Abstract
Disulfide bond (DSB) formation is catalyzed by disulfide bond proteins and is critical for the proper folding and functioning of secreted and membrane-associated bacterial proteins. Uropathogenic Escherichia coli (UPEC) strains possess two paralogous disulfide bond systems: the well-characterized DsbAB system and the recently described DsbLI system. In the DsbAB system, the highly oxidizing DsbA protein introduces disulfide bonds into unfolded polypeptides by donating its redox-active disulfide and is in turn reoxidized by DsbB. DsbA has broad substrate specificity and reacts readily with reduced unfolded proteins entering the periplasm. The DsbLI system also comprises a functional redox pair; however, DsbL catalyzes the specific oxidative folding of the large periplasmic enzyme arylsulfate sulfotransferase (ASST). In this study, we characterized the DsbLI system of the prototypic UPEC strain CFT073 and examined the contributions of the DsbAB and DsbLI systems to the production of functional flagella as well as type 1 and P fimbriae. The DsbLI system was able to catalyze disulfide bond formation in several well-defined DsbA targets when provided in trans on a multicopy plasmid. In a mouse urinary tract infection model, the isogenic dsbAB deletion mutant of CFT073 was severely attenuated, while deletion of dsbLI or assT did not affect colonization.
Disulfide bonds bridging cysteine pairs impart structural stability and protease resistance to secreted and membrane-associated proteins. Most organisms contain specific mechanisms for the formation of disulfide bonds in proteins, a process called oxidative protein folding. In bacteria, this folding process is catalyzed by the disulfide bond family of proteins (18, 22). The best-characterized bacterial disulfide bond machinery is the Escherichia coli K-12 oxidative system, which consists of two enzymes, the periplasmic DsbA and the inner-membrane DsbB (25, 35). DsbA is a monomeric protein comprising a thioredoxin (TRX) domain with an embedded helical insertion and a redox-active CPHC motif (34). This highly oxidizing protein introduces disulfide bonds into unfolded polypeptides by donating its redox-active disulfide (2, 4, 5), and as a result, the two cysteines contained in the CPHC catalytic motif become reduced. DsbB reoxidizes this cysteine pair and restores the oxidizing activity of DsbA, enabling it to assist the folding of a new substrate protein (21).
The DsbAB oxidative protein folding system plays a well-documented part in bacterial virulence. Several studies have demonstrated a direct role for both enzymes, particularly DsbA, in the biogenesis of virulence factors utilized by bacterial pathogens in various stages of the infection process (19). The protein forming the P-ring of E. coli flagella, FlgI, was one of the first DsbA substrates identified (10) and flagellum-mediated motility was subsequently demonstrated to require the presence of functional DsbA in several gram-negative pathogens, including Salmonella enterica (1), Proteus mirabilis (8), Erwinia carotovora subsp. atroseptica (9), Burkholderia cepacia (17), and Campylobacter jejuni (42). In Yersinia pestis, S. enterica, Shigella flexneri, and enteropathogenic E. coli, deletion of dsbA results in defective type III secretion, a major virulence mechanism employed by these enteric pathogens to manipulate the host during infection. The defect was shown in each case to involve the outer membrane secretin (YscC, SpiA, Spa32, and EscC, respectively), which requires a single intramolecular disulfide bond to adopt a functional conformation (23, 36, 37, 49). Fimbria-mediated adhesion is a crucial first step of the infection process as it allows host colonization by mucosal pathogens. DsbA is required for functional assembly of several types of fimbriae, including P fimbriae of uropathogenic E. coli (UPEC) (24), bundle-forming pili (Bfp) of enteropathogenic E. coli (55), mannose-resistant Proteus-like (MR/P) fimbriae of Proteus mirabilis (8), plasmid-encoded fimbriae (Pef) of Salmonella enterica (6), type IV pili of Neisseria meningitidis (47), and toxin-coregulated pili (Tcp) of Vibrio cholerae (41). A number of studies have reported that dsbA and/or dsbB mutants are attenuated in infection models (9, 16, 41, 48, 52).
The recent exponential increase in sequenced genomes has offered a first glimpse at the diversity of disulfide bond systems present in bacteria (13). In addition, it is now evident that several bacterial species encode multiple DsbA paralogues, often with demonstrated differences in substrate specificity. Neisseria meningitidis, for example, encodes three DsbA oxidoreductases: two inner membrane-associated lipoproteins (DsbA1 and DsbA2) and one periplasmic enzyme (DsbA3). While redundancy was observed in the oxidative folding of virulence-associated proteins by DsbA1 and DsbA2, DsbA3 alone was unable to restore important meningococcal virulence traits, such as type IV pilus-mediated adhesion to human endothelial cells (47). Recently, a second E. coli disulfide bond system (DsbLI) was identified in the genome-sequenced UPEC strain CFT073 and was demonstrated to be a functional paralogue of the prototypic DsbAB system (14). The oxidoreductase DsbL has the strongest oxidizing potential of all DsbA homologues characterized to date. Although the crystal structure of DsbL revealed a similar overall fold and domain architecture to DsbA, DsbL contains a longer helical insertion and deletions in the TRX domain that result in a truncated peptide binding groove. Moreover, DsbL shows different surface properties, including a distinct basic patch around the active site, which was suggested to allow stricter substrate specificity than the highly hydrophobic surface surrounding the active site of DsbA. Grimshaw and colleagues (14) demonstrated the specificity of the DsbLI system for the periplasmic enzyme arylsulfate sulfotransferase (ASST) encoded by assT, a gene found immediately upstream of dsbL and dsbI on the CFT073 chromosome. ASST belongs to a group of poorly characterized large bacterial ASSTs that are proposed to mediate detoxification of phenolic substances by catalyzing the transfer of sulfuryl groups from phenolic sulfates to phenol (26-28, 30). A reason for the specificity of DsbLI for ASST folding could be the presence of an allosteric disulfide bond, recently revealed by the enzyme's crystal structure (33). This class of disulfide bond forms between Cα atoms of cysteines in unusually close proximity (3.8 Å in the case of ASST) and has higher steric strain energy than catalytic or structural disulfide bonds, thus explaining the requirement for the stronger DsbL oxidase for its formation (33). The activity of DsbL and DsbI was studied using plasmids introduced into E. coli K-12 strains with the native DsbAB system deleted. As yet, the role of the DsbLI system in UPEC virulence has not been investigated.
