Abstract
The opportunistic pathogen Pseudomonas aeruginosa utilizes a type III secretion system (T3SS) to evade phagocytosis and damage eukaryotic cells. Transcription of the T3SS regulon is controlled by ExsA, a member of the AraC/XylS family of transcriptional regulators. These family members generally consist of an ∼100-amino acid carboxy-terminal domain (CTD) with two helix-turn-helix DNA binding motifs and an ∼200-amino acid amino-terminal domain (NTD) with known functions including oligomerization and ligand binding. In the present study, we show that the CTD of ExsA binds to ExsA-dependent promoters in vitro and activates transcription from ExsA-dependent promoters both in vitro and in vivo. Despite possessing these activities, the CTD lacks the cooperative binding properties observed for full-length ExsA at the PexsC promoter. In addition, the CTD is unaffected by the negative regulatory activity of ExsD, an inhibitor of ExsA activity. Binding studies confirm that ExsD interacts directly with the NTD of ExsA. Our data are consistent with a model in which a single ExsA molecule first binds to a high-affinity site on the PexsC promoter. Protein-protein interactions mediated by the NTD then recruit an additional ExsA molecule to a second site on the promoter to form a complex capable of stimulating wild-type levels of transcription. These findings provide important insight into the mechanisms of transcriptional activation by ExsA and inhibition of ExsA activity by ExsD.
Pseudomonas aeruginosa is an important opportunistic pathogen for patients with burn wounds, cystic fibrosis, and immunodeficiency (34, 35). An important determinant of P. aeruginosa virulence is a type III secretion system (T3SS) (1, 17, 24, 39). T3SSs are found in many gram-negative pathogens and consist of a macromolecular needle complex (injectisome) that functions by translocating toxins (effectors) into eukaryotic host cells (7, 20). Biosynthesis of the P. aeruginosa injectisome is dependent upon >30 gene products and is subject to multiple levels of transcriptional regulation (49). The primary regulator of T3SS gene expression is ExsA, a transcriptional activator of the AraC/XylS family (13, 14). ExsA-dependent transcription is coupled to type III secretory activity through a cascade of three interacting proteins, ExsC, ExsD, and ExsE. ExsD functions as an antiactivator by binding to and inhibiting ExsA-dependent transcription (28). It is unclear whether ExsD inhibits the DNA binding and/or transcriptional activation properties of ExsA. ExsC functions as an antiantiactivator by binding to and inhibiting the negative regulatory activity of ExsD (10). Finally, ExsE is a secretable regulator that binds to and inhibits ExsC activity (37, 43). Activating signals, such as growth under calcium-limiting conditions or contact of P. aeruginosa with eukaryotic cells, convert the type III export machinery to a secretion-competent state (21, 28, 44). Under those conditions, ExsE is secreted into the extracellular milieu or translocated into host cells by the T3SS (42). The corresponding decrease in intracellular ExsE releases ExsC to bind ExsD, thereby relieving ExsD-mediated inhibition of ExsA activity.
Although the ExsCDE regulatory cascade provides a mechanism for controlling ExsA activity, the mechanism of ExsA-dependent transcriptional activation is poorly understood. A total of 10 ExsA-dependent promoters are known to control T3SS gene expression. Transcriptional start sites have been mapped for 4 of the 10 promoters by primer extension (47, 48). In each case, the start sites are spaced downstream of a sequence resembling the −10 region (TATAAT) typical of σ70-dependent promoters. Each ExsA-dependent promoter also contains a −35 region similar to the consensus sequence (TTGACA) for σ70-dependent promoters. The putative −35 and −10 regions for ExsA-dependent promoters match the consensus at four or more positions out of six (4). Unlike typical σ70-dependent promoters where the −35 and −10 sites are separated by ∼17 nucleotides, the presumptive −35 and −10 sites in ExsA-dependent promoters are separated by 21 or 22 nucleotides (4). This observation has raised the question as to whether the −35 regions of ExsA-dependent promoters are authentic determinants for RNA polymerase recruitment (4).
Biochemical data indicate that ExsA is predominantly monomeric in solution and that each ExsA-dependent promoter contains two adjacent binding sites for monomeric ExsA (4). ExsA binding to the PexoT promoter is dependent upon several highly conserved nucleotides located within a region of the promoter that includes the putative −35 hexamer and extends upstream an additional 21 bp (4, 18). The current model proposes that monomeric ExsA binds to a high-affinity site (binding site 1) that overlaps the putative −35 region (4). A second ExsA monomer then binds to a lower affinity upstream site (binding site 2) that bears no obvious similarity in nucleotide sequence to binding site 1 (4). It is unclear whether ExsA binding to the two PexoT promoter sites involves cooperative interactions. The fact that occupation of site 1 by ExsA is required for efficient binding to site 2 is suggestive of cooperative binding interactions. The calculated Hill coefficient for ExsA binding to the PexoT promoter (1.1), however, is not consistent with cooperative binding (4). In contrast, ExsA binding to the PexsC promoter is suggestive of cooperative interactions (Hill coefficient, 1.9). These findings suggest that the mechanism of ExsA binding might differ between T3SS promoters.
The AraC/XylS family of transcriptional activators consists of over 800 members (11). These proteins generally consist of a distinct amino-terminal domain (NTD) and carboxy-terminal domain (CTD). The 100-amino acid CTD is most highly conserved and encodes two helix-turn-helix (HTH) DNA binding motifs (26). The NTD is poorly conserved among distantly related family members and mediates ligand binding and oligomerization (15). Binding of a small molecule ligand is a common regulatory feature of many AraC/XylS family members. The NTD of AraC, the prototypical member of the family, undergoes a conformational change upon binding to arabinose, resulting in preferential binding to adjacent half-sites on the ParaBAD promoter (38). Several AraC/XylS family proteins that regulate T3SS gene expression in mammalian pathogens, however, are known to interact with protein ligands. In some cases, the protein ligands function as coactivators (SicA in Salmonella enterica [9] and IpgC in Shigella flexneri [27]), whereas in other cases, the protein ligands function as antiactivators (OspD1 in Shigella flexneri [29] and ExsD in P. aeruginosa [28]).
The NTD and CTD of AraC/XylS family members are connected by a flexible linker region. In a number of cases, the NTD and CTD have been separated at the linker region to study domain function (2, 12, 19, 22, 23, 31, 32). In the present study, we dissect ExsA into functional NTD and CTD regions and find that the CTD binds DNA, activates transcription both in vitro and in vivo, and is immune to ExsD-mediated inhibition. Our data indicate that the ExsD interaction site is located within the NTD. Based on these data, we propose that interactions mediated by the NTD of ExsA bound to site 1 of the PexsC promoter are involved in the cooperative recruitment of a second ExsA monomer to binding site 2.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Escherichia coli strains were maintained on Luria-Bertani agar plates containing the following antibiotics as necessary: kanamycin (50 μg/ml), gentamicin (15 μg/ml), ampicillin (100 μg/ml), and tetracycline (10 μg/ml). P. aeruginosa strains were maintained on Vogel-Bonner minimal medium (45) with antibiotics as indicated (gentamicin [100 μg/ml], carbenicillin [300 μg/ml], and tetracycline [50 μg/ml]).
Strain UY241 (referred to as the Pcon-exsCEBA strain in the text) was constructed as follows. PCR-generated fragments consisting of chromosome sequences flanking the upstream (1-kb) and downstream (1.5-kb) regions of the PexsC promoter were sequentially cloned into vector pEX18Gm (16). The PexsC promoter (nucleotides −61 to +1 relative to the transcriptional start site) was replaced with a 235-bp fragment containing a derivative of the constitutive E. coli lacUV5 promoter. The resulting plasmid (p2UY36) was mobilized by conjugation to strain PA103, and allelic exchange was performed as previously described (16). The recombination was confirmed by PCR analysis using primers outside of the regions cloned on p2UY36.
Protein expression and purification.
The pET16bexsA expression construct was described previously (4). The pNTD and pCTD expression plasmids were constructed by cloning the regions of exsA encoding amino acids 1 to 180 and 159 to 278, respectively, as NdeI and BamHI restriction fragments into the corresponding sites of pET16b (EMD Biosciences, Madison, WI). E. coli Tuner (DE3) was transformed with each of the expression constructs and grown in Luria-Bertani broth containing 200 μg/ml ampicillin and 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 4 h at 30°C. Bacteria were harvested by centrifugation and suspended in 20 mM Tris (pH 8.0), 500 mM NaCl, 0.5% Tween 20, and 20 mM imidazole supplemented with a protease inhibitor cocktail (complete mini EDTA-free protease inhibitor cocktail; Roche, Indianapolis, IN). Following cell disruption via a French pressure cell, cell lysates were cleared by centrifugation (20,000 × g, 15 min, 4°C) and subjected to Ni2+-affinity chromatography as previously described (28). Peak protein fractions were pooled and dialyzed against ExsA buffer (20 mM Tris [pH 8.0], 500 mM NaCl, 1 mM dithiothreitol [DTT], and 0.5% Tween 20). ExsAHis, NTDHis, and CTDHis were purified to >90% homogeneity as determined by densitometry of silver-stained polyacrylamide gels. Each of the purified proteins was resistant to sedimentation following high-speed centrifugation (100,000 × g, 15 min, 4°C), indicating the absence of aggregates, and each preparation retained activity after prolonged storage at −80°C. Protein concentrations were determined using the Bradford protein assay and bovine serum albumin protein standards to generate a standard curve (Bio-Rad, Hercules, CA) (3).
