Abstract
In this work, the molecular basis of aerobic citrate utilization by the gram-positive bacterium Corynebacterium glutamicum was studied. Genome analysis revealed the presence of two putative citrate transport systems. The permease encoded by citH belongs to the citrate-Mg2+:H+/citrate-Ca2+:H+ symporter family, whereas the permease encoded by the tctCBA operon is a member of the tripartite tricarboxylate transporter family. The expression of citH or tctCBA in Escherichia coli enabled this species to utilize citrate aerobically, indicating that both CitH and TctABC are functional citrate transporters. Growth tests with the recombinant E. coli strains indicated that CitH is active with Ca2+ or Sr2+ but not with Mg2+ and that TctABC is active with Ca2+ or Mg2+ but not with Sr2+. We could subsequently show that, with 50 mM citrate as the sole carbon and energy source, the C. glutamicum wild type grew best when the minimal medium was supplemented with CaCl2 but that MgCl2 and SrCl2 also supported growth. Each of the two transporters alone was sufficient for growth on citrate. The expression of citH and tctCBA was activated by citrate in the growth medium, independent of the presence or absence of glucose. This activation was dependent on the two-component signal transduction system CitAB, composed of the sensor kinase CitA and the response regulator CitB. CitAB belongs to the CitAB/DcuSR family of two-component systems, whose members control the expression of genes that are involved in the transport and catabolism of tricarboxylates or dicarboxylates. C. glutamicum CitAB is the first member of this family studied in Actinobacteria.
Citrate is a ubiquitous natural compound which can be utilized as a carbon and energy source by many bacterial species. The anaerobic catabolism of citrate, which occurs, e.g., in enterobacteria like Klebsiella pneumoniae and Escherichia coli (7) and in lactic acid bacteria (14), usually involves the key enzyme citrate lyase (EC 4.1.3.6), which catalyzes the cleavage of citrate into acetate (the end product) and oxaloacetate (8, 47, 48). The subsequent catabolism of oxaloacetate can occur via different pathways, leading to, e.g., acetate and succinate as end products. Aerobic citrate utilization by bacteria possessing a complete tricarboxylic acid cycle usually requires only a citrate uptake system. Presently, at least five families of citrate transporters in bacteria have been characterized according to the classification system introduced by Saier (44): (i) the metabolite:H+ symporter family (transporter classification [TC] no. 2.A.1.6) within the major facilitator superfamily (TC no. 2.A.1), represented by CitH of K. pneumoniae (53, 54); (ii) the citrate-Mg2+:H+/citrate-Ca2+:H+ symporter (CitMHS) family (TC no. 2.A.11), represented by CitM and CitH of Bacillus subtilis (6, 30, 31); (iii) the citrate:cation symporter family (TC no. 2.A.24), represented by CitS and CitW of K. pneumoniae (24, 41); (iv) the divalent anion:Na+ symporter family (TC no. 2.A.47), represented by CitT of E. coli (42); and (v) the tripartite tricarboxylate transporter (TTT) family (TC no. 2.A.80), represented by TctABC of Salmonella enterica serovar Typhimurium (63). Whereas the transporters of the former four families consist of a single protein, the TctABC system is composed of three different subunits: two integral membrane proteins with presumably 12 (TctA) and 4 (TctB) transmembrane helices, plus a periplasmic citrate binding protein (TctC).
For most citrate transporter genes studied so far with respect to regulation, expression is induced in the presence of the substrate. In many bacteria, the transcription of genes for citrate uptake and catabolism is activated by two-component signal transduction systems (TCS) consisting of a membrane-bound histidine kinase which controls the phosphorylation status of a soluble response regulator and thereby its activity as a transcriptional regulator. Examples are the CitA-CitB TCS of K. pneumoniae and E. coli (9, 34) and the CitS-CitT TCS of B. subtilis (64), which belong to the CitAB/DcuSR family of TCS (23). The periplasmic domains of the CitA histidine kinases from K. pneumoniae and E. coli were shown to bind citrate with high specificities and affinities (between 0.1 and 10 μM at neutral pH) (22, 23). A previous mutagenesis study identified several amino acid residues critical for citrate binding, three of which are highly conserved within the CitAB/DcuSR subfamily (18). The direct involvement of these residues in citrate binding was confirmed by the first crystal structure of a periplasmic domain in a histidine kinase, i.e., that of K. pneumoniae CitA in a complex with citrate (43). More recently, a structural comparison between citrate-free and citrate-bound states of the periplasmic CitA domain became possible, revealing that ligand binding causes considerable contraction of the sensor domain. This contraction may represent the molecular switch that activates transmembrane signaling in the receptor (49).
Besides induction by citrate, the expression of citrate permease genes is sometimes also subject to carbon catabolite repression, which can at least partially override citrate induction, if other carbon sources are available. The expression of citH from B. subtilis is subject to control by the central regulator CcpA (56) and by a product of arginine metabolism (57). The expression of the tctCBA genes of S. enterica serovar Typhimurium and of citS from K. pneumoniae is controlled by the cyclic AMP receptor protein-cyclic AMP complex system (33, 62).
Whereas a wealth of information on citrate transporters and their transcriptional regulation in proteobacteria and gram-positive bacteria with low GC contents (Firmicutes) is available, not much is known about these systems in gram-positive bacteria with high GC contents (Actinobacteria). Here, we describe citrate utilization and its regulation in Corynebacterium glutamicum, a predominantly aerobic soil bacterium belonging to the Corynebacterium-Mycobacterium-Nocardia group of actinomycetes. This species is of interest due to its biotechnological importance as a producer of l-glutamate and l-lysine and its potential to serve as a nonpathogenic model organism for studying selected features common to corynebacteria and pathogenic mycobacteria. Overviews of C. glutamicum biology, genetics, physiology, and biotechnology can be found in two recent monographs (11, 15).
In an early study, Kimura (26) reported that C. glutamicum cannot grow on citrate unless the medium is supplemented with corn steep liquor. Later, Birnbaum and Demain (3) presented evidence that the function of corn steep liquor is to supply energy for the induction of a citrate transport system. They showed that resting cells of C. glutamicum pregrown on glucose with citrate take up citrate and convert it to extracellular glutamate but that cells pregrown on glucose without citrate do not or do so very slowly. Quite recently, we characterized citrate utilization in C. glutamicum by transcriptome and proteome analyses (39). In that study, it was shown that genes for two putative citrate uptake systems (citM, which has been renamed citH based on the studies presented here, and tctCBA) are strongly activated during growth on citrate and that several genes for the transport of alternative carbon sources, such as glucose, fructose, saccharose, and gluconate, show decreased mRNA levels in the presence of citrate. In addition, genes of the tricarboxylic acid cycle, of the respiratory chain, of F1Fo ATP synthase, and of gluconeogenesis show increased expression during growth on citrate (39).
In the present study, we have analyzed the molecular basis of citrate utilization by C. glutamicum in more detail. We provide evidence that both CitH (previously named CitM) and TctABC are in fact functional citrate transporters but with differing cation specificities. Citrate-dependent activation of citH and tctCBA was found to be dependent on the CitAB two-component system.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
The C. glutamicum wild-type strain ATCC 13032 and the ΔcitAB, ΔcitH, ΔtctCBA, and ΔcitH ΔtctCBA deletion mutants (Table 1) were cultivated aerobically on a rotary shaker (120 rpm) at 30°C in CGXII minimal medium (25) containing 30 mg/liter 3,4-dihydroxybenzoate as an iron chelator and carbon sources and salts as described in Results. For strain construction, plates of BHIS agar (brain heart infusion agar [Difco, Detroit, MI] with 0.5 M sorbitol) were used. Precultivation for growth experiments was usually performed in CGXII medium containing 50 mM sodium citrate, 50 mM glucose, and either 5 mM CaCl2 or 100 mM MgCl2. For cloning purposes, E. coli DHα (Bethesda Research Laboratories) was cultivated aerobically on a rotary shaker (120 rpm) at 37°C in Luria-Bertani medium (45). In order to test E. coli strains for aerobic growth on citrate as the sole carbon source, the strains were streaked onto Simmons citrate agar plates (Merck). For testing the cation specificity of the citrate transport systems CitH and TctABC, E. coli DH5α carrying either pXMJ19-citH or pXMJ19-tctCBA was cultivated in M9 minimal medium (45) supplemented with 1 g/liter Casamino Acids. For selection purposes, chloramphenicol (10 μg/ml for C. glutamicum and E. coli) and kanamycin (25 μg/ml for C. glutamicum and 50 μg/ml for E. coli) were used.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description and/or genotypea | Source or reference |
|---|---|---|
| C. glutamicum strains | ||
| ATCC 13032 | Biotin-auxotrophic wild-type strain | 1 |
| ATCC 13032 ΔcitAB | Derivative of ATCC 13032 with a citAB deletion | 27 |
| ATCC 13032 ΔcitH | Derivative of ATCC 13032 with an in-frame deletion of the citH gene | This work |
| ATCC 13032 ΔtctCBA | Derivative of ATCC 13032 with an in-frame deletion of the tctCBA operon | This work |
| ATCC 13032 ΔcitH ΔtctCBA | Derivative of ATCC 13032 with an in-frame deletion of the citH gene and in the tctCBA operon | This work |
| E. coli strains | ||
| MG1655 | Wild-type strain | 5 |
| DH5α | F− φ80dlacZΔM15 Δ(lacZYA-argF)U169 endA1 recA1 hsdR17(rK− mK+) deoR thi-1 phoA supE44 λ−gyrA96 relA1 | Invitrogen |
| BL21(DE3) | ompT hsdSB(rB− mB−) gal dcm (DE3) | Novagen |
| Plasmids | ||
| pXMJ19 | Cmr; C. glutamicum/E. coli shuttle vector for IPTG-inducible gene expression (pBL1 oriVC.g. pK18 oriVE.c. lacI Ptac) | 20 |
| pXMJ19-citAB | Cmr; pXMJ19 carrying the C. glutamicum citAB genes | This work |
| pXMJ19-citH | Cmr; pXMJ19 carrying the C. glutamicum citH gene | This work |
| pXMJ19-tctCBA | Cmr; pXMJ19 carrying the C. glutamicum tctCBA operon | This work |
| pET28b | Kmr; vector for overexpression of genes in E. coli under the control of a T7 promoter and for addition of an N-terminal or C-terminal hexahistidine tag to the synthesized protein (pBR322 oriVE.c. PT7lacI) | Novagen |
| pET28b-CgCitB-NHis6 | Kmr; pET28b derivative for overproduction of the C. glutamicum response regulator CitB with an N-terminal hexahistidine tag | This work |
| pK19mobsacB | Kmr; vector for allelic exchange in C. glutamicum (pK18 oriVE.c. sacB lacZα) | 46 |
| pK19mobsacB-ΔcitH | Kmr; pK19mobsacB derivative containing a crossover PCR product covering the regions up- and downstream of citH | This work |
| pK19mobsacB-ΔtctCBA | Kmr; pK19mobsacB derivative containing a crossover PCR product covering the region upstream of tctC and regions downstream of tctA | This work |
| pK19mobsacB-ΔcitAB | Kmr; pK19mobsacB derivative containing a crossover PCR product covering the region upstream of citA and regions downstream of citB | 27 |
oriVC.g., C. glutamicum oriV; oriVE.c., E. coli oriV.
