Abstract
The icsP promoter of Shigella spp. is repressed by H-NS and derepressed by VirB. Here, we show that an inverted repeat located between positions −1144 and −1130 relative to the icsP transcription start site is necessary for VirB-dependent derepression. The atypical location of this cis-acting site is discussed.
Shigella species are gram-negative intracellular pathogens that invade cells of the lower intestinal epithelia of humans and primates, causing bacillary dysentery. All Shigella species carry a large virulence plasmid, and many genes carried by these plasmids are thermoregulated. At the nonpermissive temperature of 30°C, the global regulator H-NS (histone-like nucleoid structuring protein) represses transcription of these genes (4, 12, 18). At the permissive temperature of 37°C, H-NS-dependent repression is relieved by temperature-induced changes in DNA topology (9, 19), VirF, or its subordinate regulator VirB (InvE) (reviewed in reference 17). The mechanism that leads to the alleviation of transcriptional repression by H-NS has been coined “antisilencing.” Antisilencing is thought to play an important role in controlling the expression of genes acquired through horizontal gene transfer and is common in bacterial pathogens in which a variety of transcription factors function to relieve repression by H-NS (reviewed in reference 29).
The icsP (sopA) gene is carried on the large virulence plasmid in all Shigella species (8, 27) and encodes an outer membrane protease, which belongs to the omptin protease family (11, 14) and cleaves the actin-tail assembly protein IcsA from the surface of Shigella (8, 27, 28). Previous studies have revealed that icsP, like other Shigella virulence plasmid genes, including virA, ospB, and phoN2, and those of the invasion locus, ipa, mxi, and spa (1, 5, 6, 25, 26, 30, 31, 33), is repressed by H-NS and derepressed by VirB (34). In this study, we identify sequences upstream of the icsP gene necessary for derepression by VirB, with a view to improve our understanding of the mechanism of transcriptional antisilencing at the icsP promoter.
Identification of sequences required for VirB-dependent regulation of the icsP promoter.
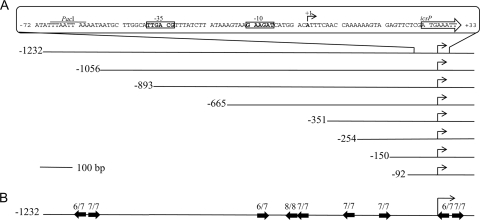
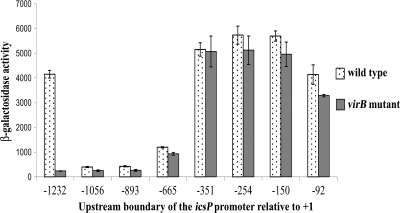
Previous work has demonstrated that VirB regulates an icsP-lacZ fusion integrated into the icsP locus on the Shigella virulence plasmid (34). To identify regions upstream of the icsP gene that mediate VirB-dependent derepression, a nested set of icsP promoter deletions was created (Fig. 1A) and cloned into a medium-copy-number lacZ reporter plasmid pHJW20 (Table 1) to replace the existing 1,232-bp icsP promoter fragment. This created eight PicsP-lacZ fusions whose upstream limits varied from positions −1232 to −92 relative to the previously annotated transcription start site of the icsP gene (8) (Fig. 1A; Table 1). Each promoter construct was introduced into wild-type Shigella strain 2457T and a mutant derivative, AWY3 (2457T virB::Tn5), and β-galactosidase production was measured using the Miller protocol (16). Of the eight promoter fragments tested, only one displayed a >2-fold increase in the presence of VirB (Fig. 2). Surprisingly, this was the longest promoter fragment (found in pHJW20). The activity of this promoter was 17-fold higher in the presence of VirB than in its absence. This increase was unlikely to be caused by sequences in the pACYC184 plasmid backbone, because these sequences would also influence the activity of the other promoter fragments. Furthermore, the increase in promoter activity was unlikely to be caused by the creation of a new VirB binding site at the boundary of the plasmid backbone and the promoter region, because two additional constructs with altered plasmid-promoter boundaries (pMIC01 and pMIC02 [Table 1]) were found to have similar activities to those displayed by pHJW20. The simplest interpretation of these data was that DNA sequences located between positions 1232 and 1056 upstream of the icsP transcription start site are required for VirB-dependent regulation of the icsP promoter.