For the copurification assay, vectors expressing ExsD alone or coexpressing ExsAHis, NTDHis, or CTDHis were generated by cloning exsD into the second multiple cloning site of pCOLADuet-1 (Novagen) as an NdeI-XhoI fragment. The coding sequences for ExsAHis, NTDHis, or CTDHis were then PCR amplified from pET16b (including the amino-terminal histidine tag) and cloned into the first multiple cloning site of pCOLADuet-1 as NcoI-SacI fragments. The entire sequences of all PCR products were confirmed by using nucleotide sequence analysis (University of Iowa DNA facility). E. coli strains expressing the indicated proteins were grown as described above for protein purification. Cell culture (1.25 ml harvested at an absorbance [A600] of 1.0) was pelleted by centrifugation and resuspended in 300 μl of binding buffer (20 mM Tris [pH 8.0], 150 mM NaCl, 0.5% Tween 20, 10% glycerol, and 10 mM imidazole). Cells were disrupted by sonication for 10 s, unbroken cells were removed by centrifugation at 4°C for 5 min, and an aliquot was saved as the unbound fraction. Supernatants were incubated with 50 μl of Talon metal-affinity resin (Clontech, Mountain View, CA) equilibrated in binding buffer. After 60 min, the resin was sedimented and washed three times with 500 μl of binding buffer containing 20 mM imidazole, and bound protein was eluted with 300 μl of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer. The unbound and bound protein samples were subjected to SDS-PAGE and analyzed by immunoblot analyses.
SDS-PAGE, immunoblots, and β-galactosidase assays.
Whole-cell lysate samples were prepared by growing cells in trypticase soy broth (TSB) to an A600 of 1.0. Samples were prepared by pelleting 1.25 ml of cell culture, suspending the pellet in 0.25 ml of SDS-PAGE sample buffer, and sonicating it for 10 s. Samples were analyzed by SDS-PAGE followed by either silver staining, Coomassie blue staining, or immunoblotting using antibodies directed against ExsA, ExsD, or the histidine epitope tag.
The activities of the PexsC-lacZ, PexsD-lacZ, and PexoT-lacZ reporters (28) were determined as follows. Strains were grown overnight on Vogel-Bonner minimal medium plates at 37°C. Bacteria were then scraped from the plate, suspended in TSB, back-diluted to an A600 of 0.1 in 10 ml TSB supplemented with 100 mM monosodium glutamate, 1% glycerol, and 2 mM EGTA as indicated, and incubated at 30°C until the A600 reached 1.0. β-Galactosidase assays were performed as previously described (28).
In vitro transcription assays.
A supercoiled transcription template was generated by cloning the PexsD promoter (nucleotides −444 to +186 relative to the transcriptional start site) as an EcoRI fragment into pOM90 (36). This vector incorporates a strong rpoC transcriptional terminator downstream of the cloning site, resulting in a terminated transcript of 261 nucleotides. Single-round transcription assays were performed by incubating ExsAHis, CTDHis, or NTDHis with supercoiled PexsD template (0.5 nM) at 25°C in 1× transcription buffer (40 mM Tris-HCl [pH 7.5], 150 mM KCl, 10 mM MgCl2, 0.01% Tween 20, and 1 mM DTT) containing 0.75 mM rATP (the initiating nucleotide of the exsD transcript) (C. A. Vakulskas and T. L. Yahr, unpublished results). After 10 min, 0.5 U (25 nM final concentration) of σ70-saturated E. coli RNA polymerase holoenzyme (Epicentre Biotechnologies, Madison, WI) was added, and open complexes were allowed to form for 5 min before the addition of the remaining unlabeled nucleotides (0.75 mM each of rUTP, rGTP, and rCTP) and 5 μCi [α32P]CTP in 1× transcription buffer containing heparin (50 μg/ml final concentration). Transcription was allowed to proceed for 10 min at 30°C before reactions (20 μl final volume) were terminated by adding an equal volume (20 μl) of stop buffer (98% formamide, 20 mM EDTA, 0.05% bromophenol blue, and 0.05% xylene cyanol). Reaction mixtures were heated to 95°C for 5 min and electrophoresed on 5% denaturing urea polyacrylamide gels. All phosphorimaging and densitometry were performed using an FLA-7000 phosphorimager (Fujifilm) and MultiGage v3.0 software (Fujifilm).
Electrophoretic mobility shift and circular permutation assays.
DNA probes containing ExsA-dependent promoters (200 bp), as well as a nonspecific fragment from the coding region of pscF (160 bp), were generated by PCR analysis and end labeled with 20 μCi [γ-32P]-ATP (PerkinElmer) and 10 U polynucleotide kinase (New England Biolabs) as previously described (4). Electrophoretic mobility shift assay (EMSA) reaction mixtures (19 μl) containing end-labeled specific and nonspecific probes (0.25 nM each), 25 ng/μl poly(2′-deoxyinosinic-2′-deoxycytidylic acid) (Sigma-Aldrich, St. Louis, MO), and ExsA DNA binding buffer (10 mM Tris [pH 7.9], 50 mM KCl, 1 mM EDTA, 1 mM DTT, 5% glycerol, and 100 μg/ml bovine serum albumin) were incubated for 5 min at 25°C. ExsAHis, NTDHis, or CTDHis were added to the indicated concentrations in a final reaction mixture volume of 20 μl and incubated at 25°C for 15 min. Samples were subjected to electrophoresis on 5% polyacrylamide glycine gels (10 mM Tris [pH 7.5], 380 mM glycine, 1 mM EDTA) at 4°C. Circular permutation assays were performed as previously described (4).
RESULTS
Functional domains of ExsA.
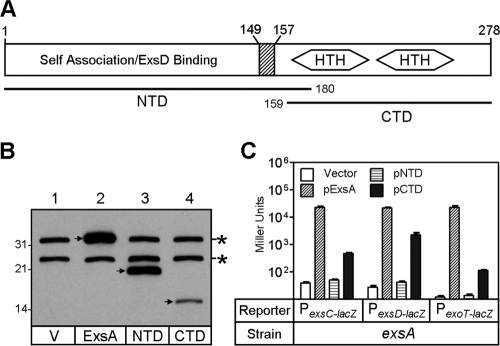
The NTD and CTD of AraC, MarA, and Rob are separated by a flexible linker sequence (25, 33, 40). Based upon an alignment of the primary amino acid sequence of ExsA with AraC, MarA, and Rob, we predicted that the NTD and CTD of ExsA are also separated by a flexible linker region located near amino acid 160 (Fig. 1A). This prediction was in agreement with a protein linker algorithm (41) that localized the linker region to amino acids 149 to 157. Based on this information, regions of exsA encoding the NTD (amino acids 1 to 180) (Fig. 1A) and CTD (amino acids 159 to 278) were cloned into an arabinose-inducible expression vector that adds an amino-terminal decahistidine tag. ExsAHis, NTDHis, and CTDHis were expressed in an exsA mutant, and expression levels were monitored by using immunoblotting with an antibody that recognizes the histidine fusion tag. Whereas the steady-state expression level of the NTD was comparable to that of the full-length ExsA, the level of the CTD was noticeably reduced (Fig. 1B). Despite the reduced steady-state level of the CTD, subsequent analyses (discussed below) found that both the NTD and CTD possess distinct activities, indicating that discrete functional domains had been defined.
FIG. 1.