E. coli CFT073 is a prototypic UPEC strain isolated from a female patient with acute pyelonephritis (38). UPEC strains are the causative agent of >80% of community-acquired urinary tract infections (UTIs) and >30% of nosocomial infections (7). The uropathogenic lifestyle of UPEC CFT073 is reflected in its genome, which contains several factors with an established role in urovirulence, including the well-studied type 1 and P fimbriae (50). Genomic comparison of CFT073—and other recently sequenced UPEC strains—with E. coli strains with distinct lifestyles (gut commensals, enteric pathogens, and avian pathogens) allows the discovery of genes unique to genomes of uropathogenic bacteria that are potentially novel urovirulence factors. One such UPEC-specific gene is assT, the gene located upstream of dsbL and dsbI in the chromosome of CFT073 (32).
Here we characterize the DsbLI system in its native genetic background of UPEC CFT073 and compare and contrast the contribution of each of the two paralogous disulfide bond systems of CFT073 in the production of UPEC-associated virulence factors and in vivo uropathogenesis. Using isogenic dsbAB and dsbLI deletion mutants of CFT073, we demonstrate that the recently identified DsbLI oxidative protein folding machinery of UPEC CFT073 plays a secondary role in the production of urovirulence factors and does not appear to contribute to virulence in the mouse infection model used in this study. We also show that in the same infection model, an isogenic assT deletion mutant of CFT073 is not attenuated.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
All strains and plasmids used in this study are listed in Table 1. E. coli strains were routinely cultured at 37°C on solid or in liquid Luria-Bertani (LB) medium supplemented with kanamycin (50 μg ml−1) or ampicillin (100 μg ml−1). Culture media were supplemented with 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) to induce expression of DsbA-DsbB and DsbL-DsbI from plasmids pDsbAB and pDsbLI, respectively. All plasmid transformations into E. coli CFT073 (wild type and deletion mutants) were carried out by electroporation. Plasmid pDsbAB was generated by PCR amplification of dsbA (primers 1147 and 1148) and dsbB (primers 1149 and 1150) genes from the chromosome of E. coli CFT073 and subsequent cloning into EcoRI-HindIII of pUC19. Plasmid pDsbLI was generated by PCR amplification of dsbL-dsbI genes (primers 1145 and 1146) from the chromosome of E. coli CFT073 and subsequent cloning into EcoRI-PstI of pGEM-T Easy (Promega). In both vectors, expression of disulfide bond enzymes was controlled by the inducible lac promoter. CFT073 mutants containing deletions of dsb genes were constructed by λ-red-mediated homologous recombination as previously described (11) and confirmed by PCR and DNA sequencing. The primers used are listed in Table S1 in the supplemental material.
TABLE 1.
E. coli strains and plasmids
| E. coli strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| CFT073 | UPEC isolate (O6:K2:H1) | 38 |
| CFT073dsbAB | CFT073dsbAdsbB::Km; Kmr | This study |
| CFT073dsbLI | CFT073dsbLI::Km; Kmr | This study |
| CFT073dsbABdsbLI | CFT073dsbAdsbB::Km dsbLI; Kmr | This study |
| CFT073assT | CFT073assT::Km; Kmr | This study |
| Plasmids | ||
| pDsbAB | dsbA and dsbB genes in pUC19; Apr | This study |
| pDsbLI | dsbL and dsbI genes in pGEM-T Easy; Apr | This study |
DNA manipulation techniques.
DNA techniques were performed as previously described (44). Chromosomal DNA was purified using the GenomicPrep cell and tissue DNA isolation kit (Amersham Pharmacia Biotech, Inc.). PCR was performed using the Expand long template PCR system according to the manufacturer's instructions (Roche, Australia). Oligonucleotides were purchased from Sigma, Australia, and are listed in Table S1 in the supplemental material. DNA sequencing of PCR products was performed using the BigDye Terminator v3.1 cycle sequencing kit according to the manufacturer's instructions (Applied Biosystems) and analyzed by the Australian Equine Genome Research Centre, Brisbane, Australia.
Purification of DsbA and DsbL, production of polyclonal sera, and immunoblotting.
The dsbA and dsbL genes were PCR amplified using primers containing ligation-independent cloning extensions. The amplified genes were subsequently inserted into a modified version of pET21a vector, which allows the introduction of an N-terminal His6 tag followed by the tobacco etch virus protease cleavage site, using ligation-independent cloning (12). Protein expression was carried out in BL21(DE3)(pLysS) cells (Invitrogen) by autoinduction (45). Briefly, cells were grown for 24 h at 30°C in minimal medium supplemented with 100 μg ml−1 ampicillin and 34 μg ml−1chloramphenicol. After lysing the cells by sonication, cell debris was removed and histidine-tagged proteins were purified using PrepEase Ni2+ affinity resin (USB). Upon His6 tag cleavage with tobacco etch virus protease, proteins were further purified by gel filtration chromatography on a Superdex S-75 column (GE Healthcare). Protein purity was then assessed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis.
Polyclonal anti-DsbA and anti-DsbL sera were raised in rabbits by the Institute of Medical and Veterinary Sciences (South Australia) following a standard protocol (http://www.imvs.sa.gov.au/). For immunoblotting, whole-cell lysates or purified protein samples were subjected to SDS-PAGE using NuPAGE Novex 10% Bis-Tris precast gels with NuPAGE MES (morpholineethanesulfonic acid)-SDS running buffer (Invitrogen) and subsequently transferred to polyvinylidene difluoride microporous membrane filters using the iBlot dry blotting system as described by the manufacturer (Invitrogen). Western blotting was performed using rabbit polyclonal anti-DsbA or anti-DsbL primary sera (1/4,000 dilution) followed by incubation with goat anti-rabbit immunoglobulin G-alkaline phosphatase-conjugated secondary antibody (1/10,000 dilution; Sigma). Signal development was performed in BCIP/NBT (5-bromo-4-chloro-3-indolylphosphate-nitroblue tetrazolium solution; Sigma).
Yeast cell and RBC agglutination.
The expression of functional type 1 and P fimbriae by E. coli CFT073 and isogenic dsb deletion mutants was investigated by their ability to agglutinate yeast (Saccharomyces cerevisiae) cells and human red blood cells (hRBCs), respectively. Agglutination was examined by mixing 10 μl of overnight E. coli culture with 10 μl of 5% (wt/vol) yeast cell suspension (for type 1 fimbriae) or 10% (vol/vol) type A hRBCs (for P fimbriae) in the presence and absence of 1% (wt/vol) mannose. E. coli strains were considered positive when visible clumping occurred within 3 min of mixing on a glass slide. All experiments were performed in triplicate.