FIG. 1.
Promoter elements of the icsP promoter and schematic representation of the truncated promoter series. (A) Angled arrows represent the icsP transcription start site (+1), determined previously (8). The hexamers at positions −10 and −35 are boxed and shown in boldface type. The translation start site is outlined by an arrow. The truncated promoters are drawn to scale, and the numbers represent the upstream boundary of the icsP promoter, relative to the transcription start site (+1). The promoter fragments represented are found in pHJW20, pJS01, pJS02, pDH01, pJS04, pHJW34, pHJW35, and pHJW36. (B) Solid black arrows represent the relative position and orientation of the nine VirB binding sites identified within the promoter fragments used in this work. In each case, the match to consensus sequence 5′-(A/G)(A/T)G(G)AAAT-3′ (30, 32) is given.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Descriptiona | Source or reference |
|---|---|---|
| Strains | ||
| S. flexneri | ||
| 2457T | S. flexneri serotype 2a | 10 |
| AWY3 | 2457T virB::Tn5; Knr | 34 |
| Plasmids | ||
| pACYC184 | Cloning vector; p15A replicon Tetr/Cmr | 24 |
| pHJW7 | icsP promoter region transcriptionally fused to lacZ in pACYC184 Cmr; carries 1,232 bp of wild-type sequence upstream of the icsP transcription start site | 34 |
| pHJW20 | pHJW7 carrying unique XbaI site upstream of lacZ gene; carries 1,232 bp of wild-type sequence upstream of the icsP transcription start site | This work |
| pJS01 | pHJW20 carrying 1,056 bp of wild-type sequence upstream of the icsP transcription start site | This work |
| pJS02 | pHJW20 carrying 893 bp of wild-type sequence upstream of the icsP transcription start site | This work |
| pDH01 | pHJW20 carrying 665 bp of wild-type sequence upstream of the icsP transcription start site | This work |
| pJS04 | pHJW20 carrying 351 bp of wild-type sequence upstream of the icsP transcription start site | This work |
| pHJW34 | pHJW20 carrying 254 bp of wild-type sequence upstream of the icsP transcription start site | This work |
| pHJW35 | pHJW20 carrying 150 bp of wild-type sequence upstream of the icsP transcription start site | This work |
| pHJW36 | pHJW20 carrying 92 bp of wild-type sequence upstream of the icsP transcription start site | This work |
| pMIC01 | pHJW20 with 33 bp deleted between the SalI and PstI sites in the multiple cloning site of pACYC184 | This work |
| pMIC02 | pHJW20 carrying 1,437 bp of wild-type sequence upstream of the icsP transcription start site | This work |
| pMIC13 | pHJW20 carrying a 7-bp mutation in box 2 of the putative VirB binding site | This work |
| pMIC17 | pHJW20 carrying a 7-bp mutation in box 1 of the putative VirB binding site | This work |
| pMIC18 | pHJW20 carrying a 14-bp mutation in both box 1 and box 2 of the putative VirB binding site | This work |
| pMIC21 | pHJW20 lacking all icsP promoter sequences | This work |
Ampr, ampicillin resistance; Tetr, tetracycline resistance; Cmr, chloramphenicol resistance; Knr, kanamycin resistance.
FIG. 2.
Activities of the truncated icsP promoter series in Shigella. Bars indicate β-galactosidase expression of the PicsP-lacZ fusions in wild-type S. flexneri (2457T) and an isogenic strain lacking virB (AWY3). β-Galactosidase activities are expressed in Miller units. Assays were run in triplicate, and the means and standard deviations of the results are shown.
Identification and site-directed mutagenesis of putative VirB binding sites responsible for VirB-dependent regulation of the icsP promoter.