Construction of C. glutamicum deletion mutants.
C. glutamicum mutants with in-frame deletions of citAB (ΔcitAB), citH (ΔcitH), tctCBA (ΔtctCBA), and citH and tctCBA (ΔcitH ΔtctCBA) were constructed via a two-step homologous recombination procedure as described previously (37). The regions up- and downstream (approximately 450 bp each) of the gene/operon to be deleted were amplified using pairs of oligonucleotides designated Delta-gene/operon-1 and Delta-gene/operon-2 and Delta-gene/operon-3 and Delta-gene/operon-4 (Table 2), respectively, and the products served as templates for crossover PCR with oligonucleotides designated Delta-gene/operon-1 and Delta-gene/operon-4. The resulting PCR product of ∼0.9 kb was digested with the restriction enzymes indicated in the primer table (Table 2) and cloned into pK19mobsacB cut with the same enzymes (46). DNA sequence analysis confirmed that the cloned PCR products did not contain spurious mutations. The transfer of the resulting plasmids, designated pK19mobsacB-Δgene/operon, into C. glutamicum and selection for the first and second recombination events were performed as described previously (37). Kanamycin-sensitive and saccharose-resistant clones were tested by colony PCR analysis with a primer pair designated Delta-gene/operon-out-fw and Delta-gene/operon-out-rv (Table 2). Clones which had the desired in-frame deletion of the gene/operon revealed a 1-kb fragment in which all nucleotides except for the first 6 codons and the last 12 codons were replaced by a 21-bp tag.
TABLE 2.
Oligonucleotides used in this study
| Oligonucleotide | Sequence (5′→3′)a | Restriction endonuclease |
|---|---|---|
| citAB-OE-fw-SalI | TAT AGT CGA CCT TCA CGC TCT GCT GAT AAT CGC | SalI |
| citAB-OE-rv-EcoRI | TAT AGA ATT CAT GCT GAC CGA GGT GGA CAT CC | EcoRI |
| citH-OE-fw-PstI | TAT ACT GCA GCA GAA CAC TCG GGA TCT CAA AGT TTC G | PstI |
| citH-OE-rv-EcoRI | TAT AGA ATT CGG GGT TTG GCG TCG AAA AGC | EcoRI |
| tct-OE-fw | TAT ATC TAG ACC CAA GCA CTT AGG CAT CAA ACA TTC | XbaI |
| tct-OE-rv | TAT ACC CGG GAT GGT CTC TAA GCG ACT TCG AGG | XmaI |
| cg-citB-for | CGC CGC CAT ATG GAT CAA ACA CTT AAA GTT TTA GTA | NdeI |
| cg-citB-rev | ATA ATA CTC GAG CTA GAG CAG TGG CTT TGA ATA | XhoI |
| citH-PE-1 | GTG GCT GAG CGT GTA GTC AGC GGC TAA TGC CG | |
| citH-PE-2 | CTT TCT GTG TTT CTC GAA ACT TTG AGA TCC CGA G | |
| citH-PE-4 | TAA TGC CGT GAC TTG TCC TGA AG | |
| tct-PE-2 | TGC GGG TTC TGC TCC TAC TTC GGC C | |
| tct-PE-3 | AAT CCC TCT CGG TCG GAC TTT TG | |
| citAB-PE-1 | AAA TAG CTA CCA CCA ACG CGA CGG TAG CCA C | |
| citAB-PE-2 | TCC ATC ATC AAA ACT GCG AAA ATT CCG GTG C | |
| Delta-citAB-1 | TAT ATC TAG AGA GCG TGT AGT CAG CGG CTA ATG | XbaI |
| Delta-citAB-2 | CCC ATC CAC TAA ACT TAA ACA CAA AAC TGC GAA AAT TCC GGT GC | |
| Delta-citAB-3 | TGT TTA AGT TTA GTG GAT GGG GGG CGA CCA GAA CAT CTA TAT TC | |
| Delta-citAB-4 | ATA TGA ATT CAC CAT GTG GTG TTG TGC GTA CC | EcoRI |
| Delta-citAB-out-fw | GAC TAC CAG GAT GCC ATC TGA G | |
| Delta-citAB-out-rv | TAT ATC CAA CCT GCA CCA AGT AC | |
| Delta-citM-1 | TAT ACT GCA GAT GAA GAT AAG CCC GAC TCC CC | XbaI |
| Delta-citM-2 | CCC ATC CAC TAA ACT TAA ACA GTA GCC CTC TTC ATC GGC GTC G | |
| Delta-citM-3 | TGT TTA AGT TTA GTG GAT GGG GAG GAT CAT GGC GAA GCC AAG | |
| Delta-citM-4 | ATA TGA ATT CAC CAT GTG GTG TTG TGC GTA CC | EcoRI |
| Delta-citM-out-fw | CGC CCG CAT ATG AAC CAA TAG C | |
| Delta-citM-out-rv | TGA ATC ACC AGT ATT CGG GTA GC | |
| Delta-tct-1 | TAT ATC TAG ACG AGC GCT ACG TGA CTT CTT TGA | XbaI |
| Delta-tct-2 | CCC ATC CAC TAA ACT TAA ACA TGC TCC TAC TTC GGC CAT TTT GGG GG | |
| Delta-tct-3 | TGT TTA AGT TTA GTG GAT GGG AAG CAC CTG ACT TCT CAG CTC GAA ACC | |
| Delta-tct-4 | TAT ACC CGG GGG TTT AGC AAG CGG GAC GCT TTC G | XmaI |
| Delta-tct-out-fw | GGG TTT GAT TTT GAG GTG ACA ACG G | |
| Delta-tct-out-rv | TTC CTA GGC TGC CCG CTT GGG CT | |
| P_citH_fw | TTT CCA GTC GGA TCC ACC AAC AG | |
| P_citH_rv | GTT TCT CGA AAC TTT GAG ATC CCG | |
| P_citH_control_fw | CGG GAT CTC AAA GTT TCG AGA AAC | |
| P_citH_control_rv | CGA AGA TGG TGG GGA CCA ACA G | |
| P_tct_fw | GAC AAC AAG GGG GAG TCC TAG C | |
| P_tct_rv | GGT TCT GCT CCT ACT TCG GCC | |
| P_cg2836_control_fw | GGT TAG CGC GCC GCC CTG TC | |
| P_cg2836_control_rv | CGA CTT CTC AAA GGC GAC TTC AC |
In some cases, oligonucleotides were designed to introduce recognition sites for restriction endonucleases (recognition sites are underlined, and the corresponding restriction endonucleases are listed) and complementary 21-mer sequences for generating crossover PCR products (indicated by italics) into the resulting PCR products.
Plasmid construction.
For the construction of pXMJ19-citAB, the entire citAB operon was amplified using oligonucleotides citAB-OE-fw-SalI and citAB-OE-rv-EcoRI. For the construction of pXMJ19-citH and pXMJ19-tctCBA, the citH gene and the tctCBA operon were amplified using the oligonucleotide pairs citH-OE-fw-PstI/citH-OE-rv-EcoRI and tct-OE-fw/tct-OE-rv, respectively. Since the plasmid pXMJ19 does not carry a ribosomal binding site, regions of 81, 52, and 70 bp upstream of citA, citH, and tctC, respectively, were also amplified with these primers. The PCR products were digested using the appropriate restriction enzymes (Table 2) and cloned into pXMJ19 (20). For the overproduction and purification of CitB with an amino-terminal histidine tag, the citB coding region was amplified using oligonucleotides that introduce an NdeI restriction site including the start codon (cg-citB-for) and an XhoI restriction site after the stop codon (cg-citB-rev). The purified 698-bp PCR product was digested with NdeI and XhoI and cloned into the expression vector pET28b cut with the same enzymes, resulting in plasmid pET28b-CgCitB-NHis6. The CitB protein encoded by this plasmid (238 amino acids; 25.6 kDa) contains a stretch of 20 additional amino acids (MGSSHHHHHHSSGLVPRGSH) at the amino terminus, including a hexahistidine tag and a thrombin cleavage site.
Overproduction and purification of CitB.
E. coli BL21(DE3) carrying plasmid pET28b-CgCitB-NHis6 was grown at 30°C in 500 ml of Luria-Bertani medium with 50 μg/ml kanamycin to an optical density at 600 nm (OD600) of ∼1 before the addition of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). After cultivation for another 3 h, cells were harvested by centrifugation, washed once, and stored at −20°C. For cell extract preparation, thawed cells were resuspended in 10 ml of TNIG5 (20 mM Tris-HCl, 300 mM NaCl, 5 mM imidazole, 5% [vol/vol] glycerol, pH 7.9). After the addition of 1 mM diisopropylfluorophosphate and 1 mM phenylmethylsulfonyl fluoride, the cells were disrupted using a French pressure cell (SLM Aminco; Spectronic Instruments, Rochester, NY). Intact cells and cell debris were removed by centrifugation (20 min at 5,000 × g and 4°C), and the cell extract was subjected to ultracentrifugation (1 h at 150,000 × g and 4°C). CitB, present in the supernatant from the ultracentrifugation step, was purified by Ni2+-chelate affinity chromatography using His-Bind resin (Novagen; Merck KGaA, Darmstadt, Germany). After the column was washed with 10 bed volumes of TNIG20 buffer and 10 bed volumes of TNIG100 buffer (the same as TNIG5 but containing 20 and 100 mM imidazole, respectively), CitB protein was eluted with TNIG400 buffer (containing 400 mM imidazole). Fractions containing CitB were pooled, and the elution buffer was exchanged for BS buffer (50 mM Tris-HCl, 50 mM KCl, 10 mM MgCl2, 0.5 mM EDTA, 10% [vol/vol] glycerol, pH 7.5) by gel filtration using PD-10 columns (Amersham Biosciences). Protein concentrations were determined with the bicinchoninic acid protein assay kit (Pierce, Rockford, IL) using bovine serum albumin as the standard. The purity of the protein preparations was assessed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and subsequent protein detection with Gel Code blue staining reagent (Pierce, Rockford, IL).
