidase (Pchr126879) and catalase (Pchr124398) are highly expressed in all media (see Table S3 in the supplemental material). In the brown rot fungus, Gloeophyllum trabeum, methanol oxidase may generate peroxide at or near the hyphal surface (7). The peroxide is hypothesized to spontaneously react with Fe2+ to form hydroxyl radicals that depolymerize cellulose. Similar nonenzymatic Fenton reactions have been implicated in lignin and cellulose depolymerization in P. chrysosporium (reviewed in reference 28). Since demethoxylation of lignin is common to white and brown rot, oxidation of methanol may be a crucial reaction. Catalase activity has been identified in the periplasmic space of P. chrysosporium grown under ligninolytic conditions (6), and recent MS studies associated the model Pchr124398 protein with membrane fractions (42).
Other expressed genes include several potentially important in posttranslational processing. Transcripts of a mannose-6-phosphatase encoding gene, mpa, showed modest increase in CLB and, as previously observed (53), the peptides were identified by LC-MS/MS (see Table S3 in the supplemental material). Evidence suggests that this enzyme dephosphorylates certain LiP isozymes (reviewed in reference 28). Proteolytic processing of peroxidases (reviewed in reference 28), cellulases (14), and CDH (12) may also play an important role in wood decay, and our results identify several candidate peptidases. Peptides corresponding to two family G1 proteases were identified solely in HBA, and the corresponding transcripts were substantially upregulated (Fig. 3). Five protease genes showed substantial accumulation in NLB relative to RB. Of these, Pchr8468 was among the most highly upregulated genes in the genome (NLB/RB ratio 10.3) and Pchr2647 was predicted to contain a GPI anchor. Likely, these observations reflect acquisition and recycling of nitrogen, but interactions with LiP cannot be ruled out. No significant (>2-fold) upregulation of proteases was observed under carbon limitation (CLB).
Not unexpectedly, we observed peptides and upregulated transcripts matching well-characterized cellulases in HBA. These included CBH2 (GH6), CBH1s (GH7s), and several GH5 endoglucanases. The potential involvement of several highly expressed GH61s in cellulose degradation merits additional study. Other genes upregulated in HBA are likely involved in hemicellulose degradation, and these include various xylanases, mannanase, α- and β-galactosidases, polygalacturonase, arabinofuranosidase, xyloglucanases, and feruloyl esterase. None of the CAZY encoding genes were upregulated in nitrogen-starved medium (NLB), although transcripts of several β-1,3-glucanase genes increased substantially under carbon limitation. Of these, lam55 (25) was among the most highly expressed (log2 15.5) and upregulated (HBA/HBG ratio of 13.2) genes in CLB (Table 2). Since hyphal cell walls contain β-1,3-glucans and chitin, GH16 and GH55 enzymes may be involved in morphogenesis and nutrient recycling.
Many of the 193 expressed hypothetical proteins observed here are undoubtedly involved in lignocellulose degradation. One-third of the sequences featured easily identifiable secretion signals, and peptides corresponding to 54 gene models were detected in extracellular filtrates. Especially intriguing among secreted proteins, Pchr138739 features a clear N-terminal cellulose-binding domain (CBM1), and transcription of the gene was substantially upregulated (ratio of 5.58) in HBA. Similarly, the gene encoding serine-rich protein Pchr138739 showed high transcript levels (log2 14.3) and upregulation (ratio of 3.80) in HBA.
Most genes encoding hypothetical proteins lack canonical secretion signals, although other structural features and expression patterns provide clues about function. Of the three genes upregulated >2-fold in both CLB and NLB, models Pchr8832, Pchr600, and Pchr1903 feature sequences supporting mitochondrial transport (TargetP, www.cbs.dtu.dk/services/TargetP/), a GPI anchor, and transmembrane location, respectively. Protein model Pchr6670 contains a centrally located transmembrane region and shows similarity to various cell surface proteins if low-complexity serine- and proline-rich regions are included in the BLASTP queries. The corresponding gene is substantially upregulated in NLB and in HBA. In this instance, as in many others mentioned above, a role in lignocellulose degradation, if any, requires additional biochemical and/or genetic analysis. Consideration of gene structure, transcriptional regulation, and protein secretion provides a filter for focusing on the more promising targets.
