Abstract
The bacterial second messenger cyclic di-GMP (c-di-GMP) regulates the transition between sessility and motility. In Salmonella enterica serovar Typhimurium, the expression of CsgD, the regulator of multicellular rdar morphotype behavior, is a major target of c-di-GMP signaling. CsgD expression is positively regulated by at least two diguanylate cyclases, GGDEF domain proteins, and negatively regulated by at least four phosphodiesterases, EAL domain proteins. Here, we show that in contrast to EAL domain proteins acting as phosphodiesterases, the EAL-like protein STM1344 regulated CsgD expression positively and motility negatively. STM1344, however, did not have a role in c-di-GMP turnover and also did not bind the nucleotide. STM1344 acted upstream of the phosphodiesterases STM1703 and STM3611, previously identified to participate in CsgD downregulation, where it repressed their expression. Consequently, although STM1344 has not retained a direct role in c-di-GMP metabolism, it still participates in the regulation of c-di-GMP turnover and has a role in the transition between sessility and motility.
Cyclic di-GMP (c-di-GMP) is a recently recognized secondary messenger which regulates the transition between sessility and motility in many bacteria (4, 27). The output of c-di-GMP signaling is determined by the temporally and spatially regulated delicate balance between the synthesis and degradation of the secondary messenger. The synthesis of c-di-GMP occurs through diguanylate cyclases which have a catalytic GGDEF domain in common (23, 35). The degradation of c-di-GMP by c-di-GMP-specific phosphodiesterases, on the other hand, is an enzymatic activity carried out by nonhomologous EAL and HD-GYP domains (32, 36). GGDEF, EAL, and HD-GYP domain proteins constitute superfamilies (7), in which subgroups of proteins can be identified (24). Although not applicable to all individual c-di-GMP-metabolizing enzymes, some general principles underlie the phenotypic output of c-di-GMP-synthesizing and -degrading activities. The expression of diguanylate cyclases leads to enhanced (local or general) intracellular c-di-GMP concentrations, which consequently promote sessility and inhibit motility. In contrast, low c-di-GMP levels promote motility and inhibit sessility. Signature motifs composed of highly conserved residues which are crucial for the c-di-GMP-metabolizing activities of GGDEF and EAL domains have been identified previously (25, 36), although the core residues required for catalytic activity are not entirely defined. Alternative functions for GGDEF and EAL domain proteins with motifs deviating from the consensus sequence have been identified (3, 6, 22, 40, 43). For example, the GGDEF-EAL domain protein LapD has been shown to bind c-di-GMP via its EAL domain (22), while CsrD has been shown to target small RNAs for degradation (40).
The transcriptional regulator CsgD is a master regulator of a multicellular/biofilm behavior in Salmonella enterica serovar Typhimurium and other Enterobacteriaceae which produces a distinct colony morphology on agar plates, the rdar (red, dry, and rough) morphotype (8). Characteristic of the rdar morphotype is the expression of the extracellular matrix components curli fimbriae and cellulose. Not only the biosynthesis of proteinaceous and polysaccharide extracellular matrix components (10, 19, 28, 47), but also the physiological adaptation to the sessile mode of growth (2) is regulated by CsgD.
CsgD and the rdar morphotype are major targets for regulation by c-di-GMP signaling (27, 33). Screens for proteins in S. Typhimurium that contribute to rdar morphotype formation revealed that at least two diguanylate cyclases (GGDEF/GGDEF-EAL domain proteins STM2123 and STM3388) and four phosphodiesterases (EAL/GGDEF-EAL domain proteins STM1703, STM1827, STM3611 [YhjH], and STM4264) are involved in the expression of CsgD in a temporally and hierarchically organized fashion (16, 37). Knockouts of STM1703 and STM4264 have more significant effects than knockouts of the other proteins, enhancing CsgD expression at least fourfold (37). STM4264 acts upstream of STM1703, as the overexpression of STM1703 downregulates CsgD expression in an STM4264 mutant while the overexpression of STM4264 in an STM1703 mutant fails to do so. Consistently, STM4264 affects a much greater fraction of the cellular c-di-GMP than STM1703. On the other hand, only one of the phosphodiesterases, STM3611, has an effect on swimming motility in the wild-type background (37), thus inversely regulating sessility and motility.