Motility assays.
The motility of E. coli CFT073 and dsb deletion mutants was assessed by inoculation of 2 μl of overnight liquid culture onto the surface of semisolid (0.25% [wt/vol]) agar. The diameter of bacterial outward growth was recorded after 18 h of incubation at 37°C. Plasmid-complemented deletion mutants were assessed in the presence and absence of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). All experiments were performed in triplicate.
Mouse model of UTI.
The mouse model of UTI was employed as previously described (43). Briefly, female C57BL/6 mice (8 to 10 weeks) were purchased from the University of Queensland Animal Facility and housed in sterile cages with ad libitum access to sterile water. Preinoculation urine was examined microscopically and by culture, and animals with a combination of >5 × 102 CFU/ml and >2 × 105 leukocytes in urine were excluded. Mice were transurethrally catheterized with ∼5 × 108 CFU (20 μl bacterial phosphate-buffered saline suspension) under isoflurane anesthesia. Mice were euthanized at 18 h postinfection by cervical dislocation. Bacterial loads in urine and bladder homogenates of individual mice were determined by viable counts performed in triplicate. Data are expressed as mean log CFU per ml of urine or 0.1 g of bladder tissue ± the standard error of the mean. A minimum of six mice were included in each group. Equality of group medians of log-transformed data was tested using the Kruskal-Wallis nonparametric test in the Minitab15 statistical package. P values of <0.05 were considered significant.
RESULTS
Sequence comparison of DsbA and DsbL paralogues of E. coli CFT073.
The two genes of the prototypic DsbAB system (dsbA and dsbB) are located at separate loci on the CFT073 chromosome (c4804 and c1633, respectively), while the dsbL and dsbI genes lie adjacent to one another at a distinct chromosomal location (c3786 and c3787). Despite sharing a similar overall fold and domain architecture (14), DsbA and DsbL display low amino acid identity (Fig. 1A). Amino acid sequence divergence, as well as insertions and deletions, is observed in key regions of the protein, including the CXXC active site (CPHC and CPFC in DsbA and DsbL, respectively), the hydrophobic area proximal to the active site, and the target binding groove (positively charged in DsbL) (Fig. 1A and B). As these differences are found in areas involved in substrate recognition and binding, they may account for differences in substrate specificity between DsbA and DsbL.
FIG. 1.
(A) Structure-based sequence alignment of DsbA (Protein Data Bank no. 1FVK [15]) and DsbL (Protein Data Bank no. 3C7M [14]) using STRAP (http://www.charite.de/bioinf/strap/). Residues belonging to the TRX and helical domain are indicated by a gray bar and dark blue bar, respectively. Identical residues are shown in red. Regions in DsbL that differ from DsbA are highlighted in boxes (the CXXC catalytic site and the deletion at the C terminus that shortens the peptide binding groove) or underlined (positively charged residues in DsbL generate a basic patch where DsbA has a hydrophobic patch). Arrows indicate the second cysteine in the CXXC motif and the conserved cis-proline, residues utilized to measure the length of the inserted alpha-helical domain in the TRX fold. (B) Ribbon representations of E. coli K-12 DsbA (Protein Data Bank no. 1FVK [15]) incorporating a TRX fold (gray) with a helical insertion (dark blue). The active-site disulfide is shown in a space-filling representation (yellow). Arrows point at areas that differ between DsbA and DsbL (also highlighted in panel A). (C) Physical map demonstrating the genomic location of the dsbL gene in selected genera from Gammaproteobacteria (pink box), Deltaproteobacteria (blue box), and Epsilonproteobacteria (green box). Genes and intergenic regions are drawn to scale.
Prevalence and genomic organization of the dsbLI genes in bacteria.
In order to investigate the distribution of dsbL in bacteria, we searched 404 complete bacterial genomes deposited in the J. Craig Venter Institute public database (Comprehensive Microbial Resource) using the amino acid sequence of DsbL from E. coli CFT073. Proteins with significant sequence identity to DsbL (P < 0.0002) were considered DsbL homologues if (i) their sequence identity to DsbL was higher than their sequence identity to DsbA from E. coli CFT073 and (ii) they contained an identical active site to DsbL (CPFC) or if the length of the inserted alpha-helical domain in the TRX fold (counting from the catalytic CXXC domain to the conserved cis-proline residue [Fig. 1A], which is present in most TRX-like proteins) was ≥118 residues (minimum insert length present in CPFC-containing DsbL homologues). Using this set of criteria, 23 DsbL proteins were identified from 17 genomes of Gammaproteobacteria, Deltaproteobacteria, and Epsilonproteobacteria of the following genera: Escherichia, Salmonella, Campylobacter, Geobacter, and Shewanella (see Table S2 in the supplemental material). Protein sequences satisfying the above criteria were also identified in other Proteobacteria (Yersinia frederiksenii, Enterobacter amnigenus, Klebsiella sp. strain K-36, Citrobacter freundii, and Edwardsiella tarda) upon interrogation of the NCBI database with the amino acid sequence of DsbL from E. coli CFT073 (see Table S2 in the supplemental material). The genomic location of dsbL was examined, and a dsbI gene was present downstream of dsbL in all genomes (Fig. 1C; and see Table S2 in the supplemental material). The genomes of Campylobacter curvus, Campylobacter fetus, and Campylobacter concisus contained a second copy of dsbL carried at a different position on the chromosome without a dsbI gene in close proximity, while the second copy of dsbL in the genome of Camplobacter jejuni was carried immediately downstream of dsbL, dsbI, and a degenerate copy of assT (Fig. 1C; and see Table S2 in the supplemental material). The assT gene, encoding the periplasmic substrate of DsbL in E. coli CFT073, was colocated with the dsbL and dsbI genes in all but one genome (Fig. 1C).
Construction and characterization of dsbA-dsbB and dsbL-dsbI deletion mutants in E. coli CFT073.
We employed λ-red-mediated homologous recombination to delete the dsbA-dsbB and dsbL-dsbI genes in E. coli CFT073. As the dsbA and dsbB genes are located in different positions on the CFT073 chromosome, deletion of these genes required two successive rounds of mutagenesis. The CFT073 dsbA-dsbB deletion mutant (CFT073dsbAB) was nonmotile (Fig. 2B). In contrast, deletion of the dsbL-dsbI genes (CFT073dsbLI) did not affect motility compared to that of wild-type CFT073 (Fig. 2C and A). This suggested that either DsbL is not expressed at levels sufficient to restore the loss of motility in the dsbA-dsbB mutant or that FlgI—the flagella P-ring protein targeted by DsbA—is not a substrate of DsbL.
