Abstract
A gram-positive anaerobic pathogen, Clostridium perfringens, causes clostridial myonecrosis or gas gangrene in humans by producing numerous extracellular toxins and enzymes that act in concert to degrade host tissue. The agr system is known to be important for the regulation of virulence genes in a quorum-sensing manner in Staphylococcus aureus. A homologue for S. aureus agrBD (agrBDSa) was identified in the C. perfringens strain 13 genome, and the role of C. perfringens agrBD (agrBDCp) was examined. The agrBDCp knockout mutant did not express the theta-toxin gene, and transcription of the alpha- and kappa-toxin genes was also significantly decreased in the mutant strain. The mutant strain showed a recovery of toxin production after the addition of the culture supernatant of the wild-type strain, indicating that the agrBDCp mutant lacks a signal molecule in the culture supernatant. An agr-virR double-knockout mutant was constructed to examine the role of the VirR/VirS two-component regulatory system, a key virulence regulator, in agrBDCp-mediated regulation of toxin production. The double-mutant strain could not be stimulated for toxin production with the wild-type culture supernatant. These results indicate that the agrBDCp system plays an important role in virulence regulation and also suggest that VirR/VirS is required for sensing of the extracellular signal and activation of toxin gene transcription in C. perfringens.
Clostridium perfringens is a gram-positive, spore-forming, anaerobic bacterium. C. perfringens is the causative agent of several human and animal diseases, including clostridial myonecrosis, or gas gangrene (7). C. perfringens produces various extracellular enzymes and toxins, including alpha-, theta-, and kappa-toxins encoded by plc, pfoA, and colA, respectively (21). These toxin genes are positively regulated by the two-component VirR/VirS system (25) that is a major regulator of virulence in C. perfringens. The VirS is a sensor histidine kinase, and VirR is a response regulator. When VirS senses specific stimuli in the environment, VirS autophosphorylates at a histidine residue and then transfers the phosphate to VirR. Once VirR is activated by phosphorylation, it regulates gene expression. The genomic sequence of C. perfringens strain 13 was determined in 2002, and it was found that the genome contains only five genes, including pfoA and VR-RNA, that have VirR-binding sites on their promoter regions (24). VR-RNA is known to be a small regulatory RNA and positively regulates colA and plc transcription (26). Recent microarray analysis suggested that many other genes are regulated by the VirRS-VR-RNA cascade. Thus, a number of virulence-related genes and also some housekeeping genes are included in the VirRS-VR-RNA-regulon (K. Ohtani et al., unpublished data). The C. perfringens genome contains many genes for toxins or for enzymes that can degrade host tissue, while the genome lacks many genes related to the synthesis of amino acids. Under infectious conditions, C. perfringens might secrete these toxins and enzymes in order to degrade the host tissue. It may then import the resulting amino acids, using them to survive in the host tissue. The VirR/VirS system is therefore very important for the activation of toxin production that results in the degradation of host cells and is critical for the survival of C. perfringens, especially within the host. However, it is still unclear what the signal of VirS is and how this signaling system effectively stimulates toxin production.
Many bacteria regulate gene expression in response to cell population density, a phenomenon known as “quorum sensing” (4). Quorum sensing involves the production of extracellular signaling molecules (autoinducers). In general, many known autoinducers of gram-positive bacteria are actively secreted peptides that are processed from larger propeptides. These peptide autoinducers function as ligands for signal receptors such as the two-component sensor histidine kinase (17). In gram-negative bacteria, the N-acylhomoserine lactones (AHLs) are well known as autoinducers (14). They diffuse freely in and out of cells and interact directly with intracellular regulatory proteins. AHL accumulates as cells grow, and when it reaches a certain threshold, AHL can efficiently regulate the expression of many genes. In Vibrio fischeri, the LuxR protein binds to AHL, and this complex regulates the lux operon and many other genes at the transcriptional level (14). Moreover, the luxS gene is responsible for the production of another kind of autoinducer, autoinducer 2 (30). Highly conserved luxS homologues have been identified in both gram-positive and gram-negative bacteria (2). These quorum-sensing systems play important roles in the regulation of virulence factors and in biofilm formation in various pathogenic bacteria (6, 28, 30).
In C. perfringens, the possibility that cell-cell signaling exists has been suggested (8). In a previous report, two types of toxin-negative strains were cross-streaked on a blood agar plate, and one toxin-negative strain recovered its toxin production just after the crossing point of the two strains on the plate (8, 10). This experiment suggested that there is a signal molecule (called substance A) (9) that stimulates toxin production from outside of the cell. In 2002, cell-cell signaling mediated by luxS was reported, and it was concluded that the signal produced actually regulated the transcription of toxin genes (18). However, the mutant strain of luxS still retained toxin production; therefore, it was concluded that the luxS signaling system might be different from that mediated by substance A and thus that there may be a different cell-cell signaling system in C. perfringens.
In gram-positive bacteria, a secreted peptide regulates gene expression in the quorum-sensing manner described above (17). In the case of S. aureus, the autoinducer propeptide (AIP) acts as a signal to stimulate gene expression. This peptide contains an intramolecular thiolactone ring. The agrD gene is a structural gene for AIP, and AgrB is a protein that is required for modification of the AgrD propeptide. In the genome of S. aureus, the genes of a two-component system, agrA and agrC, lie next to the agrBD genes. The AgrA protein is a response regulator, and AgrC is a sensor histidine kinase. The AIP, synthesized from the AgrD protein, is secreted and accumulates in the supernatant. Once AIP reaches a certain threshold level, it activates its receptor, AgrC sensor histidine kinase, which then activates AgrA by phosphotransfer. Finally, AgrA activates the transcription of the regulatory RNA, RNAIII, that regulates the expression of various virulence genes of S. aureus (5, 15, 17). This signaling system is highly conserved among many gram-positive bacteria (12, 16, 20).
In the present study we identified an agrBD gene in C. perfringens (agrBDCp) that is homologous to the agrBD gene of S. aureus (agrBDSa). Functional genetic analysis revealed that agrBDCp is involved in the positive regulation of alpha-, kappa-, and theta-toxin genes through a cell-cell signaling mechanism that involves a two-component VirR/VirS system.
