Abstract
The genome of an Escherichia coli MC4100 strain with a λ placMu50 fusion revealed numerous regulatory differences from MG1655, including one that arose during laboratory storage. The 194 mutational differences between MC4100(MuLac) and other K-12 sequences were mostly allocated to specific lineages, indicating the considerable mutational divergence between K-12 strains.
Strains of Escherichia coli K-12 commonly used in various laboratories were derived from a common ancestor, but different lineages have been exposed to various forms of mutagenesis, as well as recombinational crosses involving conjugation and transduction (1). Some K-12 strains were also recipients in crosses involving E. coli B donors, as happened with the common araD139 mutation from an E. coli B/r strain (7). Laboratories in earlier eras also used different culture and storage conditions, also potentially impacting genomic integrity, especially in the movement of insertion sequences and in polymorphisms arising during storage (20, 22). Here, we used genomics to analyze the chromosomal characteristics of a commonly used K-12 lineage with a history different from that of reference K-12 strains MG1655 and W3110 (13) and pieced together its derivation by using the origins of single-nucleotide polymorphisms (SNPs) and indels as markers.
Strain MC4100 [genotype according to the E. coli Genetic Stock Center: F− (araD139) Δ(argF-lac)169 λ− e14− flhD5301 Δ(fruK-yeiR)725(fruA25) relA1 rpsL150(Strr) rbsR22 Δ(fimB-fimE)632(::IS1) deoC1] was obtained in a series of strain constructions (4) from an HfrC-derived MO strain of S. Brenner (genotype according to the E. coli Genetic Stock Center: F− λ− e14− relA1 rspL150 spoT1) (J. Beckwith, personal communication; 1, 6). Strain MC4100 has been widely adopted following studies involving lacZ reporter gene fusions in the Beckwith laboratory (4, 30, 31). MC4100 is an E. coli K-12 strain frequently used in fundamental studies of gene regulation and protein export (30) and bacterial growth and physiology, including cell division (33), DNA replication (16), metabolism (26), and stationary-phase regulation (18). MC4100 is also being used in systems biology approaches to defining E. coli (15) and as a starting strain in laboratory evolution experiments (21).
The genome of strain MC4100 has been previously compared to that of reference strain MG1655 by restriction mapping (14) and using microarrays based on the MG1655 sequence (25). There are substantial band differences between MG1655 and MC4100 as determined by pulsed-field electrophoresis (14), and several deletions have been defined by microarray analysis, followed by PCR analysis of the flanking regions (25). The microarrays did not reveal differences other than deletions, but there remain differences between MC4100 and MG1655 that are unexplained by the known genotypes. Differences in the positions of insertion sequences in MG1655 and MC4100 influence anaerobic gene regulation (29), and another far-reaching difference is the level of sigma factor σS in the two widely used strains (17). There also appear to be differences in central metabolism between the K-12 strains (26), and a recent unexpected finding was the presence of a spoT1 mutation in MC4100 not previously defined in its widely cited genotype (32). Clearly, a full genome sequence of MC4100 would greatly benefit the interpretation of a wide range of fundamental studies.
The strain of MC4100 sequenced here contains an additional element, a λ placMu50 operon fusion (3) in the malEFG operon (24). According to citations, this transposable reporter construct has been used in more than 100 studies of gene regulation but has not been fully sequenced. λ placMu50 was introduced into MC4100 to generate MC4100(MuLac) strain BW2952, the ancestor strain in experimental evolution experiments, because mal expression is a useful marker for detecting an assortment of regulatory mutations in evolving cultures (9, 23).
Major characteristics of the genome.
The sequence of MC4100(MuLac) strain BW2952 (24) was fully assembled by using the hybrid Sanger-pyrosequencing strategy described previously (10). For the full details of the sequencing and annotation strategies used, see the supplemental material.