FIG. 2.
Motility analysis for wild-type CFT073 (A), CFT073dsbAB (B), CFT073dsbLI (C), CFT073dsbABdsbLI (D), CFT073dsbABdsbLI(pDsbAB) (E), CFT073dsbABdsbLI(pDsbLI) (F), CFT073dsbABdsbLI(pUC19) (G), and CFT073dsbABdsbLI(pGEM-T Easy) (H).
The DsbAB and DsbLI systems can catalyze oxidative folding of the FlgI flagella P-ring protein.
To compare the specificity of the DsbAB and DsbLI systems, we constructed a dsbA-dsbB dsbL-dsbI double deletion mutant in E. coli CFT073 (CFT073dsbABdsbLI) and transiently expressed each disulfide bond system in this genetic background. The lack of motility in CFT073dsbABdsbLI (Fig. 2D) was restored upon complementation with both a dsbA-dsbB-carrying plasmid (pDsbAB) and a dsbL-dsbI-carrying plasmid (pDsbLI) (Fig. 2E and F). Transformation of CFT073dsbABdsbLI with the vector control did not affect the motility phenotype (Fig. 1G and H). While complementation with pDsbAB restored motility of CFT073dsbABdsbLI to wild-type levels, pDsbLI restored only partial motility. These results suggest that DsbL-DsbI can direct the correct folding of FlgI in the absence of DsbA-DsbB when expressed at high levels, albeit with reduced efficiency.
DsbA and DsbL expression in E. coli CFT073.
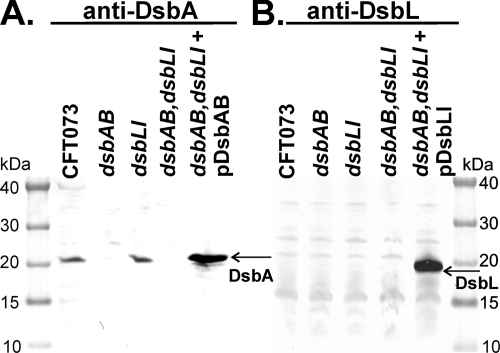
The loss of motility in CFT073dsbAB and the complementation by multicopy dsbL-dsbI suggest that DsbL might be expressed at low levels in E. coli CFT073. To investigate whether both oxidoreductase paralogues are expressed in CFT073, we developed rabbit polyclonal antisera against DsbA and DsbL. The specificity of each antiserum was checked using purified recombinant DsbA and DsbL, and no cross-reactivity was observed (data not shown). A band of the correct size for DsbA (∼21 kDa) was observed in whole-cell lysates of CFT073 and CFT073dsbLI but was absent from CFT073dsbAB and CFT073dsbABdsbLI (Fig. 3A). Complementation of CFT073dsbABdsbLI with pDsbAB restored DsbA expression (Fig. 3A). No expression was detected for DsbL in CFT073 and all three isogenic mutants under the conditions tested (Fig. 3B). However, a band corresponding to DsbL was detected upon complementation of CFT073dsbABdsbLI with pDsbLI (Fig. 3B). No DsbL expression was detected following growth of CFT073 in human urine (data not shown).
FIG. 3.
Western blot analysis of DsbA expression in wild-type CFT073, CFT073dsbAB, CFT073dsbLI, CFT073dsbABdsbLI, and CFT073dsbABdsbLI(pDsbAB) whole-cell lysates (A) and DsbL expression in wild-type CFT073, CFT073dsbAB, CFT073dsbLI, CFT073dsbABdsbLI, and CFT073dsbABdsbLI(pDsbLI) whole-cell lysates (B).
Role of DsbA and DsbL in the production of P and type 1 fimbriae by E. coli CFT073.
We investigated the contribution of each disulfide bond system to the production of functional P and type 1 fimbriae by testing the ability of CFT073 single and double dsb deletion mutants to agglutinate hRBCs (indicative of P fimbria expression) and yeast cells (indicative of type 1 fimbria expression). The ability of CFT073 to cause hemagglutination (HA) of hRBCs required the DsbAB but not the DsbLI system (Table 2). However, a positive HA phenotype was restored in CFT073dsbAB and CFT073dsbABdsbLI following complementation with either pDsbAB or pDsbLI (Table 2). In contrast, mannose-sensitive yeast agglutination (YA) was still observed with CFT073 deletion mutants for either or both disulfide bond systems (Table 2). To investigate this further, we measured the exact time required by each strain to produce YA (Table 2). While CFT073dsbLI agglutinated yeast cells as fast as wild-type CFT073, CFT073dsbAB displayed a small (10 to 15 s) delay, which was partially restored upon complementation of CFT073dsbAB and CFT073dsbABdsbLI with either pDsbAB or pDbsLI. Taken together, these results suggest that both disulfide bond systems can catalyze the biogenesis of functional P fimbriae in CFT073, with DsbAB primarily contributing to the process, while functional assembly of type 1 fimbriae can occur independently of both disulfide bond systems.
TABLE 2.
Production of P and type 1 fimbriae by E. coli CFT073 and isogenic disulfide bond deletion mutants
| E. coli strain | HA (P fimbriae) | YA (type 1 fimbriae) | Avg YA time (s) ± SD |
|---|---|---|---|
| CFT073 | + | + | 3 ± 1 |
| CFT073dsbLI | + | + | 3.7 ± 0.6 |
| CFT073dsbAB | − | + | 21.7 ± 1.5 |
| CFT073dsbAB(pDsbAB) | + | + | 13.3 ± 1.2 |
| CFT073dsbAB(pUC19) | − | + | 23 ± 1.7 |
| CFT073dsbAB(pDsbLI) | + | + | 14.7 ± 2.5 |
| CFT073dsbAB(pGEM-T Easy) | − | + | 19 ± 2.6 |
| CFT073dsbABdsbLI | − | + | 17.7 ± 3.1 |
| CFT073dsbABdsbLI(pDsbAB) | + | + | 7.3 ± 0.6 |
| CFT073dsbABdsbLI(pUC19) | − | + | 18 ± 1 |
| CFT073dsbABdsbLI(pDsbLI) | + | + | 7.7 ± 0.6 |
| CFT073dsbABdsbLI(pGEM-T Easy) | − | + | 19 ± 2.6 |
Contribution of the DsbAB and DsbLI systems to colonization of the mouse urinary tract.
