Abstract
The activities of the Vsr and MutH endonucleases of Escherichia coli are stimulated by MutL. The interaction of MutL with each enzyme is enhanced in vivo by 2-aminopurine treatment and by inactivation of the mutY gene. We hypothesize that MutL recruits the endonucleases to sites of DNA damage.
The Escherichia coli Dcm protein methylates the second C of CCWGG sites (W = A or T). Deamination of 5-methylcytosine converts CG base pairs to T/G mismatches, causing CCWGG-to-CTWGG transition mutations. Very-short-patch (VSP) repair minimizes these mutations (2). Repair is initiated by a sequence- and mismatch-specific endonuclease, Vsr, which cleaves the DNA 5′ of the T. DNA polymerase I removes the T along with a few 3′ nucleotides and resynthesizes the missing bases, restoring the CG base pair. Vsr is both necessary and sufficient for initiating VSP repair. However, two other proteins, MutS and MutL, enhance VSP repair of deamination damage (1).
MutS and MutL are best known for their roles in postreplication mismatch repair (MMR) (9, 11). MutL couples mismatch recognition by MutS to the activation of MutH, an endonuclease that cleaves the unmethylated strand of GATC sequences that are transiently hemimethylated following DNA replication. The nicked strand, containing the erroneous base, is removed by the UvrD helicase and one of several exonucleases to beyond the mismatch and then resynthesized by DNA polymerase III.
MutL stimulates the endonuclease activities of both Vsr and MutH in vitro (8, 17). The requirements for stimulation are the same: a mismatch, MutS, and ATP hydrolysis by MutL (8, 8a). Cross-linking studies showed that MutH and Vsr interact with the same region in the N-terminal domain of MutL (Heinze et al., submitted). Competition of Vsr with MutH for access to MutL explains the ability of Vsr to inactivate MMR in vivo when overexpressed (6, 13). Thus, the interactions of the two repair endonucleases with MutL are structurally and functionally very similar.
In contrast to MMR, where the cleavage site for MutH may be several kilobases away from the mismatch, VSP repair requires that mismatch recognition and endonucleolytic cleavage occur at the same C(T/G)WGG site. How MutS and MutL stimulate VSP repair if MutS and Vsr compete for the same mismatch remains unknown (2, 12). We hypothesized that MutS binds the mismatch first and that a MutS-MutL complex then recruits Vsr. If so, then the MMR proteins would initially mask the mismatch, making the interaction of Vsr with MutL independent of lesion identity.
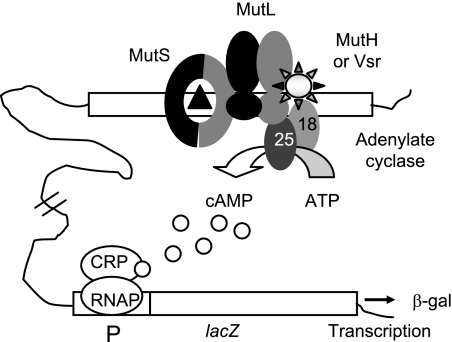
To test this hypothesis, we studied the interaction of MutL with Vsr and with MutH in response to two types of mismatch by using a bacterial two-hybrid assay (10). This assay detects all known interactions among the Mut proteins: homodimerization of MutS and MutL, interaction of MutL with MutS and with MutH, and interaction of Vsr with the N-terminal domain of MutL (15). We found no false positives or false negatives. Furthermore, since the assay relies on reconstitution of a soluble protein (adenylate cyclase), the DNA repair proteins are free to interact with the DNA (Fig. 1).
FIG. 1.
Known interactions among repair proteins as detected by the bacterial two-hybrid assay. The T18 and T25 subunits of CyaA are fused to any two repair proteins (illustrated here by MutL and Vsr), allowing measurement of all pairwise interactions as units of β-galactosidase (β-gal). T25 fusions are repair proficient. CRP, cyclic AMP (cAMP) receptor protein; P, lac operon promoter; RNAP, RNA polymerase.