Characterization of ExsA domains. (A) Regions corresponding to the NTD and CTD constructs used in this work are indicated with solid lines. The HTH motifs of ExsA as well as the predicted linker region (hatched box) are indicated. Amino acids are numbered with respect to the initiating methionine. (B) Steady-state expression levels of ExsAHis, NTDHis, and CTDHis in P. aeruginosa. A vector control (pJN105) (lane 1) (V) or arabinose-inducible plasmids expressing either full-length ExsAHis (lane 2), NTDHis (lane 3), or CTDHis (lane 4) were introduced into an exsA mutant. Cells were grown in T3SS-inducing medium in the presence of arabinose (0.5%), and cell lysate samples were subjected to immunoblot analyses using an anti-histidine monoclonal antibody. The expressed products are indicated with arrows, and cross-reactive bands that serve as loading controls are indicated with asterisks. (C) Transcriptional activation by ExsA, NTD, and CTD. A vector control or plasmids expressing full-length ExsA, NTD, or CTD were introduced into an exsA mutant carrying the PexsC-lacZ, PexsD-lacZ, or PexoT-lacZ transcriptional reporters. Strains were grown under inducing conditions for T3SS gene expression in the presence of arabinose (0.5%) and assayed for β-galactosidase activity. Note that the y axis is plotted on a logarithmic scale. Reported values (Miller units) are the averages from three independent experiments, and error bars indicate the standard errors of the means.
The CTD activates T3SS promoters in vivo and in vitro.
To determine whether the NTD or CTD could activate transcription in vivo, expression plasmids were introduced into an exsA mutant carrying one of three previously described transcriptional reporters for T3SS gene expression (PexsC-lacZ, PexsD-lacZ, or PexoT-lacZ) (28). The resulting strains were grown under conditions permissive for T3SS gene expression (Ca2+ limiting) and assayed for β-galactosidase activity. The NTD, lacking the DNA binding domain, was unable to stimulate expression of the lacZ reporters (Fig. 1C). The CTD, however, activated expression of each of the reporters, although to significantly lower levels than the expression levels seen with full-length ExsA. Given the reduced steady-state levels of the CTD compared to those of ExsA (Fig. 1B), it was unclear whether the decreased activity of the CTD resulted from a difference in expression levels or a defect in DNA binding and/or transcriptional activation.
To more quantitatively compare the transcriptional activation properties of ExsA to the CTD, we employed in vitro transcription assays. ExsAHis, NTDHis, and CTDHis were purified from E. coli as histidine-tagged fusion proteins to >90% homogeneity as judged by silver-stained SDS-polyacrylamide gels (Fig. 2A). Single-round transcription assays were performed using a plasmid template carrying the PexsD promoter cloned upstream of a strong transcriptional terminator. RNA polymerase alone (Fig. 2B, lane 1) or in the presence of 20 nM NTDHis (data not shown) was unable to generate a transcript from the PexsD promoter. The addition of either ExsAHis (Fig. 2B, lanes 2 to 7) or CTDHis (Fig. 2B, lanes 8 to 13), however, generated a terminated transcript of the expected size. Neither ExsAHis nor CTDHis had an effect on a constitutive transcript produced from a plasmid-borne promoter (Fig. 2B). Compared to ExsA, approximately fourfold-higher concentrations of CTDHis were required to generate an equivalent amount of terminated transcript. We were unable to determine the concentration of CTDHis required for half-maximal activation because protein concentrations higher than those utilized in Fig. 2B nonspecifically inhibit the in vitro transcription assay (data not shown). These data indicate that most if not all of the transcriptional activation properties of ExsA are located within the CTD.
FIG. 2.
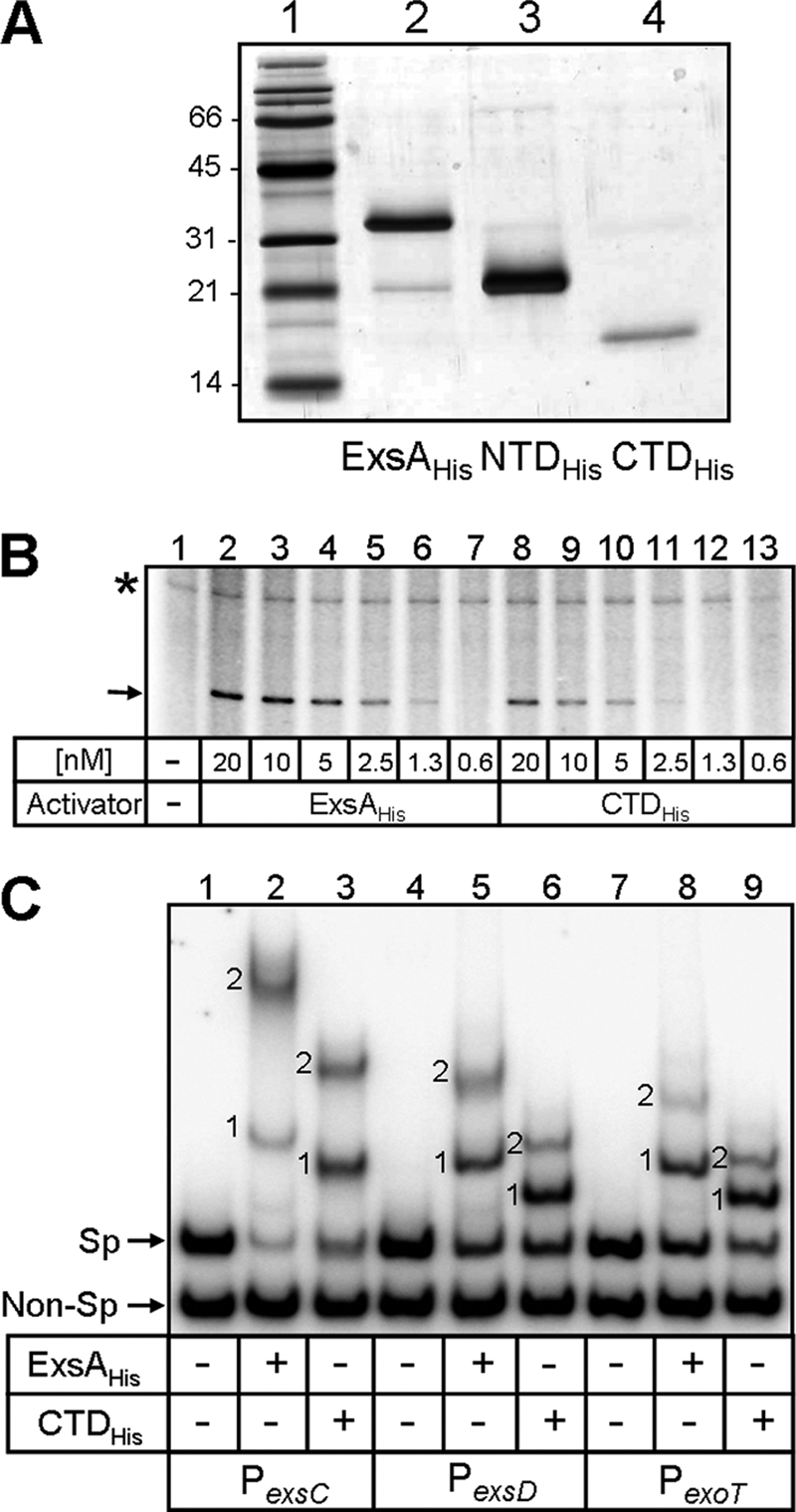
DNA binding and transcriptional activation properties of ExsAHis, NTDHis, and CTDHis. (A) ExsAHis, NTDHis, and CTDHis were purified from E. coli by using Ni2+-affinity chromatography, and samples (1 μg) were subjected to SDS-PAGE and detected by silver staining. Molecular mass standards (in kDa) are indicated on the left. (B) A plasmid template carrying the PexsD promoter followed by a strong transcriptional terminator was incubated alone (lane 1) or in the presence of ExsAHis (lanes 2 to 7) or CTDHis (lanes 8 to 13) at the indicated concentrations for 10 min with rATP. RNA polymerase was then added for 5 min to allow for open complex formation. A single round of elongation (10 min at 30°C) was initiated by adding heparin, ribonucleotides, and [α-P32]rCTP. Reaction mixtures were analyzed by electrophoresis, followed by phosphorimaging. ExsA-dependent (arrow) and ExsA-independent (asterisk) transcripts are indicated. (C) EMSA comparing the DNA binding properties of ExsAHis and CTDHis. Nonspecific (Non-Sp) and specific (Sp) promoter fragments (PexsC, PexsD, and PexoT) were incubated in the absence (−) or presence (+) of ExsAHis (74 nM) or CTDHis (280 nM) for 15 min. Samples were subjected to electrophoresis and phosphorimaging. Shift products 1 and 2 for each of the promoter fragments are indicated.
DNA binding properties of the CTD.