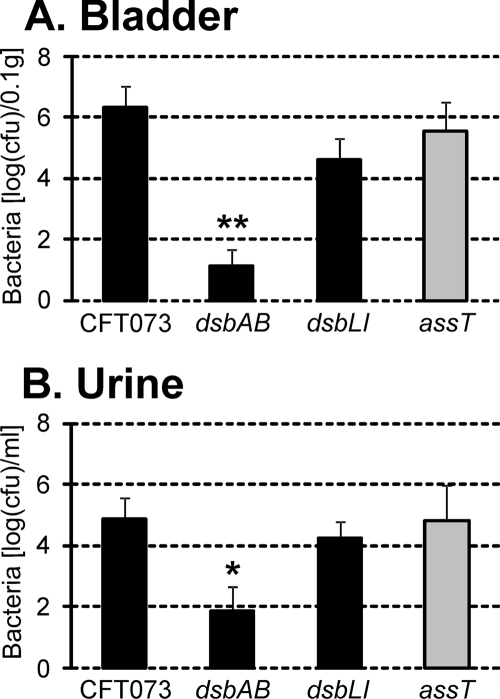
To study the role of both disulfide bond systems in virulence, we examined the ability of CFT073, CFT073dsbAB, and CFT073dsbLI to survive in the mouse urinary tract. CFT073dsbAB was severely attenuated in its ability to colonize the bladder (P < 0.001) despite the fact that it still produced type 1 fimbriae (Fig. 4A). Significantly lower numbers of CFT073dsbAB cells were also recovered from the urine of these mice compared to wild-type CFT073 (Fig. 4B). In contrast, no significant difference in the colonization of the bladder or urine was observed for CFT073dsbLI compared to CFT073. No significant colonization of the kidneys was observed for any of the strains (data not shown); this is consistent with previous data from our laboratory using C57BL/6 mice.
FIG. 4.
Bacterial load in mouse bladders (A) and urine (B) 18 h postinfection with CFT073, CFT073dsbAB, CFT073dsbLI, and CFT073assT. The average log CFU per 0.1 g bladder or ml urine is plotted ± the standard error of the mean. *, P < 0.05; **, P < 0.001.
Disruption of the assT gene does not affect colonization of the mouse bladder.
ASST, the periplasmic enzyme specifically oxidized by DsbL, has been implicated as a virulence factor of UPEC (32, 33). However, in our study CFT073dsbLI colonized the mouse bladder at wild-type levels. To examine the possibility that DsbA is responsible for functional ASST production in the CFT073dsbLI mutant and to test whether ASST is required for UPEC virulence, we constructed an assT deletion mutant in CFT073 (CFT073assT) and tested its ability to colonize the urinary tract of C57BL/6 mice. There was no significant difference in the abilities of CFT073 and CFT073assT to colonize the mouse bladder following transurethral infection (Fig. 4A). Similar numbers of each strain were also recovered from the urine of infected mice (Fig. 4B); neither strain was isolated from the mouse kidney (data not shown).
DISCUSSION
UPEC strains are successful extraintestinal pathogens equipped with a mosaic genome encoding a plethora of factors contributing to their uropathogenic lifestyle, such as surface adhesins and secreted toxins. Membrane and secreted protein folding in UPEC CFT073 is assisted by two distinct disulfide catalytic systems, the prototypic DsbAB system also found in E. coli K-12 and the recently identified DsbLI system. The DsbAB system introduces disulfide bonds into unfolded polypeptides in the periplasm and has broad substrate specificity. In contrast, the DsbLI system is proposed to be highly specific for oxidizing the periplasmic enzyme ASST. Here we demonstrated that when overexpressed, the DsbLI system can catalyze disulfide bond formation in two targets of the DsbAB system in E. coli CFT073: the flagellar motor component FlgI and the P fimbrial chaperone PapD. Additionally, we constructed isogenic dsb mutants in CFT073 and assessed the relative contribution of each disulfide bond system in UPEC virulence by using a mouse UTI model.
Flagella-mediated motility is important for the dissemination of pathogenic bacteria in different niches within the host. In the case of UPEC, this is required for bacterial ascent from the bladder to the kidneys via the ureters against the flow of urine (31). FlgI forms the P-ring of the flagellar structure and P-ring assembly requires the introduction of a disulfide bond by DsbAB in E. coli K-12 (10). The difference observed in the ability of DsbLI and DsbAB to fully restore motility in CFT073dsbABdsbLI in this study is most likely due to the different biochemical properties of the two oxidases. This is consistent with a previous study that examined DsbLI overexpression in an E. coli K-12 dsbA-dsbB deletion mutant (14).
An important first step in UPEC infection is adhesion to the host mucosa. Adhesion mediated by P fimbriae has been extensively studied and shown to be required for the early establishment of UPEC in the human urinary tract (51). P fimbria biogenesis follows the chaperone-usher pathway of protein secretion, where the periplasmic chaperone PapD binds the unfolded P fimbrial subunits emerging through the inner membrane in a Sec-dependent manner and targets them to the outer membrane usher PapC, which in turn directs their ordered assembly and secretion to the bacterial cell surface to form the fimbrial fiber (46). PapD requires the formation of an intramolecular disulfide bond in order to adopt its native conformation in vivo, and this allows it to interact with P fimbrial subunit proteins (24). Overexpression of DsbL in the absence of DsbA restored the production of functional P fimbriae in E. coli CFT073dsbABdsbLI, demonstrating the capacity of DsbL to catalyze disulfide bond formation in PapD. Other members of the PapD-like chaperone superfamily have been described from E. coli and other organisms (20, 40). For example, the Caf1M chaperone of capsule in Yersinia pestis is the prototypic member of the FGL subfamily of molecular chaperones and contains two invariable cysteine residues that form a disulfide bond in the subunit binding pocket (53, 54). Formation of this disulfide bond is catalyzed by DsbA and is required for the appropriate folding of Caf1M and for binding to the Caf1 capsule subunit of Y. pestis (53). Based on our results, it is possible that DsbL may also be capable of catalyzing disulfide bond formation in Caf1M and other members of the FGL subfamily of chaperones.
In contrast to P fimbriae, the production of functional type 1 fimbriae (as measured by the ability to mediate yeast cell agglutination) was not completely abolished in CFT073dsbABdsbLI. This difference is most likely attributed to the lack of cysteine residues in the FimC chaperone. These results also suggest that disulfide bond formation within the major (FimA) and minor (FimF, FimG, and FimH) components of type 1 fimbriae can occur independently of DsbA and DsbL. Our data are in agreement with a previous study which demonstrated that removal of the cysteine bond in the mannose-binding domain of FimH does not affect FimH-mannose binding under static or low-shear conditions (39). This is in contrast to high-flow conditions, where FimH-mediated adhesion requires the formation of a cysteine bond for the stabilization of its mannose binding pocket.