MATERIALS AND METHODS
Strains, media, and culture conditions.
The C. perfringens strains 13 (13) and TS133 (23), as well as the other strains used in the present study (Table 1), were cultured in Gifu anaerobic medium (GAM) or TSF (tryptone, 40 g/liter; soytone, 4 g/liter; fructose, 5 g/liter [pH 5.7]) (9) medium at 37°C under anaerobic conditions as described previously (23). Escherichia coli strain DH5α was cultured under standard conditions (22). The plasmid pUC19 was used for general cloning in E. coli, and pJIR418 (27) was used as an E. coli-C. perfringens shuttle vector. The plasmid pTS405 was used as a complementation vector for virR/virS genes (19).
TABLE 1.
C. perfringens strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Source |
|---|---|---|
| C. perfringens strains | ||
| Strain 13 | Wild-type strain (type A) | |
| TS133 | Strain 13 virR::Tetr | |
| TS230 | Strain 13 ΔagrBD::Emr | This study |
| TS231 | Strain 13 ΔagrBD-ΔvirR::Tetr Emr | This study |
| Plasmids | ||
| pJIR418 | E. coli -C. perfringens shuttle vector, Cmr Emr; pJIR418 Ω (PstI 4.3-kb strain 13 genomic library) | |
| pTS405 | virR+virS+ complementation vector; Cmr | T. Shimizu, unpublished data |
| pTS1301 | pJIR418 Ω(Δpromoter CPE1562-CPE1561-agrD-CPE1560) | This study |
| pTS1302 | pJIR418 Ω(Δpromoter agrD-CPE1560) | This study |
| pTS1303 | pJIR418 Ω(CPE1563-CPE1562-CPE1561-agrD -CPE1560) including promoter | This study |
| pTS1304 | pJIR418 Ω(CPE1563-CPE1562-CPE1561-agrD) including promoter | This study |
| pTS1305 | pJIR418 Ω(Δpromoter CPE1562-CPE1561-agrD) | This study |
| pTS1306 | pJIR418 Ω(Δpromoter CPE1561-agrD) | This study |
| pTS1307 | pJIR418 Ω(Δpromoter agrD) | This study |
| pTS1308 | pJIR418 Ω(CPE1562-CPE1561-agrD) | This study |
| pTS1309 | pJIR418 Ω(CPE1561-agrD) | This study |
| pTS1310 | pJIR418 Ω(agrD) | This study |
| pTS1311 | pJIR418 Ω(CPE1563-agrD) | This study |
| pTS1312 | pJIR418 Ω(CPE1563-CPE1562-agrD) | This study |
| pTS1313 | pJIR418 Ω(CPE1562-agrD) | This study |
| pTS1314 | pJIR418 Ω(CPE1562-CPE1561-agrD) | This study |
Cmr, chloramphenicol resistance; Emr, erythromycin resistance; Tetr, tetracycline resistance.
DNA manipulation.
General recombinant DNA techniques were performed as described in Sambrook et al. (22) unless otherwise noted. C. perfringens strains were transformed by an electroporation-mediated transformation as previously described (23). Deletion endpoints were confirmed by nucleotide sequencing using reverse or universal primers, a BigDye terminator reaction kit, and an ABI 310 sequencer (Applied Biosystems, Tokyo, Japan).
Northern and Southern hybridization.
Total RNA from C. perfringens was extracted according to a method described previously (1). Northern hybridization was also carried out as described previously (3, 11) except that DNA fragments were labeled with an AlkPhos-direct kit (Amersham Pharmacia Biotech, Buckinghamshire, United Kingdom), and signals were detected by CDPstar chemiluminescence. Southern hybridization was also performed using the same labeling and detection procedures.
Culture supernatant replacement experiments.
C. perfringens was cultured in TSF medium to stationary phase (optical density at 600 nm of 5.0) as a primary culture, and then these bacteria were inoculated at 5% concentration. The culture was continued to various growth stages and centrifuged at 15,000 rpm for 5 min, and then the culture supernatant was collected. To prepare recipient cells, C. perfringens was cultured in TSF medium for 5 h at 37°C and centrifuged at 15,000 rpm for 5 min, followed by removal of the supernatant from the cells. Recipient cells were suspended with the appropriate culture supernatant and incubated at 37°C for 15 min. Total RNA was then isolated from the incubated cells.
Construction of plasmids for allelic replacements.
Specific mutants were constructed by using PCR, and the sequences of all primers used for PCR are shown in Table 2. The fragment upstream of the agr region was amplified by PCR using primers 1 and 2 and inserted into the HincII site of pUC118. The fragment downstream of the agr region was amplified by PCR using primers 23 and 25 and inserted into the SmaI site of the plasmid containing the upstream region. Reverse PCR was performed using primers 2 and 23 and the erythromycin resistance gene was cloned into the region deleted by reverse PCR.
TABLE 2.
Primers used in this study
| Primer | Sequence | Description |
|---|---|---|
| 1 | GAACATATGTTTGCATGGAGG | To make plasmid for allelic replacement |
| 2 | CAAGCTCTGGGGCACTAGTT | To make plasmid for allelic replacement |
| 3 | ATTGTAAAGAGTGAAGGGAG | To construct pTS1303,1304 |
| 4 | AAAGTTGGACAATCTATCCTA | To construct pTS1301 |
| 5 | AGGATAGATTGTCCAACTTT | To construct pTS1308-1310 |
| 6 | TTTATGGGTAACTATGATGT | To construct pTS1305 |
| 7 | ACTTGTTCCTATCATATGTA | To make CPE1563 probe |
| 8 | ATTCTTCCTCCGCTGTCACT | To construct pTS1314 |
| 9 | AGTGACAGCGGAGGAAGAAT | To make CPE1562 probe |
| 10 | ATGGTATTCATACAATATTG | To construct pTS1306 |
| 11 | TTTAAACCTTCACATAAA | To make CPE1562 probe |
| 12 | TAGGTATTCCATCTACTAT | Sequence primer |
| 13 | TTTTTCAGCTATTAACTTCGA | To construct pTS1313,1314 |
| 14 | TTTACAGCAAGCATACTTA | To construct pTS1310,1311, to make CPE1561 probe |
| 15 | TTCTGGAGGAGCACATTCAG | To construct pTS1302 |
| 16 | TCCTTAGAGTCATACATTGC | To make CPE1561 probe |
| 18 | TTGTTAAAAACTATAGATTCTT | To check mutation, agrD Northern |
| 19 | GGCCGGTTTAAAACCTACCT | To check mutation, agrD Northern |
| 20 | TATACTAGATTAGAGAGGGAGAAT | To make CPE1560 probe |
| 21 | CTCTTCTCCTCCATATCTAGC | To make CPE1560 probe |
| 22 | ACTTCAGCTAAGCTATGCTG | To construct pTS1302 |
| 23 | AAGGTCATAGGTGTTGTATAGC | To make plasmid for allelic replacement |
| 24 | TAACAGTACGTGTTCCAAAC | To construct pTS1301,1303 |
| 25 | AGATGGGGCGGTAGACGTAG | To make plasmid for allelic replacement |
Construction of deletion strains.