Previous analysis of the icsB, spa15, and virA promoters of Shigella sonnei established a consensus binding site for VirB (30). Our analysis of sequences upstream of the icsP gene identified nine sites with a match greater than 6/7 to the consensus, 5′-(A/G)(A/T)G(G)AAAT-3′ (Fig. 1B). Three of these sites are located between positions −665 and −351. The location of these sites may explain why small yet significant increases in VirB-dependent promoter activity were observed in wild-type Shigella flexneri carrying promoter constructs with upstream boundaries of −665, −893, and −1056 relative to the icsP transcription start site (+1) (Fig. 2). Two other putative VirB binding sites are located immediately downstream of the transcription start site. Although the location of these sites may explain the small increase in VirB-dependent promoter activity associated with the shortest promoter fragment used in our studies, our data suggest that these sites alone play no significant role in the presence of upstream promoter sequences.
Interestingly, two of the nine sites identified upstream of the icsP gene are located between positions −1144 and −1130 and are organized as an inverted repeat (Fig. 1B). Since our truncation analysis indicates that sequences between positions −1232 and −1056 are essential for a 17-fold increase in promoter activity in the presence of VirB (Fig. 2), we chose to analyze these sites further. Seven base pair substitutions were made in either the upstream site (box 2), the downstream site (box 1) or both, using a PCR site-directed mutagenesis method described by Lie and Leigh (15) (Table 2). Each mutated promoter fragment was then introduced into the lacZ reporter plasmid pHJW20 to replace the existing wild-type sites, the resulting plasmids pMIC13, pMIC17, pMIC18, and pHJW20 and a promoterless control, pMIC21 (Table 1), were introduced into wild-type Shigella strain 2457T and the virB mutant (AWY3), and β-galactosidase levels were measured (Table 2). Our data revealed that complete mutagenesis of the upstream binding site (box 2), the downstream binding site (box 1), or both, resulted in complete loss of VirB-dependent regulation of the icsP promoter. Furthermore, these results were not an artifact of the lacZ reporter constructs, because similar patterns of expression were observed when the icsP gene was fused to each of our promoter constructs and IcsP levels were measured by Western blotting (data not shown). These data strongly suggest that VirB regulates the icsP promoter from sequences located more than 1 kb upstream of the icsP transcription start site. To our knowledge, this is the first evidence that VirB can influence promoter activity from such distal sites.
TABLE 2.
Summary of mutations introduced into the two boxes that form the upstream inverted repeat and activities of wild-type and mutated icsP promoter fragments
| Fragment description | Sequencea | β-Galactosidase activityb
|
|
|---|---|---|---|
| virB+ | virB mutant | ||
| WT box 1 and 2 | CGGGGATTTCAGTATGAAATGAAGTA | 4,412 ± 80 | 388 ± 10 |
| Mutated box 1 | CGGGGATTTCAGTCGACCCGGAAGTA | 307 ± 13 | 326 ± 75 |
| Mutated box 2 | CGGGGGCCCAGCTATGAAATGAAGTA | 309 ± 27 | 345 ± 22 |
| Mutated box 1 and 2 | CGGGGGCCCAGCTCGACCCGGAAGTA | 297 ± 18 | 341 ± 15 |
| Promoterless lacZ | 284 ± 13 | 388 ± 25 | |
5′→3′ DNA sequences of the wild-type and mutated boxes that form the upstream inverted repeat. Sequences lie between positions −1144 and −1130 relative to the annotated transcription start site of icsP (+1) (8). Underlined sequences are box 2 (left) and box 1 (right) sequences.
All promoter fragments were fused to lacZ, and β-galactosidase activities were measured in wild-type S. flexneri (2457T) and the isogenic strain that lacks virB (AWY3). The parent cloning vector with a promoterless lacZ gene was included as a negative control. β-Galactosidase activities are expressed in Miller units. Assays were run in triplicate, and the means and standard deviations of the results are shown.
Conserved sequence and location of the two distal VirB binding sites in all known Shigella sequences and in EIEC strain HN280.