We next examined the DNA binding activity of the CTD by EMSA. For these assays, ExsAHis, NTDHis, and CTDHis were incubated with a specific promoter probe (PexsC, PexsD, or PexoT) and an equivalent amount of a nonspecific probe. Samples were then analyzed by PAGE and phosphorimaging. As expected, NTDHis lacked DNA binding activity (data not shown), whereas ExsAHis (74 nM) specifically retarded the mobility of the PexsC, PexsD, and PexoT promoter probes, resulting in the appearance of two distinct shift products (1 and 2) (4) (Fig. 2C, lanes 2, 5, and 8). A previous study found that shift products 1 and 2 result from the binding of an ExsA monomer to binding site 1 followed by occupation of site 2, respectively (4) (Fig. 3A). The CTD (280 nM) also specifically bound each of the promoter probes, generating shift products 1 and 2 (Fig. 2C, lanes 3, 6, and 9). The noticeable difference in migration of the shift products generated by ExsAHis and CTDHis presumably reflects the difference in molecular masses of CTDHis (17 kDa) and ExsAHis (34 kDa). Formation of similar amounts of shift products 1 and 2 at each of the promoters required a fourfold-higher concentration of CTDHis relative to that of ExsAHis. These data suggest that the CTD has a lower binding affinity for T3SS promoters than ExsAHis or that the specific activity of purified CTDHis is lower than that of ExsAHis.
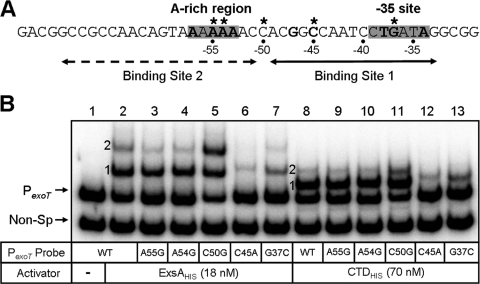
FIG. 3.
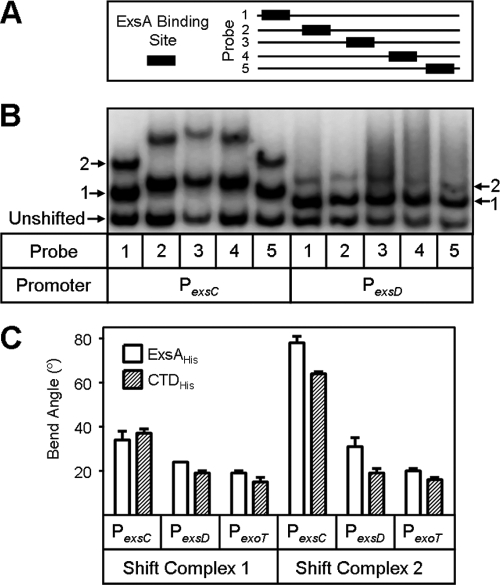
ExsAHis and CTDHis preferentially bind to site 1 on the promoter PexoT. (A) Diagram of the promoter PexoT. The putative −35 consensus and adenine-rich regions are indicated and highlighted with gray boxes. Nucleotides highly conserved in all ExsA-dependent promoters are bold, and those changed by site-directed mutagenesis are indicated with asterisks. ExsA binding sites 1 (solid arrow) and 2 (dashed arrow) are indicated. (B) EMSA comparing the DNA binding of ExsAHis and CTDHis to PexoT promoter mutants. PexoT probes with the indicated point mutations and a nonspecific (Non-Sp) probe were incubated in the absence (−) or presence (+) of 18 nM ExsAHis or 70 nM CTDHis for 15 min. Samples were subjected to electrophoresis and phosphorimaging. Shift products 1 and 2 for each of the promoter fragments are indicated.
ExsAHis and CTDHis preferentially bind to site 1 on the PexoT promoter.
Maximal formation of shifts products 1 and 2 at the PexoT promoter is dependent upon several highly conserved nucleotides (Fig. 3A) that constitute the ExsA consensus-binding site (4). To determine whether the same nucleotides are required for binding by the CTD, PexoT promoter probes carrying mutations within binding site 1 (C45A or G37C), binding site 2 (A55G or A54G), or a nonconserved position (C50G) were subjected to EMSA in the presence of ExsAHis or CTDHis. As previously reported, mutations within binding site 2 have little effect on the formation of shift product 1 by ExsAHis (Fig. 3B, lanes 3 and 4), while binding site 1 mutations result in a significant reduction in both shift products 1 and 2 (4) (Fig. 3B, lanes 6 to 7). Similar results were seen when using the CTDHis (Fig. 3B). These findings indicate that the binding of ExsA and CTD to site 1 is required for maximal formation of shift products 1 and 2 and further suggest that binding to site 1 precedes the occupation of site 2.
The NTD is required for cooperative interaction at the PexsC promoter.
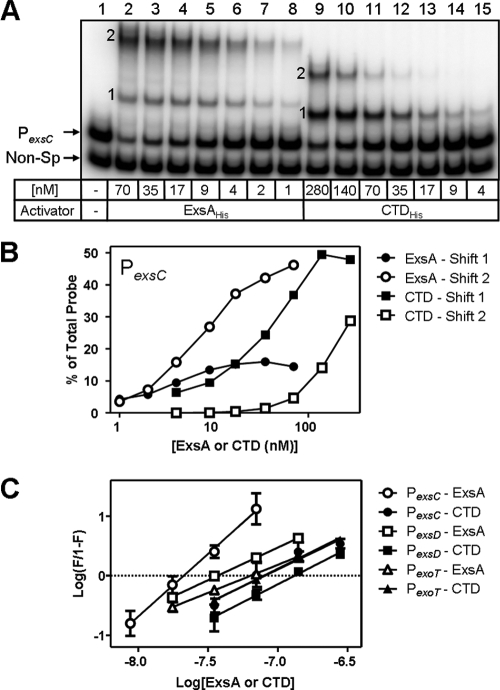
The previously determined Hill coefficient of 1.9 ± 0.2 for ExsAHis binding to the PexsC promoter is suggestive of cooperative binding (4). Since the NTDs of some AraC/XylS family members are involved in self-interactions and cooperative binding (15), we tested whether CTDHis binding was also cooperative. EMSA experiments using the PexsC promoter probe were performed over a range of ExsAHis or CTDHis concentrations. As noted previously (4), low concentrations of ExsAHis result in the simultaneous appearance of shift products 1 and 2, whereas higher concentrations result in the preferential accumulation of shift product 2 with no net increase in the amount of shift product 1 (4) (Fig. 4A, lanes 2 to 8). In contrast, however, titrations with CTDHis resulted in the preferential formation of shift product 1, followed by the eventual appearance of shift product 2 at higher concentrations of CTDHis (Fig. 4A, lanes 9 to 15). In addition, at protein concentrations sufficient to shift 50% of the probe, the ratio of shift product 2 to shift product 1 (P2/P1) generated by ExsAHis was approximately 3:1, whereas this ratio is reversed (ca. 1:3) for products generated by CTDHis. These findings indicate that CTDHis lacks the cooperative binding properties seen for ExsAHis binding to the PexsC promoter. In support of this conclusion, the calculated Hill coefficients for binding of ExsAHis and CTDHis to the PexsC promoter were 2.1 ± 0.3 and 1.2 ± 0.2, respectively (Fig. 4C). These data indicate that cooperative binding of ExsAHis to the PexsC promoter is dependent upon the NTD.
FIG. 4.
The CTD lacks cooperative binding to the PexsC promoter. (A) The PexsC promoter and a nonspecific (Non-Sp) DNA fragment were incubated in the absence (lane 1) or presence of the indicated concentrations of ExsAHis (lanes 2 to 8) or CTDHis (lanes 9 to 15) for 15 min at 25°C, followed by electrophoresis and phosphorimaging. Shift products 1 and 2 are indicated. (B) Quantitation of shift products 1 and 2 formed by ExsAHis and CTDHis. The reported values represent the percentage of shift products 1 and 2 generated (y axis) relative to that of the unshifted probe (A, lane 1) plotted as a function of ExsAHis or CTDHis concentration (x axis). (C) Hill plots used to determine the Hill coefficient for ExsAHis and CTDHis binding to the PexsC, PexsD, and PexoT promoters. The amount of probe shifted in EMSA experiments (y axis; log[F/1 − F]), where F equals the fraction of probe shifted) was plotted as a function of ExsAHis or CTDHis concentration (x axis; log[ExsAHis or CTDHis]). The Hill coefficients were calculated as previously described (4), and the reported values are the averages from three independent experiments, with error bars representing the standard errors of the means.