The MC4100(MuLac) genome is smaller (4,578,159 bp) than that of MG1655 due to the deletions considered below. The number of genes (see Table S1 in the supplemental material) is 4,098, compared to the 4,305 protein coding sequences of MG1655, the 4,227 of W3110 (accession number AP009048), and the 4,058 of DH10B (accession number CP000948), which also has a number of constructed deletions (6, 13). There are 57 genes belonging to the λ placMu50 element integrated into the chromosome. An alignment shows the expected high level of similarity and colinearity over extensive stretches of the four K-12 genome sequences (Fig. 1).
FIG. 1.
Alignment of the four sequenced E. coli K-12 genomes. Scales are in megabase pairs. The gray- and yellow-shaded regions represent segments present in all strains, with inverted segments shaded in yellow. Red and green boxes within the gray and yellow areas represent major indels (insertions and deletions, respectively) (see Table S4 in the supplemental material), and the event numbering is also shown (see Table S4 in the supplemental material). R indicates the E. coli B segment gained by recombination. Green lines (no numbering) represent small deletions treated as mutations, and red lines represent insertions (see Table S5 in the supplemental material). Full-height blue lines represent nonsynonymous substitutions, half-height blue lines represent synonymous substitutions, and gray lines represent substitutions in noncoding regions. For an expanded version of this figure showing individual genes, see Fig. S1 in the supplemental material.
The major differences between the genomes are shown in Fig. 1 (for details, see Table S2 in the supplemental material). The locations of the four large deletions detected by Peters et al. (25) are confirmed by the genomic sequence. Prophage e14, present in MG1655, is absent from MC4100(MuLac) and DH10B, and prophage CP4-6 is absent from MC4100(MuLac) as part of the large (argF-lac)169 deletion (event 02; see Table S2 in the supplemental material). Altogether, there are 42 IS elements in BW2952, similar to the 43 in MG1655 but considerably fewer than the 63 found in DH10B, which is more prone to IS150 transpositions (6). There are several positional differences of insertion sequences between MG1655 and BW2952; two of the MG1655-unique IS1 elements were lost together with CP4-6, and one IS1 element between MG1655 genes b1893 and b1894 and two IS3 elements in b0298 and b0299, respectively, are absent in MC4100(MuLac). Eight additional IS elements found in the MC4100(MuLac) genome include three IS1B elements (in genes BWG_0877, BWG_1061, and BWG_4010), three IS2 elements (in genes BWG_2520-2521, BWG_3106-3108, and BWG_3445-3446), one IS5 element (in gene BWG_1166), and one IS186 element (in gene BWG_1071). All are present in MG1655 at other locations within the chromosome.
The rRNA and tRNA coding sequences were found to be highly conserved, and known small RNAs in MG1655 are also all present in MC4100(MuLac) (see Table S3 in the supplemental material). Minor differences include a tandem duplication of lysQ lysine tRNA in MC4100(MuLac) (Table 1) and changes in the early part of gltT in glutamate tRNA.
TABLE 1.
Insertions unique to MC4100(MuLac)
| Event no. | Genome position
|
Region length (bp) | Insertion location | |
|---|---|---|---|---|
| Start | End | |||
| 07 | 683643 | 683850 | 208 | Duplication of lysQ |
| 09 | 994588 | 995363 | 776 | IS1 in pgaA-ycdT noncoding region |
| 15 | 1178208 | 1178984 | 777 | IS1 in rssB |
| 18 | 1190846 | 1192196 | 1,351 | IS186 in oppB |
| 20 | 1288468 | 1289677 | 1,210 | IS5 in fnr-ogt noncoding region |
| 41 | 3431760 | 3433096 | 1,336 | IS2 in gntT |
| 46 | 3825129 | 3826464 | 1,336 | IS2 in rbsR |
| 51 | 4131098 | 4180256 | 49,158 | λ placMu in malG |
| 58 | 4477679 | 4478446 | 768 | IS1 Δ(fimB-fimE) |
For the mutational differences between strain MC4100(MuLac) and strains MG1655, W3110, and DH10B, see Table S4 in the supplemental material. Mutational changes (mostly SNPs) and major indels are distinguished for convenience, but the boundary is arbitrary and indels of up to 19 bp are treated as mutational, leaving the smallest treated as a major indel at 110 bp. For the distribution of the 194 mutational differences, see Table S5 in the supplemental material. For the mutations, allocated to lineages as discussed below, see Fig. S1 in the supplemental material.