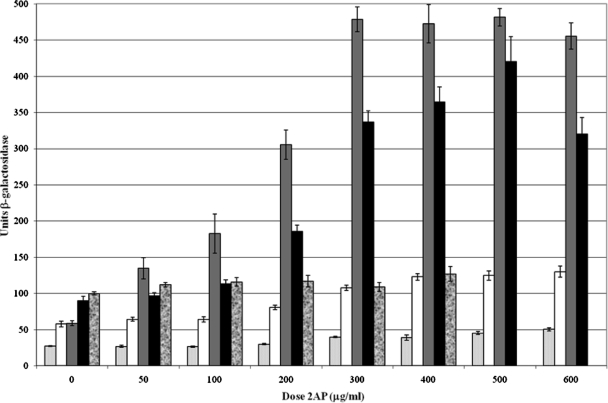
2-Aminopurine (2AP) mispairs with C during DNA replication, causing transition and frameshift mutations (5). The transitions are due primarily to the mismatch itself; the frameshifts are due to saturation of MMR, which leaves slipped-strand intermediates caused by DNA replication errors unrepaired (19). MutS and MutL bind to 2AP/C lesions (22), although the lesions may not be subject to MMR (19). As shown in Fig. 2, treatment with 2AP causes a dose-dependent increase in the interaction of MutL with both Vsr and MutH; dimerization of MutL and interaction of MutL with MutS are somewhat increased.
FIG. 2.
Effect of 2AP treatment on protein-protein interactions in the bacterial two-hybrid assay. Results in units of β-galactosidase ± standard errors of the means (n = 9) are shown for BTH101(F galE15 ga1K16 rpsL1 hsdR2 mcrA1 mcrB1 cyaA-99) cells treated with 2AP as described previously (5, 19). Cells were cotransformed with pT18 and pT25 vectors (light gray bars), pT18-mutS and pT25-mutL (white bars), pT18-vsr and pT25-mutL (gray bars), pT18-mutH and pT25-mutL (black bars), or pT18-mutL and pT25-mutL (mottled bars). (NB: The dose-response curve for the pT18-mutS pT25-mutS transformants is similar to that of the pT18-mutL pT25-mutL transformants; it has been omitted for graphical clarity since the MutS-MutS interaction gives very high units of β-galactosidase activity [15]).
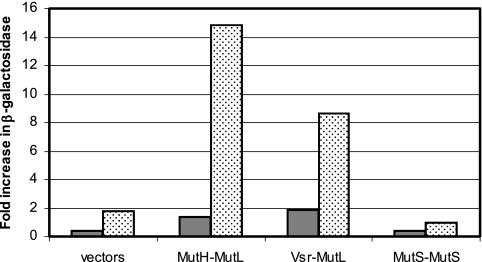
The MutY adenine glycosylase removes A's which have mispaired with oxidized guanine (8-oxoG) during DNA replication. Cells with a deletion of mutY have an elevated frequency of CG-to-AT transversion mutations (18); these are reduced by excess MutS, suggesting that 8-oxoG/A mismatches are also subject to MMR (23). As shown in Fig. 3, the interactions between Vsr and MutL and between MutH and MutL increase in a mutY cell (stippled bars). Other interactions, such as MutS dimerization, are unaffected (not shown).
FIG. 3.
Effects of mutY and mutT deletions on protein-protein interactions in the bacterial two-hybrid assay. Results are in units of β-galactosidase, relative to the level in the wild type, in mutT (solid) and mutY (stippled) derivatives of BTH101 cotransformed with pT18 and pT25 vectors, pT18-mutH and pT25-mutL, pT18-vsr and pT25-mutL, or pT18-mutS and pT25-mutS (n = 3).
8-OxoG/A mismatches also arise by incorporation of oxidized dGTP opposite A during DNA replication. The MutT nuclease minimizes this by removing oxidized dGTP from the nucleotide pool. The high frequency of AT-to-CG mutations in mutT strains is unaffected by the status of the MMR system (7, 21, 23), possibly because these 8-oxoG/A mispairs are in a conformation that MutS does not recognize. As shown in Fig. 3, neither the interaction between MutL and Vsr nor that between MutL and MutH is elevated in a mutT strain (solid bars).