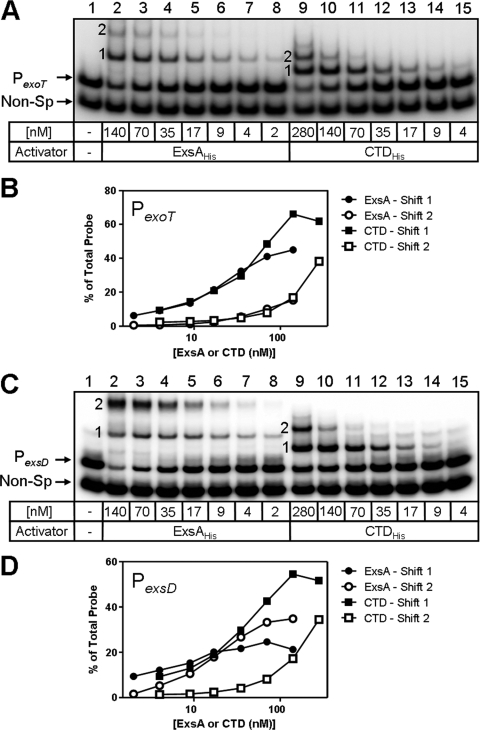
Although ExsA likely binds cooperatively to the PexsC promoter, the manner in which it binds to the PexoT and PexsD promoters is less clear. EMSA experiments using the PexoT promoter demonstrate that when 50% of the probe is bound, the ratio of shift product 2 to shift product 1 is similar (P2/P1, ∼1:1) for both ExsAHis and the CTDHis (Fig. 5A and B). In addition, the Hill coefficients for binding of ExsAHis and CTDHis to the PexoT (1.0 ± 0.1 and 1.1 ± 0.1, respectively) are nearly identical (Fig. 4C). These data suggest a lack of cooperative binding and indicate that the NTD has little effect on ExsA binding to the PexoT promoter.
FIG. 5.
EMSA experiments demonstrating the binding of ExsAHis and CTDHis to the PexoT and PexsD promoters. (A and C) EMSA experiments using radiolabeled PexoT (A) or PexsD (C) promoter probes and a nonspecific (Non-Sp) DNA fragment. Probes were incubated in the absence (lane 1) or presence of the indicated concentrations of ExsAHis (lanes 2 to 8) or CTDHis (lanes 9 to 15) as indicated for 15 min at 25°C, followed by electrophoresis and phosphorimaging. Shift products 1 and 2 are indicated. (B and D) Quantitation of shift products 1 and 2 from Fig. 5A (PexoT) and Fig. 5C (PexsD) was performed as described in legend to Fig. 4.
EMSA experiments using the PexsD promoter probe reveal that shift products 2 and 1 are generated at the same rate by ExsAHis, demonstrating a P2/P1 ratio of 1:1 (Fig. 5C and D). In contrast, CTDHis preferentially generates shift product 1 followed by delayed formation of shift product 2 at elevated CTDHis concentrations (P2/P1, ∼1:4). In this regard, binding of CTDHis to the PexsD and PexsC promoters appears to be quite similar. A notable difference, however, is that the Hill coefficients for binding of ExsAHis and CTDHis to the PexsD promoter (1.1 ± 0.1 and 1.2 ± 0.2) are nearly identical and not supportive of cooperativity. These findings suggest that the mechanisms by which ExsA interacts with the PexsC, PexoT, and PexsD promoters are different.
DNA bending at T3SS promoters is mediated by the CTD.
A previous study found that ExsA induces moderate bending of the PexsD and PexoT promoters and more pronounced bending of the PexsC promoter (4). To test whether the CTD is capable of inducing bending, we used circular permutation assays. These assays are based on the fact that DNA fragments with a bend near the middle migrate more slowly in an EMSA than an unbent fragment of the same size or fragments having a bend near the ends (8). Promoter probes were generated by positioning the ExsA binding site from the PexsC, PexsD, or PexoT promoters at regular intervals along an ∼200-bp DNA fragment (Fig. 6A). The probes were then incubated with CTDHis and subjected to EMSA. Similar to previous findings with ExsA (4), CTDHis induced pronounced DNA bending of the PexsC promoter probe and slight bending of the PexsD (Fig. 6B) and PexoT (data not shown) promoter probes. Importantly, the probes showed no evidence of bending in the absence of ExsAHis (4) or CTDHis. The degree of bending induced by CTDHis was similar to that seen previously with ExsAHis for each of the promoters (4) (Fig. 6C). Based on these data, we conclude that the NTD and, by inference, cooperative DNA binding between ExsA monomers do not play significant roles in DNA bending.
FIG. 6.
DNA bending properties of the CTD. (A) The position of the ExsA binding site (70 bp) within each EMSA probe (196 bp) is indicated (black boxes). (B) Promoter fragments (PexsC or PexsD; PexoT not shown) were incubated with 280 nM CTDHis for 15 min, followed by electrophoresis and phosphorimaging. Unshifted probe and shift products 1 and 2 are indicated. (C) Calculated bend angles generated by CTDHis for the PexsC, PexsD, and PexoT promoters. Bending values for ExsAHis were previously reported (4). Values for the CTD are the averages from three independent experiments, and error bars represent the standard errors of the means.
The NTD directly interacts with ExsD.
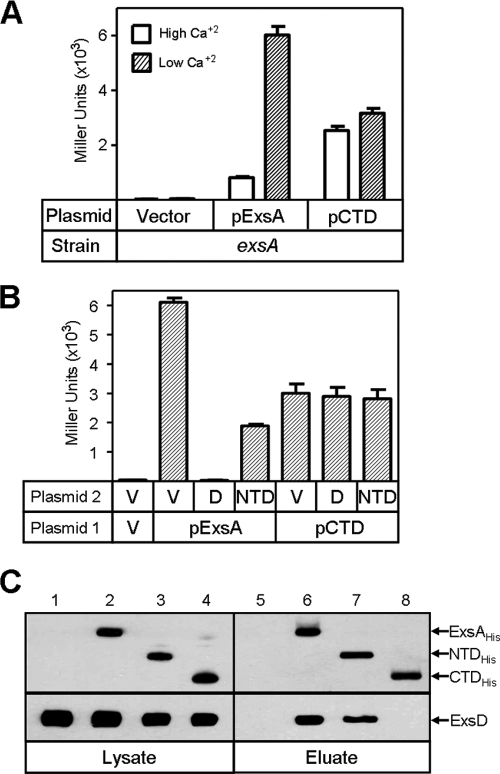
Under noninducing (Ca2+-replete) conditions for T3SS gene expression, ExsD binds to ExsA and inhibits transcription (28). To determine whether transcriptional activation by the CTD is sensitive to ExsD-dependent inhibition, the pExsA and pCTD expression plasmids were introduced into an exsA mutant carrying a PexsD-lacZ transcriptional reporter. Transformants were cultured under noninducing and inducing conditions for T3SS gene expression and assayed for β-galactosidase activity. Under noninducing conditions, ExsA-dependent expression of the PexsD-lacZ reporter is inhibited due to the activity of ExsD (Fig. 7A). Activation of the reporter by the CTD, however, was similar under both noninducing and inducing conditions for T3SS gene expression. This finding indicates that the CTD is insensitive to the inhibitory activity of ExsD. To further test this idea, the strains described above were transformed with a second plasmid expressing ExsD or a vector control. Whereas overexpression of ExsD completely inhibited ExsA-dependent expression of the PexsD-lacZ reporter, activation of the reporter by the CTD was unaffected by overexpressed ExsD (Fig. 7B). Similar results were obtained using the PexsC-lacZ and PexoT-lacZ reporters (data not shown). These findings suggest that the binding of ExsD to ExsA is mediated through the NTD. Interestingly, when the strains described above were transformed with a plasmid expressing the NTD, ExsA-dependent transcription decreased approximately threefold, while CTD-dependent transcription was unaffected (discussed below).
FIG. 7.
The NTD interacts with ExsD. (A) Vector control or a plasmid expressing either full-length ExsA or CTD was introduced into an exsA mutant carrying the PexsD-lacZ transcriptional reporter. Strains were grown in the presence of arabinose (0.5%) under Ca2+-replete (white bars) or Ca2+-limiting (hatched bars) conditions and assayed for β-galactosidase activity. (B) The CTD is immune to inhibition by ExsD and to the dominant negative activity of the NTD. An exsA mutant carrying a vector control (V), pExsA, or pCTD was transformed with a second plasmid [vector control, pExsD (D), or pNTD]. Cells were cultured in the presence of arabinose (0.5%) under conditions permissive for T3SS gene expression and assayed for expression of the PexsD-lacZ reporter. The reported values (Miller units) in panels A and B are the averages from three independent experiments, and error bars represent the standard errors of the means. (C) The NTD directly interacts with ExsD. Cell lysates were prepared from E. coli expressing ExsD alone (lanes 1 and 5) or coexpressing ExsD with ExsAHis (lanes 2 and 6), NTDHis (lanes 3 and 7), or CTDHis (lanes 4 and 8). Lysates were incubated with Talon metal-affinity resin and washed, and bound protein was eluted with SDS-PAGE sample buffer. The starting lysate (lanes 1 to 4) and eluate (lanes 5 to 8) samples were subjected to SDS-PAGE and immunoblotted using antibody against ExsA or ExsD as indicated.