In S. Typhimurium, bioinformatic analyses previously detected the presence of five GGDEF, seven EAL, and seven GGDEF-EAL domain proteins. Among them, the EAL-like protein STM1344, highly similar to the phosphodiesterase STM3611, was identified (26). STM1344 behaves unconventionally, as it is required for rdar morphotype and CsgD expression but downregulates motility. Although STM1344 affects the c-di-GMP pool in vivo, biochemical analysis showed that STM1344 neither degrades nor synthesizes c-di-GMP and also does not bind it. However, STM1344 is integrated into the c-di-GMP network, as it works independently of the phosphodiesterases STM4264 and STM1827 to affect the expression of the phosphodiesterases STM1703 and STM3611. Therefore, STM1344 exerts a novel function in aiding the transition between sessility and motility.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. For cloning purposes, Escherichia coli and S. Typhimurium were grown at 37°C on Luria-Bertani (LB) agar plates supplemented with the appropriate antibiotics. For analysis, bacteria were precultured on LB agar plates at 37°C overnight and directly inoculated onto LB agar plates without salt at 28 or 37°C for the times indicated below. Cells were grown on agar plates containing LB medium without salt, supplemented with Congo red (40 μg ml−1) and Coomassie brilliant blue G-250 (20 μg ml−1) or calcofluor white (fluorescence brightener 28; 50 μg ml−1) when required. For the expression of genes, 0.1% arabinose or 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) was used. The antibiotics employed were ampicillin (100 μg ml−1), chloramphenicol (20 μg ml−1), kanamycin (30 μg ml−1), and tetracycline (20 μg ml−1).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or descriptiona | Reference or source |
|---|---|---|
| S. Typhimurium strains | ||
| ATCC 14028 | Wild type | |
| UMR1 | ATCC 14028-1s Nalr | 28 |
| MAE46 | UMR1 ΔompR101::Ampr | 28 |
| MAE52 | UMR1 PcsgD1 | 31 |
| MAE420 | UMR1 STM3611::Cmr | 37 |
| MAE424 | UMR1 STM1344::Cmr | 37 |
| MAE282 | UMR1 STM1703::Cmr | 16 |
| MAE425 | UMR1 STM4264::Cmr | 37 |
| MAE422 | UMR1 STM1827::Cmr | 37 |
| MAE130 | UMR1 STM3611-SPA Kmr | This study |
| MAE131 | UMR1 STM1703-SPA Kmr | This study |
| MAE132 | UMR1 STM1344-SPA Kmr | This study |
| MAE440 | UMR1 STM1344::Cm ΔSTM1703::101 | This study |
| MAE1251 | UMR1 STM1344::Cm ΔSTM1827::101 | This study |
| MAE451 | UMR1 STM1344::Cm ΔSTM3611::101 | This study |
| MAE439 | UMR1 STM1344::Cm ΔSTM4264::101 | This study |
| MAE1253 | UMR1 STM1344::Cm STM1703-SPA Kmr | This study |
| MAE1252 | UMR1 STM1344::Cm STM3611-SPA Kmr | This study |
| LT2 | Wild type | |
| LB5010 | metA22 metE551 ilv-452 leu-3121 trpO2 xyl-404 galE856 hsdLT6 hsdSA29 hsdSB121 rpsL120 | 31 |
| E. coli strains | ||
| K-12 DH5α | endA1 hsdR17 supE44 thi-1 recA1 gyrA relA1 Δ(lacZYA-argF)U169 (φ80lacZΔM15) | Laboratory collection |
| BL21(DE3) | F−ompT hsdSB(rB− mB−) gal dcm (DE3) | Novagen |
| BL21(DE3)(pLysS) | F−ompT hsdSB(rB− mB−) gal dcm (DE3) pLysS | Novagen |
| Plasmids | ||
| pBAD30 | Arabinose-regulated promoter; Ampr | 11 |
| pKD46 | Expresses λ Red recombinase system; Ampr; temp-sensitive replicon | 5 |
| pET23a(+) | Expression vector with T7 promoter; Ampr | Novagen |
| pRGS40 (pSTM1344) | pBAD30::STM1344-His6 Ampr | This study |
| pRGS42 | pBAD30::STM1344(N85A)-His6 Ampr | This study |
| pRGS44 | pBAD30::STM1344(F144A)-His6 Ampr | This study |
| pRGS45 | pBAD30::STM1344(D165A)-His6 Ampr | This study |
| pRGS46 | pBAD30::STM1344(F168A)-His6 Ampr | This study |
| pRGS47 | pET23a::STM1344; expresses C-terminal His6 tag; Ampr | This study |
| pRGS48 | pET23a::STM3611; expresses N-terminal FLAG tag; Ampr | This study |
| pMMB-PA3702 | pMMB::PA3702 Ampr | 18 |
| pEZ-YcgR | pET23a::K-12ycgR; expresses C-terminal His6 tag | 34 |
| pJL148 | Expresses SPA tag (calmodulin binding peptide, tobacco etch virus protease cleavage site, and three FLAG tags); Kmr | 46 |
Nalr, nalidixic acid resistant; Ampr, ampicillin resistant; Cmr, chloramphenicol resistant; Kmr, kanamycin resistant.