Supplementary Material
Acknowledgments
This study was supported by National Research Initiative of the USDA Cooperative State Research, Education and Extension Service, grant 2007-35504-18257 to the Forest Products Laboratory; by U.S. Department of Energy grant DE-FG02-87ER13712 to the University of Wisconsin-Madison; and by National Institutes of Health grant GM060201 to the University of New Mexico.
Footnotes
Published ahead of print on 17 April 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Abbas, A., H. Koc, F. Liu, and M. Tien. 2004. Fungal degradation of wood: initial proteomic analysis of extracellular proteins of Phanerochaete chrysosporium grown on oak substrate. Curr. Genet. 47:49-56. [DOI] [PubMed] [Google Scholar]
- 2.Bao, W., E. Lymar, and V. Renganathan. 1994. Optimization of cellobiose dehydrogenase and β-glucosidase production by cellulose-degrading cultures of Phanerochaete chrysosporium. Appl. Biochem. Biotechnol. 42:642-646. [Google Scholar]
- 3.Benjdia, M., E. Rikirsch, T. Muller, M. Morel, C. Corratge, S. Zimmermann, M. Chalot, W. B. Frommer, and D. Wipf. 2006. Peptide uptake in the ectomycorrhizal fungus Hebeloma cylindrosporum: characterization of two di- and tripeptide transporters (HcPTR2A and B). New Phytol. 170:401-410. [DOI] [PubMed] [Google Scholar]
- 4.Brazma, A., P. Hingamp, J. Quackenbush, G. Sherlock, P. Spellman, C. Stoeckert, J. Aach, W. Ansorge, C. A. Ball, H. C. Causton, T. Gaasterland, P. Glenisson, F. C. Holstege, I. F. Kim, V. Markowitz, J. C. Matese, H. Parkinson, A. Robinson, U. Sarkans, S. Schulze-Kremer, J. Stewart, R. Taylor, J. Vilo, and M. Vingron. 2001. Minimum information about a microarray experiment (MIAME)—toward standards for microarray data. Nat. Genet. 29:365-371. [DOI] [PubMed] [Google Scholar]
- 5.Cantarel, B. L., P. M. Coutinho, C. Rancurel, T. Bernard, V. Lombard, and B. Henrissat. 2009. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37:D233-D238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daniel, G., P. Binarova, J. Volc, and E. Kubatova. 1995. Cytochemical and immunocytochemical studies on sites of H2O2 production and catalase activity in hyphae of Phanerochaete chyrsosporium grown on liquid culture and on wood, p. 583-587. In E. Srebotnik and K. Messner (ed.), Proceedings of the Sixth International Conference on Biotechnology in the Pulp and Paper Industry. Facultas Universitatsverlag, Vienna, Austria.