One segment of 28 kb [from strain MC4100(MuLac) sites 65594 to 94277] had a much higher frequency of SNPs. There were 331 SNPs with an average spacing of 90 bp, compared with an average spacing of 56 kb for the remainder of the chromosome. This region included araD with the araD139 mutation that was transferred from E. coli B by transduction (7). It seems clear that the 28-kb region was transferred from E. coli B in this event, and the SNPs involved are not included in the discussion of mutations.
Assignment of mutations to specific lineages.
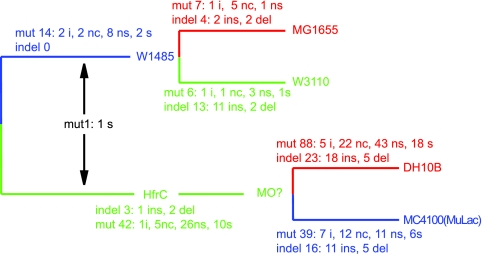
Two of the published genome strains, MG1655 and W3110, are from the W1485 lineage and are quite early isolates (1), while the other, DH10B, is a derivative of HfrC (6), as is MC4100. Thus, the relationships are known to be as shown in Fig. 2. In addition, an alignment was used to attribute events to the W1485 common lineage, the HfrC common lineage, or one of the individual strain lineages. For this analysis, events allocated to the HfrC lineage will include any events that occurred prior to the divergence of MC4100(MuLac) and DH10B, and while the HfrC lineage is well documented as far as the isolation of HfrC, the manipulations in the generation of DH10B and MC1000 are less well defined. Major events in the derivation of DH10B from HfrC were summarized by Durfee et al. (6). The plot of the four-genome alignment (see Fig. S1 in the supplemental material) and the locations of putative recombinant segments were generated by methods used previously for Vibrio cholerae (8) and collectively known as GA-Plot (Genome Alignment Plot) (see the supplemental material).
FIG. 2.
Tree showing the relationships of strains W3310 and MG1655 and strains MC4100(MuLac) and DH10B and the events allocated to the various lineages. The tree topography for HfrC, W1485, MG1655, and W3110 is taken from reference 1. Note that the W1485 lineage discussed in the text extends from the ancestral K-12 strain through W1485 to the divergence between W3310 and MG1655 and includes any events that occurred after W1485 was isolated but before the generation of W3310 and MG1655. The same is true of the HfrC lineage, which for the present purposes extends to the divergence of MC4100(MuLac) and DH10B. For each lineage, the number of mutations (mut) and insertion or deletion events (indel) is shown, first as a total and then for mutations as intergenic (i), other noncoding (nc), nonsynonymous (ns), or synonymous (s) and for indels as insertions (ins) or deletions (del). One mutation allocated to the divergence between W1485 and HfrC but not to either lineage is shown between the two lineages.
For 140 sites, one allele was present in only one of the strains, and the mutations were allocated to that strain. For the other 57 sites, one allele was present in W3110 and MG1655, the other was present in MC4100(MuLac) and DH10B, and all were attributed to events that occurred during the divergence of the W1485 and HfrC lineages.