Electrophoretic mobility shift assays (EMSAs).
For testing the binding of CitB to putative target promoters, purified CitB protein (0 to 1.4 μM) was mixed with 100-ng DNA fragments (219 to 336 bp; final concentrations, 22 to 35 nM) in a total volume of 20 μl. The binding buffer contained 50 mM Tris-HCl (pH 7.5), 50 mM KCl, 10 mM MgCl2, 0.5 mM EDTA, and 10% (vol/vol) glycerol. The reaction mixture was incubated at room temperature for 30 min and then loaded onto a native 15% polyacrylamide gel. Electrophoresis and DNA detection with Sybr green I were performed as described before (59). The DNA fragments were generated by PCR, purified with a PCR purification kit (Qiagen, Hilden, Germany), and eluted in EB buffer (10 mM Tris-HCl, pH 8.5).
Analysis of glucose and citrate.
The quantitation of glucose and citrate in the supernatants of cultures was performed by high-performance liquid chromatography (HPLC) using a Merck Hitachi HPLC system equipped with an Aminex HPX-87H column (300 by 7.8 mm; Bio-Rad). Isocratic elution was performed with 6 mM H2SO4 at 80°C for 20 min at a flow rate of 0.6 ml/min. Citric acid was detected with a LaChrom L-7400 UV detector (Merck Hitachi) at 215 nm. Glucose was detected with a LaChrom L-7490 refractive index detector (Merck Hitachi). Quantification was performed using calibration curves obtained with external standards.
Primer extension analysis.
Nonradioactive primer extension analysis was performed as described previously (16). Each transcriptional start site was determined using at least two different digoxigenin-labeled oligonucleotides: citH-PE-1, citH-PE-2, and citH-PE-4 for citH, tct-PE-2 and tct-PE-3 for tctCBA, and citAB-PE-1 and citAB-PE-2 for citAB (Table 2).
DNA microarray analysis.
The preparation of RNA and the synthesis of fluorescently labeled cDNA were carried out as described previously (35). Custom-made DNA microarrays printed with 70-mer oligonucleotides for C. glutamicum ATCC 13032 were obtained from Operon (Cologne, Germany) and are based on the genome sequence entry with accession no. NC_006958 (21). Hybridization and stringent washing of the microarrays were performed according to the instructions of the supplier. Hybridization was carried out for 16 to 18 h at 42°C by using a microarray user interface hybridization system (BioMicro Systems, Salt Lake City, UT). After being washed, the microarrays were dried by centrifugation (5 min at 1,600 × g) and the levels of fluorescence at 532 nm (for Cy3-dUTP) and 635 nm (for Cy5-dUTP) were determined with 10-μm resolution by using a GenePix 6000 laser scanner (Axon Instruments, Sunnyvale, CA). Quantitative image analysis was carried out using GenePix image analysis software (GenePix Pro 6.0; Axon Instruments), and the results were saved as GenePix results files. For data normalization, GenePix results files were processed using the Bioconductor/R packages limma and marray (http://www.bioconductor.org). Processed and normalized data, as well as experimental details (minimum information about a microarray experiment) (10), were stored in the in-house microarray database for further analysis (40).
RESULTS
Use of E. coli to test whether the C. glutamicum genes citH and tctCBA encode functional citrate transporters.
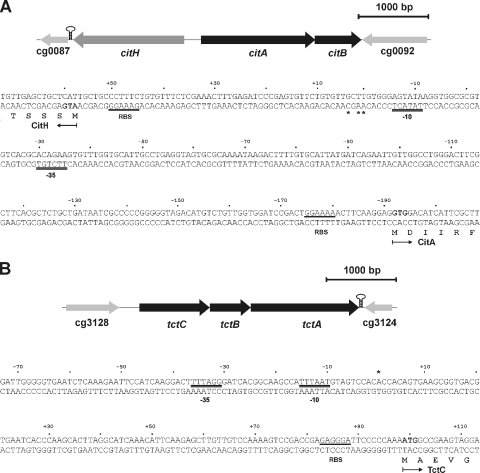
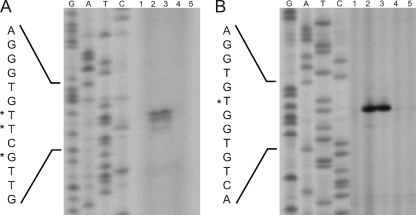
Our previous study on citrate utilization by C. glutamicum had shown that the genes for two putative citrate transport systems, citH (cg0088; previously designated citM) and tctCBA (cg3127, cg3126, and cg3125), were the ones most strongly induced in the presence of citrate in the medium (39). CitH belongs to the CitMHS family (TC no. 2.A.11), whereas TctABC is a member of the TTT family (TC no. 2.A.80) (63). The CitH protein shows 42 and 43% sequence identity to the B. subtilis citrate transporters CitM and CitH (6, 30). The first of several putative start codons for citH (Fig. 1A) leads to a protein consisting of 526 amino acid residues. The tctCBA genes (Fig. 1B) presumably form an operon, since the last nucleotides of the tctC and tctB stop codons correspond to the first nucleotides of the start codons of tctB and tctA, respectively. Downstream of tctA, a Rho-independent transcriptional terminator structure (ΔG° = −128.5 kJ/mol) was identified. TctA (510 amino acid residues) and TctB (188 residues) are integral membrane proteins with 12 and 4 predicted transmembrane helices, respectively. TctC (334 residues) is presumably a periplasmic citrate binding protein that contains a putative transmembrane helix (residues 19 to 39) in the N-terminal region. This transmembrane helix may either tether the protein to the cytoplasmic membrane or be cleaved off by signal peptidase. TctA, TctB, and TctC from C. glutamicum show 38, 29, and 27% sequence identity to the corresponding proteins from S. enterica serovar Typhimurium, the first and most carefully studied members of the TTT transporter family (50-52, 60-62).
FIG. 1.
Physical map of the cit locus of C. glutamicum and DNA sequence of the citA-citH intergenic region (A) and physical map of the tct locus and DNA sequence of the tctC promoter region (B). Transcriptional start sites as determined by primer extension analyses are indicated by asterisks. For citH, three transcriptional start sites were identified. The one resulting in the longest transcript and having the most intense signal was set as +1. The derived −10 and −35 regions of the promoters are indicated by double lines. The proposed start codons of citH, citA, and tctC are indicated in bold, and the putative ribosome binding sites (RBS) are underlined. The proposed N-terminal amino acid residues of CitH, CitA, and TctC are also shown. Downstream of citH and tctA, Rho-independent transcriptional terminator structures are indicated by stem-loop structures.
In order to test whether the C. glutamicum CitH and TctABC proteins are in fact citrate transporters, the corresponding genes were cloned and expressed in the heterologous host E. coli MG1655. This strain does not synthesize a citrate transporter under aerobic conditions and consequently cannot grow on citrate. It is therefore a suitable host to test the functions of putative citrate transporters (42). The recombinant strains were tested for their abilities to utilize citrate as the sole carbon source under aerobic conditions by being streaked onto Simmons citrate agar supplemented with 10 μg/ml chloramphenicol. The formation of colonies and a color change of the pH indicator bromthymol blue from green to blue indicated that MG1655/pXMJ19-citH and MG1655/pXMJ19-tctCBA, but not the control strain, MG1655/pXMJ19, were able to utilize citrate (data not shown). The color change is observed only with cells that are able to metabolize citrate, which leads to the alkalinization of the medium. These results clearly indicate that both CitH and the TctABC complex of C. glutamicum are functional citrate transporters.
Cation dependency of CitH and TctABC tested in the E. coli system.
CitH belongs to the CitMHS family of transporters. Representatives of this family transport citrate in symport with different cations, such as Mg2+ (CitM of B. subtilis), Ca2+ (CitH of B. subtilis), and Fe3+ (CitH of Streptococcus mutans) (4, 29, 30). Since the metal ion specificity cannot be deduced from sequence comparisons, the identity of the metal ion cotransported by C. glutamicum CitH is not yet known. The second citrate transport system of C. glutamicum, TctABC, belongs to the TTT family (63), for which the cation dependency is also not yet clear. Thus, further studies to clarify the cation dependency of the two C. glutamicum citrate transporters were performed.
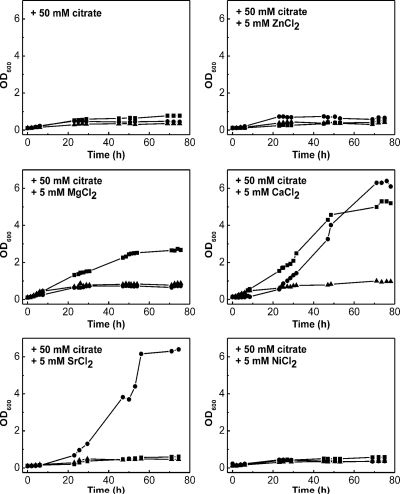
The growth of the above-mentioned recombinant E. coli MG1655 strains in M9 minimal medium supplemented with 1 g/liter Casamino Acids in order to allow some basic growth and 50 mM sodium citrate as the major carbon source was monitored under aerobic conditions. The medium was supplemented with a 5 mM concentration of either CaCl2, MgCl2, NiCl2, SrCl2, or ZnCl2. As shown in Fig. 2, MG1655/pXMJ19-citH can grow on citrate in the presence of Ca2+ and Sr2+ but not in the presence of Mg2+, Ni2+, or Zn2+. On the other hand, MG1655/pXMJ19-tctCBA can grow on citrate in the presence of Ca2+ and Mg2+ but not in the presence of Sr2+, Ni2+, or Zn2+. The control strain MG1655/pXMJ19 did not grow on citrate under any tested conditions. These results indicate that both CitH and TctABC of C. glutamicum transport citrate in a complex with Ca2+. CitH can also transport an Sr2+-citrate complex, and TctABC can also transport an Mg2+-citrate complex. It cannot be ruled out that the two transporters are able to take up citrate in a complex with Ni2+, as the concentration used also did not allow growth with 50 mM glucose and thus is toxic for the cells (data not shown). In contrast, growth with 50 mM glucose and 5 mM Zn2+ was possible, indicating that nongrowth with citrate was due to the situation that neither CitH nor TctABC is active with Zn2+.