- 7.Daniel, G., J. Volc, L. Filonova, O. Plihal, E. Kubatova, and P. Halada. 2007. Characteristics of Gloeophyllum trabeum alcohol oxidase, an extracellular source of H2O2 in brown rot decay of wood. Appl. Environ. Microbiol. 73:6241-6253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daniel, G., J. Volc, and E. Kubatova. 1994. Pyranose oxidase, a major source of H2O2 during wood degradation by Phanerochaete chrysosporium, Trametes versicolor, and Oudemansiella mucida. Appl. Environ. Microbiol. 60:2524-2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Decelle, B., A. Tsang, and R. Storm. 2004. Cloning, functional expression and characterization of three Phanerochaete chrysosporium endo-1,4-β-xylanases. Curr. Genet. 46:166-175. [DOI] [PubMed] [Google Scholar]
- 10.de Koker, T. H., M. D. Mozuch, D. Cullen, J. Gaskell, and P. J. Kersten. 2004. Pyranose 2-oxidase from Phanerochaete chrysosporium: isolation from solid substrate, protein purification, and characterization of gene structure and regulation. Appl. Environ. Microbiol. 70:5794-5800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duranova, M., S. Spanikova, H. A. Wosten, P. Biely, and R. P. de Vries. 2009. Two glucuronoyl esterases of Phanerochaete chrysosporium. Arch. Microbiol. 191:133-140. [DOI] [PubMed] [Google Scholar]
- 12.Eggert, C., N. Habu, U. Temp, and K.-E. L. Eriksson. 1996. Cleavage of Phanerochaete chrysosporium cellobiose dehydrogenase (CDH) by three endogenous proteases, p. 551-554. In E. Srebotnik and K. Messner (ed.), Biotechnology in the pulp and paper industry. Fakultas Universitatsverlag, Vienna, Austria.
- 13.Eriksson, K.-E., and S. G. Hamp. 1978. Regulation of endo-1,4-β-glucanase production in Sporotrichum pulverulentum. Eur. J. Biochem. 90:183-190. [DOI] [PubMed] [Google Scholar]
- 14.Eriksson, K.-E., and B. Pettersson. 1982. Purification and partial characterization of two acidic proteases from the white rot fungus Sporotrichium pulverulentum. Eur. J. Biochem. 124:635-642. [DOI] [PubMed] [Google Scholar]
- 15.Eriksson, K.-E. L., R. A. Blanchette, and P. Ander. 1990. Microbial and enzymatic degradation of wood and wood components. Springer-Verlag, Berlin, Germany.
- 16.Hammel, K. E., and D. Cullen. 2008. Role of fungal peroxidases in biological ligninolysis. Curr. Opin. Plant Biol. 11:349-355. [DOI] [PubMed] [Google Scholar]
- 17.Hart, D. O., S. He, C. J. Chany, 2nd, S. G. Withers, P. F. Sims, M. L. Sinnott, and H. Brumer III. 2000. Identification of Asp-130 as the catalytic nucleophile in the main α-galactosidase from Phanerochaete chrysosporium, a family 27 glycosyl hydrolase. Biochemistry 39:9826-9836. [DOI] [PubMed] [Google Scholar]
- 18.Henriksson, G., A. Nutt, H. Henriksson, B. Pettersson, J. Stahlberg, G. Johansson, and G. Pettersson. 1999. Endoglucanase 28 (Cel12A), a new Phanerochaete chrysosporium cellulase. Eur. J. Biochem. 259:88-95. [DOI] [PubMed] [Google Scholar]
- 19.Henriksson, G., L. Zhang, J. Li, P. Ljungquist, T. Reitberger, G. Pettersson, and G. Johansson. 2000. Is cellobiose dehydrogenase from Phanerochaete chrysosporium a lignin degrading enzyme? Biochim. Biophys. Acta 1480:83-91. [DOI] [PubMed] [Google Scholar]
- 20.Highley, T. L. 1973. Influence of carbon source on cellulase activity of white rot and brown rot fungi. Wood Fiber 5:50-58. [Google Scholar]
- 21.Irizarry, R. A., B. Hobbs, F. Collin, Y. D. Beazer-Barclay, K. J. Antonellis, U. Scherf, and T. P. Speed. 2003. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4:249-264. [DOI] [PubMed] [Google Scholar]