A few of the mutation events allocated to the HfrC/W1485 divergence can be allocated to a specific lineage by historical information, but a more comprehensive allocation can be done by reference to other E. coli strains. It is usual to use an outgroup strain for this purpose, but within a species subject to recombination, such as E. coli (11), variation in regions that undergo recombination will be much greater than the variation within a lineage such as K-12, and this can confuse the analysis. There is also the problem that a significant part of the genome is found only in some strains and not in others (27), so some segments are expected to be missing in any given outgroup strain. We previously overcame these difficulties for V. cholerae by using a set of strains as a “virtual” outgroup, using only the bases involved in variation within the lineage for the analysis (8). This is expected to work, as the difference between strains of E. coli averages between 1 and 2% (12, 34) and the likelihood that any specific site is affected by recombination is very low.
The 17 E. coli strains with a complete genome sequence available were used to generate the virtual outgroup. The strain MC4100(MuLac) genome was compared with these genomes by using Blastn. The regions including mutation sites were retrieved, and local alignments of the 4 E. coli K-12 genomes and 17 reference genomes were then generated by using Mauve (5) and joined to form a consensus alignment. For the base (if any) present in each genome at the 194 mutation sites, see Table S5 in the supplemental material. For most mutational sites, one base predominated, and it was also one of the two bases present in E. coli K-12, making it the putative ancestral base in the original K-12 strain and the other base the outcome of mutation. Strains 8739 and HS were the most closely related to K-12, and in cases of ambiguity these were given priority in allocation.
We looked first at the 140 mutations present in only one K-12 strain and found excellent agreement between the allocation based on distribution within K-12 and the inference from the outgroup analysis. Of the 140 allocations, 131 were confirmed by the virtual outgroup analysis, 4 were not contradicted (essentially no outgroup data), and for only 5 was there a contradiction between the two approaches. For details and the criteria used to estimate the level of support, see Table S5 in the supplemental material.
This outcome provides excellent support for the use of the virtual outgroup approach, which thus allowed for allocation of the mutations in the main divergence to either the W1485 or the HfrC lineage. Of the 57 mutations allocated to the divergence between the HfrC and W1485 lineages, 56 could be allocated to a specific lineage, leaving only 1 with no basis for allocation.
In summary, analysis of the mutations showed a much higher level of mutation in MC4100(MuLac) (n = 39) and DH10B (n = 88) than in W1485 (n = 6) or MG1655 (n = 7), presumably reflecting the greater number of generations during the many additional construction events. This difference is also present in the two shared components of the lineages, with 13 mutations attributed to the W1485 lineage and 42 attributed to the HfrC lineage. Some, of course, are mutations added by design, and others may be due to recombination with more-divergent K-12 strains.
Deletions and SNPs unique to BW2952 and the known MC4100 genotype.
The insertions in strain MC4100(MuLac) absent from the other K-12 strains are listed in Table 1. Several of the IS changes are insertions into genes functional in MG1655, including the regulatory genes considered below. SNPs not present in the other K-12 strains are shown in Table 2. Some, like the frameshift in flhD, are consistent with the nonchemotactic nature of strain MC4100. Some of the amino acid changes may influence function, so regulation (fabR) and metabolism (tktB) are possibly affected, and the other changes listed in Table 2 could also influence function. Relatively few of the changes unique to MC4100(MuLac) were synonymous substitutions, as in yjeS, yjgL, and amiB. A further 12 unique base changes were in intergenic regions (see Table S4 in the supplemental material).
TABLE 2.