To examine how well conserved DNA sequences located upstream of the icsP promoter are among other Shigella strains, species, and other enterics, a 2-kb sequence upstream of the icsP gene in Shigella flexneri 2457T was subjected to BLAST analysis. All known Shigella virulence plasmid sequences and the virulence plasmid of the enteroinvasive Escherichia coli (EIEC) strain HN280 contained nearly identical sequences (99 to 100% identity) over the entire 2-kb sequence upstream of the icsP gene. Furthermore, the upstream inverted repeat identified by our studies was 100% identical in all strains and located in exactly the same position relative to the annotated transcription start site identified in S. flexneri. These findings strongly suggest that icsP genes found in all Shigella spp. and the EIEC strain HN280 are likely to be regulated by VirB from a binding site located more than 1 kb upstream of the gene.
VirB is structurally homologous to plasmid partitioning proteins, which can influence transcription from distances of several kilobases.
While it is unusual for transcription factors to influence transcription from distances greater than 200 bp upstream or downstream of the transcription start site in bacteria (2, 7), some examples exist. For example, the enhancer of the Bacillus subtilis rocG gene is located 1.5 kb downstream of the promoter and, beyond the end of the rocG coding region (3) and the two NtrC binding sites required for the transcriptional activation of E. coli σ54-regulated glnA promoter, can still function when placed as far as 3 kb from the promoter (20). Furthermore, bacterial plasmid partitioning factors, while not typically considered transcription factors, have also been shown to silence the promoters of genes in the vicinity of their cis-acting binding sites from distances of several kilobase pairs (13, 21-23, 35). One of these proteins, the P1 ParB protein, displays structural homology to VirB and has bridging capabilities—the ability to interact with other ParB monomers located at binding sites further up- or downstream. It is therefore possible that the seven other sites with close matches to the VirB consensus binding site play an important role in bridging by VirB and that the resulting DNA topology is central to the alleviation of H-NS-dependent repression of the icsP promoter, although this needs to be tested.
In summary, although transcription factors typically bind to sequences located within 200 bp upstream or downstream of the transcription start site (2, 7), here we provide strong evidence that VirB has the capacity to alleviate H-NS-dependent repression of the icsP promoter from sites located more than 1 kb upstream of the transcription start site. This raises two important questions. (i) Are other Shigella virulence plasmid genes regulated from remote VirB binding sites? (ii) Is it common for transcriptional antisilencing mechanisms to employ distal regulator binding sites? Future studies will address these questions and elucidate the molecular mechanism of H-NS-dependent repression and VirB-mediated derepression of the icsP promoter.
Acknowledgments
We thank J. Dodsworth for critical reading of the manuscript.
This study was supported by the NIH grant P20 RR-016464 from the INBRE Program of the National Center for Research Resources, by the NSF grant 0649267, and by UNLV start-up funds to H.J.W. J.M.S. was a recipient of an NSF REU scholarship (NSF 0649267) in 2007; K.M.L. is a recipient of a Barry Goldwater scholarship; D.J.H. is an LT MSC USN microbiologist, NAVMED MPT&E.
The views expressed in this article are those of the authors and do not necessarily reflect the official policy or position of the Navy, Department of Defense, or the U.S. government.
Footnotes
Published ahead of print on 10 April 2009.