- 22.Ishida, T., K. Yaoi, A. Hiyoshi, K. Igarashi, and M. Samejima. 2007. Substrate recognition by glycoside hydrolase family 74 xyloglucanase from the basidiomycete Phanerochaete chrysosporium. FEBS J. 274:5727-5736. [DOI] [PubMed] [Google Scholar]
- 23.Jensen, K. A., K. M. Evans, T. K. Kirk, and K. E. Hammel. 1994. Biosynthetic pathway for veratryl alcohol in the ligninolytic fungus Phanerochaete chrysosporium. Appl. Environ. Microbiol. 60:709-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karkehabadi, S., H. Hansson, S. Kim, K. Piens, C. Mitchinson, and M. Sandgren. 2008. The first structure of a glycoside hydrolase family 61 member, Cel61B from Hypocrea jecorina, at 1.6 Å resolution. J. Mol. Biol. 383:144-154. [DOI] [PubMed] [Google Scholar]
- 25.Kawai, R., K. Igarashi, and M. Samejima. 2006. Gene cloning and heterologous expression of glycoside hydrolase family 55 β-1,3-glucanase from the basidiomycete Phanerochaete chrysosporium. Biotechnol. Lett. 28:365-371. [DOI] [PubMed] [Google Scholar]
- 26.Kawai, R., K. Igarashi, M. Yoshida, M. Kitaoka, and M. Samejima. 2006. Hydrolysis of β-1,3/1,6-glucan by glycoside hydrolase family 16 endo-1,3(4)-β-glucanase from the basidiomycete Phanerochaete chrysosporium. Appl. Microbiol. Biotechnol. 71:898-906. [DOI] [PubMed] [Google Scholar]
- 27.Kersten, P., and D. Cullen. 1993. Cloning and characterization of a cDNA encoding glyoxal oxidase, a peroxide-producing enzyme from the lignin-degrading basidiomycete Phanerochaete chrysosporium. Proc. Natl. Acad. Sci. USA 90:7411-7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kersten, P., and D. Cullen. 2007. Extracellular oxidative systems of the lignin-degrading basidiomycete Phanerochaete chrysosporium. Fungal Genet. Biol. 44:77-87. [DOI] [PubMed] [Google Scholar]
- 29.Kersten, P. J. 1990. Glyoxal oxidase of Phanerochaete chrysosporium: its characterization and activation by lignin peroxidase. Proc. Natl. Acad. Sci. USA 87:2936-2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kersten, P. J., and T. K. Kirk. 1987. Involvement of a new enzyme, glyoxal oxidase, in extracellular H2O2 production by Phanerochaete chrysosporium. J. Bacteriol. 169:2195-2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kirk, T. K., E. Schultz, W. J. Conners, L. F. Lorentz, and J. G. Zeikus. 1978. Influence of culture parameters on lignin metabolism by Phanerochaete chrysosporium. Arch. Microbiol. 117:277-285. [Google Scholar]
- 32.Kroon, P. A., G. Williamson, N. M. Fish, D. B. Archer, and N. J. Belshaw. 2000. A modular esterase from Penicillium funiculosum which releases ferulic acid from plant cell walls and binds crystalline cellulose contains a carbohydrate binding module. Eur. J. Biochem. 267:6740-6752. [DOI] [PubMed] [Google Scholar]
- 33.Kurek, B., and P. Kersten. 1995. Physiological regulation of glyoxal oxidase from Phanerochaete chrysosporium by peroxidase systems. Enzymol. Microb. Technol. 17:751-756. [Google Scholar]
- 34.Levi, M. P., and E. B. Cowling. 1969. Role of nitrogen in wood deterioration. VII. Physiological adaptation of wood-destroying and other fungi to substrates deficient in nitrogen. Phytopathology 59:460-468. [Google Scholar]