To determine if the NTD binds to ExsD, we employed copurification assays. Untagged ExsD was expressed alone or coexpressed with ExsAHis, NTDHis, or CTDHis in E. coli. Cell lysates were then mixed with Talon metal-affinity resin, the resin was washed, and bound histidine-tagged proteins were eluted. Although ExsD was present in each of the whole-cell lysate samples, ExsD was detected only in the eluate samples when coexpressed with either ExsAHis or NTDHis (Fig. 7C). These data indicate that the NTD of ExsA is sufficient for binding ExsD.
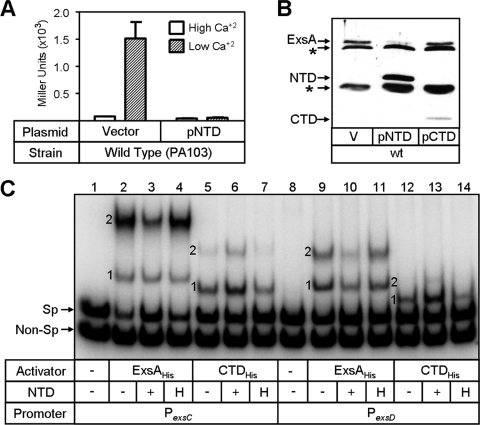
The NTD has dominant negative activity.
Based on the finding that the NTD interacts with ExsD, we predicted that overexpression of the NTD in a wild-type cell might deregulate T3SS gene expression by sequestering ExsD and thereby allowing chromosomally encoded ExsA to constitutively activate transcription even under Ca2+-replete conditions. To test this idea, wild-type PA103 carrying the PexsD-lacZ reporter was transformed with the pNTD expression plasmid and assayed for β-galactosidase activity. Contrary to our expectations, expression of the NTD had a dominant negative effect on T3SS gene expression irrespective of whether cells were cultured under Ca2+-replete or Ca2+-limiting conditions (Fig. 8A). Because transcription of the exsCEBA operon is autoregulated by ExsA, it was unclear whether the dominant negative activity of the NTD was occurring at the transcriptional or posttranscriptional levels. To address this question, we generated a strain (Pcon-exsCEBA) in which the ExsA-dependent promoter (PexsC) driving expression of the exsCEBA operon on the chromosome was replaced with a constitutive derivative of the lac promoter (Pcon). Expression of the NTD in this strain also inhibited transcription of the PexsD-lacZ reporter (data not shown), and immunoblotting revealed that coexpression of the NTD decreased the steady-state expression levels of ExsA. In contrast, coexpression of the CTD had no effect on ExsA levels (Fig. 8B). Since transcription of the exsCEBA operon in the Pcon-exsCEBA strain is independent of ExsA (data not shown), the dominant negative activity of the NTD is likely occurring at the posttranscriptional level. These data suggest that the NTD interacts with full-length ExsA and affects ExsA translation and/or stability.
FIG. 8.
Inhibitory activity of the NTD. (A) Wild-type P. aeruginosa carrying a vector control or a plasmid expressing the NTD was cultured in the presence of arabinose (0.5%) under Ca2+-replete (white bars) or Ca2+-limiting (hatched bars) conditions and assayed for expression of the PexsD-lacZ reporter. (B) Overexpression of the NTD in P. aeruginosa reduces the steady-state expression levels of ExsA. Cell lysates harvested from P. aeruginosa Pcon-exsCEBA carrying a vector control (V) or a plasmid expressing either the NTD (pNTD) or CTD (pCTD) were subjected to immunoblot analysis using ExsA antibody. Bands corresponding to ExsA, the NTD, the CTD, and cross-reactive proteins (asterisks) are indicated. wt, wild type. (C) Inhibition of ExsA-DNA complex formation by the NTD. Specific (Sp) and nonspecific (Non-Sp) promoter fragments were incubated in the absence (−) or presence of ExsAHis (18 nM or 36 nM for PexsC and PexsD, respectively) or CTDHis (70 nM) for 5 min. An equivalent volume of buffer (−), NTD (1.7 nM) (+), or heat-inactivated NTD (1.7 nM) (H) was then added and incubated for 10 min, followed by electrophoresis and phosphorimaging.
We hypothesized that binding of the NTD to full-length ExsA might inhibit DNA binding activity in vitro. In support of this, coincubation of the NTD with ExsAHis reduced the amount of shift product 2 for both the PexsC and PexsD promoter probes compared to ExsAHis alone (Fig. 8C, lane 2 versus lane 3 and lane 9 versus lane 10). Inhibition by NTDHis appears to be specific as the activity was heat sensitive (Fig. 8C, lanes 4 and 11). If the inhibitory effect of the NTD on generation of the shift product 2 resulted from interactions between the NTD and the amino terminus of full-length ExsA, then DNA binding by the CTD should be immune to the NTD. Two pieces of data confirm that this is indeed the case. First, the addition of NTDHis does not inhibit binding of CTDHis to the PexsC or PexsD promoters (Fig. 8C, compare lane 5 to lane 6 and lane 12 to lane 13) but rather seems to enhance generation of both shift products 1 and 2, and this enhancement was heat labile (Fig. 8C, lanes 7 and 14). Second, although expression of the NTD reduced ExsA-dependent expression of the PexsD-lacZ reporter approximately threefold in vivo, there was no effect of the NTD on CTD-dependent expression of the reporter (Fig. 7B).
DISCUSSION
AraC/XylS family members are prominent in regulatory pathways controlling T3SS gene expression in a number of mammalian pathogens (5). Although few of these proteins have been studied in detail, the best characterized share the following properties: (i) allosteric regulation by protein ligands rather than small molecules (9, 28-30), (ii) binding to long, poorly defined degenerate DNA sequences (4), and (iii) a domain structure consisting of an NTD followed by a DNA-binding CTD. Being amenable to purification and biochemical analyses, ExsA serves as a model system to better understand AraC/XylS family members involved in controlling T3SS gene expression. In the present study, we find that the NTD is highly stable in vitro (Fig. 2A) and can be maintained at relatively high concentrations (>1 mg/ml) in the absence of detergent without losing solubility (E. D. Brutinel and T. L. Yahr, unpublished data). In contrast, ExsA and the CTD are prone to aggregation at moderate protein concentrations (>0.5 mg/ml), and maintenance of solubility is dependent upon the presence of detergent (0.5% Tween 20). These findings indicate that the CTD is the primary contributor to the poor solubility of full-length ExsA at protein concentrations >0.5 mg/ml under the conditions tested herein.
High concentrations of ExsA (74 nM) or the CTD (280 nM) in DNA binding assays generate shift products 1 and 2 for the PexsC, PexsD, and PexoT promoter probes (Fig. 2B). A previous study concluded that shift products 1 and 2 result from the sequential binding of monomeric ExsA to binding sites 1 and 2 on the PexoT promoter, respectively (4). The CTD likely interacts with the PexoT promoter in the same manner because point mutants within binding site 2 have little effect formation of shift product 1, while point mutants within binding site 1 significantly decrease binding to sites 1 and 2 (Fig. 3B). Whether binding of the two ExsA monomers to the PexsC, PexsD, or PexoT promoter involves cooperative interactions has been an open question. For the PexsC promoter, we now conclude that ExsA binding is cooperative based upon the following data. First, titration of ExsA into DNA binding assays results in the preferential accumulation of shift product 2, while the amount of shift product 1 remains steady (Fig. 4A). This observation indicates that the second binding event is intimately linked to the first binding event. Second, titrations with the CTD result in the preferential formation of shift product 1 and a delay in the formation of shift product 2. This suggests that the NTD facilitates binding of a second ExsA monomer to the PexsC promoter. Finally, whereas the Hill coefficient for ExsA binding to the PexsC promoter (2.1 ± 0.3) is indicative of positive cooperativity, the Hill coefficient for the CTD (1.2 ± 0.2) is not. These combined data are consistent with a model in which monomeric ExsA binds to the PexsC promoter and, through interactions mediated by the NTD, recruits a second ExsA molecule to the promoter.
Two additional pieces of data are also suggestive of a role for the NTD in mediating dimeric interactions between ExsA monomers. First, overexpression of the NTD results in a dominant negative phenotype due to degradation of ExsA (Fig. 8B). Because the CTD is immune to NTD-dependent degradation, the NTD is likely interacting with the NTD of full-length ExsA to promote degradation. Second, the NTD inhibits the DNA binding activity of ExsAHis but has no effect on binding by the CTDHis (Fig. 8C). It remains unclear whether the dominant negative activity seen for the NTD in vivo results from degradation, inhibition of DNA binding, or a combination of both. Although these combined data suggest that ExsA monomers are capable of interacting with one another, ExsA purified from E. coli is largely monomeric in solution (4). We propose that interactions between ExsA monomers are dependent upon the NTD and occur minimally in solution but are enhanced when ExsA monomers are bound to the PexsC promoter.