Despite production of functional type 1 fimbriae (albeit at a reduced level), deletion of the dsbAB system, but not the dsbLI system, resulted in severe attenuation of E. coli CFT073 in a mouse UTI model. This is likely due to a combined effect of the dsbAB deletion on flagellum-mediated motility and the production of P and type 1 fimbriae. Additionally, disruption of the DsbAB system could have affected several other potential targets among the >150 exported proteins that contain a cysteine pair in E. coli (13). Thus, the severe attenuation of CFT073dsbAB may result from the cumulative contribution of direct and pleiotropic effects. In this regard, we note that the growth of CFT073dsbAB and CFT073dsbABdsbLI did not differ from wild-type CFT073 in LB broth and minimal media.
DsbLI is the proposed specialized oxidase system for the periplasmic enzyme ASST (14). The genomic location of DsbL homologues identified in this study supports this proposition, as an assT gene is found in close proximity to dsbL and dsbI in all but one of the examined genomes. The only exception is Campylobacter concisus, where one of the two assT genes present on the chromosome is located 17 genes apart from dsbL and dsbI. In our bioinformatic search, we applied a series of stringent cutoffs for the identification of DsbL homologues. This resulted in the selection of less than 40 proteins distributed across 10 genera of Proteobacteria (see Table S2 in the supplemental material). Our set of DsbL homologues was congruent with the list of DsbL homologues identified in 33 assT-containing genomes by Grimsahw et al. (14) and included additional DsbL homologues from other Proteobacteria (Yersinia, Citrobacter, Edwardsiella, Enterobacter, and Klebsiella). In a recent comprehensive study of bacterial DsbA homologues (19), we also identified CPFC-containing DsbA proteins in Firmicutes (Listeria monocytogenes), Spirochaetes (Leptospira interrogans and Leptospira innocua), and Actinobacteria (Actinomyces naeslundii, Streptomyces coelicolor, Corynebacterium glutamicum, and Mycobacterium sp.). However, these proteins were not included in the current data set due to their low sequence identity to DsbL (P > 0.0002) and lack of biochemical data confirming that they have similar redox properties to DsbL.
In this study, we demonstrate functional redundancy between two paralogous oxidative protein folding systems from E. coli CFT073 upon overexpression of DsbAB and DsbLI in an isogenic double-dsb deletion mutant. Lack of DsbL expression in wild-type CFT073 could be due to the limited sensitivity of detection by the Western blotting assay employed. Alternatively, the optimal conditions for expression of the DsbLI system were not reproduced in our experiments. In the chromosome of E. coli CFT073, the dsbL and dsbI genes lie immediately downstream of the assT gene and the three genes form a tricistronic operon (50). ASST is a large periplasmic enzyme whose function is dependent upon the formation of a single disulfide bond by the DsbLI dithiol oxidase system (14). Therefore, it is likely that the DsbLI system is specifically expressed under ASST-inducing conditions. ASST enzymes have been characterized from various intestinal bacteria and are proposed to function as detoxifying catalysts, converting toxic phenolic compounds and antibiotics into nontoxic substances (3, 26-30). Thus, although we were unable to demonstrate a role for assT in colonization of the mouse urinary tract, we cannot rule out the possibility that the precise conditions required to activate assT expression were not reproduced in this experiment. The precise environmental cues that regulate assT gene expression remain to be elucidated.
Supplementary Material
Acknowledgments
This work was supported by grants from the Australian Research Council to M.A.S. (DP666852) and B.H. (DP0556707) and a grant from the National Health and Medical Research Council to M.A.S. (455914).
Footnotes
Published ahead of print on 17 April 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Agudo, D., M. T. Mendoza, C. Castañares, C. Nombela, and R. Rotger. 2004. A proteomic approach to study Salmonella typhi periplasmic proteins altered by a lack of the DsbA thiol: disulfide isomerase. Proteomics 4355-363. [DOI] [PubMed] [Google Scholar]
- 2.Akiyama, Y., S. Kamitani, N. Kusukawa, and K. Ito. 1992. In vitro catalysis of oxidative folding of disulfide-bonded proteins by the Escherichia coli dsbA (ppfA) gene product. J. Biol. Chem. 26722440-22445. [PubMed] [Google Scholar]
- 3.Baek, M. C., S. K. Kim, D. H. Kim, B. K. Kim, and E. C. Choi. 1996. Cloning and sequencing of the Klebsiella K-36 astA gene, encoding an arylsulfate sulfotransferase. Microbiol. Immunol. 40531-537. [DOI] [PubMed] [Google Scholar]
- 4.Bardwell, J. C., J. O. Lee, G. Jander, N. Martin, D. Belin, and J. Beckwith. 1993. A pathway for disulfide bond formation in vivo. Proc. Natl. Acad. Sci. USA 901038-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bardwell, J. C. A., K. McGovern, and J. Beckwith. 1991. Identification of a protein required for disulfide bond formation in vivo. Cell 67581-589. [DOI] [PubMed] [Google Scholar]
- 6.Bouwman, C. W., M. Kohli, A. Killoran, G. A. Touchie, R. J. Kadner, and N. L. Martin. 2003. Characterization of SrgA, a Salmonella enterica Serovar Typhimurium virulence plasmid-encoded paralogue of the disulfide oxidoreductase DsbA, essential for biogenesis of plasmid-encoded fimbriae. J. Bacteriol. 185991-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bouza, E., R. San Juan, P. Munoz, A. Voss, and J. Kluytmans. 2001. A European perspective on nosocomial urinary tract infections II. Report on incidence, clinical characteristics and outcome (ESGNI-004 study). Clin. Microbiol. Infect. 7532-542. [DOI] [PubMed] [Google Scholar]