- 35.Li, B., S. R. Nagalla, and V. Renganathan. 1996. Cloning of a cDNA encoding cellobiose dehydrogenase, a hemoflavoenzyme from Phanerochaete chrysosporium. Appl. Environ. Microbiol. 62:1329-1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lilly, W. W., J. E. Stajich, P. J. Pukkila, S. K. Wilke, N. Inoguchi, and A. C. Gathman. 2008. An expanded family of fungalysin extracellular metallopeptidases of Coprinopsis cinerea. Mycol. Res. 112:389-398. [DOI] [PubMed] [Google Scholar]
- 37.Lucic, E., C. Fourrey, A. Kohler, F. Martin, M. Chalot, and A. Brun-Jacob. 2008. A gene repertoire for nitrogen transporters in Laccaria bicolor. New Phytol. 180:343-364. [DOI] [PubMed] [Google Scholar]
- 38.Martinez, D., L. F. Larrondo, N. Putnam, M. D. Sollewijn Gelpke, K. Huang, J. Chapman, K. G. Helfenbein, P. Ramaiya, J. C. Detter, F. Larimer, P. M. Coutinho, B. Henrissat, R. Berka, D. Cullen, and D. Rokhsar. 2004. Genome sequence of the lignocellulose degrading fungus Phanerochaete chrysosporium strain RP78. Nat. Biotechnol. 22:695-700. [DOI] [PubMed] [Google Scholar]
- 39.Ravalason, H., G. Jan, D. Molle, M. Pasco, P. M. Coutinho, C. Lapierre, B. Pollet, F. Bertaud, M. Petit-Conil, S. Grisel, J. C. Sigoillot, M. Asther, and I. Herpoel-Gimbert. 2008. Secretome analysis of Phanerochaete chrysosporium strain CIRM-BRFM41 grown on softwood. Appl. Microbiol. Biotechnol. 80:719-733. [DOI] [PubMed] [Google Scholar]
- 40.Reiser, J., A. Muheim, M. Hardegger, G. Frank, and A. Feichter. 1994. Aryl-alcohol dehydrogenase from the white-rot fungus Phanerochaete chrysosporium: gene cloning, sequence analysis, expression and purification of recombinant protein. J. Biol. Chem. 269:28152-28159. [PubMed] [Google Scholar]
- 41.Sato, S., F. Liu, H. Koc, and M. Tien. 2007. Expression analysis of extracellular proteins from Phanerochaete chrysosporium grown on different liquid and solid substrates. Microbiology 153:3023-3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shary, S., A. N. Kapich, E. A. Panisko, J. K. Magnuson, D. Cullen, and K. E. Hammel. 2008. Differential expression in Phanerochaete chrysosporium of membrane-associated proteins relevant to lignin degradation. Appl. Environ. Microbiol. 74:7252-7257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shimizu, M., N. Yuda, T. Nakamura, H. Tanaka, and H. Wariishi. 2005. Metabolic regulation at the tricarboxylic acid and glyoxylate cycles of the lignin-degrading basidiomycete Phanerochaete chrysosporium against exogenous addition of vanillin. Proteomics 5:3919-3931. [DOI] [PubMed] [Google Scholar]
- 44.Sims, A. H., N. S. Dunn-Coleman, G. D. Robson, and S. G. Oliver. 2004. Glutamic protease distribution is limited to filamentous fungi. FEMS Microbiol. Lett. 239:95-101. [DOI] [PubMed] [Google Scholar]
- 45.Singh, D., and S. Chen. 2008. The white-rot fungus Phanerochaete chrysosporium: conditions for the production of lignin-degrading enzymes. Appl. Microbiol. Biotechnol. 81:399-417. [DOI] [PubMed] [Google Scholar]
- 46.Smith, T. F., and M. S. Waterman. 1981. Identification of common molecular subsequences. J. Mol. Biol. 147:195-197. [DOI] [PubMed] [Google Scholar]