The role of cooperativity in binding of ExsA to the PexoT and PexsD promoters remains poorly defined. A number of observations are suggestive of cooperative binding to the PexoT promoter, as follows: (i) promoter mapping studies indicate that ExsA binding to site 1 is required for occupation of site 2 (4); (ii) mutations within binding site 1 have a significant effect on ExsA binding to site 2, whereas mutations within binding site 2 have only a minimal effect on binding to site 1 (4); and (iii) DNase I footprinting studies reveal no evidence of preferential filling of binding sites 1 or 2, suggesting that occupation of site 1 results in the immediate occupation of binding site 2 (4). At odds with these data, however, are the findings that the NTD has little to no effect on ExsAHis binding to the PexoT promoter (Fig. 5A and B) and that the Hill coefficients for ExsAHis and CTDHis binding are similar and not supportive of cooperativity. If binding of ExsA to the PexoT promoter does not involve cooperative interactions, however, it is difficult to envision how mutations in binding site 1 would affect binding to site 2. Data regarding the question of cooperative binding of ExsA to the PexsD promoter are more limited. While the Hill coefficients for ExsAHis and CTDHis binding are not supportive of cooperativity (Fig. 4C), the binding properties of ExsAHis and CTDHis differ significantly and indicate a role for the NTD in binding to the PexsD promoter (Fig. 5C and D). While no solid conclusions can be made from these data, it is evident that the manner in which ExsA interacts with the PexsC, PexsD, and PexoT promoters is different in each case and requires further investigation.
The CTD activates transcription from T3SS promoters in vitro; however, a fourfold-higher concentration of CTDHis relative to ExsAHis is required to yield similar levels of single-round transcription products (Fig. 2B). This difference is in close agreement with the approximately fourfold-reduced binding affinity of the CTD measured by EMSA, suggesting that the CTD contains all of the necessary determinants for transcriptional activation and that once bound to the promoter, the CTD is able to activate transcription to the same level seen with full-length ExsA. Previous studies have reported the DNA binding and transcriptional activation properties for the CTD of several AraC/XylS family members. For AraC, which displays cooperative binding, the CTD is unable to activate transcription of the ParaBAD promoter because the CTD binds only one of the necessary half sites (araI1). The addition of a heterologus dimerization domain to the CTD, however, results in the occupation of both half sites (araI1 and araI2) (6). The CTD of MelR, the transcriptional activator of the melAB operon in E. coli also binds only one half site (sites 1 and 2 but not sites 1′ and 2′) and is unable to activate transcription (19). In contrast, the CTDs of RhaS (46) and XylS (22) constitutively activate transcription in the absence of their respective activating ligands (l-rhamnose and meta-toluate). Like RhaS and XylS, the CTD of ExsA is constitutively active, but in this case the constitutive activity results from a loss of interaction with the ExsD antiactivator.
The CTD induces DNA bending to a similar degree as full-length ExsA (Fig. 6). These data suggest that bending of the PexsC promoter does not result from interactions between two promoter-bound ExsA monomers because the NTD is required for those interactions. This conclusion is supported by our previous observation that shift product 1, which represents a single molecule of bound ExsA, also exhibits DNA bending (4). The crystal structure of the AraC/XylS family protein MarA complexed with DNA suggests a mechanism for ExsA-induced DNA bending. MarA is a somewhat atypical member of the AraC/XylS family in that it binds DNA as a monomer. The two HTH motifs of MarA insert into adjacent major grooves of the DNA and are separated by a rigid α helix. The length of the α helix is shorter than the space between the major grooves, and the DNA is forced to bend to accommodate the binding of both HTH motifs (33). Although these data suggest how monomeric ExsA might bend DNA, the mechanism for increased bending of the promoter PexsC compared to that for the promoters PexsD and PexoT is unclear (4).
Our data demonstrate that ExsD binds to the NTD of ExsA and that the NTD is required for cooperative binding to the PexsC promoter. Although the primary determinants for DNA binding and transcriptional activation are located within the CTD, binding of ExsD to the NTD could sterically interfere with CTD function or induce/prevent a conformational change in the CTD involved in cooperativity, DNA binding, or transcriptional activation. The PexsC promoter controls transcription of the exsCEBA operon in an ExsA-dependent manner and in theory could represent the sole target for ExsD-mediated inhibition of T3SS gene expression. Under such a scenario, ExsD might function by interfering with the cooperative recruitment of ExsA to the PexsC promoter. Alternatively, ExsD could have a general effect on ExsA activity, thereby resulting in inhibition of all T3SS promoters. Potential models accounting for a general inhibition of ExsA activity by binding of ExsD to the NTD include (i) inhibiting the DNA binding activity of ExsA, (ii) preventing ExsA from interacting with or recruiting RNA polymerase, or (iii) promoting degradation of ExsA.
ExsA and ExsD homologs are also likely involved in regulation of T3SS gene expression in Vibrio parahaemolyticus, Aeromonas spp., and Photorhabdus spp. (43). The ExsA and ExsD homologs in V. parahaemolyticus are known to function as an activator and inhibitor of T3SS gene expression, respectively, suggesting that the functions of ExsA and ExsD might be conserved in each of these organisms (50). Protein ligands are also involved in regulation of other AraC/XylS family members that regulate T3SS gene expression. The most relevant is OspD1, as it functions as an antiactivator of MxiE in Shigella flexneri (29). Neither OspD1 nor the NTD of MxiE shares obvious sequence similarity with ExsD or the NTD of ExsA, however, making it difficult to draw parallels between the two systems. Future work is necessary to define the mechanistic basis for negative regulation by ExsD and other protein ligands that modulate the activities of AraC/XylS family members.
Acknowledgments
We thank Mark Urbanowski and Gary Gussin for their suggestions regarding this work.
This study was supported by the National Institutes of Health (RO1-AI055042-06).
Footnotes
Published ahead of print on 17 April 2009.
REFERENCES
- 1.Apodaca, G., M. Bomsel, R. Lindstedt, J. Engel, D. Frank, K. E. Mostov, and J. Wiener-Kronish. 1995. Characterization of Pseudomonas aeruginosa-induced MDCK cell injury: glycosylation-defective host cells are resistant to bacterial killing. Infect. Immun. 631541-1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Basturea, G. N., M. D. Bodero, M. E. Moreno, and G. P. Munson. 2008. Residues near the amino terminus of Rns are essential for positive autoregulation and DNA binding. J. Bacteriol. 1902279-2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72248-254. [DOI] [PubMed] [Google Scholar]
- 4.Brutinel, E. D., C. A. Vakulskas, K. M. Brady, and T. L. Yahr. 2008. Characterization of ExsA and of ExsA-dependent promoters required for expression of the Pseudomonas aeruginosa type III secretion system. Mol. Microbiol. 68657-671. [DOI] [PubMed] [Google Scholar]
- 5.Brutinel, E. D., and T. L. Yahr. 2008. Control of gene expression by type III secretory activity. Curr. Opin. Microbiol. 11128-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bustos, S. A., and R. F. Schleif. 1993. Functional domains of the AraC protein. Proc. Natl. Acad. Sci. USA 905638-5642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cornelis, G. R., and F. Van Gijsegem. 2000. Assembly and function of type III secretory systems. Annu. Rev. Microbiol. 54735-774. [DOI] [PubMed] [Google Scholar]
- 8.Crothers, D. M., M. R. Gartenberg, and T. E. Shrader. 1991. DNA bending in protein-DNA complexes. Methods Enzymol. 208118-146. [DOI] [PubMed] [Google Scholar]
- 9.Darwin, K. H., and V. L. Miller. 2000. The putative invasion protein chaperone SicA acts together with InvF to activate the expression of Salmonella typhimurium virulence genes. Mol. Microbiol. 35949-960. [DOI] [PubMed] [Google Scholar]
- 10.Dasgupta, N., G. L. Lykken, M. C. Wolfgang, and T. L. Yahr. 2004. A novel anti-anti-activator mechanism regulates expression of the Pseudomonas aeruginosa type III secretion system. Mol. Microbiol. 53297-308. [DOI] [PubMed] [Google Scholar]