The resulting plasmid for allelic replacement of agr operon was transformed into wild-type strain 13. Transformants were screened on a blood agar plate containing erythromycin (25 μg/ml). A hemolysis-negative colony was picked up and Southern analysis was performed to confirm the null mutation of the agr region in TS230.
To construct double-knockout mutants, an internal PCR fragment of the virR gene was inserted into pUC18 containing the tetA gene. The resulting plasmid was transformed into TS230 and screened on an agar plate containing 25 μg of erythromycin/ml and 2.5 μg of tetracycline/ml. The single-crossover mutation of virR in TS231 was confirmed by PCR using the appropriate primer set.
Construction of deletion mutants.
To construct the pTS1304 deletion mutant containing the genomic fragment stretching from CPE1563 to agrDCp, PCR was performed using the primers listed in Table 2. This PCR fragment was inserted into the HincII site of pUC118, and the resulting plasmid was used as a template for further PCR. Each fragment amplified by PCR was self-ligated and transformed into E. coli DH5α. The inserted fragments containing various agr genomic regions were then subcloned into pJIR418. To construct the pTS1313 deletion mutant, PCR was performed using pTS1312 as a template.
RESULTS AND DISCUSSION
Identification of an agrBD homologue in C. perfringens.
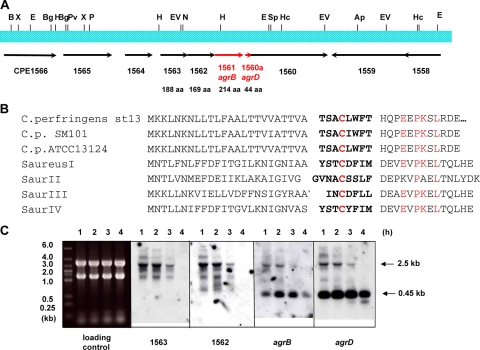
The agr operon of S. aureus is known to mediate a quorum-sensing system (17). It has been reported that there is a homologue of this agr system in C. perfringens SM101 and ATCC 13124 genomes (29). However, the function of the agr system in C. perfringens has not been determined. To investigate the function of the agr system in C. perfringens, we searched for homologues of the agr operon in the genome of C. perfringens strain 13. We found that the amino acid sequence deduced from CPE1561 showed a 29% identity and 50% similarity with the AgrBSa protein of S. aureus. The agrB gene encodes an integral membrane protein that modifies the AIP produced by AgrD protein. Downstream of CPE1561 (agrBCp), there was a small open reading frame (ORF) that was not assigned as an ORF when the C. perfringens genome sequence was determined (Fig. 1A). The protein from this ORF (designated CPE1560a) was similar to the AgrD peptide of S. aureus (32% identity and 46% similarity in a 43-amino-acid-overlap region), which is a propeptide for AIP. Next to the agrBDSa gene in S. aureus, there are genes for a two-component system (agrA and agrC) that can act as a receptor for AIP and induce gene expression. However, in the C. perfringens strain 13 genome, a similar two-component system could not be found in the vicinity of the agrBD gene (Fig. 1A). AIPs in S. aureus show a variety of amino acid sequences, but the central cysteine, which is important for the formation of a thiolactone ring with the C-terminal amino acid, is conserved in all of them (15). The amino acid sequence of the C. perfringens AgrD (AgrDCp) is completely different from that of AIPs, with the exception that this same central cysteine is conserved (Fig. 1B). However, the predicted peptide sequences are conserved in all three C. perfringens whose genome sequences are available (Fig. 1B).
FIG. 1.
Analysis of the agr region in C. perfringens. (A) Gene map of the agr region in C. perfringens. (B) Alignment of the deduced amino acid sequence of AgrDCp in C. perfringens and S. aureus AIPs. Conserved residues are indicated in red, and the deduced sequence of the mature peptides is in boldface. (C) Northern analysis of the agrBDCp region. RNA was isolated from strain 13 after culture for 1, 2, 3, and 4 h.
To examine the mRNA corresponding to the agrBDCp region, Northern analysis was performed using gene probes for agrDCp, CPE1561 (agrBCp), CPE1562, CPE1563, CPE1564, and CPE1560. The mRNA obtained from the CPE1561 region was ∼2.5 kb in length (Fig. 1C). This length is consistent with the total length of the CPE1561 operon calculated from genome information. Thus, the CPE1561 operon encodes agrDCp, CPE1562, and CPE1563. These data also suggest that CPE1564 and CPE1560 must be independently transcribed, since mRNAs of different sizes were detected by Northern hybridization using gene probes for CPE1564 and CPE1560 (data not shown). The agrDCp gene is included in the operon, but a second, small independent mRNA was also identified that corresponds to the agrDCp gene (Fig. 1C). This mRNA is transcribed at a high level up to the stationary phase of growth (data not shown). The length of the agrDCp mRNA was calculated as 0.45 kb (Fig. 1C). This 0.45-kb mRNA was also detected by using the CPE1561 (agrBCp) probe, probably because the transcription start site of this mRNA exists in the coding region of CPE1561.
Effect of agrBDCp on toxin gene expression.