General molecular biology methods.
Common molecular biological procedures such as the isolation of plasmid and chromosomal DNA, PCR analyses, and the transformation of cells with plasmids, as well as restriction digestion, were carried out using standard protocols (1). PCR amplifications were performed using GeneCraft Taq polymerase (Biostar). PCR products were cleaned using the QIAquick PCR purification kit (Qiagen), and DNA fragments were recovered from agarose gels by using the QIAquick elute gel purification minikit (Qiagen). Primer sequences are provided in Table 2.
TABLE 2.
Primers used in this study
| Purpose and name | Sequencea | Restriction site and/or tag |
|---|---|---|
| Construction of SP4-tagged proteins | ||
| STM1344-SPA-F | TGGCCTCCGGTGCCGGTAAGCCAGCTCATTAAACTCGTTCAGCGATCCATGGAAAAGAGAAG | |
| STM1344-SPA-R | AGTGTAGACGGTTAATCACCGGTTAAACACCGGCAAACAGAAAGGCATATGAATATCCTCCTTAG | |
| STM3611-SPA-F | CGACCCGTGCCGCTGATATCGCTCGAAGAGGTGATTCTGACCCTGTCCATGGAAAAGAGAAG | |
| STM3611-SPA-R | ATGACCGCGCCTCGTAATACCACGTATTACGGGAACAGTCTGGCGCATATGAATATCCTCCTTAG | |
| STM1703-SPA-F | TCTTTTGAACGCTGGTACAAACGTTATCAGACGAAAAAAATGCGTTCCATGGAAAAGAGAAG | |
| STM1703-SPA-R | GGCTATGCCTGTCTGCACAGGCTGTTCTCCCTATTATCGTCGCCACATATGAATATCCTCCTTAG | |
| SP4-tag control | ||
| STM1344_SPA_control | TGCACGCTATTGTTGCCCA | |
| STM1344_Control_Reverse | GGGCAGTAAAAGACAGGGT | |
| STM3611_SPA_control | TGCTGCTGCAACTGATGAAC | |
| STM3611_Control_Reverse | TGTGATGGCGGCGGTTATT | |
| STM1703_SPA_Control | AGGCGCTAAATTTACAAGTG | |
| STM1703_Control_Reverse | AAATTGATTGTTGTCGGGAGT | |
| Cloning | ||
| STM1344_pET23a_Start | CGTGGCTAGCATTGCTTCACTTGATGAGCTT | NheI |
| STM1344_pET23a_Stop | CCTAAGCTTTCGCTGAACGAGTTTAATGA | HindIII |
| STM3611_pET23a_N-FLAG_Start | AGCCATATGGACTACAAAGACGATGACGACAAGGGTATAAAGCAGGTTATCCAGCA | NdeI, FLAG tag |
| STM3611_pET23a_N-Tag_Stop | AGCAAGCTTTTACAGGGTCAGAATCACCTCT | HindIII |
| Construction of variant STM1344 | ||
| E29A_forward2 | TTGGTTTAGCAATTATTGCAACGT | |
| E29A_reverse2 | GTTGCAATAATTGCTAAACCAACAA | |
| N85A_forward | CATGGCTTGCATTACCGCCTGCAATTAGTGA | |
| N85A_reverse | GCGGTAATGCAAGCCATGCAATAAGTTTATGC | |
| F144A_forward | CTGGCAAATGCTGGGGCAGGCGAGGCG | |
| F144A_reverse | TGCCCCAGCATTTGCCAGCATTAATGGGA | |
| D165A_forward | GTCATGTTGGCTAAAAATTTTATTCAGCAGCGAG | |
| D165A_reverse | TAAAATTTTTAGCCAACATGACTCGTTTGAAAA | |
| F168A_forward | TTGGATAAAAATGCTATTCAGCAGC | |
| F168A_reverse | CTGCTGAATAGCATTTTTATCCAAC | |
| RT-PCR | ||
| recA_forward | GGCGAAATCGGCGACTCT | |
| recA_reverse | CATACGGATCTGGTTGATGAAAATC | |
| STM1703_forward | GGCGCAGGAGCTCTCTTT | |
| STM1703_reverse | GCAATGACTGAGTTGGCTTCTG | |
| STM3611_forward | CTGCTGCTGCAACTGATGAAC | |
| STM3611_reverse | CTCCACGCCCTCGACAAT | |
| csgD_forward | ACGCTACTGAAGACCAGGAAC | |
| csgD_reverse | GCATTCGCCACGCAGAATA | |
| Sequencing | ||
| pBAD_MCS_Start | ATCGCAACTCTCTACTGTTTC | |
| pBAD_MCS_Stop | CTGATTTAATCTGTATCAGGC |
SPA tag-specific priming sequences are underlined only, the FLAG tag sequence is in bold and underlined, restriction sites are shown in italics, and nucleotide exchanges leading to amino acid replacement are shown in bold.