FIG. 2.
Testing the cation dependency of the citrate transporters CitH and TctABC of C. glutamicum by using the growth of E. coli on citrate as the indicator. E. coli MG1655 transformed with pXMJ19-citH (•), pXMJ19-tctCBA (▪), or pXMJ19 (▴) was grown in a 500-ml Erlenmeyer flask containing 50 ml of M9 minimal medium supplemented with 1 g/liter Casamino Acids, 50 mM sodium citrate, and none or one of the following salts: 5 mM CaCl2, 5 mM MgCl2, 5 mM ZnCl2, 5 mM NiCl2, and 5 mM SrCl2. The cultures were incubated at 37°C on a rotary shaker at 120 rpm. The growth curves shown here are representative of those from three independent growth experiments with comparable results.
Cation dependency of the growth of the C. glutamicum wild type on citrate as the sole carbon and energy source.
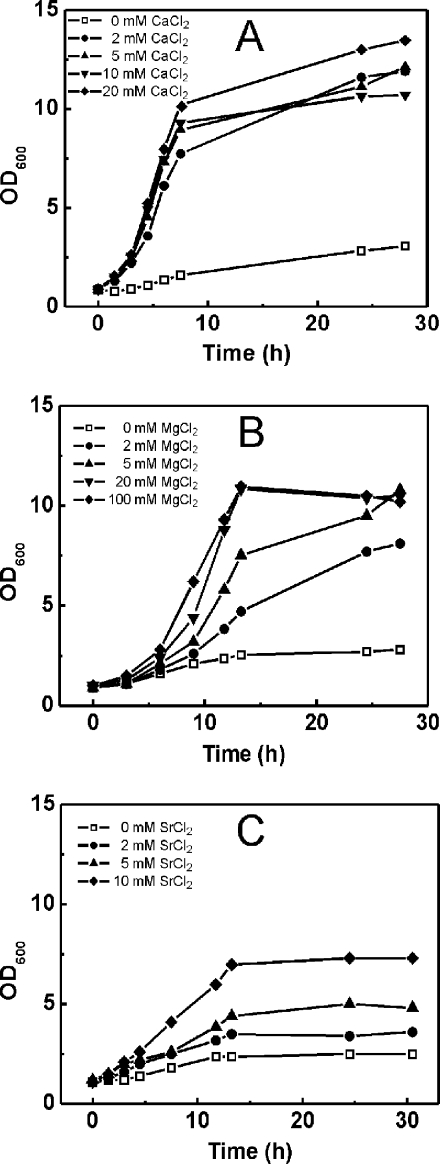
Based on the fact that C. glutamicum possesses two functional citrate transporters, we tested whether the organism is able to grow on citrate as the sole carbon and energy source. In initial studies, wild-type cells pregrown in standard CGXII minimal medium with 50 mM glucose and 50 mM sodium citrate showed only minimal growth (from an OD600 of 0.9 to an OD600 of 3) when they were subsequently cultivated in standard CGXII minimal medium containing 50 mM sodium citrate as the sole carbon source (Fig. 3). Standard CGXII minimal medium contains 0.09 mM Ca2+ and 1 mM Mg2+, indicating that the poor growth may be due to insufficient concentrations of the cations that were found to be required for citrate utilization via CitH and TctABC in E. coli. Therefore, we supplemented the standard CGXII medium with additional CaCl2, MgCl2, or SrCl2. As shown in Fig. 3A, C. glutamicum grew at a rate of 0.31 h−1 to an OD600 of about 12 on citrate as the sole carbon source with 2 mM CaCl2. Increased CaCl2 concentrations up to 20 mM did not further stimulate either the growth rate or the growth yield. Even higher Ca2+ concentrations led to the precipitation of salt, presumably as calcium phosphate. Growth on citrate was accompanied by an increase of the pH of the medium from an initial pH of 7 to 8.5 to 8.7 at the end of growth (data not shown). Growth behavior in the presence of MgCl2 was different, as the maximal growth rate (0.20 h−1) was obtained only with 100 mM (Fig. 3B) and then decreased with even higher concentrations (data not shown). In the case of SrCl2, the maximal growth rate (0.14 h−1) and cell yield were obtained with 10 mM (Fig. 3C). Higher concentrations of dissolved SrCl2 could not be achieved. In the subsequent growth experiments with C. glutamicum on citrate, the medium was usually supplemented with 5 mM CaCl2.
FIG. 3.
Influence of Ca2+ (A), Mg2+ (B), or Sr2+ (C) on the growth of the C. glutamicum wild type with citrate as the sole carbon and energy source. Cells pregrown in CGXII minimal medium with 50 mM glucose, 50 mM citrate, and 20 mM CaCl2, 50 mM MgCl2, or 5 mM SrCl2, respectively, were used to inoculate 50 ml of CGXII medium (containing 0.09 mM CaCl2, 1 mM MgSO4, and 0 mM SrCl2) containing 50 mM citrate as the sole carbon and energy source. The cultures were incubated at 30°C on a rotary shaker at 120 rpm. The growth curves shown here are representative of those from three independent growth experiments with comparable results.
Testing the roles and cation specificities of CitH and TctABC for the growth of C. glutamicum on citrate.
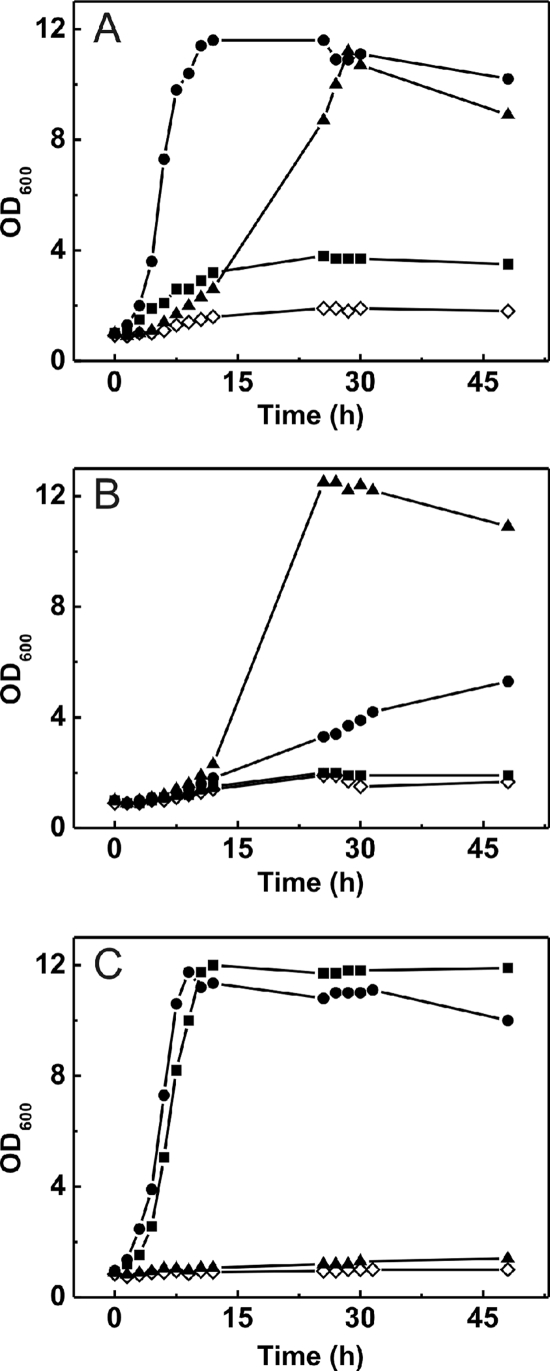
Since C. glutamicum possesses two functional citrate transporters, the question arises of whether both are required for growth on citrate or if each one alone is sufficient for growth. Moreover, the cation specificities determined for the two transporters in E. coli should also be tested in C. glutamicum. For this purpose, three deletion mutants of C. glutamicum, the ΔcitH, ΔtctCBA, and ΔcitH ΔtctCBA strains, were constructed as described in Materials and Methods. The three strains and, for comparison, the wild type were cultivated in CGXII minimal medium with 50 mM citrate as the sole carbon and energy source, with and without the addition of 5 mM CaCl2, MgCl2, or SrCl2. In the case of MgCl2 and SrCl2, the concentration of 5 mM is below the concentration required for maximal growth but still sufficient to allow growth.
As shown in Fig. 4A, the wild type grew best in the presence of Ca2+, followed by Mg2+ and Sr2+. In the presence of Sr2+, the wild type reached only about one-third of the OD600 obtained with calcium or magnesium ions (a final OD600 of 4 compared to a final OD600 of 12). The ΔcitH mutant grew well in the presence of magnesium ions, worse with additional calcium ions, and not at all with added strontium ions (Fig. 4B). The ΔtctCBA mutant grew equally well in the presence of calcium and strontium ions but did not grow in the presence of magnesium ions (Fig. 4C). This behavior reflects the cation specificity determined for the TctCBA system (Ca2+ and Mg2+) and CitH (Ca2+ and Sr2+) in E. coli. A remarkable result becomes obvious by a comparison of Fig. 4A and C. In the absence of TctABC, Sr2+ allows growth on citrate at the same rate and to the same final OD600 as Ca2+ (Fig. 4C). In the presence of TctABC, however, both the growth rate and the final OD600 obtained with Sr2+ are much lower than those obtained with Ca2+ (Fig. 4A). This result indicates that the simultaneous presence of TctABC and Sr2+ ions inhibits growth on citrate in a yet unknown manner. The much better growth of the ΔcitH strain with Mg2+ than with Ca2+ may indicate that Mg2+ is the preferred cation for citrate transport by the TctCBA system.
FIG. 4.
Cation dependency of the growth of the C. glutamicum wild type (A), the ΔcitH strain (B), and the ΔtctCBA strain (C) in CGXII medium with 50 mM citrate as the sole carbon and energy source. The medium contained either no additional salts (⋄) or 5 mM CaCl2 (•), 5 mM MgCl2 (▴), or 5 mM SrCl2 (▪). Cells were pregrown in CGXII minimal medium with 50 mM glucose and 50 mM citrate. The cultures were incubated at 30°C on a rotary shaker at 120 rpm. The growth curves shown here are representative of those from three independent growth experiments with comparable results.