Unique SNPs in MC4100(MuLac) not present in the other three K-12 strains
| Site in MG1655 | Codon | Site in MC4100(MuLac) | Codon | Gene | Amino acid change |
|---|---|---|---|---|---|
| 14172 | GCT | 14172 | GTT | dnaJ | A2V |
| 1976107 | −1 | 1868167 | Frameshift | flhD | |
| 2479422 | CTG | 2365227 | CAG | emrY | L249Q |
| 2577965 | GGC | 2463770 | GAC | tktB | G103D |
| 4111874 | CAT | 4001543 | TAT | fpr | H208Y |
| 4141400 | TCA | 4031069 | TTA | frwC | S283L |
| 4159271 | GGC | 4048940 | GTC | fabR | G42V |
| 4280350 | −1 | 4219196 | Frameshift | yjcF | |
| 4291804 | GGC | 4230650 | GAT | nrfG | G80D |
| 4291805 | GGC | 4230651 | GAT | nrfG | G80D |
| 4323667 | AGC | 4262402 | AAC | yjdN | S33N |
| 4391020 | GTC | 4329755 | GTT | yjeS | |
| 4391575 | TTC | 4330310 | TTT | yjeS | |
| 4394396 | ACC | 4333131 | ACT | amiB | |
| 4394795 | TTT | 4333530 | TTC | amiB | |
| 4471923 | GCA | 4410658 | AGA | yjgI | G52R |
| 4473516 | TTT | 4412251 | TTC | yjgL |
The strain MC4100 lineage retained the relA1 mutation present in the ancestor (strain MO, genotype F− λ− e14− relA1 rspL150 spoT1), as confirmed by genomic sequencing. As shown recently, and confirmed in the genomic sequence, MC4100 also contains a spoT1 mutation resulting in elevated basal levels of ppGpp and higher sigma factor RpoS levels than found with wild-type spoT (32). This spoT1 mutation is not recorded in the MC4100 genotype but is inherent in this lineage (1).
Several further disruptions affect global regulation in MC4100. First, one of the intentionally introduced deletions contributing to the genotype is now shown to have the effect of disrupting fruR (cra) between codons 82 and 83. FruR regulates multiple genes in central metabolic pathways (28) and so would be expected to affect metabolic regulation. The fruR defect can explain the observed lower expression of the glyoxylate cycle in strain MC4100(MuLac) than in strain MG1655 (26) because it lacks the activation of aceBAK caused by FruR (28). Also documented and confirmed in the genome is the presence of an IS5 element upstream from fnr that differentially affects anaerobic regulation in strain MC4100 (29).
Finally, an additional unexpected mutation in strain MC4100(MuLac) is an insertion sequence integrated into the first codons of rssB, which encodes a response regulator involved in RpoS turnover (2, 35). A reduced function of RssB lowers the proteolytic breakdown of RpoS and hence increases the level of this sigma factor in the cell. This rssB mutation, together with spoT1, accounts for the previously unexplained observation that strain MC4100(MuLac) has a higher endogenous RpoS level while having the same rpoS sequence as strain MG1655 (17, 32). Interestingly, as shown in the PCR-based screening (see Fig. S2 in the supplemental material), the IS1 insertion in rssB is present in glycerol stocks of strain MC4100 and MC4100(MuLac) strain BW2952 (constructed in November 1992) from the Ferenci laboratory but absent from earlier freeze-dried stocks of MC4100 from 1984. IS1 is also absent from strain MC4100 lineages received in the Ferenci laboratory from seven outside laboratories (see Fig. S2 in the supplemental material), so it probably arose during the preparation of the MC4100 glycerol stocks in the early 1990s. The mutation does not have an obvious phenotype on rich or glucose minimal media, so it was not detected until now.
Complete sequence of the incorporated fusion vector λ placMu50.
The vector λ placMu50 was found to be integrated into the ninth codon of malG, consistent with its earlier transductional mapping (24). The structure of the fusion vector was remarkably accurately described by Bremer et al. (3), who undertook the in vivo recombinational construction of the λ-Mu hybrid. The only surprise is that the construct contains six genes in the middle of the λ backbone probably derived from the cryptic lambdoid DLP12 phage, also found at 12 min in the E. coli chromosome (19) and present in the MC4100(MuLac) genome. The sequences of the corresponding BWG_0429-0434 and BWG_3716-3721 genes have several base differences in each gene pair, suggesting that the DLP12 genes in the λ-Mu hybrid originated before the construction of strain BW2952. As expected, the fusion vector also contains genes for the aminoglucoside phosphotransferase conferring kanamycin resistance and the promoterless lacZY genes after a trp gene fragment.
Conclusions.