These data show that mismatches which attract MutS and MutL increase the interaction of MutL with MutH in vivo. Although these mismatches are not subject to VSP repair, they also increase the interaction between MutL and Vsr. The simplest interpretation is that a MutS-MutL complex recruits MutH and Vsr to the DNA independent of the identity of the mismatch. MutS and MutL could then clear the mismatch, delivering the (activated) endonuclease to its specific target site, no matter how far away it is.
Interaction of MutL with MutH, leading to MMR, is probably the default option. However, the MutS-MutL complex may recruit other repair proteins, such as Vsr or UvrB (20), to lesions that are poorly processed by MMR. The T/G mismatch in hemimethylated CTWGG sequences may be one such site. Vsr is expressed at very low levels in growing cells (14), so this recruitment would enhance VSP repair. However, recruitment of Vsr to other lesions would reduce VSP repair. For example, recruitment of Vsr by MutL to 2AP/C lesions (Fig. 2) could explain why CCWGG sites are hotspots for 2AP-induced mutations (4, 19).
We have argued that Vsr is kept at low levels while DNA is replicating to avoid interference with MMR (14). However, if, as we suggest here, MutS and MutL are needed to recruit scarce Vsr to its target sequence, this argument loses its merit. It seems more likely that Vsr levels are kept low to avoid CTWGG-to-CCWGG mutations; Vsr creates these mutations by converting T/G mismatches formed at CTAGG sites by errors in DNA replication to CG (3, 6, 16). Vsr levels rise in nongrowing cells (14), when mutagenesis is no longer a risk. Under these circumstances, it is likely that MutS and MutL are no longer required for efficient VSP repair.
Acknowledgments
This work was supported by a grant to C.G.C. from the Canadian Institutes for Health Research. J.M. was supported in part by an undergraduate research award from the Faculty of Science, University of Victoria.
Footnotes
Published ahead of print on 17 April 2009.
REFERENCES
- 1.Bell, D. C., and C. G. Cupples. 2001. Very-short-patch repair in Escherichia coli requires the dam adenine methylase. J. Bacteriol. 1833631-3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhagwat, A. S., and M. Lieb. 2002. Cooperation and competition in mismatch repair: very short-patch repair and methyl-directed mismatch repair in Escherichia coli. Mol. Microbiol. 441421-1428. [DOI] [PubMed] [Google Scholar]
- 3.Bhagwat, A. S., and M. McClelland. 1992. DNA mismatch correction by Very Short Patch repair may have altered the abundance of oligonucleotides in the E. coli genome. Nucleic Acids Res. 201663-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coulondre, C., J. H. Miller, P. J. Farabaugh, and W. Gilbert. 1978. Molecular basis of base substitution hotspots in Escherichia coli. Nature 274775-780. [DOI] [PubMed] [Google Scholar]
- 5.Cupples, C. G., M. Cabrera, C. Cruz, and J. H. Miller. 1990. A set of lacZ mutations in Escherichia coli that allow rapid detection of specific frameshift mutations. Genetics 125275-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doiron, K. M., S. Viau, M. Koutroumanis, and C. G. Cupples. 1996. Overexpression of vsr in Escherichia coli is mutagenic. J. Bacteriol. 1784294-4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fowler, R. G., S. J. White, C. Koyama, S. C. Moore, R. L. Dunn, and R. M. Schaaper. 2003. Interactions among the Escherichia coli mutT, mutM, and mutY damage prevention pathways. DNA Repair (Amsterdam) 2159-173. [DOI] [PubMed] [Google Scholar]
- 8.Hall, M. C., and S. W. Matson. 1999. The Escherichia coli MutL protein physically interacts with MutH and stimulates the MutH-associated endonuclease activity. J. Biol. Chem. 2741306-1312. [DOI] [PubMed] [Google Scholar]