- 8.Burall, L. S., J. M. Harro, X. Li, C. V. Lockatell, S. D. Himpsl, J. R. Hebel, D. E. Johnson, and H. L. T. Mobley. 2004. Proteus mirabilis genes that contribute to pathogenesis of urinary tract infection: identification of 25 signature-tagged mutants attenuated at least 100-fold. Infect. Immun. 722922-2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coulthurst, S. J., K. S. Lilley, P. E. Hedley, H. Liu, I. K. Toth, and G. P. C. Salmond. 2008. DsbA plays a critical and multi-faceted role in the production of secreted virulence factors by the phytopathogen, Erwinia carotovora subsp. atroseptica. J. Biol. Chem. 28323739-23753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dailey, F. E., and H. C. Berg. 1993. Mutants in disulfide bond formation that disrupt flagellar assembly in Escherichia coli. Proc. Natl. Acad. Sci. USA 901043-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 976640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Donnelly, M. I., M. Zhou, C. S. Millard, S. Clancy, L. Stols, W. H. Eschenfeldt, F. R. Collart, and A. Joachimiak. 2006. An expression vector tailored for large-scale, high-throughput purification of recombinant proteins. Protein Expr. Purif. 47446-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dutton, R. J., D. Boyd, M. Berkmen, and J. Beckwith. 2008. Bacterial species exhibit diversity in their mechanisms and capacity for protein disulfide bond formation. Proc. Natl. Acad. Sci. USA 10511933-11938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grimshaw, J. P. A., C. U. Stirnimann, M. S. Brozzo, G. Malojcic, M. G. Grütter, G. Capitani, and R. Glockshuber. 2008. DsbL and DsbI form a specific dithiol oxidase system for periplasmic arylsulfate sulfotransferase in uropathogenic Escherichia coli. J. Mol. Biol. 380667-680. [DOI] [PubMed] [Google Scholar]
- 15.Guddat, L. W., J. C. A. Bardwell, R. Glockshuber, M. Huber-Wunderlich, T. Zander, and J. L. Martin. 1997. Structural analysis of three His32 mutants of DsbA: support for an electrostatic role of His32 in DsbA stability. Protein Sci. 61893-1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ha, U.-H., Y. Wang, and S. Jin. 2003. DsbA of Pseudomonas aeruginosa is essential for multiple virulence factors. Infect. Immun. 711590-1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayashi, S., M. Abe, M. Kimoto, S. Furukawa, and T. Nakazawa. 2000. The dsbA-dsbB disulfide bond formation system of Burkholderia cepacia is involved in the production of protease and alkaline phosphatase, motility, metal resistance, and multi-drug resistance. Microbiol. Immunol. 4441-50. [DOI] [PubMed] [Google Scholar]
- 18.Heras, B., M. Kurz, S. R. Shouldice, and J. L. Martin. 2007. The name's bond…disulfide bond. Curr. Opin. Struct. Biol. 17691-698. [DOI] [PubMed] [Google Scholar]
- 19.Heras, B., S. R. Shouldice, M. Totsika, M. J. Scanlon, M. A. Schembri, and J. L. Martin. 2009. DSB proteins and bacterial pathogenicity. Nat. Rev. Microbiol. 7215-225. [DOI] [PubMed] [Google Scholar]
- 20.Hung, D. L., S. D. Knight, R. M. Woods, J. S. Pinkner, and S. J. Hultgren. 1996. Molecular basis of two subfamilies of immunoglobulin-like chaperones. EMBO J. 153792-3805. [PMC free article] [PubMed] [Google Scholar]
- 21.Inaba, K., S. Murakami, M. Suzuki, A. Nakagawa, E. Yamashita, K. Okada, and K. Ito. 2006. Crystal structure of the DsbB-DsbA complex reveals a mechanism of disulfide bond generation. Cell 127789-801. [DOI] [PubMed] [Google Scholar]
- 22.Ito, K., and K. Inaba. 2008. The disulfide bond formation (Dsb) system. Curr. Opin. Struct. Biol. 18450-458. [DOI] [PubMed] [Google Scholar]
- 23.Jackson, M. W., and G. V. Plano. 1999. DsbA is required for stable expression of outer membrane protein YscC and for efficient Yop secretion in Yersinia pestis. J. Bacteriol. 1815126-5130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jacob-Dubuisson, F., J. Pinkner, Z. Xu, R. Striker, A. Padmanhaban, and S. J. Hultgren. 1994. PapD chaperone function in pilus biogenesis depends on oxidant and chaperone-like activities of DsbA. Proc. Natl. Acad. Sci. USA 9111552-11556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kadokura, H., F. Katzen, and J. Beckwith. 2003. Protein disulfide bond formation in prokaryotes. Annu. Rev. Biochem. 72111-135. [DOI] [PubMed] [Google Scholar]
- 26.Kang, J. W., Y. J. Jeong, A. R. Kwon, H. J. Yun, D. H. Kim, and E. C. Choi. 2001. Cloning, sequence analysis, and characterization of the astA gene encoding an arylsulfate sulfotransferase from Citrobacter freundii. Arch. Pharm. Res. 24316-322. [DOI] [PubMed] [Google Scholar]
- 27.Kang, J. W., A. R. Kwon, D. H. Kim, and E. C. Choi. 2001. Cloning and sequencing of the astA gene encoding arylsulfate sulfotransferase from Salmonella typhimurium. Biol. Pharm. Bull. 24570-574. [DOI] [PubMed] [Google Scholar]
- 28.Kim, B., Y. J. Hyun, K. S. Lee, K. Kobashi, and D. H. Kim. 2007. Cloning, expression and purification of arylsulfate sulfotransferase from Eubacterium A-44. Biol. Pharm. Bull. 3011-14. [DOI] [PubMed] [Google Scholar]
- 29.Kwon, A. R., and E. C. Choi. 2005. Role of disulfide bond of arylsulfate sulfotransferase in the catalytic activity. Arch. Pharm. Res. 28561-565. [DOI] [PubMed] [Google Scholar]
- 30.Kwon, A. R., T. G. Oh, D. H. Kim, and E. C. Choi. 1999. Molecular cloning of the arylsulfate sulfotransferase gene and characterization of its product from Enterobacter amnigenus AR-37. Protein Expr. Purif. 17366-372. [DOI] [PubMed] [Google Scholar]
- 31.Lane, M. C., C. J. Alteri, S. N. Smith, and H. L. T. Mobley. 2007. Expression of flagella is coincident with uropathogenic Escherichia coli ascension to the upper urinary tract. Proc. Natl. Acad. Sci. USA 10416669-16674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lloyd, A. L., D. A. Rasko, and H. L. T. Mobley. 2007. Defining genomic islands and uropathogen-specific genes in uropathogenic Escherichia coli. J. Bacteriol. 1893532-3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malojcic, G., R. L. Owen, J. P. A. Grimshaw, M. S. Brozzo, H. Dreher-Teo, and R. Glockshuber. 2008. A structural and biochemical basis for PAPS-independent sulfuryl transfer by aryl sulfotransferase from uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 10519217-19222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martin, J. L., J. C. A. Bardwell, and J. Kuriyan. 1993. Crystal structure of the DsbA protein required for disulphide bond formation in vivo. Nature 365464-468. [DOI] [PubMed] [Google Scholar]