To examine the role of the agrBDCp region in detail, an agrBDCp-null mutant strain and its complement strain were constructed as described in Materials and Methods. The resulting mutant strain (TS230) lacked PfoA-hemolytic activity on blood agar plates (see Fig. 4). Transcription of agrDCp was completely absent in TS230 but was recovered in the strains that carry pTS1303 and pTS1304 (Fig. 2). In the TS230/pTS1304, an extra band was detected above the agrDCp mRNA; this band presumably originated from a readthrough transcription occurring in the recombinant plasmid (Fig. 2). The transcription of pfoA in TS230/pJIR418 was very low, and the plc and colA mRNA levels were significantly decreased (Fig. 2). In the TS230 strain that was complemented with a plasmid containing the intact 2.5-kb agrBDCp operon (TS230/pTS1304), transcription of the toxin genes increased to almost the same level as that in the wild-type strain (Fig. 2). Since the level of toxin gene transcription was practically the same between the TS230/pTS1304 strain complemented with the 2.5-kb operon and the TS230/pTS1303 strain complemented with the 2.5-kb operon and the downstream CPE1560 (Fig. 2), it was concluded that CPE1560 does not have a significant effect on toxin gene expression. From these data it was concluded that the agrBDCp operon is responsible for the transcriptional activation of toxin genes in C. perfringens.
FIG. 4.
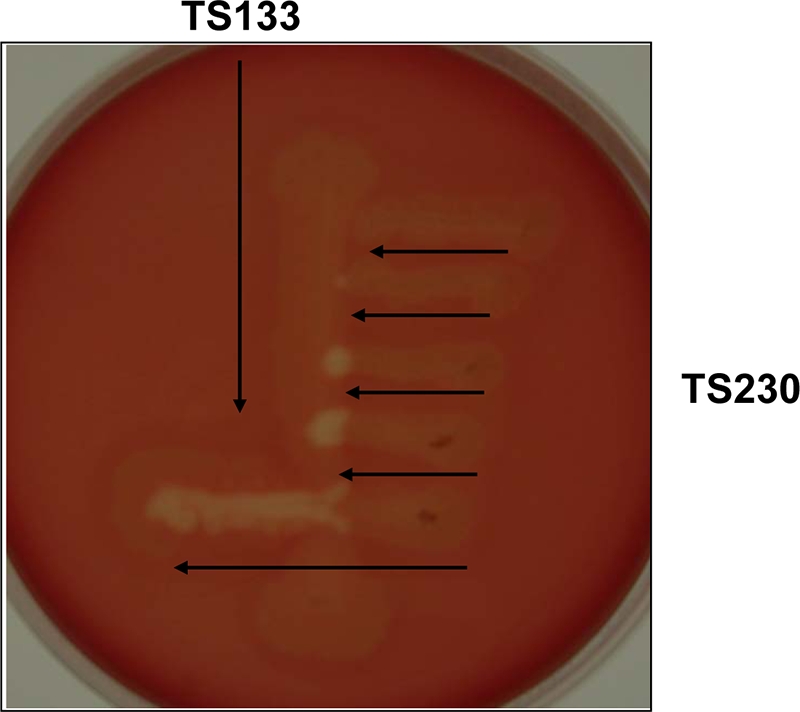
Cross-streaking of TS230 and TS133. The virR mutant strain, TS133, was streaked onto a blood agar plate, and then several streaks of TS230 were made at a right angle to TS133. The distance between the two strains was decreased with each successive streak.
FIG. 2.
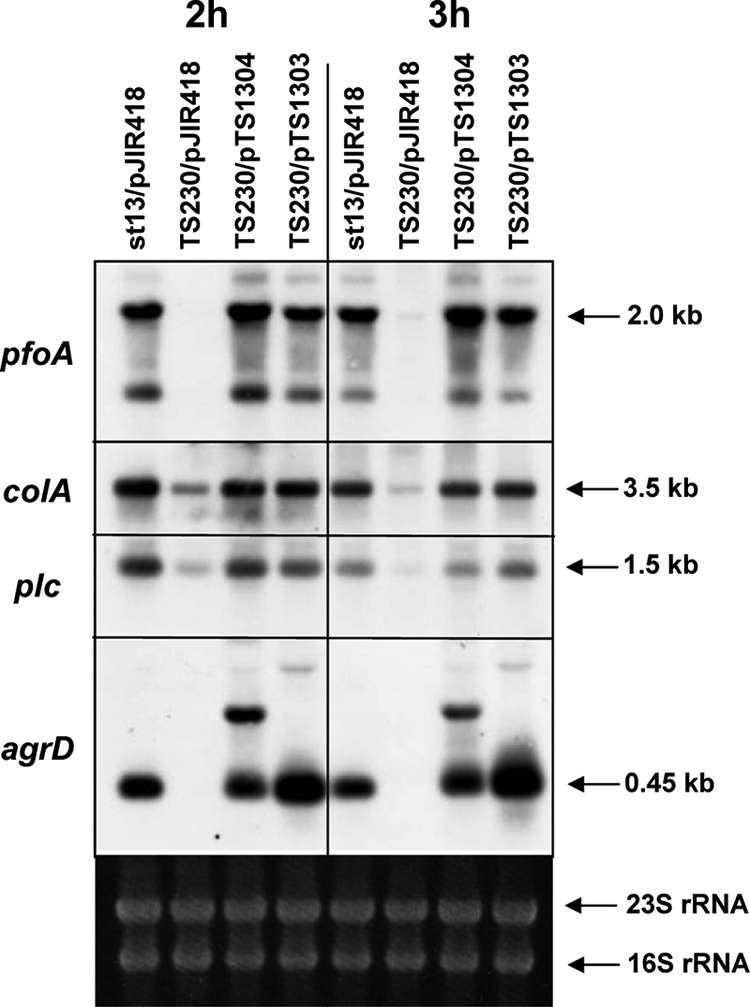
Northern analysis of the agrBDCp mutant and complemented strains. An agrBDCp-null mutant (TS230) was constructed by a double-crossing-over method, and the agrBDCp region was complemented by transformation with pTS1304 and pTS1303. Total RNA was prepared from 2- and 3-h-cultured cells, and 10 μg of total RNA was used for Northern analysis. The internal regions of pfoA, colA, plc, and agrD were used as probes.