- 47.Stewart, P., and D. Cullen. 1999. Organization and differential regulation of a cluster of lignin peroxidase genes of Phanerochaete chrysosporium. J. Bacteriol. 181:3427-3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stewart, P., P. Kersten, A. Vanden Wymelenberg, J. Gaskell, and D. Cullen. 1992. The lignin peroxidase gene family of Phanerochaete chrysosporium: complex regulation by carbon and nitrogen limitation, and the identification of a second dimorphic chromosome. J. Bacteriol. 174:5036-5042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:2552-2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tien, M., and T. K. Kirk. 1984. Lignin-degrading enzyme from Phanerochaete chrysosporium: purification, characterization, and catalytic properties of a unique H2O2-requiring oxygenase. Proc. Natl. Acad. Sci. USA 81:2280-2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Uzcategui, E., G. Johansson, B. Ek, and G. Pettersson. 1991. The 1,4-d-glucan glucanohydrolases from Phanerochaete chrysosporium. Re-assessment of their significance in cellulose degradation mechanisms. J. Biotechnol. 21:143-160. [DOI] [PubMed] [Google Scholar]
- 52.Uzcategui, E., A. Ruiz, R. Montesino, G. Johansson, and G. Pettersson. 1991. The 1,4-β-d-glucan cellobiohydrolase from Phanerochaete chrysosporium. I. A system of synergistically acting enzymes homologous to Trichoderma reesei. J. Biotechnol. 19:271-286. [DOI] [PubMed] [Google Scholar]
- 53.Vanden Wymelenberg, A., P. Minges, G. Sabat, D. Martinez, A. Aerts, A. Salamov, I. Grigoriev, H. Shapiro, N. Putnam, P. Belinky, C. Dosoretz, J. Gaskell, P. Kersten, and D. Cullen. 2006. Computational analysis of the Phanerochaete chrysosporium v2.0 genome database and mass spectrometry identification of peptides in ligninolytic cultures reveals complex mixtures of secreted proteins. Fungal Genet. Biol. 43:343-356. [DOI] [PubMed] [Google Scholar]
- 54.Vanden Wymelenberg, A., G. Sabat, D. Martinez, A. S. Rajangam, T. T. Teeri, J. Gaskell, P. J. Kersten, and D. Cullen. 2005. The Phanerochaete chrysosporium secretome: database predictions and initial mass spectrometry peptide identifications in cellulose-grown medium. J. Biotechnol. 118:17-34. [DOI] [PubMed] [Google Scholar]
- 55.Vanden Wymelenberg, A., G. Sabat, M. D. Mozuch, P. Kersten, D. Cullen, and R. A. Blanchette. 2006. Structure, organization, and transcriptional regulation of a family of copper radical oxidase genes in the lignin-degrading basidiomycete Phanerochaete chrysosporium. Appl. Environ. Microbiol. 72:4871-4877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Varela, E., J. M. Martinez, and A. T. Martinez. 2000. Aryl-alcohol oxidase protein sequence: a comparison with glucose oxidase and other FAD oxidoreductases. Biochim. Biophys. Acta 1481:202-208. [DOI] [PubMed] [Google Scholar]
- 57.Wariishi, H., K. Valli, and M. H. Gold. 1992. Mn oxidation by manganese peroxidase from Phanerochaete chrysosporium: kinetic mechanisms and role of chelators. J. Biol. Chem. 267:23688-23695. [PubMed] [Google Scholar]
- 58.Whittaker, M. M., P. J. Kersten, N. Nakamura, J. Sanders-Loehr, E. S. Schweizer, and J. W. Whittaker. 1996. Glyoxal oxidase from Phanerochaete chrysosporium is a new radical-copper oxidase. J. Biol. Chem. 271:681-687. [DOI] [PubMed] [Google Scholar]
- 59.Wood, J. D., and P. M. Wood. 1992. Evidence that cellobiose:quinone oxidoreductase from Phanerochaete chrysosporium is a breakdown product of cellobiose oxidase. Biochim. Biophys. Acta 1119:90-96. [DOI] [PubMed] [Google Scholar]
- 60.Yoshida, M., K. Igarashi, M. Wada, S. Kaneko, N. Suzuki, H. Matsumura, N. Nakamura, H. Ohno, and M. Samejima. 2005. Characterization of carbohydrate-binding cytochrome b562 from the white-rot fungus Phanerochaete chrysosporium. Appl. Environ. Microbiol. 71:4548-4555. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.