Plasmid construction.
For complementation, the STM1344 gene was cloned into pBAD30 with a sequence expressing a C-terminal His tag. Mutant STM1344 alleles were constructed by PCR-based mutagenesis using overlapping primers (Table 2) and subsequently cloned into pBAD30.
For the expression of STM1344 and STM3611 proteins in E. coli strain BL21(DE3), the corresponding genes were cloned into the expression vector pET23a(+) with sequences encoding a C-terminal His tag and an N-terminal FLAG tag, respectively.
Primers used to construct the recombinant plasmids are listed in Table 2. E. coli DH5α was transformed with the plasmids, which were passaged through the restriction-deficient strain S. Typhimurium LB5010 before being introduced into S. Typhimurium UMR1 derivatives by electroporation.
Construction of mutants.
Epitope tagging of STM1344, STM1703, and STM3611 was performed by the λ Red recombination method (5). The tag was a sequential peptide affinity (SPA) tag (consisting of a calmodulin binding peptide, a tobacco etch virus protease cleavage site, and three FLAG tags) PCR amplified from plasmid pJL148 (46). Briefly, approximately 300 ng of processed PCR product was introduced by electroporation into S. Typhimurium UMR1 containing pKD46, which expresses λ Red recombinase. Recovered colonies were purified at least twice on LB medium containing appropriate antibiotics.
Phage transduction was carried out with phage P22 HT105/1 int-201, a high-frequency generalized transducing mutant of phage P22. Transductants were colony purified twice on LB agar plates containing EGTA (10 mM) and appropriate antibiotics. All constructed mutants were verified by PCR with control primers flanking the deleted open reading frame or the inserted DNA fragment. Gene fusions were sequenced to ensure the in-frame insertion of the SPA tag sequence, leading to the production of a fusion protein.
Protein techniques.
For Western blot analysis, 5 mg (wet weight) of cells was harvested, resuspended in sample buffer, and heated to 95°C for 10 min. The protein content was analyzed by staining with a Coomassie blue solution (20% methanol, 10% acetic acid, 0.1% Coomassie brilliant blue G). Equal amounts of protein were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using 15% resolving gel with 4% stacking gel and were transferred onto a polyvinylidene difluoride membrane (Immobilon P; Millipore). The detection of CsgD was carried out as described previously (30) using the polyclonal anti-CsgD peptide antibody (1:5,000) as the primary antibody and goat anti-rabbit immunoglobulin G conjugated with horseradish peroxidase (1:2,000; Jackson ImmunoResearch Laboratories Inc.) as the secondary antibody. The detection of FLAG tags was done using anti-FLAG antibody (1:2,000) and anti-mouse immunoglobulin G conjugated with horseradish peroxidase (1:2,000; Jackson ImmunoResearch Laboratories Inc.) as the secondary antibody. Chemiluminescence from the Lumi-Light WB substrate (Roche) was recorded using the LAS-1000 system (Fujifilm) and quantified using ImageQuant software (version 5.2).
The enrichment of curli fibers was performed as described previously (29). CsgA subunit expression was detected by colloidal Coomassie blue staining after depolymerization of the curli fibers and separation on gels (4% stacking gel and 15% separating gel).
Phenotypic evaluation. (i) Congo red and calcofluor white binding assays.
Samples of 5 μl of an overnight culture suspended in water (to an optical density at 600 nm [OD600] of 5) were spotted onto LB agar plates lacking NaCl and supplemented with Congo red (40 μg ml−1) and Coomassie brilliant blue (20 μg ml−1) or calcofluor white (fluorescence brightener 28; 50 μg ml−1). Plates were incubated at 28°C for 48 h or at 37°C for 24 h. The development of the colony morphology and dye binding were analyzed over time.
(ii) Analysis of swimming motility.
Swimming motility on 0.3% LB agar plates at 37°C was observed for 6 h. The plates were inoculated with 5-μl aliquots of an overnight culture resuspended at an OD600 of 5. The diameter of the area covered by swimming bacteria was measured after 1 to 6 h.
(iii) Analysis of swarming motility.