The ΔcitH ΔtctCBA double mutant did not grow on citrate under any tested conditions during the first 10 h of incubation; however, in a few cases, growth up to an OD600 of 10 to 12 after 20 to 30 h was observed (data not shown).
The growth of C. glutamicum on citrate is not subject to catabolite repression.
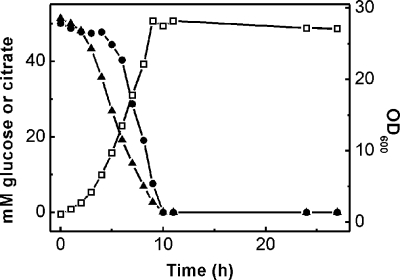
Previous studies (3, 19, 55) had indicated that citrate and glucose are catabolized simultaneously by C. glutamicum. To confirm this hypothesis, we monitored the consumption of the substrates by a culture growing with 50 mM citrate and 50 mM glucose (supplemented with 5 mM CaCl2). As shown in Fig. 5, the substrates were consumed simultaneously, indicating that the genes required for citrate utilization are not subject to catabolite repression. Interestingly, citrate consumption started before glucose consumption, indicating that citrate initially had some inhibitory effect on glucose utilization. Nevertheless, the two substrates were consumed after the same time period of about 10 h.
FIG. 5.
Simultaneous consumption of carbon sources during the growth (□) of C. glutamicum on citrate and glucose. The wild type was grown in CGXII medium containing 50 mM glucose, 50 mM sodium citrate, and 5 mM CaCl2. Cells were precultivated in CGXII minimal medium with 50 mM glucose and 50 mM citrate. The consumption of citrate (▴) and glucose (•) was determined via HPLC as described in Materials and Methods. The curves shown here are representative of those from three independent growth experiments with comparable results.
The CitAB two-component system is required for citrate utilization by C. glutamicum.
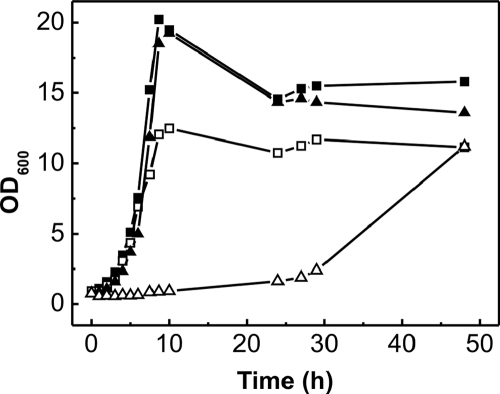
C. glutamicum possesses genes for 13 TCS (27). One of these TCS, encoded by the citAB genes, belongs to the CitAB/DcuSR subfamily (23). The C. glutamicum sensor kinase CitA (551 amino acid residues) contains two putative transmembrane helices separated by an extracytoplasmic domain extending from residues 48 to 188. It displays 24 and 29% overall sequence identity to the CitA histidine kinases from K. pneumoniae and E. coli, respectively, which were previously shown to function as citrate receptors (22, 23). The C. glutamicum response regulator CitB (218 amino acid residues) shows 32 and 35% sequence identity to the CitB proteins from K. pneumoniae and E. coli, respectively. The affiliation with the CitAB/DcuSR subfamily and the fact that the citAB genes are located immediately upstream of and divergent from the citH gene (Fig. 1A) suggested an involvement of the C. glutamicum CitAB TCS in the regulation of citrate uptake genes. This suggestion was confirmed by the fact that a C. glutamicum mutant lacking the citAB genes (the ΔcitAB strain) was unable to grow with citrate as the sole carbon and energy source within the first 20 h of cultivation but grew like the wild type on glucose (Fig. 6) or pyruvate (data not shown). The growth defect could be abolished by the transformation of the deletion mutant with the citAB expression plasmid pXMJ19-citAB (data not shown). Interestingly, the growth of the ΔcitAB mutant on citrate occurred after prolonged incubation (about 30 h in the experiment with results shown in Fig. 6). When such cultures were used to inoculate fresh citrate minimal medium, cells grew either immediately or with a shortened lag phase, indicating that a suppressor mutation had presumably allowed the expression of either citH or tctCBA.
FIG. 6.
Growth of the C. glutamicum wild type (squares) and the mutant strain ΔcitAB (triangles) in CGXII minimal medium containing either 50 mM glucose (filled symbols) or 50 mM citrate (hollow symbols) as the sole carbon and energy source. Cells were pregrown in CGXII minimal medium with 50 mM glucose and 50 mM citrate. The medium was supplemented with 5 mM CaCl2. The cultures were incubated at 30°C on a rotary shaker at 120 rpm. The growth curves shown here are representative of those from three independent growth experiments with comparable results.
Transcriptome comparisons reveal that CitAB is responsible for the activation of citH and tctCBA.
To analyze the influence of the citAB deletion on genomewide gene expression in the presence of citrate, the transcriptomes of the ΔcitAB mutant and the wild type were compared using DNA microarrays. For this purpose, the cells were grown in CGXII minimal medium with 50 mM citrate, 50 mM glucose, and 50 mM MgCl2. Four sets of comparisons, each starting with independent cultures, were made. Sixty genes showed alterations in mRNA levels of more than threefold in at least three of four experiments (P ≤ 0.05). Thirty-six of these genes showed decreases and 29 genes showed increases in mRNA levels in the ΔcitAB mutant compared to those in the wild type. As shown in Table 3, the functions of the encoded proteins are highly diverse, and only those genes showing the most drastic changes in mRNA levels will be dealt with here. The citH and tctCBA mRNA levels in the ΔcitAB mutant were 25- to 100-fold lower than those in the wild type, indicating that the CitAB TCS is responsible for the activation of these genes. Only two other genes, namely, cg2380 and ssuC, showed mRNA levels that were changed to the same extent and in the same manner (Table 3). The former gene encodes a protein of 14,084 Da that contains a single transmembrane helix extending from amino acid residue 65 to 85, with the N-terminal region located extracytoplasmically. Homologs are present in many other corynebacteria, mycobacteria, and other genera of the actinomycetes; however, the function is still unknown. The ssuC gene is part of the ssuD1CBA operon encoding a sulfonatase (ssuD1) and an ABC transporter for aliphatic sulfonates (28). The other genes of this operon, as well as further genes involved in sulfonate metabolism (ssuD2, ssuI, and seuA), also showed altered mRNA levels (Table 3). The reason for the changed expression patterns of these genes in the comparison of the ΔcitAB mutant and the wild type is unclear. In contrast to the genes mentioned above, cg3195, encoding a flavin-containing monooxygenase, showed a 27-fold increase in the mRNA level in the ΔcitAB mutant compared to that in the wild type. Again, an obvious reason for this change is not apparent.
TABLE 3.
Genes with ≥3-fold change in mRNA levelsa
| cg designation | NCgl designation | Gene name | Description of (putative) product | Avg mRNA ratio (ΔcitAB strain mRNA/wt mRNA) |
|---|---|---|---|---|
| cg0089 | NCgl0067 | citA | Sensor histidine kinase | 0.00 |
| cg0090 | NCgl0068 | citB | Response regulator | 0.00 |
| cg3125 | NCgl2724 | tctA | Tricarboxylate transport membrane protein | 0.04 |
| cg3126 | NCgl2725 | tctB | Tricarboxylate transport membrane protein | 0.01 |
| cg3127 | NCgl2726 | tctC | Tricarboxylate binding protein | 0.02 |
| cg2380 | NCgl2088 | 14-kDa membrane protein | 0.03 | |
| cg0088 | NCgl0066 | citH | Citrate transporter | 0.04 |
| cg1376 | NCgl1173 | ssuD1 | Sulfonatase (alkanesulfonate monooxygenase) | 0.10 |
| cg1377 | NCgl1174 | ssuC | Aliphatic sulfonate transmembrane ABC transporter protein | 0.04 |
| cg1379 | NCgl1175 | ssuB | Aliphatic sulfonate ATP binding ABC transporter protein | 0.10 |
| cg1380 | NCgl1176 | ssuA | Aliphatic sulfonate binding protein | 0.20 |
| cg3226 | NCgl2816 | l-Lactate permease | 0.05 | |
| cg3227 | NCgl2817 | lldD | Quinone-dependent l-lactate dehydrogenase | 0.15 |
| cg3308 | NCgl2881 | rpsF | 30S ribosomal protein S6 | 0.09 |
| cg0810 | NCgl0676 | Hypothetical protein | 0.09 | |
| cg1628 | NCgl1383 | Hydrolase of the α/β superfamily | 0.12 | |
| cg1581 | NCgl1341 | argJ | Bifunctional ornithine acetyltransferase/N-acetylglutamate synthase protein | 0.14 |
| cg1582 | NCgl1342 | argB | N-Acetylglutamate kinase | 0.18 |
| cg1583 | NCgl1343 | argD | Acetylornithine aminotransferase | 0.27 |
| cg3026 | NCgl2635 | mrpD | Subunit D of multisubunit Na+/H+ antiporter | 0.19 |
| cg3027 | NCgl2636 | mrpE | Subunit E of multisubunit Na+/H+ antiporter | 0.15 |
| cg3028 | NCgl2637 | mrpF | Subunit F of multisubunit Na+/H+ antiporter | 0.24 |
| cg3029 | NCgl2638 | mrpG | Subunit G of multisubunit Na+/H+ antiporter | 0.23 |
| cg1156 | NCgl0975 | ssuD2 | Monooxygenase for sulfonate utilization | 0.19 |
| cg1451 | NCgl1235 | serA | Phosphoglycerate dehydrogenase | 0.22 |
| cg0576 | NCgl0471 | rpoB | Subunit β of DNA-directed RNA polymerase | 0.23 |
| cg2894 | NCgl2523 | Drug resistance-related transcriptional repressor | 0.24 | |
| cg1364 | NCgl1161 | atpF | Subunit B of ATP synthase | 0.26 |
| cg2157 | NCgl1892 | terC | Tellurium resistance membrane protein | 0.30 |
| cg1151 | NCgl0972 | seuA | Monooxygenase for sulfonate ester utilization | 0.30 |
| cg1386 | NCgl1182 | fixA | Subunit β of putative electron transfer flavoprotein | 0.30 |
| cg0653 | NCgl0538 | rpsK | 30S ribosomal protein S11 | 0.31 |
| cg1147 | NCgl0968 | ssuI | Flavin mononucleotide binding protein required for sulfonate and sulfonate ester utilization | 0.32 |
| cg1349 | NCgl1147 | Membrane protein containing CBS domain | 0.33 | |
| cg0825 | NCgl0689 | fabG | 3-Ketoacyl-(acyl carrier protein) reductase | 0.35 |
| cg0882 | NCgl0738 | Hypothetical protein | 0.36 | |
| cg3420 | NCgl2983 | sigM | RNA polymerase σ factor of ECF subfamily | 2.37 |
| cg2069 | NCgl1774 | psp1 | Putative secreted protein | 2.50 |
| cg3351 | NCgl2920 | nagI | Gentisate 1,2-dioxygenase | 2.56 |
| cg2630 | NCgl2314 | pcaG | Subunit α of protocatechuate dioxygenase | 2.57 |