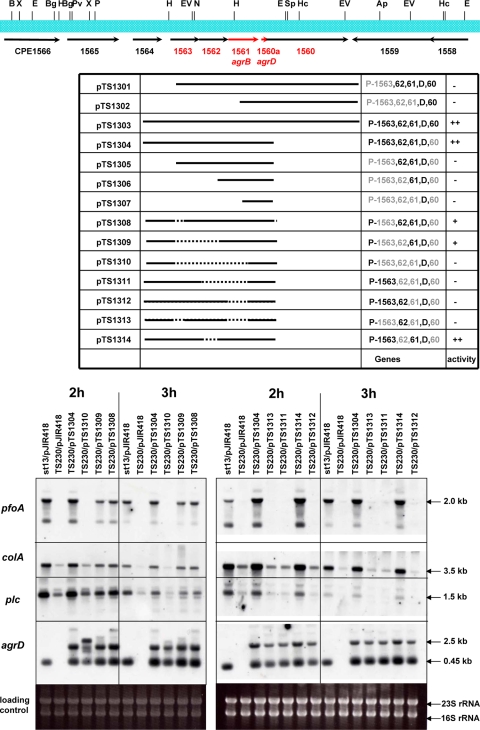
Function of each gene in the operon.
In S. aureus, agrBDSa and a two-component regulatory system are all included in a single operon. However, in the case of C. perfringens there is no apparent two-component system in the vicinity of agrBDCp in the genome. Instead, two other hypothetical genes exist upstream of CPE1561 (agrBCp) and compose a 2.5-kb operon together with agrBDCp (Fig. 1A). It was therefore considered a possibility that these genes might also have a regulatory effect on toxin gene expression. To analyze the effect of these genes on toxin transcription, plasmids encoding various deletions in these genes were constructed and transformed into the agrBDCp mutant TS230 (Fig. 3). Deletion plasmids containing both an intact agrDCp and the CPE1561 gene (pTS1303, pTS1308, pTS1309, and pTS1314) could restore transcription of the toxin genes, whereas plasmids that do not contain the CPE1561 gene (pTS1302, pTS1307, pTS1310, pTS1311, pTS1312, and pTS1313) could not recover toxin gene transcription even when the plasmids contained an intact agrDCp gene (Fig. 3). Plasmids that contain both the agrDCp and CPE1561 genes but that do not contain a potential promoter region located upstream of CPE1563 (pTS1301, pTS1302, pTS1305, and pTS1306) also could not activate transcription of the toxin genes (Northern blot data not shown). These experiments suggest that at least CPE1561 (agrBCp) and agrDCp appear to be essential to the regulatory function of this operon and that transcription is started from a position upstream of CPE1563. Interestingly, in TS230/pTS1308 and TS230/pTS1309 (the plasmids that contain CPE1561 and agrDCp but not CPE1563) toxin genes are transcribed, but the level of transcription is much weaker than that in TS230/pTS1304 (the plasmid containing all of the genes). However, transcription of the toxin genes in the mutant strain with pTS1314 (ΔCPE1562) was at almost the same level as that in the TS230/pTS1304 strain. These data indicated that CPE1561 (agrBCp) and agrDCp are essential genes for toxin gene activation but that CPE1563 is required for complete activation.
FIG. 3.
Deletion analysis of the agr region. To determine the role of each gene in the operon, deletion plasmids were constructed and transformed into the agrBDCp-null mutant, TS230. Each strain was cultured, and RNA was isolated after 2 and 3 h of culture. The RNA was used for Northern analysis of the indicated toxin genes. In the deletion table, “−” indicates no activity, “++” indicates the plasmid has activity to induce the expression of toxin genes, and “+” indicates the plasmids have activity but that the activity is lower than that of pTS1304. The internal regions of pfoA, colA, plc, and agrD were used as probes.
Activation of toxin production by the toxin-negative strain TS133.
We examined whether TS230 can recover its hemolytic activity by exposure to a signal molecule produced from another toxin-negative virR mutant strain, TS133. First, TS133 was streaked on a blood agar plate, and then TS230 was streaked at a right angle to TS133 at various distances (Fig. 4). As the two strains became closer, hemolysis from TS230 became stronger. This finding suggested that TS133 secreted a signal molecule that activated toxin production and that TS230 recovered its toxin production by absorbing this molecule from TS133. However, this signal molecule did not appear to diffuse widely in the agar medium because hemolysis of TS230 only occurred when the distance between TS230 and TS133 was quite short (Fig. 4).
Production of the signal molecule and its putative sensor protein.
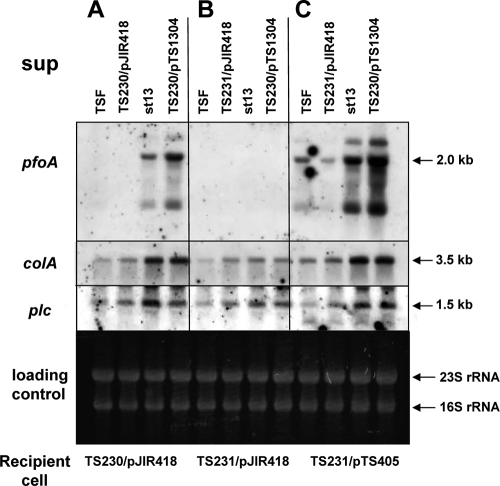
In S. aureus, AIP is produced from the agrBDSa region and is secreted from the cell into the culture supernatant, where it regulates gene expression via a two-component system consisting of AgrA and AgrC (17). To determine whether agrBDCp is related to the signaling component that is secreted from C. perfringens cells, we assayed the ability of agrBDCp to modulate toxin expression. The culture supernatant was collected from the wild-type C. perfringens strain 13 or the agrBDCp mutant TS230 at early log phase (optical density at 600 nm of 0.5) and was then added to TS230 cells. The cells were incubated at 37°C for 15 min, and total RNA was prepared and analyzed by Northern analysis. The transcription of toxin genes was significantly increased in the TS230 cells only when the wild-type supernatant was added (Fig. 5A), suggesting that the TS230 cells lacked the ability to produce the signal molecule and release it into the supernatant. To further confirm that the signal molecule in the supernatant of strain 13 was produced from the agrBDCp region, the supernatant was collected from a TS230 mutant strain that had been complemented with an intact agrBDCp (TS230/pTS1304). When this supernatant was tested on TS230 cells, the expression of toxin genes, especially that of pfoA, was strongly induced (Fig. 5A). These data clearly indicate that the agrBDCp gene is responsible for the production of an extracellular autoinducible signal molecule that controls the expression of toxin genes in C. perfringens.