Swarming motility on 0.5% LB agar plates supplemented with 0.5% glucose was analyzed. The plates were inoculated with 5-μl aliquots of an overnight culture (OD600 = 5) and incubated at 37°C. The radius from the point of inoculation to the edge of the swarming zone was measured every hour for up to 6 h.
RNA extraction.
The extraction of total RNA was performed using the SV total RNA isolation system according to the protocol of the manufacturer (Promega) with minor modifications. Prior to RNA extraction, the bacterial cells were incubated for ≥30 min in a mixture of ice-cold 5% (vol/vol) phenol and 95% (vol/vol) ethanol on ice to stabilize the RNA. The samples were centrifuged at 4,000 rpm for 10 min, and the supernatant was discarded. Subsequently, the pellet was resuspended in 100 μl of lysis buffer (50 mg/ml lysozyme in Tris-EDTA buffer) and incubated for 5 min at room temperature. The following steps of RNA purification, including on-column DNase I treatment, were performed according to the instructions of the RNA isolation system manufacturer. RNA concentrations were determined using the NanoDrop system (Thermo Scientific), and the quality of the RNA was assessed via gel electrophoresis or similar methods.
Quantitative real-time RT-PCR.
The expression of target genes was evaluated by two-step real-time reverse transcription (RT)-PCR using the Power Sybr green PCR master mix (Applied Biosystems) and the 7500 real-time PCR system (Applied Biosystems). First-strand cDNA synthesis from RNA samples (1 μg) was performed with the high-capacity cDNA RT kit (Applied Biosystems). Relative transcript abundance was determined by the 2−ΔΔCt method (20) using 7500 SDS software v1.3.1 (Applied Biosystems). The recA gene was used as an endogenous control for internal normalization. All experiments were performed as biological and technical replicates using the mean expression from quadruplicates per real-time PCR assay relative to a calibrator value (the value for S. Typhimurium UMR1). Statistical significance levels (P values) were determined with a paired t test using GraphPad Prism software.
Quantification of c-di-GMP.
Nucleotide extracts were prepared essentially as described previously (39). Bacteria were suspended in 0.19% ice-cold formalin, heated to 95°C for 10 min, and extracted by ethanol treatment. Nucleotide extracts equivalent to 10 mg (wet weight) were subjected to high-performance liquid chromatography (HPLC) separation using a Hypersil octadecyl silane (ODS) reversed-phase column (5-μm particle size; Hypersil-Keystone). Runs were carried out with a multistep gradient using 0.1 M triethylammonium acetate (pH 6.0) at 1 ml min−1 with increasing concentrations of acetonitrile. Relevant fractions were collected, lyophilized, and resuspended in 10 μl of water. Fractions containing c-di-GMP were pinpointed by matrix-assisted laser desorption ionization-time of flight analysis and pooled. Synthetic c-di-AMP was added to the pooled fractions at a concentration suitable for use as an internal standard. A standard curve was established using fractions spiked with known amounts of c-di-GMP, with a fixed amount of synthetic c-di-AMP as an internal control. The peak areas for c-di-GMP and c-di-AMP were calculated, and the ratio was determined. c-di-GMP measurements were done in triplicate in two independent experiments.
Purification of proteins.
STM1344, YcgR (34), and WspR*, constitutively active WspR (18), were purified as soluble C-terminal His6 tag fusions by nickel affinity chromatography essentially according to the protocol of the chromatography system manufacturer (Qiagen). STM1344 was eluted in 50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 10 mM MgCl2, and 250 mM imidazole. Soluble STM3611 with an N-terminal FLAG tag fusion was purified using monoclonal anti-FLAG M2 affinity gel according to the protocol of the manufacturer (Sigma-Aldrich). Buffer exchange and the concentration of proteins were done by centrifugation in Amicon ultrafiltration units (Millipore) with a cutoff of 10 kDa. STM1344 and STM3611 were concentrated to 1 mg/ml in a solution of 50 mM Tris-HCl, pH 8.0, 150 mM NaCl, and 10 mM MgCl2 and used directly in assays or stored for up to 2 days at 4°C. Protein concentrations were determined using a bicinchoninic acid protein assay kit (Pierce).
c-di-GMP binding assays. (i) UV cross-linking of c-di-[32P]GMP.