| cg2966 | NCgl2588 | Phenol 2-monooxygenase | 2.71 | |
| cg1255 | NCgl1060 | HNH endonuclease | 2.87 | |
| cg0638 | NCgl0524 | HD superfamily hydrolase | 3.09 | |
| cg3216 | NCgl2808 | gntP | Gluconate permease | 3.10 |
| cg2504 | NCgl2201 | Hypothetical protein | 3.14 | |
| cg2181 | NCgl1915 | Secreted component of ABC-type peptide transport system | 3.26 | |
| cg2182 | NCgl1916 | Permease component of ABC-type peptide transport system | 3.46 | |
| cg2183 | NCgl1917 | Permease component of ABC-type peptide transport system | 3.71 | |
| cg2184 | NCgl1918 | ATPase component of ABC-type peptide transport system; contains duplicated ATPase domains | 4.41 | |
| cg3151 | NCgl2131 | tnp2b | Transposase | 3.56 |
| cg0759 | prpD2 | Methylcitrate dehydratase | 3.63 | |
| cg0760 | NCgl0629 | prpB2 | Methylisocitrate lyase | 9.11 |
| cg0762 | NCgl0630 | prpC2 | Methylcitrate synthase | 11.05 |
| cg2938 | NCgl2563 | Permease component of ABC-type dipeptide/oligopeptide/nickel transport system | 3.93 | |
| cg2940 | NCgl2565 | ATPase component of ABC-type transport system; contains duplicated ATPase domains | 3.69 | |
| cg2836 | NCgl2476 | sucD | Subunit α of succinyl-CoA synthetase | 4.92 |
| cg2837 | NCgl2477 | sucC | Subunit β of succinyl-CoA synthetase | 3.82 |
| cg3047 | NCgl2656 | ackA | Acetate kinase | 4.00 |
| cg3048 | NCgl2657 | pta | Phosphotransacetylase | 5.43 |
| cg0961 | NCgl0805 | metA | Homoserine O-acetyltransferase | 4.89 |
| cg2426 | tnp2d | Transposase | 5.32 | |
| cg1612 | NCgl1368 | Acetyltransferase | 5.69 | |
| cg2610 | NCgl2294 | Secreted component of ABC-type dipeptide/oligopeptide/nickel transport system | 6.19 | |
| cg3107 | NCgl2709 | adhA | Zn-dependent alcohol dehydrogenase | 9.21 |
| cg3195 | NCgl2787 | Flavin-containing monooxygenase | 26.82 |
For the genes listed, the mRNA level in the ΔcitAB strain was altered ≥3-fold compared to the mRNA level in the wild type (wt) in at least three of four independent experiments after the growth of the strains in CGXII minimal medium containing 50 mM citrate and 50 mM glucose as carbon sources. The table includes those genes for which the mean mRNA ratio showed at least a twofold change (increase or decrease) and had a P value of ≤0.05. The mRNA ratios shown are mean values from three to four independent DNA microarray experiments starting from independent cultures. mRNA was prepared from cells in the exponential growth phase. CBS, cystathionine-β-synthase; ECF, extracytoplasmic function; succinyl-CoA, succinyl coenzyme A.
Requirement of citrate for CitAB-dependent activation of citH and tctCBA.
In order to confirm the microarray data and to test whether the presence of citrate is required for citH and tctCBA induction, primer extension experiments were performed. As shown in Fig. 7, primer extension products were detectable for citH and tctC RNAs of the wild type cultivated on citrate or citrate plus glucose, but not for RNAs of the wild type cultivated on glucose or the ΔcitAB mutant cultivated on glucose or glucose plus citrate. From these results, three conclusions can be drawn: (i) the expression of citH and tctCBA is activated in the presence of citrate, and in the absence of citrate, there is no or very weak expression; (ii) glucose does not inhibit the expression of citH and tctCBA, since the primer extension products for RNAs from cells grown on citrate and those grown on citrate plus glucose were of equal strengths; and (iii) citrate-inducible expression of both citH and tctCBA is strictly dependent on the CitAB TCS.
FIG. 7.
Expression analyses and determination of the transcriptional start sites of the C. glutamicum citH gene (A) and tctCBA operon (B) by primer extension analyses with the oligonucleotides citH-PE-1 and tct-PE-3. Results for genes from the following strains cultivated under the indicated conditions are shown: lanes 1, wild type grown on glucose; lanes 2, wild type grown on citrate; lanes 3, wild type grown on citrate and glucose; lanes 4, ΔcitAB mutant grown on glucose; and lanes 5, ΔcitAB mutant grown on citrate and glucose. RNAs from the strains were isolated after cultivation in CGXII minimal medium with 50 mM MgCl2 and a 25 mM concentration of the indicated carbon source. In all primer extension experiments for which results are shown, equivalent amounts of total RNA (20 μg) were used. The transcriptional start sites are indicated by asterisks.
In the case of citH, one major and two minor primer extension products were observed. The major transcription start site was located 56 bp upstream of the first putative start codon of citH, and the minor ones were located 55 and 53 bp upstream of the start codon (Fig. 1A and 7). The same start sites were also obtained with a second primer annealing at a different position (data not shown). The −10 region (Fig. 1A) deduced from the transcription start site (5′-TATACT-3′) is very similar to the −10 consensus core hexamer reported previously for C. glutamicum, 5′-TA(C/T)AAT-3′ (38). In the case of tctC, a single primer extension product, which ended 98 bp upstream of the predicted start codon of tctC, was identified (Fig. 1B and 7). Again, the same transcription start site was identified with a second primer annealing at a different position (data not shown), and the sequence of the deduced −10 region (5′-TTTAAT-3′) was similar to the consensus sequence listed above.
Binding of the response regulator CitB to the promoter regions of citH and tctCBA.
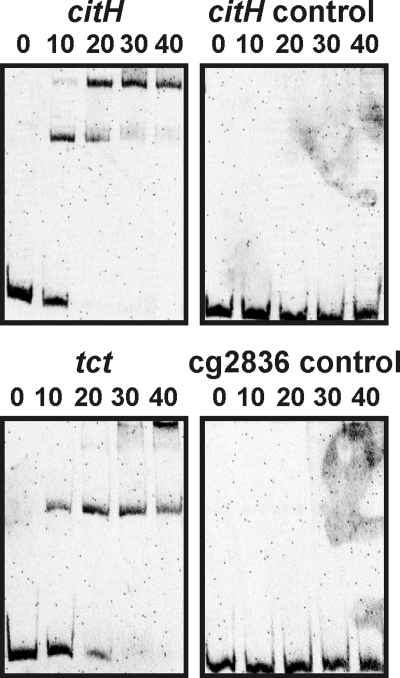
To support the assumption that CitB functions as a direct transcriptional activator of citH and tctCBA, EMSAs were performed with purified CitB and the promoter regions of citH and tctC. For this purpose, the CitB protein containing an amino-terminal hexahistidine tag was overproduced in E. coli BL21(DE3) and purified to apparent homogeneity by affinity chromatography (data not shown). DNA fragments covering the promoter regions of citH (bp −179 to +41 with respect to the major citH transcriptional start site, defined as bp +1) and tctC (bp −216 to +120 with respect to the tctC transcriptional start site) were incubated with four different concentrations of CitB and separated by native polyacrylamide gel electrophoresis. As negative controls, a fragment extending from bp +17 to +248 with respect to the major citH transcriptional start site and a promoter fragment of the arbitrarily chosen gene cg2836 (bp −557 to −226 with respect to the translational start site of cg2836) were tested. As shown in Fig. 8, the citH and tct promoter fragments in the presence of only a 10-fold molar excess of CitB were partially shifted, and complete retardation in the presence of a 20- or 30-fold molar excess of CitB was observed. No shift in the two DNA fragments serving as negative controls was observed, even at the highest CitB concentration used. In the case of the citH and tct promoter regions, a second protein-DNA complex was formed with increasing CitB concentrations, indicating either the presence of a second binding site or the binding of further CitB molecules to the initial protein-DNA complex. An inspection of the citH and tctC promoter regions did not reveal a highly conserved sequence motif that might represent the CitB binding site.
FIG. 8.
Binding of CitB to the promoter regions of citH and tctCBA. DNA fragments (219 to 336 bp; final concentrations, 22 to 35 nM) covering the promoter regions of citH and tctCBA and two DNA fragments serving as negative controls (see the text) were incubated for 30 min at room temperature without CitB (far left lanes) or with a 10- to 40-fold molar excess of purified CitB protein, as indicated, before separation by native polyacrylamide (15%) gel electrophoresis and staining with Sybr green I.
DISCUSSION
In this study, we have unraveled the molecular basis for citrate utilization and its regulation in C. glutamicum. As the expression of the C. glutamicum genes citH and tctCBA in E. coli MG1655 enabled growth on citrate, both CitH and TctCBA must be able to catalyze citrate uptake into the cell (Fig. 2). However, the two transporters differ in their cation dependency. CitH is active with Ca2+ and Sr2+ but not with Mg2+, and TctABC is active with Ca2+ and Mg2+ but not with Sr2+. In B. subtilis, the citrate transporters CitM and CitH, which show 42 and 43% sequence identity to C. glutamicum CitH, have complementary metal ion specificities. CitM of B. subtilis transports citrate in complexes with Mg2+, Ni2+, Mn2+, Co2+, and Zn2+ and CitH in complexes with Ca2+, Sr2+, and Ba2+ (30). As the metal ion specificity of the C. glutamicum homolog clearly resembles that of B. subtilis CitH, the original designation of CitM was changed to CitH. For B. subtilis CitH, apparent Km values of 33 and 40 μM for the uptake of Ca-citrate and Sr-citrate complexes into E. coli cells were determined previously (30), and similar values can be expected for C. glutamicum CitH. In the case of the TctABC transporter, a number of physiological and biochemical studies were performed with the system of S. enterica serovar Typhimurium (reviewed in reference 63). However, no clear picture of the cation dependency of this system exists. Our results suggest that Ca2+ or Mg2+ is required for citrate transport catalyzed by C. glutamicum TctABC. Since high concentrations of these cations were required for growth on citrate by using the Tct system, a different cation may be used by this transporter in the natural habitat.