MC4100(MuLac) and its parent MC4100 contain many genomic alterations that affect gene regulation compared to MG1655, indicative of numerous mutational differences within K-12 laboratory strains. Its HfrC-derived genome is traceably different from the MG1655-like genomes derived from W1485. A salutary lesson of the ease with which laboratory-acquired mutations can be incorporated into stored strains is the IS1 transposition into the rssB gene in MC4100(MuLac), which arose in an MC4100 colony in the 1980s prior to storage in glycerol. Speculatively, this mutation was selected by the storage of streaked plates at 4°C before inoculation, as was common in the Ferenci laboratory in the 1980s. A high level of RpoS, as would result from the rssB mutation, improves survival in the cold (32), as well as recovery from −80°C stocks. The advantage that this confers provides a greater chance of being picked from cold-stored streaked plates. It would not be surprising if other laboratory stocks contained unexpected mutations selected by laboratory culture and storage.
There are 194 mutations found in one or more of the four E. coli K-12 genomes, far more than accounted for by the known construction events. Most of these have no predictable effect on phenotype but should be taken into account when comparing these strains. There are 81 in MC4100(MuLac) and 130 in DH10B, of which 42 are shared, compared with 19 in W3110 and 20 in MG165, of which 13 are shared. The higher number in MC4100(MuLac) and DH10B may be due to a longer time in culture during the many construction events. In addition, the known introduction of the araD139 mutation into the ancestor of MC4100(MuLac) and DH10B by transduction from E. coli B appears to have introduced an additional 331 SNPs not present in the ancestral K-12 strain. Altogether, MC4100 is not as closely related to MG1655 as proposed by Peters et al. (25).
Nucleotide sequence accession number.
The genome sequence of MC4100(MuLac) strain BW2952 has been deposited at GenBank/EMBL/DDBJ under accession number CP001396.
Supplementary Material
Acknowledgments
We thank J. Beckwith for information on the source of the MC4100 strain. E. Bremer, R. Misra, M. Villarejo, W. Boos, T. Silhavy, B. Spira, and R. Hengge are gratefully acknowledged for providing strains. We thank Tianjin Biochip Corporation for assistance with sequencing and analysis.
We thank the Australian Research Council for grant support to T.F. This work was supported by Tianjin Municipal Special Fund for Science and Technology Innovation grant 05FZZDSH00800, National Natural Science Foundation of China (NSFC) Key Programs grants 30530010 and 20536040, NSFC General Program grant 30670038, National 863 Program of China grants 2006AA020703 and 2006AA06Z409, National 973 Program of China grant 2009CB522603, and National Key Programs for Infectious Diseases of China grants 2008ZX10004-002 and 2008ZX10004-009.
Footnotes
Published ahead of print on 17 April 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Bachmann, B. J. 1996. Derivations and genotypes of some mutant derivatives of Escherichia coli, p. 2460-2488. In R. Curtiss III, F. C. Neidhardt, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. American Society for Microbiology, Washington, DC.