FIG. 5.
Effect of the wild-type supernatant on the expression of toxin genes in TS230 and TS231. The culture supernatant was collected from strain 13 and added to the indicated strains to check the effect of the supernatant on sensor protein activity. The supernatants were collected from the wild-type strain, strain 13/pJIR418, and strain TS230/pTS1304 after culture for 1.5 h. Total RNA was prepared 15 min after addition of the supernatant. (A) The supernatant (sup) was added onto the agr-null mutant, TS230. (B) The supernatant (sup) was added onto the agr-null virR mutant, TS231/pJIR418. (C) The supernatant (sup) was added onto TS231 that contains an intact virR/virS, TS231/pTS405.
In C. perfringens, the VirR/VirS-VR-RNA system is known as a global regulator and can regulate the expression of many toxin genes, including plc, pfoA, and colA; however, the signal that activates the sensor protein VirS has not been identified. Since the agrBDCp locus controls the expression of a subset of toxin genes similar to that of the VirR/VirS-VR-RNA system, it seemed highly probable that VirS is a sensor protein for the signal molecule produced from the agrBDCp region. To examine this hypothesis, an agrBDCp-virR/virS double-knockout mutant was constructed (designated TS231), and the effect of the wild-type supernatant on toxin transcription in the double mutant was examined. The transcription of pfoA in the TS231 mutant was not activated by the wild-type supernatant (Fig. 5B). In contrast, when TS231 was complemented with the plasmid pTS405, which contains the intact virR/virS genes, the resulting strain (TS231/pTS405) could sense the extracellular signal, and the transcription of toxin genes was significantly induced by the addition of wild-type or TS230/pTS1304 supernatants (Fig. 5C). In addition, the transcription of plc and colA in TS231/pJIR418 or TS231/pTS1304 was also upregulated by addition of the wild-type supernatant (Fig. 5C). It was suggested from these data that VirR/VirS is important for sensing of the extracellular signal and activation of toxin gene transcription in C. perfringens. However, it remains possible that another two-component system or another protein plays a role in the sensing of this signal, and thus further experiments are needed to elucidate the relationship between the signal molecule from agrBDCp and the VirS sensor protein.
Regulation between agr and virR/virS.
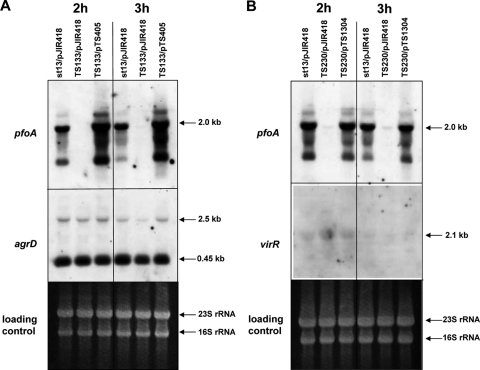
In S. aureus, the agr signaling system results in a positive-feedback loop, and the expression of both agrBDSa for AIP production and agrA/agrC for AIP sensing are positively regulated in an operon (15). To examine the regulatory mechanism of the agr system in C. perfringens, Northern analysis was performed by using TS133 and TS230. At first, RNA was isolated from the wild-type strain (strain 13), TS133, and its complement strain TS133/pTS405, which were cultured for 2 h and 3 h. As in previous experiments, transcription of pfoA was absent in TS133 but recovered in TS133/pTS405 (Fig. 6A). In contrast, the transcriptional levels of agrDCp and the 2.5-kb operon in the three strains were almost the same at 2 h under a virR/virS-negative background (Fig. 6A), although the level of agrDCp transcript was slightly decreased in TS133/pJIR418 at 3 h, which was thought to be not significant.
FIG. 6.
Regulatory relationship between agrBDCp and virR/virS. (A) Regulation of agrBDCp by virR/virS. Total RNA was isolated from 2- and 3-h-cultured strain 13/pJIR418, TS133/pJIR418, and TS133/pTS405. (B) Regulation of virR/virS by agrBDCp. Total RNA was isolated from strain 13/pJIR418, TS230/pJIR418, and TS230/pTS1304. A 10-μg portion of total RNA was used for Northern analysis.
Next, Northern analysis was performed using strains 13/pJIR418, TS230/pJIR418, and TS230/pTS1304 to check the virR/virS transcription under agrBDCp-negative conditions. As shown in Fig. 6B, the transcription of the virR/virS operon was too faint to confirm its regulation, but the mRNA level was almost the same in all three strains. These results suggested that the agr regulatory system involving the agrBDCp and virR/virS operons in C. perfringens is not completely analogous to the agr regulation system in S. aureus.
Effect of a stationary culture supernatant on pfoA transcription.