REFERENCES
- 1.Adler, B., C. Sasakawa, T. Tobe, S. Makino, K. Komatsu, and M. Yoshikawa. 1989. A dual transcriptional activation system for the 230 kb plasmid genes coding for virulence-associated antigens of Shigella flexneri. Mol. Microbiol. 3627-635. [DOI] [PubMed] [Google Scholar]
- 2.Balleza, E., L. N. Lopez-Bojorquez, A. Martinez-Antonio, O. Resendis-Antonio, I. Lozada-Chavez, Y. I. Balderas-Martinez, S. Encarnacion, and J. Collado-Vides. 2009. Regulation by transcription factors in bacteria: beyond description. FEMS Microbiol. Rev. 33133-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belitsky, B. R., and A. L. Sonenshein. 1999. An enhancer element located downstream of the major glutamate dehydrogenase gene of Bacillus subtilis. Proc. Natl. Acad. Sci. USA 9610290-10295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beloin, C., and C. J. Dorman. 2003. An extended role for the nucleoid structuring protein H-NS in the virulence gene regulatory cascade of Shigella flexneri. Mol. Microbiol. 47825-838. [DOI] [PubMed] [Google Scholar]
- 5.Beloin, C., S. McKenna, and C. J. Dorman. 2002. Molecular dissection of VirB, a key regulator of the virulence cascade of Shigella flexneri. J. Biol. Chem. 27715333-15344. [DOI] [PubMed] [Google Scholar]
- 6.Berlutti, F., M. Casalino, C. Zagaglia, P. A. Fradiani, P. Visca, and M. Nicoletti. 1998. Expression of the virulence plasmid-carried apyrase gene (apy) of enteroinvasive Escherichia coli and Shigella flexneri is under the control of H-NS and the VirF and VirB regulatory cascade. Infect. Immun. 664957-4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collado-Vides, J., B. Magasanik, and J. D. Gralla. 1991. Control site location and transcriptional regulation in Escherichia coli. Microbiol. Rev. 55371-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Egile, C., H. d'Hauteville, C. Parsot, and P. J. Sansonetti. 1997. SopA, the outer membrane protease responsible for polar localization of IcsA in Shigella flexneri. Mol. Microbiol. 231063-1073. [DOI] [PubMed] [Google Scholar]
- 9.Falconi, M., B. Colonna, G. Prosseda, G. Micheli, and C. O. Gualerzi. 1998. Thermoregulation of Shigella and Escherichia coli EIEC pathogenicity. A temperature-dependent structural transition of DNA modulates accessibility of virF promoter to transcriptional repressor H-NS. EMBO J. 177033-7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Formal, S. B., G. J. Dammin, E. H. LaBrec, and H. Schneider. 1958. Experimental Shigella infections: characteristics of a fatal infection produced in guinea pigs. J. Bacteriol. 75604-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hritonenko, V., and C. Stathopoulos. 2007. Omptin proteins: an expanding family of outer membrane proteases in gram-negative Enterobacteriaceae. Mol. Membr. Biol. 24395-406. [DOI] [PubMed] [Google Scholar]
- 12.Hromockyj, A. E., S. C. Tucker, and A. T. Maurelli. 1992. Temperature regulation of Shigella virulence: identification of the repressor gene virR, an analogue of hns, and partial complementation by tyrosyl transfer RNA (tRNA1(Tyr)). Mol. Microbiol. 62113-2124. [DOI] [PubMed] [Google Scholar]
- 13.Kim, S. K., and J. C. Wang. 1999. Gene silencing via protein-mediated subcellular localization of DNA. Proc. Natl. Acad. Sci. USA 968557-8561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kukkonen, M., and T. K. Korhonen. 2004. The omptin family of enterobacterial surface proteases/adhesins: from housekeeping in Escherichia coli to systemic spread of Yersinia pestis. Int. J. Med. Microbiol. 2947-14. [DOI] [PubMed] [Google Scholar]
- 15.Lie, T. J., and J. A. Leigh. 2007. Genetic screen for regulatory mutations in Methanococcus maripaludis and its use in identification of induction-deficient mutants of the euryarchaeal repressor NrpR. Appl. Environ. Microbiol. 736595-6600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller, J. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 17.Porter, M. E., and C. J. Dorman. 1997. Differential regulation of the plasmid-encoded genes in the Shigella flexneri virulence regulon. Mol. Gen. Genet. 25693-103. [DOI] [PubMed] [Google Scholar]