- 35.Messens, J., and J.-F. Collet. 2006. Pathways of disulfide bond formation in Escherichia coli. Int. J. Biochem. Cell Biol. 381050-1062. [DOI] [PubMed] [Google Scholar]
- 36.Miki, T., N. Okada, and H. Danbara. 2004. Two periplasmic disulfide oxidoreductases, DsbA and SrgA, target outer membrane protein SpiA, a component of the Salmonella pathogenicity island 2 type III secretion Syst. J. Biol. Chem. 27934631-34642. [DOI] [PubMed] [Google Scholar]
- 37.Miki, T., N. Okada, Y. Kim, A. Abe, and H. Danbara. 2008. DsbA directs efficient expression of outer membrane secretin EscC of the enteropathogenic Escherichia coli type III secretion apparatus. Microb. Pathog. 44151-158. [DOI] [PubMed] [Google Scholar]
- 38.Mobley, H. L., D. M. Green, A. L. Trifillis, D. E. Johnson, G. R. Chippendale, C. V. Lockatell, B. D. Jones, and J. W. Warren. 1990. Pyelonephritogenic Escherichia coli and killing of cultured human renal proximal tubular epithelial cells: role of hemolysin in some strains. Infect. Immun. 581281-1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nilsson, L. M., O. Yakovenko, V. Tchesnokova, W. E. Thomas, M. A. Schembri, V. Vogel, P. Klemm, and E. V. Sokurenko. 2007. The cysteine bond in the Escherichia coli FimH adhesin is critical for adhesion under flow conditions. Mol. Microbiol. 651158-1169. [DOI] [PubMed] [Google Scholar]
- 40.Nuccio, S.-P., and A. J. Baumler. 2007. Evolution of the chaperone/usher assembly pathway: fimbrial classification goes Greek. Microbiol. Mol. Biol. Rev. 71551-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peek, J. A., and R. K. Taylor. 1992. Characterization of a periplasmic thiol:disulfide interchange protein required for the functional maturation of secreted virulence factors of Vibrio cholerae. Proc. Natl. Acad. Sci. USA 896210-6214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raczko, A. M., J. M. Bujnicki, M. Pawlowski, R. Godlewska, M. Lewandowska, and E. K. Jagusztyn-Krynicka. 2005. Characterization of new DsbB-like thiol-oxidoreductases of Campylobacter jejuni and Helicobacter pylori and classification of the DsbB family based on phylogenomic, structural and functional criteria. Microbiology 151219-231. [DOI] [PubMed] [Google Scholar]
- 43.Roos, V., G. C. Ulett, M. A. Schembri, and P. Klemm. 2006. The asymptomatic bacteriuria Escherichia coli strain 83972 outcompetes uropathogenic E. coli strains in human urine. Infect. Immun. 74615-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 45.Studier, F. W. 2005. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 41207-234. [DOI] [PubMed] [Google Scholar]
- 46.Thanassi, D. G., and S. J. Hultgren. 2000. Assembly of complex organelles: pilus biogenesis in Gram-negative bacteria as a model system. Methods 20111-126. [DOI] [PubMed] [Google Scholar]
- 47.Tinsley, C. R., R. Voulhoux, J.-L. Beretti, J. Tommassen, and X. Nassif. 2004. Three homologues, including two membrane-bound proteins, of the disulfide oxidoreductase DsbA in Neisseria meningitidis: effects on bacterial growth and biogenesis of functional type IV pili. J. Biol. Chem. 27927078-27087. [DOI] [PubMed] [Google Scholar]
- 48.Tomb, J. F. 1992. A periplasmic protein disulfide oxidoreductase is required for transformation of Haemophilus influenzae Rd. Proc. Natl. Acad. Sci. USA 8910252-10256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Watarai, M., T. Tobe, M. Yoshikawa, and C. Sasakawa. 1995. Disulfide oxidoreductase activity of Shigella flexneri is required for release of Ipa proteins and invasion of epithelial cells. Proc. Natl. Acad. Sci. USA 924927-4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Welch, R. A., V. Burland, G. Plunkett III, P. Redford, P. Roesch, D. Rasko, E. L. Buckles, S. R. Liou, A. Boutin, J. Hackett, D. Stroud, G. F. Mayhew, D. J. Rose, S. Zhou, D. C. Schwartz, N. T. Perna, H. L. Mobley, M. S. Donnenberg, F. R. Blattner, D. M. Green, A. L. Trifillis, D. E. Johnson, G. R. Chippendale, C. V. Lockatell, B. D. Jones, and J. W. Warren. 2002. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 9917020-17024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wullt, B., G. Bergsten, H. Connell, P. Rollano, N. Gebretsadik, R. Hull, and C. Svanborg. 2000. P fimbriae enhance the early establishment of Escherichia coli in the human urinary tract. Mol. Microbiol. 38456-464. [DOI] [PubMed] [Google Scholar]
- 52.Yu, J. 1998. Inactivation of DsbA, but not DsbC and DsbD, affects the intracellular survival and virulence of Shigella flexneri. Infect. Immun. 663909-3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zav'yalov, V. P., T. V. Chernovskaya, D. A. Chapman, A. V. Karlyshev, S. MacIntyre, A. V. Zavialov, A. M. Vasiliev, A. I. Denesyuk, G. A. Zav'yalova, I. V. Dudich, T. Korpela, and V. M. Abramov. 1997. Influence of the conserved disulphide bond, exposed to the putative binding pocket, on the structure and function of the immunoglobulin-like molecular chaperone Caf1M of Yersinia pestis. Biochem. J. 324571-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zav'yalov, V. P., G. A. Zav'yalova, A. I. Denesyuk, and T. Korpela. 1995. Modelling of steric structure of a periplasmic molecular chaperone Caf1M of Yersinia pestis, a prototype member of a subfamily with characteristic structural and functional features. FEMS Immunol. Med. Microbiol. 1119-24. [DOI] [PubMed] [Google Scholar]
- 55.Zhang, H.-Z., and M. S. Donnenberg. 1996. DsbA is required for stability of the type IV pilin of enteropathogenic Escherichia coli. Mol. Microbiol. 21787-797. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.