Purified protein (10 μM) was incubated with c-di-GMP (100 μM) in binding buffer 1 (50 mM sodium phosphate, pH 7.4, 300 mM NaCl, 0.5 mM EDTA, 10% glycerol) (34) or binding buffer 2 (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 10 mM MgCl2) for 20 min at room temperature. Samples were irradiated at 254 nm for 5 to 15 min on ice in a Hoefer UVC 500 UV cross-linker with a UVG-54 mineral light lamp (UVP) at a distance of 3 cm (intensity, ∼14,000 μW cm−2). After irradiation, samples were treated with 2× SDS-PAGE sample buffer (40% glycerol, 8% SDS, 2% β-mercaptoethanol, 40 mM EDTA, 0.05% bromophenol blue, 250 mM Tris-HCl, pH 6.8) and boiled for 5 min. Proteins were separated by SDS-PAGE (with 15% separating gel) and detected by storage phosphorimaging with a Typhoon scanner (Amersham Bioscience) and Coomassie blue staining.
(ii) Equilibrium dialysis.
c-di-GMP binding to STM1344 was assessed using the rapid equilibrium dialysis (RED) device disposable chambers and plate according to the instructions of the manufacturer (Pierce). In short, at the start of the assay, 200 pmol of the protein to be tested was added to the binding buffer in the RED sample chamber, which resulted in a 2 μM concentration of protein. Two hundred picomoles of c-di-GMP was added to 300 μl of binding buffer in the control chamber to give a starting c-di-GMP concentration of 667 nM. The chambers were covered with parafilm and incubated for 16 h at room temperature to allow equilibrium between the two chambers to be reached. The c-di-GMP concentrations in both chambers were determined and normalized against the c-di-GMP concentrations in the chambers of the RED device insert in which no protein had been added to the sample chamber. The amount of bound c-di-GMP was calculated according to the protocol of the manufacturer, as follows: % bound c-di-GMP = 100 × (1 − [c-di-GMP]Buffer chamber/[c-di-GMP]Sample chamber), where [c-di- GMP]Buffer chamber is the concentration of c-di-GMP in the control chamber and [c-di-GMP]Sample chamber is the concentration of c-di-GMP in the sample chamber.
Synthesis of c-di-[32P]GMP.
c-di-[32P]GMP was synthesized essentially as described previously (21) by incubating 50 μg of purified His6-WspR* (18) with 200 μCi of [32P]GTP (Amersham Biosciences) at room temperature in 100 μl of reaction buffer (75 mM Tris-HCl, pH 8.0, 250 mM NaCl, 25 mM KCl, 10 mM MgCl2). Residual [32P]GTP was dephosphorylated by the addition of 5 U of calf intestinal phosphatase (New England Biolabs) to the reaction mixture, followed by 15 min of incubation at room temperature. The protein was precipitated by heating the sample to 95°C for 5 min and centrifuging at 16,000 × g. The c-di-[32P]GMP was passed through an Ultrafree-MC column (Millipore) with a cutoff of 5 kDa by centrifugation for 30 min at 5,000 × g. The conversion of [32P]GTP to c-di-[32P]GMP was analyzed by thin-layer chromatography on polyethyleneimine-cellulose plates (Sigma) using 1.5 M KH2PO4, pH 3.65, as the mobile phase. The 32P-labeled nucleotides were visualized and quantified by storage phosphorimaging with a Typhoon scanner (Amersham Bioscience).
Enzymatic assay.
The c-di-GMP-synthesizing activity was tested by incubating 5 μM protein and 2 μM GTP in 100 μl of reaction buffer (75 mM Tris-HCl, pH 8.0, 250 mM NaCl, 25 mM KCl, 10 mM MgCl2) for 1 h. Phosphodiesterase activity was assayed by incubating 1.0 μM protein and 100 μM c-di-GMP in 100 μl of reaction buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 10 mM MgCl2) for 1 h. The reactions were stopped by heating the sample to 95°C for 5 min, and protein was precipitated by centrifugation at 16,000 × g for 5 min. The supernatant was analyzed by reversed-phase HPLC on a Hypersil ODS C18 column with the dimensions 50 by 4.6 mm and a 3-μm particle size and a Hypersil ODS guard column (10 by 4 mm; 5-μm particle size). The mobile phase was buffer A (0.1 M trimethylammonium acetate, pH 6.0) and buffer B (80% acetonitrile and 20% buffer A) at a flow rate of 1 ml/min in a gradient from 0 to 10% buffer B in 5 min. c-di-GMP eluted at 3.35 min.
RESULTS
Identification of STM1344 as an unconventional EAL domain protein.
Recently, a knockout library of genes in the S. Typhimurium chromosome known to encode EAL domain proteins was created (37). The bioinformatic analysis identified STM1344 (also called YdiV) as a single-domain EAL domain protein with a high overall degree of similarity (43%) to the phosphodiesterase STM3611 yet missing several consensus residues required for phosphodiesterase activity (Fig. 1) (26).
FIG. 1.