- 18.Porter, M. E., and C. J. Dorman. 1994. A role for H-NS in the thermo-osmotic regulation of virulence gene expression in Shigella flexneri. J. Bacteriol. 1764187-4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prosseda, G., P. A. Fradiani, M. Di Lorenzo, M. Falconi, G. Micheli, M. Casalino, M. Nicoletti, and B. Colonna. 1998. A role for H-NS in the regulation of the virF gene of Shigella and enteroinvasive Escherichia coli. Res. Microbiol. 14915-25. [DOI] [PubMed] [Google Scholar]
- 20.Reitzer, L. J., and B. Magasanik. 1986. Transcription of glnA in E. coli is stimulated by activator bound to sites far from the promoter. Cell 45785-792. [DOI] [PubMed] [Google Scholar]
- 21.Rine, J. 1999. On the mechanism of silencing in Escherichia coli. Proc. Natl. Acad. Sci. USA 968309-8311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodionov, O., M. Lobocka, and M. Yarmolinsky. 1999. Silencing of genes flanking the P1 plasmid centromere. Science 283546-549. [DOI] [PubMed] [Google Scholar]
- 23.Rodionov, O., and M. Yarmolinsky. 2004. Plasmid partitioning and the spreading of P1 partition protein ParB. Mol. Microbiol. 521215-1223. [DOI] [PubMed] [Google Scholar]
- 24.Rose, R. E. 1988. The nucleotide sequence of pACYC184. Nucleic Acid Res. 16355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sakai, T., C. Sasakawa, and M. Yoshikawa. 1988. Expression of four virulence antigens of Shigella flexneri is positively regulated at the transcriptional level by the 30 kiloDalton virF protein. Mol. Microbiol. 2589-597. [DOI] [PubMed] [Google Scholar]
- 26.Santapaola, D., F. Del Chierico, A. Petrucca, S. Uzzau, M. Casalino, B. Colonna, R. Sessa, F. Berlutti, and M. Nicoletti. 2006. Apyrase, the product of the virulence plasmid-encoded phoN2 (apy) gene of Shigella flexneri, is necessary for proper unipolar IcsA localization and for efficient intercellular spread. J. Bacteriol. 1881620-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shere, K. D., S. Sallustio, A. Manessis, T. G. D'Aversa, and M. B. Goldberg. 1997. Disruption of IcsP, the major Shigella protease that cleaves IcsA, accelerates actin-based motility. Mol. Microbiol. 25451-462. [DOI] [PubMed] [Google Scholar]
- 28.Steinhauer, J., R. Agha, T. Pham, A. W. Varga, and M. B. Goldberg. 1999. The unipolar Shigella surface protein IcsA is targeted directly to the bacterial old pole: IcsP cleavage of IcsA occurs over the entire bacterial surface. Mol. Microbiol. 32367-377. [DOI] [PubMed] [Google Scholar]
- 29.Stoebel, D. M., A. Free, and C. J. Dorman. 2008. Anti-silencing: overcoming H-NS-mediated repression of transcription in gram-negative enteric bacteria. Microbiology 1542533-2545. [DOI] [PubMed] [Google Scholar]
- 30.Taniya, T., J. Mitobe, S. Nakayama, Q. Mingshan, K. Okuda, and H. Watanabe. 2003. Determination of the InvE binding site required for expression of IpaB of the Shigella sonnei virulence plasmid: involvement of a ParB boxA-like sequence. J. Bacteriol. 1855158-5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tobe, T., S. Nagai, N. Okada, B. Adler, M. Yoshikawa, and C. Sasakawa. 1991. Temperature-regulated expression of invasion genes in Shigella flexneri is controlled through the transcriptional activation of the virB gene on the large plasmid. Mol. Microbiol. 5887-893. [DOI] [PubMed] [Google Scholar]
- 32.Turner, E. C., and C. J. Dorman. 2007. H-NS antagonism in Shigella flexneri by VirB, a virulence gene transcription regulator that is closely related to plasmid partition factors. J. Bacteriol. 1893403-3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Uchiya, K., T. Tobe, K. Komatsu, T. Suzuki, M. Watarai, I. Fukuda, M. Yoshikawa, and C. Sasakawa. 1995. Identification of a novel virulence gene, virA, on the large plasmid of Shigella, involved in invasion and intercellular spreading. Mol. Microbiol. 17241-250. [DOI] [PubMed] [Google Scholar]
- 34.Wing, H. J., A. W. Yan, S. R. Goldman, and M. B. Goldberg. 2004. Regulation of IcsP, the outer membrane protease of the Shigella actin tail assembly protein IcsA, by virulence plasmid regulators VirF and VirB. J. Bacteriol. 186699-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yarmolinsky, M. 2000. Transcriptional silencing in bacteria. Curr. Opin. Microbiol. 3138-143. [DOI] [PubMed] [Google Scholar]