- 8a.Heinze, R., L. Giron-Monzon, A. Solovyova, S. Elliot, S. Geisler, C. Cupples, B. Connolly, and P. Friedhoff. Physical and functional interactions between Escherichia coli MutL and the Vsr repair endonuclease. Nucleic Acids Res., in press. [DOI] [PMC free article] [PubMed]
- 9.Jun, S. H., T. G. Kim, and C. Ban. 2006. DNA mismatch repair system. Classical and fresh roles. FEBS J. 2731609-1619. [DOI] [PubMed] [Google Scholar]
- 10.Karimova, G., J. Pidoux, A. Ullmann, and D. Ladant. 1998. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc. Natl. Acad. Sci. USA 955752-5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li, G. M. 2008. Mechanisms and functions of DNA mismatch repair. Cell Res. 1885-98. [DOI] [PubMed] [Google Scholar]
- 12.Lieb, M., S. Rehmat, and A. S. Bhagwat. 2001. Interaction of MutS and Vsr: some dominant-negative mutS mutations that disable methyladenine-directed mismatch repair are active in very-short-patch repair. J. Bacteriol. 1836487-6490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Macintyre, G., K. M. Doiron, and C. G. Cupples. 1997. The Vsr endonuclease of Escherichia coli: an efficient DNA repair enzyme and a potent mutagen. J. Bacteriol. 1796048-6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Macintyre, G., P. Pitsikas, and C. G. Cupples. 1999. Growth phase-dependent regulation of Vsr endonuclease may contribute to 5-methylcytosine mutational hot spots in Escherichia coli. J. Bacteriol. 1814435-4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mansour, C. A., K. M. Doiron, and C. G. Cupples. 2001. Characterization of functional interactions among the Escherichia coli mismatch repair proteins using a bacterial two-hybrid assay. Mutat. Res. 485331-338. [DOI] [PubMed] [Google Scholar]
- 16.Merkl, R., M. Kroger, P. Rice, and H. J. Fritz. 1992. Statistical evaluation and biological interpretation of nonrandom abundance in the E. coli K-12 genome of tetra- and pentanucleotide sequences related to VSP DNA mismatch repair. Nucleic Acids Res. 201657-1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Monastiriakos, S. K., K. M. Doiron, M. I. Siponen, and C. G. Cupples. 2004. Functional interactions between the MutL and Vsr proteins of Escherichia coli are dependent on the N-terminus of Vsr. DNA Repair (Amsterdam) 3639-647. [DOI] [PubMed] [Google Scholar]
- 18.Nghiem, Y., M. Cabrera, C. G. Cupples, and J. H. Miller. 1988. The mutY gene: a mutator locus in Escherichia coli that generates G.C→T.A transversions. Proc. Natl. Acad. Sci. USA 852709-2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pitsikas, P., J. M. Patapas, and C. G. Cupples. 2004. Mechanism of 2-aminopurine-stimulated mutagenesis in Escherichia coli. Mutat. Res. 55025-32. [DOI] [PubMed] [Google Scholar]
- 20.Pitsikas, P., Y. Y. Polosina, and C. G. Cupples. 2009. Interaction between the mismatch repair and nucleotide excision repair pathways in the prevention of 5-azacytidine-induced CG-to-GC mutations in Escherichia coli. DNA Repair (Amsterdam) 8354-359. [DOI] [PubMed] [Google Scholar]
- 21.Schaaper, R. M., B. I. Bond, and R. G. Fowler. 1989. A.T→C.G transversions and their prevention by the Escherichia coli mutT and mutHLS pathways. Mol. Gen. Genet. 219256-262. [DOI] [PubMed] [Google Scholar]
- 22.Smith, B. T., A. D. Grossman, and G. C. Walker. 2001. Visualization of mismatch repair in bacterial cells. Mol. Cell 81197-1206. [DOI] [PubMed] [Google Scholar]
- 23.Zhao, J., and M. E. Winkler. 2000. Reduction of GC → TA transversion mutation by overexpression of MutS in Escherichia coli K-12. J. Bacteriol. 1825025-5028. [DOI] [PMC free article] [PubMed] [Google Scholar]