Comparison of the amino acid sequence of the EAL-like protein STM1344 to that of the most similar EAL domain protein, the phosphodiesterase STM3611 in S. Typhimurium UMR1. Both proteins have only an EAL domain, with the name-giving EAL motif changed to EII in STM1344 and ELL in STM3611. Amino acid alignment was performed using the FFAS03 pairwise alignment server (14) and manual adjustment. Amino acids conserved in STM1344 and STM3611 are shown with a black background. The consensus motifs for functional phosphodiesterases are displayed below the alignment and shown with a gray background. The asterisks indicate alanine replacement of amino acids in STM1344, whereby only F168 was shown to have a functional role.
In order to elucidate the function of STM1344, if any, in rdar morphotype expression, an STM1344 knockout mutant of S. Typhimurium UMR1 was created. Surprisingly, rdar morphotype expression in the ΔSTM1344 mutant was downregulated compared with that in UMR1 (Fig. 2A). In line with the downregulation of the rdar morphotype, CsgD, the major regulator of rdar morphotype expression (9), was downregulated (Fig. 2D), as were extracellular matrix components cellulose and curli fimbriae (Fig. 2B and C; also data not shown), which are commonly activated by CsgD (26).
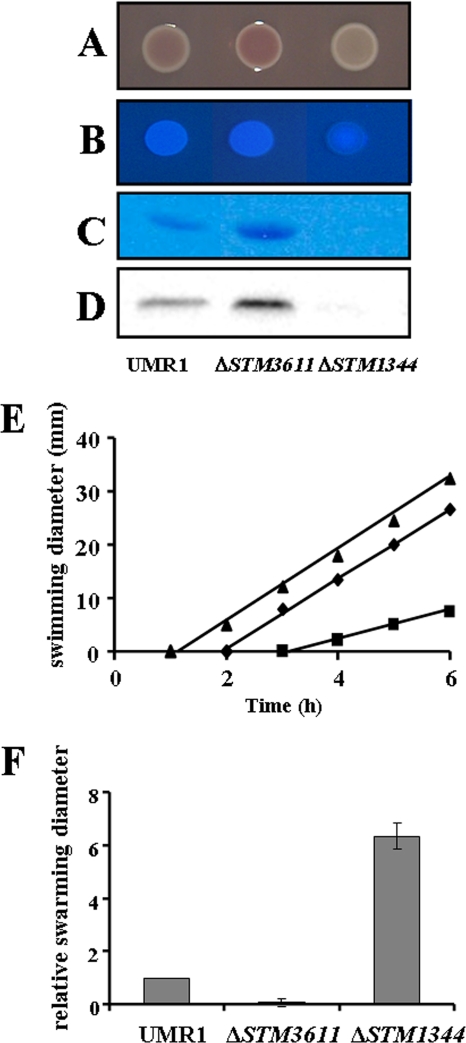
FIG. 2.
Phenotypes of the STM1344 knockout mutant in comparison with those of wild-type S. Typhimurium UMR1 and the c-di-GMP-specific phosphodiesterase STM3611 knockout mutant. (A to D) The expression of the rdar morphotype (A), calcofluor white binding capability (indicative of cellulose expression) (B), extracellular matrix component curli fimbriae (C), and the master regulator of rdar morphotype expression, CsgD (D), in the ΔSTM1344 mutant was downregulated in comparison to that in wild-type S. Typhimurium UMR1. Cells were grown on plates of LB agar without salt supplemented with Congo red (A) or calcofluor white (B) at 28°C for 20 h. The expression of CsgD was evaluated by Western blot analysis. Curli fimbriae were visualized by Coomassie brilliant blue staining of protein gels. (E and F) The swimming (E) and swarming (F) motilities of the ΔSTM1344 mutant were upregulated in comparison to those of wild-type S. Typhimurium UMR1. Diameters of the zones covered by swimming and swarming bacteria were measured. The regulation of all phenotypes of the ΔSTM1344 mutant was opposite that of the phenotypes of the STM3611 knockout mutant. Plates were incubated at 37°C for up to 6 h. In panel E, diamonds represent the wild-type strain UMR1, squares represent the STM3611 mutant, and triangles represent the STM1344 mutant.
Motility is another major phenotype regulated by c-di-GMP signaling (38), and wild-type S. Typhimurium cells show swimming and swarming motilities. The knockout of STM1344 created a mutant that swam at the same speed as, but started to swim earlier than, the wild type (Fig. 2E). On the other hand, the swarming motility of the STM1344 knockout mutant was highly enhanced compared to that of the wild type (Fig. 2F). In summary, the phenotypes of the STM1344 knockout mutant are opposite to the phenotypes of the c-di-GMP-specific phosphodiesterase STM3611 knockout mutant, which shows upregulation of rdar morphotype expression and downregulation of motility (Fig. 2) (37).