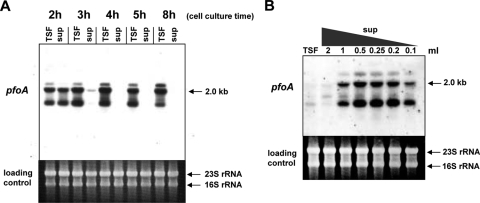
To further analyze the mechanism by which the extracellular signal in the culture supernatant of C. perfringens regulates toxin gene expression, the effect of addition of the C. perfringens culture supernatant on pfoA expression was examined in more detail by Northern analysis. Although the expression of plc and colA was also partially regulated by the extracellular signal molecule in the supernatant, we focused on the regulation of pfoA in this analysis, since pfoA appears to be the main target of this system. First, the supernatant was removed from wild-type strain 13 cells that were cultured to various growth stages (Fig. 7A, 2 to 8 h). These cells were used as recipient cells and were resuspended in fresh TSF medium. As a control, cells were resuspended in the supernatant that had been removed. After 15 min of incubation in the added medium or supernatant, total RNA was prepared from the recipient cells. In the control experiment (see the “sup” lane in Fig. 7A), maximum transcription of pfoA was observed when the supernatant from a 2-h cell culture was added. However, pfoA transcription in the recipient cells was clearly observed within 15 min after the supernatant was replaced with fresh TSF medium (Fig. 7A, lane TSF). Surprisingly, the transcription of pfoA occurred even in the 8-h-cultured recipient cells after replacement of the supernatant with fresh medium (Fig. 7A). Furthermore, the transcription of pfoA in the 3- to 8-h-cultured recipient cells (lane TSF; 3 to 8 h of culture) was at a much higher level than that observed in the recipient cells cultured for 2 h in the presence of a 2-h culture supernatant (lane 2h sup). These data suggest that there is another signaling molecule in the supernatant that negatively controls pfoA expression, especially at the stationary phase, because removal of the culture supernatant and re-addition of fresh medium leads to activation of pfoA transcription in the 3-h (mid log)- to 8-h (stationary)-cultured recipient cells. Furthermore, these data presumably suggest that the amount of signal molecule that binds to recipient cells is sufficient to activate pfoA transcription. Moreover, through the removal of the stationary-phase supernatant, the concentration of the inhibitory substance might decrease, and the remaining activator bound to cells could stimulate pfoA transcription.
FIG. 7.
Effect of the supernatant on toxin gene expression. (A) The supernatant was removed from the various time points of the culture. The cells from each time point were incubated with TSF at 37°C. As a control, the removed supernatant was added again to the same cells. RNA was isolated after 15 min of incubation. Lanes: TSF, TSF control; sup, culture supernatant. (B) The supernatant from strain 13 after 6 h of culture was diluted with TSF medium and added to TS230 cells. RNA was isolated after a 15-min incubation.
To confirm this hypothesis, the supernatant from the stationary phase was diluted with TSF medium and added to TS230 recipient cells. As predicted, diluted supernatant from the stationary phase could activate pfoA transcription, with a maximum activation observed at a fourfold dilution (Fig. 7B). These data suggest that there may be an inhibitory molecule in the supernatant from the stationary phase that represses pfoA expression but that this inhibition is abrogated when the hypothetical inhibitor is diluted. The proportions of activator concentration and inhibitory molecule might be important for determining the transcriptional level of the pfoA gene. Thus, in C. perfringens, a gradual accumulation of the inhibitor might occur over the culture period and, when the concentration of the inhibitor reaches a certain threshold, it may completely stop the transcription of pfoA. This mechanism could explain the decrease in toxin production at the stationary phase of growth in C. perfringens.
In the present study, we examined novel regulatory genes (agrBDCp) for toxin production in C. perfringens. These genes are highly similar to the agr system in S. aureus, and we have shown that the agrBDCp locus is responsible for the production of an extracellular signal molecule that stimulates the expression of toxin genes in C. perfringens. We also found that the two-component VirR/VirS system appears to be required for the regulation by the signaling molecule produced by agrBDCp.
In C. perfringens the functions of agrBDCp, the VirR/VirS system, and VR-RNA seem to be quite similar to those of S. aureus agrBDSa, AgrA/AgrC, and RNAIII, respectively. Consequently, the two bacteria might have evolved similar regulatory systems to control their pathogenicity toward humans. However, the genes involved in the regulation of toxin genes are scattered around the genome of C. perfringens, whereas the genes involved in the agr system are located in a cluster on the S. aureus chromosome (17).
It is noteworthy that toxin gene expression in C. perfringens reaches a maximum during the log phase of growth and completely stops at the stationary phase, whereas in many other pathogenic bacteria, toxin gene expression commonly starts at the stationary phase. Induction of toxin gene expression at the stationary phase is mainly mediated by a quorum-sensing mechanism. In contrast, the agrBDCp system of C. perfringens induces the expression of toxin genes in the early stages of cell growth. For this expression pattern, there may be other unique systems that ensure the specific expression of toxin genes at the early stages of cell growth. From the data in the present study, we predict that there might exist in C. perfringens a system whereby inhibitory molecules are secreted into the medium. However, these molecules would stop toxin gene expression only upon reaching a critical level at the stationary phase. The balance between the agrBDCp activator system and a second, as-yet-undefined inhibitory system may be important for the proper control of gene expression in C. perfringens.
The unique regulation of toxin expression in C. perfringens is consistent with the requirement of C. perfringens to secrete various tissue-degrading toxins and enzymes at an early stage of infection. These secreted products enable the organism to acquire essential nutrients from the host (resulting in gas gangrene) that are required for the survival and growth of the bacteria. Genomic analysis has shown that C. perfringens lacks many genes related to amino acid biosynthesis, with the exception of genes for the three amino acids cysteine, serine, and glycine. Thus, in order to survive, especially in a host environment, C. perfringens may require a well-coordinated system to secrete numerous toxins and enzymes for the degradation of host cells and for the effective import of nutrients from the environment. Therefore, it is very important to precisely elucidate how these extracellular regulatory systems control the virulence of C. perfringens. Elucidation of these regulatory systems may lead to an understanding of the relationship between C. perfringens and other bacteria that coexist in the intestine or in wounds and, furthermore, to the identification of new therapeutic targets for the treatment of life-threatening diseases caused by C. perfringens.
Acknowledgments
This study was supported by a KAKENHI (Grant-in-Aid for Scientific Research) on the Priority Area “Applied Genomics” from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Footnotes
Published ahead of print on 10 April 2009.