- 2.Becker, G., E. Klauck, and R. Hengge-Aronis. 2000. The response regulator RssB, a recognition factor for σS proteolysis in Escherichia coli, can act like an anti-σS factor. Mol. Microbiol. 35657-666. [DOI] [PubMed] [Google Scholar]
- 3.Bremer, E., T. J. Silhavy, and G. M. Weinstock. 1985. Transposable λ placMu phages for creating lacZ operon fusions and kanamycin resistance insertions in Escherichia coli. J. Bacteriol. 1621092-1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casadaban, M. J. 1976. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J. Mol. Biol. 104541-555. [DOI] [PubMed] [Google Scholar]
- 5.Darling, A. C. E., B. Mau, F. R. Blattner, and N. T. Perna. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 141394-1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Durfee, T., R. Nelson, S. Baldwin, G. Plunkett, V. Burland, B. Mau, J. F. Petrosino, X. Qin, D. M. Muzny, M. Ayele, R. A. Gibbs, B. Csorgo, G. Posfai, G. M. Weinstock, and F. R. Blattner. 2008. The complete genome sequence of Escherichia coli DH10B: insights into the biology of a laboratory workhorse. J. Bacteriol. 1902597-2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Englesberg, E., R. L. Anderson, R. Weinberg, N. Lee, P. Hoffee, G. Huttenhauer, and H. Boyer. 1962. l-Arabinose-sensitive, l-ribulose 5-phosphate 4-epimerase-deficient mutants of Escherichia coli. J. Bacteriol. 84137-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feng, L., P. R. Reeves, R. Lan, Y. Ren, C. Gao, Z. Zhou, Y. Ren, J. Cheng, W. Wang, J. Wang, W. Qian, D. Li, and L. Wang. 2008. A recalibrated molecular clock and independent origins for the cholera pandemic clones. PLoS ONE 3e4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferenci, T. 2008. Bacterial physiology, regulation and mutational adaptation in a chemostat environment. Adv. Microb. Physiol. 53169-229. [DOI] [PubMed] [Google Scholar]
- 10.Goldberg, S. M. D., J. Johnson, D. Busam, T. Feldblyum, S. Ferriera, R. Friedman, A. Halpern, H. Khouri, S. A. Kravitz, F. M. Lauro, K. Li, Y. H. Rogers, R. Strausberg, G. Sutton, L. Tallon, T. Thomas, E. Venter, M. Frazier, and J. C. Venter. 2006. A Sanger/pyrosequencing hybrid approach tor the generation of high-quality draft assemblies of marine microbial genomes. Proc. Natl. Acad. Sci. USA 10311240-11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guttman, D. S., and D. E. Dykhuizen. 1994. Clonal divergence in Escherichia coli as a result of recombination, not mutation. Science 2661380-1383. [DOI] [PubMed] [Google Scholar]
- 12.Hall, B. G., and P. M. Sharp. 1992. Molecular population genetics of Escherichia coli: DNA sequence diversity at the celC, crr and gutB loci of natural isolates. Mol. Biol. Evol. 9654-665. [DOI] [PubMed] [Google Scholar]
- 13.Hayashi, K., N. Morooka, Y. Yamamoto, K. Fujita, K. Isono, S. Choi, E. Ohtsubo, T. Baba, B. L. Wanner, H. Mori, and T. Horiuchi. 2006. Highly accurate genome sequences of Escherichia coli K-12 strains MG1655 and W3110. Mol. Syst. Biol. 22006.0007. doi: 10.1038/msb4100049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heath, J. D., J. D. Perkins, B. Sharma, and G. M. Weinstock. 1992. NotI genomic cleavage map of Escherichia coli K-12 strain MG1655. J. Bacteriol. 174558-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ishihama, Y., T. Schmidt, J. Rappsilber, M. Mann, F. U. Hartl, M. Kerner, and D. Frishman. 2008. Protein abundance profiling of the Escherichia coli cytosol. BMC Genomics 9102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khodursky, A. B., B. J. Peter, M. B. Schmid, J. DeRisi, D. Botstein, P. O. Brown, and N. R. Cozzarelli. 2000. Analysis of topoisomerase function in bacterial replication fork movement: use of DNA microarrays. Proc. Natl. Acad. Sci. USA 979419-9424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.King, T., A. Ishihama, A. Kori, and T. Ferenci. 2004. A regulatory trade-off as a source of strain variation in the species Escherichia coli. J. Bacteriol. 1865614-5620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lange, R., and R. Hengge-Aronis. 1991. Identification of a central regulator of stationary-phase gene expression in Escherichia coli. Mol. Microbiol. 549-59. [DOI] [PubMed] [Google Scholar]