- 11.Egan, S. M. 2002. Growing repertoire of AraC/XylS activators. J. Bacteriol. 1845529-5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eustance, R. J., S. A. Bustos, and R. F. Schleif. 1994. Reaching out. Locating and lengthening the interdomain linker in AraC protein. J. Mol. Biol. 242330-338. [DOI] [PubMed] [Google Scholar]
- 13.Frank, D. W. 1997. The exoenzyme S regulon of Pseudomonas aeruginosa. Mol. Microbiol. 26621-629. [DOI] [PubMed] [Google Scholar]
- 14.Frank, D. W., and B. H. Iglewski. 1991. Cloning and sequence analysis of a trans-regulatory locus required for exoenzyme S synthesis in Pseudomonas aeruginosa. J. Bacteriol. 1736460-6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gallegos, M. T., R. Schleif, A. Bairoch, K. Hofmann, and J. L. Ramos. 1997. Arac/XylS family of transcriptional regulators. Microbiol. Mol. Biol. Rev. 61393-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoang, T. T., R. R. Karkhoff-Schweizer, A. J. Kutchma, and H. P. Schweizer. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 21277-86. [DOI] [PubMed] [Google Scholar]
- 17.Holder, I. A., A. N. Neely, and D. W. Frank. 2001. Type III secretion/intoxication system important in virulence of Pseudomonas aeruginosa infections in burns. Burns 27129-130. [DOI] [PubMed] [Google Scholar]
- 18.Hovey, A. K., and D. W. Frank. 1995. Analyses of the DNA-binding and transcriptional activation properties of ExsA, the transcriptional activator of the Pseudomonas aeruginosa exoenzyme S regulon. J. Bacteriol. 1774427-4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Howard, V. J., T. A. Belyaeva, S. J. Busby, and E. I. Hyde. 2002. DNA binding of the transcription activator protein MelR from Escherichia coli and its C-terminal domain. Nucleic Acids Res. 302692-2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hueck, C. J. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iglewski, B. H., J. Sadoff, M. J. Bjorn, and E. S. Maxwell. 1978. Pseudomonas aeruginosa exoenzyme S: an adenosine diphosphate ribosyltransferase distinct from toxin A. Proc. Natl. Acad. Sci. USA 753211-3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaldalu, N., U. Toots, V. de Lorenzo, and M. Ustav. 2000. Functional domains of the TOL plasmid transcription factor XylS. J. Bacteriol. 1821118-1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kolin, A., V. Jevtic, L. Swint-Kruse, and S. M. Egan. 2007. Linker regions of the RhaS and RhaR proteins. J. Bacteriol. 189269-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kurahashi, K., O. Kajikawa, T. Sawa, M. Ohara, M. A. Gropper, D. W. Frank, T. R. Martin, and J. P. Wiener-Kronish. 1999. Pathogenesis of septic shock in Pseudomonas aeruginosa pneumonia. J. Clin. Investig. 104743-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kwon, H. J., M. H. Bennik, B. Demple, and T. Ellenberger. 2000. Crystal structure of the Escherichia coli Rob transcription factor in complex with DNA. Nat. Struct. Biol. 7424-430. [DOI] [PubMed] [Google Scholar]
- 26.Martin, R. G., and J. L. Rosner. 2001. The AraC transcriptional activators. Curr. Opin. Microbiol. 4132-137. [DOI] [PubMed] [Google Scholar]
- 27.Mavris, M., A. L. Page, R. Tournebize, B. Demers, P. Sansonetti, and C. Parsot. 2002. Regulation of transcription by the activity of the Shigella flexneri type III secretion apparatus. Mol. Microbiol. 431543-1553. [DOI] [PubMed] [Google Scholar]
- 28.McCaw, M. L., G. L. Lykken, P. K. Singh, and T. L. Yahr. 2002. ExsD is a negative regulator of the Pseudomonas aeruginosa type III secretion regulon. Mol. Microbiol. 461123-1133. [DOI] [PubMed] [Google Scholar]
- 29.Parsot, C., E. Ageron, C. Penno, M. Mavris, K. Jamoussi, H. d'Hauteville, P. Sansonetti, and B. Demers. 2005. A secreted anti-activator, OspD1, and its chaperone, Spa15, are involved in the control of transcription by the type III secretion apparatus activity in Shigella flexneri. Mol. Microbiol. 561627-1635. [DOI] [PubMed] [Google Scholar]
- 30.Pilonieta, M. C., and G. P. Munson. 2008. The chaperone IpgC copurifies with the virulence regulator MxiE. J. Bacteriol. 1902249-2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poore, C. A., C. Coker, J. D. Dattelbaum, and H. L. Mobley. 2001. Identification of the domains of UreR, an AraC-like transcriptional regulator of the urease gene cluster in Proteus mirabilis. J. Bacteriol. 1834526-4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prouty, M. G., C. R. Osorio, and K. E. Klose. 2005. Characterization of functional domains of the Vibrio cholerae virulence regulator ToxT. Mol. Microbiol. 581143-1156. [DOI] [PubMed] [Google Scholar]
- 33.Rhee, S., R. G. Martin, J. L. Rosner, and D. R. Davies. 1998. A novel DNA-binding motif in MarA: the first structure for an AraC family transcriptional activator. Proc. Natl. Acad. Sci. USA 9510413-10418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Richards, M. J., J. R. Edwards, D. H. Culver, and R. P. Gaynes. 2000. Nosocomial infections in combined medical-surgical intensive care units in the United States. Infect. Control Hosp. Epidemiol. 21510-515. [DOI] [PubMed] [Google Scholar]
- 35.Richards, M. J., J. R. Edwards, D. H. Culver, and R. P. Gaynes, and the National Nosocomial Infections Surveillance System. 1999. Nosocomial infections in medical intensive care units in the United States. Crit. Care Med. 27887-892. [DOI] [PubMed] [Google Scholar]
- 36.Richet, E., and L. Sogaard-Andersen. 1994. CRP induces the repositioning of MalT at the Escherichia coli malKp promoter primarily through DNA bending. EMBO J. 134558-4567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rietsch, A., I. Vallet-Gely, S. L. Dove, and J. J. Mekalanos. 2005. ExsE, a secreted regulator of type III secretion genes in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 1028006-8011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saviola, B., R. Seabold, and R. F. Schleif. 1998. Arm-domain interactions in AraC. J. Mol. Biol. 278539-548. [DOI] [PubMed] [Google Scholar]
- 39.Sawa, T., T. L. Yahr, M. Ohara, K. Kurahashi, M. A. Gropper, J. P. Wiener-Kronish, and D. W. Frank. 1999. Active and passive immunization with the Pseudomonas V antigen protects against type III intoxication and lung injury. Nat. Med. 5392-398. [DOI] [PubMed] [Google Scholar]
- 40.Soisson, S. M., B. MacDougall-Shackleton, R. Schleif, and C. Wolberger. 1997. Structural basis for ligand-regulated oligomerization of AraC. Science 276421-425. [DOI] [PubMed] [Google Scholar]
- 41.Tanaka, T., S. Yokoyama, and Y. Kuroda. 2006. Improvement of domain linker prediction by incorporating loop-length-dependent characteristics. Biopolymers 84161-168. [DOI] [PubMed] [Google Scholar]
- 42.Urbanowski, M. L., E. D. Brutinel, and T. L. Yahr. 2007. Translocation of ExsE into Chinese hamster ovary cells is required for transcriptional induction of the Pseudomonas aeruginosa type III secretion system. Infect. Immun. 754432-4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Urbanowski, M. L., G. L. Lykken, and T. L. Yahr. 2005. A secreted regulatory protein couples transcription to the secretory activity of the Pseudomonas aeruginosa type III secretion system. Proc. Natl. Acad. Sci. USA 1029930-9935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vallis, A. J., T. L. Yahr, J. T. Barbieri, and D. W. Frank. 1999. Regulation of ExoS production and secretion by Pseudomonas aeruginosa in response to tissue culture conditions. Infect. Immun. 67914-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vogel, H. J., and D. M. Bonner. 1956. Acetylornithinase of Escherichia coli: partial purification and some properties. J. Biol. Chem. 21897-106. [PubMed] [Google Scholar]
- 46.Wickstrum, J. R., J. M. Skredenske, A. Kolin, D. J. Jin, J. Fang, and S. M. Egan. 2007. Transcription activation by the DNA-binding domain of the AraC family protein RhaS in the absence of its effector-binding domain. J. Bacteriol. 1894984-4993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yahr, T. L., and D. W. Frank. 1994. Transcriptional organization of the trans-regulatory locus which controls exoenzyme S synthesis in Pseudomonas aeruginosa. J. Bacteriol. 1763832-3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yahr, T. L., A. K. Hovey, S. M. Kulich, and D. W. Frank. 1995. Transcriptional analysis of the Pseudomonas aeruginosa exoenzyme S structural gene. J. Bacteriol. 1771169-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yahr, T. L., and M. C. Wolfgang. 2006. Transcriptional regulation of the Pseudomonas aeruginosa type III secretion system. Mol. Microbiol. 62631-640. [DOI] [PubMed] [Google Scholar]
- 50.Zhou, X., D. H. Shah, M. E. Konkel, and D. R. Call. 2008. Type III secretion system 1 genes in Vibrio parahaemolyticus are positively regulated by ExsA and negatively regulated by ExsD. Mol. Microbiol. 69747-764. [DOI] [PMC free article] [PubMed] [Google Scholar]