REFERENCES
- 1.Aiba, H., S. Adhya, and B. de Crombrugghe. 1981. Evidence for two functional gal promoters in intact Escherichia coli cells. J. Biol. Chem. 25611905-11910. [PubMed] [Google Scholar]
- 2.Bassler, B. L. 1999. How bacteria talk to each other: regulation of gene expression by quorum sensing. Curr. Opin. Microbiol. 2582-587. [DOI] [PubMed] [Google Scholar]
- 3.Ba-Thein, W., M. Lyristis, K. Ohtani, I. T. Nisbet, H. Hayashi, J. I. Rood, and T. Shimizu. 1996. The virR/virS locus regulates the transcription of genes encoding extracellular toxin production in Clostridium perfringens. J. Bacteriol. 1782514-2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fuqua, W. C., S. C. Winans, and E. P. Greenberg. 1994. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J. Bacteriol. 176269-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.George, E. A., and T. W. Muir. 2007. Molecular mechanisms of agr quorum sensing in virulent staphylococci. Chembiochem. 8847-855. [DOI] [PubMed] [Google Scholar]
- 6.Gobbetti, M., M. De Angelis, R. Di Cagno, F. Minervini, and A. Limitone. 2007. Cell-cell communication in food related bacteria. Int. J. Food Microbiol. 12034-45. [DOI] [PubMed] [Google Scholar]
- 7.Hatheway, C. L. 1990. Toxigenic clostridia. Clin. Microbiol. Rev. 366-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Higashi, Y., M. Chazono, K. Inoue, Y. Yanagase, T. Amano, and K. Shimada. 1973. Complementation of theta toxinogenecity between mutants of two groups of Clostridium perfringens. Biken J. 161-9. [PubMed] [Google Scholar]
- 9.Imagawa, T., and Y. Higashi. 1992. An activity which restores theta toxin activity in some theta toxin-deficient mutants of Clostridium perfringens. Microbiol. Immunol. 36523-527. [DOI] [PubMed] [Google Scholar]
- 10.Imagawa, T., T. Tatsuki, Y. Higashi, and T. Amano. 1981. Complementation characteristics of newly isolated mutants from two groups of strains of Clostridium perfringens. Biken J. 2413-21. [PubMed] [Google Scholar]
- 11.Kobayashi, T., T. Shimizu, and H. Hayashi. 1995. Transcriptional analysis of the β-galactosidase gene (pbg) in Clostridium perfringens. FEMS Microbiol. Lett. 13365-69. [DOI] [PubMed] [Google Scholar]
- 12.Lyon, G. J., and R. P. Novick. 2004. Peptide signaling in Staphylococcus aureus and other gram-positive bacteria. Peptides 251389-1403. [DOI] [PubMed] [Google Scholar]
- 13.Mahony, D. E., and T. J. Moore. 1976. Stable l-forms of Clostridium perfringens and their growth on glass surfaces. Can. J. Microbiol. 22953-959. [DOI] [PubMed] [Google Scholar]
- 14.Miller, M. B., and B. L. Bassler. 2001. Quorum sensing in bacteria. Annu. Rev. Microbiol. 55165-199. [DOI] [PubMed] [Google Scholar]
- 15.Muir, T. W. 2003. Turning virulence on and off in staphylococci. J. Pept. Sci. 9612-619. [DOI] [PubMed] [Google Scholar]
- 16.Nakayama, J., A. D. Akkermans, and W. M. De Vos. 2003. High-throughput PCR screening of genes for three-component regulatory system putatively involved in quorum sensing from low-G+C gram-positive bacteria. Biosci. Biotechnol. Biochem. 67480-489. [DOI] [PubMed] [Google Scholar]
- 17.Novick, R. P., and T. W. Muir. 1999. Virulence gene regulation by peptides in staphylococci and other gram-positive bacteria. Curr. Opin. Microbiol. 240-45. [DOI] [PubMed] [Google Scholar]
- 18.Ohtani, K., H. Hayashi, and T. Shimizu. 2002. The luxS gene is involved in cell-cell signaling for toxin production in Clostridium perfringens. Mol. Microbiol. 44171-179. [DOI] [PubMed] [Google Scholar]
- 19.Okumura, K., K. Ohtani, H. Hayashi, and T. Shimizu. 2008. Characterization of genes regulated directly by the VirR/VirS system in Clostridium perfringens. J. Bacteriol. 1907719-7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rieu, A., S. Weidmann, D. Garmyn, P. Piveteau, and J. Guzzo. 2007. Agr system of Listeria monocytogenes EGD-e: role in adherence and differential expression pattern. Appl. Environ. Microbiol. 736125-6133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rood, J. I. 1998. Virulence genes of Clostridium perfringens. Annu. Rev. Microbiol. 52333-360. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook, J., E. F. Fritch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 23.Shimizu, T., W. Ba-Thein, M. Tamaki, and H. Hayashi. 1994. The virR gene, a member of a class of two-component response regulators, regulates the production of the perfringolysin O, collagenase, and hemagglutinin in Clostridium perfringens. J. Bacteriol. 1761616-1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shimizu, T., K. Ohtani, H. Hirakawa, K. Ohshima, A. Yamashita, T. Shiba, N. Ogasawara, M. Hattori, S. Kuhara, and H. Hayashi. 2002. Complete genome sequence of Clostridium perfringens, an anaerobic flesh-eater. Proc. Natl. Acad. Sci. USA 99996-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shimizu, T., A. Okabe, and J. I. Rood. 1997. Regulation of toxin production in Clostridium perfringens, p. 451-470. In J. I. Rood, G. Songer, B. A. McClane, and R. W. Titball (ed.), The clostridia: molecular biology and pathogenesis. Academic Press, London, England.
- 26.Shimizu, T., H. Yaguchi, K. Ohtani, S. Banu, and H. Hayashi. 2002. Clostridial VirR/VirS regulon involves a regulatory RNA molecule for expression of toxins. Mol. Microbiol. 43257-265. [DOI] [PubMed] [Google Scholar]
- 27.Sloan, J., T. A. Warner, P. T. Scott, T. L. Bannam, D. I. Berryman, and J. I. Rood. 1992. Construction of a sequenced Clostridium perfringens-Escherichia coli shuttle plasmid. Plasmid 27207-219. [DOI] [PubMed] [Google Scholar]
- 28.Waters, C. M., and B. L. Bassler. 2005. Quorum sensing: cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 21319-346. [DOI] [PubMed] [Google Scholar]
- 29.Wuster, A., and M. M. Babu. 2008. Conservation and evolutionary dynamics of the agr cell-to-cell communication system across firmicutes. J. Bacteriol. 190743-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xavier, K. B., and B. L. Bassler. 2003. LuxS quorum sensing: more than just a numbers game. Curr. Opin. Microbiol. 6191-197. [DOI] [PubMed] [Google Scholar]