- 19.Lindsey, D. F., D. A. Mullin, and J. R. Walker. 1989. Characterization of the cryptic lambdoid prophage DLP12 of Escherichia coli and overlap of the DLP12 integrase gene with the tRNA gene argU. J. Bacteriol. 1716197-6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu, G. R., K. Edwards, A. Eisenstark, Y. M. Fu, W. Q. Liu, K. E. Sanderson, R. N. Johnston, and S. L. Liu. 2003. Genomic diversification among archival strains of Salmonella enterica serovar Typhimurium LT7. J. Bacteriol. 1852131-2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maharjan, R., S. Seeto, L. Notley-McRobb, and T. Ferenci. 2006. Clonal adaptive radiation in a constant environment. Science 313514-517. [DOI] [PubMed] [Google Scholar]
- 22.Naas, T., M. Blot, W. M. Fitch, and W. Arber. 1994. Insertion sequence-related genetic variation in resting Escherichia coli K-12. Genetics 136721-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Notley-McRobb, L., and T. Ferenci. 1999. The generation of multiple coexisting mal-regulatory mutations through polygenic evolution in glucose-limited populations of Escherichia coli. Environ. Microbiol. 145-52. [DOI] [PubMed] [Google Scholar]
- 24.Notley, L., and T. Ferenci. 1995. Differential expression of mal genes under cAMP and endogenous inducer control in nutrient stressed Escherichia coli. Mol. Microbiol. 16121-129. [DOI] [PubMed] [Google Scholar]
- 25.Peters, J. E., T. E. Thate, and N. L. Craig. 2003. Definition of the Escherichia coli MC4100 genome by use of a DNA array. J. Bacteriol. 1852017-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prasad Maharjan, R., P. L. Yu, S. Seeto, and T. Ferenci. 2005. The role of isocitrate lyase and the glyoxylate cycle in Escherichia coli growing under glucose limitation. Res. Microbiol. 156178-183. [DOI] [PubMed] [Google Scholar]
- 27.Rasko, D. A., M. J. Rosovitz, G. S. A. Myers, E. F. Mongodin, W. F. Fricke, P. Gajer, J. Crabtree, M. Sebaihia, N. R. Thomson, R. Chaudhuri, I. R. Henderson, V. Sperandio, and J. Ravel. 2008. The pangenome structure of Escherichia coli: comparative genomic analysis of E. coli commensal and pathogenic isolates. J. Bacteriol. 1906881-6893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saier, M. H., Jr., and T. M. Ramseier. 1996. The catabolite repressor/activator (Cra) protein of enteric bacteria. J. Bacteriol. 1783411-3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sawers, R. G. 2005. Expression of fnr is constrained by an upstream IS5 insertion in certain Escherichia coli K-12 strains. J. Bacteriol. 1872609-2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Silhavy, T. J., and J. R. Beckwith. 1985. Uses of lac fusions for the study of biological problems. Microbiol. Rev. 49398-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Silhavy, T. J., M. J. Casadaban, H. A. Shuman, and J. R. Beckwith. 1976. Conversion of β-galactosidase to a membrane-bound state by gene fusion. Proc. Natl. Acad. Sci. USA 733423-3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spira, B., X. Hu, and T. Ferenci. 2008. Strain variation in ppGpp concentration and RpoS levels in laboratory strains of Escherichia coli K-12. Microbiology 1542887-2895. [DOI] [PubMed] [Google Scholar]
- 33.Wientjes, F. B., and N. Nanninga. 1989. Rate and topography of peptidoglycan synthesis during cell division in Escherichia coli: concept of a leading edge. J. Bacteriol. 1713412-3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wirth, T., D. Falush, R. T. Lan, F. Colles, P. Mensa, L. H. Wieler, H. Karch, P. R. Reeves, M. C. J. Maiden, H. Ochman, and M. Achtman. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol. 601136-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou, Y. N., S. Gottesman, J. R. Hoskins, M. R. Maurizi, and S. Wickner. 2001. The RssB response regulator directly targets òS for degradation by ClpXP. Genes Dev. 15627-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.