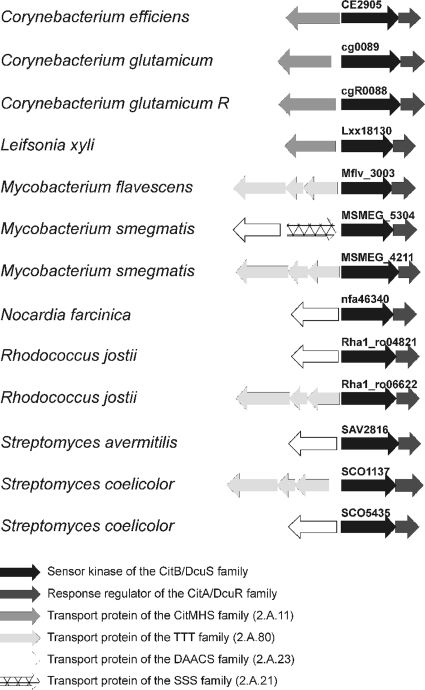
Birnbaum and Demain (3) suggested that citrate transport by C. glutamicum is inducible. The findings from our previous DNA microarray analyses (39) and the primer extension studies in this work clearly show that both citH and tctCBA are expressed only when citrate is present in the medium, and the transcriptome data indicate that the tctCBA operon is activated much more strongly than citH. The reason for this difference and its physiological meaning are not yet clear. The activation of citH and tctCBA in the presence of citrate is strictly dependent on the CitAB TCS, and a ΔcitAB deletion mutant of C. glutamicum was unable to grow on citrate as the sole carbon and energy source within the first ∼30 h of cultivation (see below). These results indicate (i) that the histidine kinase CitA may function as a citrate sensor, as shown previously for the orthologous CitA proteins from K. pneumoniae (23) and E. coli (22), and (ii) that the response regulator CitB presumably functions as a transcriptional activator of citH and the tctCBA operon. The latter hypothesis was supported by results from EMSAs, which showed that purified CitB binds to the promoter regions of citH and tctC. According to these results, the transcriptional control of citrate permease genes by a TCS of the CitAB/DcuSR family occurs not only in gram-negative bacteria and low-GC-content gram-positive bacteria, but also in high-GC-content gram-positive bacteria and thus is distributed in all major lines of the eubacteria. BLAST searches (2) with completed genome sequences of Actinobacteria revealed homologs of CitA/DcuS and CitB/DcuR in many members of this class of bacteria, such as Mycobacterium smegmatis, Nocardia farcinica, and Streptomyces coelicolor, and in nearly all cases, the corresponding genes were localized next to genes for a tricarboxylate or dicarboxylate transporter (Fig. 9).
FIG. 9.
Colocalization of genes encoding two-component systems of the CitAB/DcuSR family with genes for tricarboxylate or dicarboxylate transporters in selected Actinobacteria. Gene identifiers for all sensor kinases are indicated. DAACS, dicarboxylate/amino acid:cation (Na+ or H+) symporter; SSS, solute:sodium symporter.
Whereas citrate utilization is subject to catabolite repression in many bacteria (see the introduction), C. glutamicum uses citrate and glucose simultaneously (Fig. 5). In accord with this observation, the activation of citH and tctCBA was not influenced by the presence of glucose in the growth medium (39) (Fig. 7). Cometabolism of different carbon sources with glucose is typical for C. glutamicum and was shown previously for, e.g., fructose (13), acetate (58), propionate (12), serine (36), vanillate and protocatechuate (32), and gluconate (17).
Footnotes
Published ahead of print on 17 April 2009.
REFERENCES
- 1.Abe, S., K. Takayama, and S. Kinoshita. 1967. Taxonomical studies on glutamic acid producing bacteria. J. Gen. Appl. Microbiol. 13279-301. [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. H. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 253389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Birnbaum, J., and A. L. Demain. 1969. Conversion of citrate to extracellular glutamate by penicillin-treated resting cells of Corynebacterium glutamicum. Agric. Biol. Chem. 331169-1173. [Google Scholar]
- 4.Blancato, V. S., C. Magni, and J. S. Lolkema. 2006. Functional characterization and Me2+ ion specificity of a Ca2+-citrate transporter from Enterococcus faecalis. FEBS J. 2735121-5130. [DOI] [PubMed] [Google Scholar]
- 5.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 2771453-1474. [DOI] [PubMed] [Google Scholar]
- 6.Boorsma, A., M. E. van der Rest, J. S. Lolkema, and W. N. Konings. 1996. Secondary transporters for citrate and the Mg2+-citrate complex in Bacillus subtilis are homologous proteins. J. Bacteriol. 1786216-6222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bott, M. 1997. Anaerobic citrate metabolism and its regulation in enterobacteria. Arch. Microbiol. 16778-88. [PubMed] [Google Scholar]
- 8.Bott, M., and P. Dimroth. 1994. Klebsiella pneumoniae genes for citrate lyase and citrate lyase ligase: localization, sequencing, and expression. Mol. Microbiol. 14347-356. [DOI] [PubMed] [Google Scholar]
- 9.Bott, M., M. Meyer, and P. Dimroth. 1995. Regulation of anaerobic citrate metabolism in Klebsiella pneumoniae. Mol. Microbiol. 18533-546. [DOI] [PubMed] [Google Scholar]
- 10.Brazma, A., P. Hingamp, J. Quackenbush, G. Sherlock, P. Spellman, C. Stoeckert, J. Aach, W. Ansorge, C. A. Ball, H. C. Causton, T. Gaasterland, P. Glenisson, F. C. Holstege, I. F. Kim, V. Markowitz, J. C. Matese, H. Parkinson, A. Robinson, U. Sarkans, S. Schulze-Kremer, J. Stewart, R. Taylor, J. Vilo, and M. Vingron. 2001. Minimum information about a microarray experiment (MIAME)—toward standards for microarray data. Nat. Genet. 29365-371. [DOI] [PubMed] [Google Scholar]
- 11.Burkovski, A. (ed.). 2008. Corynebacteria: genomics and molecular biology. Caister Academic Press, Norfolk, United Kingdom.
- 12.Claes, W. A., A. Puhler, and J. Kalinowski. 2002. Identification of two prpDBC gene clusters in Corynebacterium glutamicum and their involvement in propionate degradation via the 2-methylcitrate cycle. J. Bacteriol. 1842728-2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dominguez, H., M. Cocaign-Bousquet, and N. D. Lindley. 1997. Simultaneous consumption of glucose and fructose from sugar mixtures during batch growth of Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 47600-603. [Google Scholar]
- 14.Drider, D., S. Bekal, and H. Prevost. 2004. Genetic organization and expression of citrate permease in lactic acid bacteria. Genet. Mol. Res. 3273-281. [PubMed] [Google Scholar]
- 15.Eggeling, L., and M. Bott (ed.). 2005. Handbook of Corynebacterium glutamicum. CRC Press, Taylor & Francis Group, Boca Raton, FL.
- 16.Engels, S., J. E. Schweitzer, C. Ludwig, M. Bott, and S. Schaffer. 2004. clpC and clpP1P2 gene expression in Corynebacterium glutamicum is controlled by a regulatory network involving the transcriptional regulators ClgR and HspR as well as the ECF sigma factor σH. Mol. Microbiol. 52285-302. [DOI] [PubMed] [Google Scholar]
- 17.Frunzke, J., V. Engels, S. Hasenbein, C. Gätgens, and M. Bott. 2008. Co-ordinated regulation of gluconate catabolism and glucose uptake in Corynebacterium glutamicum by two functionally equivalent transcriptional regulators, GntR1 and GntR2. Mol. Microbiol. 67305-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gerharz, T., S. Reinelt, S. Kaspar, L. Scapozza, and M. Bott. 2003. Identification of basic amino acid residues important for citrate binding by the periplasmic receptor domain of the sensor kinase CitA. Biochemistry 425917-5924. [DOI] [PubMed] [Google Scholar]
- 19.Guillouet, S., A. A. Rodal, G. An, P. A. Lessard, and A. J. Sinskey. 1999. Expression of the Escherichia coli catabolic threonine dehydratase in Corynebacterium glutamicum and its effect on isoleucine production. Appl. Environ. Microbiol. 653100-3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jacoby, M., C. E. Nguouoto-Nkili, and A. Burkovski. 1999. Construction and application of new Corynebacterium glutamicum vectors. Biotechnol. Techniques 13437-441. [Google Scholar]
- 21.Kalinowski, J., B. Bathe, D. Bartels, N. Bischoff, M. Bott, A. Burkovski, N. Dusch, L. Eggeling, B. J. Eikmanns, L. Gaigalat, A. Goesmann, M. Hartmann, K. Huthmacher, R. Krämer, B. Linke, A. C. McHardy, F. Meyer, B. Möckel, W. Pfefferle, A. Pühler, D. A. Rey, C. Rückert, O. Rupp, H. Sahm, V. F. Wendisch, I. Wiegrabe, and A. Tauch. 2003. The complete Corynebacterium glutamicum ATCC 13032 genome sequence and its impact on the production of l-aspartate-derived amino acids and vitamins. J. Biotechnol. 1045-25. [DOI] [PubMed] [Google Scholar]
- 22.Kaspar, S., and M. Bott. 2002. The sensor kinase CitA (DpiB) of Escherichia coli functions as a high-affinity citrate receptor. Arch. Microbiol. 177313-321. [DOI] [PubMed] [Google Scholar]
- 23.Kaspar, S., R. Perozzo, S. Reinelt, M. Meyer, K. Pfister, L. Scapozza, and M. Bott. 1999. The periplasmic domain of the histidine autokinase CitA functions as a highly specific citrate receptor. Mol. Microbiol. 33858-872. [DOI] [PubMed] [Google Scholar]
- 24.Kästner, C. N., K. Schneider, P. Dimroth, and K. M. Pos. 2002. Characterization of the citrate/acetate antiporter CitW of Klebsiella pneumoniae. Arch. Microbiol. 177500-506. [DOI] [PubMed] [Google Scholar]
- 25.Keilhauer, C., L. Eggeling, and H. Sahm. 1993. Isoleucine synthesis in Corynebacterium glutamicum: molecular analysis of the ilvB-ilvN-ilvC operon. J. Bacteriol. 1755595-5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kimura, K. 1964. Utilization of organic acids for the glutamic acid production by Micrococcus glutamicus. J. Gen. Appl. Microbiol. 1023-31. [Google Scholar]
- 27.Kocan, M., S. Schaffer, T. Ishige, U. Sorger-Herrmann, V. F. Wendisch, and M. Bott. 2006. Two-component systems of Corynebacterium glutamicum: deletion analysis and involvement of the PhoS-PhoR system in the phosphate starvation response. J. Bacteriol. 188724-732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koch, D. J., C. Rückert, D. A. Rey, A. Mix, A. Pühler, and J. Kalinowski. 2005. Role of the ssu and seu genes of Corynebacterium glutamicum ATCC 13032 in utilization of sulfonates and sulfonate esters as sulfur sources. Appl. Environ. Microbiol. 716104-6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Korithoski, B., K. Krastel, and D. G. Cvitkovitch. 2005. Transport and metabolism of citrate by Streptococcus mutans. J. Bacteriol. 1874451-4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krom, B. P., J. B. Warner, W. N. Konings, and J. S. Lolkema. 2000. Complementary metal ion specificity of the metal-citrate transporters CitM and CitH of Bacillus subtilis. J. Bacteriol. 1826374-6381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li, H., and A. M. Pajor. 2002. Functional characterization of CitM, the Mg2+-citrate transporter. J. Membr. Biol. 1859-16. [DOI] [PubMed] [Google Scholar]
- 32.Merkens, H., G. Beckers, A. Wirtz, and A. Burkovski. 2005. Vanillate metabolism in Corynebacterium glutamicum. Curr. Microbiol. 5159-65. [DOI] [PubMed] [Google Scholar]
- 33.Meyer, M., P. Dimroth, and M. Bott. 2001. Catabolite repression of the citrate fermentation genes in Klebsiella pneumoniae: evidence for involvement of the cyclic AMP receptor protein. J. Bacteriol. 1835248-5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meyer, M., P. Dimroth, and M. Bott. 1997. In vitro binding of the response regulator CitB and of its carboxy-terminal domain to A + T-rich DNA target sequences in the control region of the divergent citC and citS operons of Klebsiella pneumoniae. J. Mol. Biol. 269719-731. [DOI] [PubMed] [Google Scholar]
- 35.Möker, N., M. Brocker, S. Schaffer, R. Krämer, S. Morbach, and M. Bott. 2004. Deletion of the genes encoding the MtrA-MtrB two-component system of Corynebacterium glutamicum has a strong influence on cell morphology, antibiotics susceptibility and expression of genes involved in osmoprotection. Mol. Microbiol. 54420-438. [DOI] [PubMed] [Google Scholar]
- 36.Netzer, R., P. Peters-Wendisch, L. Eggeling, and H. Sahm. 2004. Cometabolism of a nongrowth substrate: l-serine utilization by Corynebactetium glutamicum. Appl. Environ. Microbiol. 707148-7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Niebisch, A., and M. Bott. 2001. Molecular analysis of the cytochrome bc1-aa3 branch of the Corynebacterium glutamicum respiratory chain containing an unusual diheme cytochrome c1. Arch. Microbiol. 175282-294. [DOI] [PubMed] [Google Scholar]
- 38.Patek, M., J. Nesvera, A. Guyonvarch, O. Reyes, and G. Leblon. 2003. Promoters of Corynebacterium glutamicum. J. Biotechnol. 104311-323. [DOI] [PubMed] [Google Scholar]
- 39.Polen, T., D. Schluesener, A. Poetsch, M. Bott, and V. F. Wendisch. 2007. Characterization of citrate utilization in Corynebacterium glutamicum by transcriptome and proteome analysis. FEMS Microbiol. Lett. 273109-119. [DOI] [PubMed] [Google Scholar]
- 40.Polen, T., and V. F. Wendisch. 2004. Genomewide expression analysis in amino acid-producing bacteria using DNA microarrays. Appl. Biochem. Biotechnol. 118215-232. [DOI] [PubMed] [Google Scholar]
- 41.Pos, K. M., and P. Dimroth. 1996. Functional properties of the purified Na+-dependent citrate carrier of Klebsiella pneumoniae: evidence for asymmetric orientation of the carrier protein in proteoliposomes. Biochemistry 351018-1026. [DOI] [PubMed] [Google Scholar]
- 42.Pos, K. M., P. Dimroth, and M. Bott. 1998. The Escherichia coli citrate carrier CitT: a member of a novel eubacterial transporter family related to the 2-oxoglutarate/malate translocator from spinach chloroplasts. J. Bacteriol. 1804160-4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reinelt, S., E. Hofmann, T. Gerharz, M. Bott, and D. R. Madden. 2003. The structure of the periplasmic ligand-binding domain of the sensor kinase CitA reveals the first extracellular PAS domain. J. Biol. Chem. 27839189-39196. [DOI] [PubMed] [Google Scholar]
- 44.Saier, M. H., Jr. 2000. A functional-phylogenetic classification system for transmembrane solute transporters. Microbiol. Mol. Biol. Rev. 64354-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 46.Schäfer, A., A. Tauch, W. Jäger, J. Kalinowski, G. Thierbach, and A. Pühler. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 14569-73. [DOI] [PubMed] [Google Scholar]
- 47.Schneider, K., P. Dimroth, and M. Bott. 2000. Biosynthesis of the prosthetic group of citrate lyase. Biochemistry 399438-9450. [DOI] [PubMed] [Google Scholar]
- 48.Schneider, K., P. Dimroth, and M. Bott. 2000. Identification of triphosphoribosyl-dephospho-CoA as precursor of the citrate lyase prosthetic group. FEBS Lett. 483165-168. [DOI] [PubMed] [Google Scholar]
- 49.Sevvana, M., V. Vijayan, M. Zweckstetter, S. Reinelt, D. R. Madden, R. Herbst-Irmer, G. M. Sheldrick, M. Bott, C. Griesinger, and S. Becker. 2008. A ligand-induced switch in the periplasmic domain of sensor histidine kinase CitA. J. Mol. Biol. 377512-523. [DOI] [PubMed] [Google Scholar]
- 50.Somers, J. M., and W. W. Kay. 1983. Genetic fine structure of the tricarboxylate transport (tct) locus of Salmonella typhimurium. Mol. Gen. Genet. 19020-26. [DOI] [PubMed] [Google Scholar]
- 51.Sweet, G. D., C. M. Kay, and W. W. Kay. 1984. Tricarboxylate-binding proteins of Salmonella typhimurium: purification, crystallization, and physical properties. J. Biol. Chem. 2591586-1592. [PubMed] [Google Scholar]
- 52.Sweet, G. D., J. M. Somers, and W. W. Kay. 1979. Purification and properties of a citrate-binding transport component, the C-protein of Salmonella typhimurium. Can. J. Biochem. 57710-715. [DOI] [PubMed] [Google Scholar]
- 53.van der Rest, M. E., T. Abee, D. Molenaar, and W. N. Konings. 1991. Mechanism and energetics of a citrate-transport system of Klebsiella pneumoniae. Eur. J. Biochem. 19571-77. [DOI] [PubMed] [Google Scholar]
- 54.van der Rest, M. E., E. Schwarz, D. Oesterhelt, and W. N. Konings. 1990. DNA sequence of a citrate carrier of Klebsiella pneumoniae. Eur. J. Biochem. 189401-407. [DOI] [PubMed] [Google Scholar]
- 55.von der Osten, C. H., C. Gioannetti, and A. J. Sinskey. 1989. Design of a defined medium for growth of Corynebacterium glutamicum in which citrate facilitates iron uptake. Biotechnol. Lett. 1111-16. [Google Scholar]
- 56.Warner, J. B., B. P. Krom, C. Magni, W. N. Konings, and J. S. Lolkema. 2000. Catabolite repression and induction of the Mg2+-citrate transporter CitM of Bacillus subtilis. J. Bacteriol. 1826099-6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Warner, J. B., C. Magni, and J. S. Lolkema. 2003. CcpA-independent regulation of expression of the Mg2+-citrate transporter gene citM by arginine metabolism in Bacillus subtilis. J. Bacteriol. 185854-859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wendisch, V. F., A. A. De Graaf, H. Sahm, and B. J. Eikmanns. 2000. Quantitative determination of metabolic fluxes during coutilization of two carbon sources: comparative analyses with Corynebacterium glutamicum during growth on acetate and/or glucose. J. Bacteriol. 1823088-3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wennerhold, J., A. Krug, and M. Bott. 2005. The AraC-type regulator RipA represses aconitase and other iron proteins from Corynebacterium under iron limitation and is itself repressed by DtxR. J. Biol. Chem. 28040500-40508. [DOI] [PubMed] [Google Scholar]
- 60.Widenhorn, K. A., W. Boos, J. M. Somers, and W. W. Kay. 1988. Cloning and properties of the Salmonella typhimurium tricarboxylate transport operon in Escherichia coli. J. Bacteriol. 170883-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Widenhorn, K. A., J. M. Somers, and W. W. Kay. 1988. Expression of the divergent tricaryboxylate transport operon (tctI) of Salmonella typhimurium. J. Bacteriol. 1703223-3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Widenhorn, K. A., J. M. Somers, and W. W. Kay. 1989. Genetic regulation of the tricarboxylate transport operon (tctI) of Salmonella typhimurium. J. Bacteriol. 1714436-4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Winnen, B., R. N. Hvorup, and M. H. Saier, Jr. 2003. The tripartite tricarboxylate transporter (TTT) family. Res. Microbiol. 154457-465. [DOI] [PubMed] [Google Scholar]
- 64.Yamamoto, H., M. Murata, and J. Sekiguchi. 2000. The CitST two-component system regulates the expression of the Mg-citrate transporter in Bacillus subtilis. Mol. Microbiol. 37898-912. [DOI] [PubMed] [Google Scholar]