Complementation analysis of ΔSTM1344.
The ΔSTM1344 mutant was complemented by STM1344 expressed from a plasmid as a C-terminally His-tagged protein. Complementation studies revealed that, at 28°C, STM1344-His6 overexpression enhanced rdar morphotype expression slightly over wild-type levels and downregulated swimming and swarming motilities (Fig. 3A and C; also data not shown). The temperature regulation of rdar morphotype expression was not overcome by the overexpression of STM1344-His6 since no rdar morphotype expression at 37°C was observed (data not shown).
FIG. 3.
Complementation of an S. Typhimurium UMR1 ΔSTM1344 mutant with wild-type and variant STM1344-His6 proteins. The complementation of ΔSTM1344 with a pBAD30 vector expressing STM1344-His6 (pSTM1344) leads to the upregulation of the rdar morphotype. (A) Calcofluor white binding and CsgD expression. (B) Complementation of ΔSTM1344 with STM1344-His6 variants. Only the F168A mutant of STM1344 did not complement CsgD expression. The N85A STM1344 variant produced a hypercomplementation phenotype. For Western blot analysis (A and B), cells were grown for 20 h on plates of LB agar without salt at 28°C. Pictures of plate-grown cells were taken after 48 h to make morphotype changes strongly visible. (C) Repression of swarming motility by the overexpression of STM1344-His6 and selected variants. While the N85A variant of STM1344 repressed swarming motility as did wild-type STM1344, the F164 variant of STM1344 had no effect. VC, pBAD30 vector control.
To elucidate which amino acids are critical for the function of STM1344, alanine replacement of selected amino acids was carried out. Amino acids highly conserved in experimentally confirmed c-di-GMP-dependent phosphodiesterases (25, 36) were chosen, in particular E29, N85, F144, D165, and F168. The F168A replacement had an effect on the function of STM1344, as the altered protein did not complement the expression of CsgD and the rdar morphotype and did not repress swimming and swarming motility compared to the wild-type STM1344 protein (Fig. 3B and C). On the other hand, the N85A variant showed a gain of function (Fig. 3B and C). While the E29A replacement in STM1344 could not be obtained without secondary mutations, F144A and D165A did not have an effect on STM1344 functionality (Fig. 3B; also data not shown). All proteins were expressed at the same level as wild-type STM1344-His6, with the exception of the N85A variant, which was expressed at a lower level (data not shown).
Expression of STM1344 and effect of the knockout on CsgD expression.
The analysis of CsgD expression in the ΔSTM1344 mutant during growth revealed that CsgD was downregulated approximately 50% during the entire time period from 10 to 48 h (Fig. 4). The growth phase-dependent pattern of CsgD expression, with a characteristic peak at 24 h, remained. As ompR is required for csgD transcription (28), the ompR mutant served as a negative control. We investigated whether the expression of STM1344 parallels the expression of CsgD. STM1344 was expressed at similar levels throughout the growth phase (data not shown).
FIG. 4.
Western blot analysis of growth phase-dependent expression of CsgD in the ΔSTM1344 mutant of S. Typhimurium UMR1. As in wild-type S. Typhimurium UMR1, there is growth phase-dependent expression of CsgD in the ΔSTM1344 mutant. The expression of CsgD in the ΔSTM1344 mutant is reduced approximately 50% compared to that in the wild type. The reference was S. Typhimurium UMR1 or the ΔSTM1344 mutant grown for 16 h; the negative control was S. Typhimurium MAE46 (ΔompR) grown for 24 h.
Effect of STM1344 on csgD transcription.
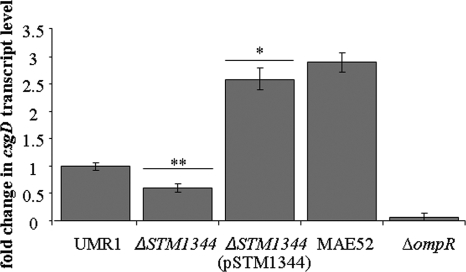
CsgD protein levels are downregulated in the ΔSTM1344 mutant. To investigate on which level the regulation of CsgD expression occurs, the transcription of csgD was measured by real-time RT-PCR analysis. csgD transcript levels in the ΔSTM1344 mutant were downregulated to 60% compared to those in the wild type after 20 h of growth, while the overexpression of STM1344-His6 led to a 2.6-fold increase in csgD transcript levels compared to wild-type levels (Fig. 5).
FIG. 5.
Effect of STM1344 on csgD transcription in S. Typhimurium UMR1. The deletion of the STM1344 gene downregulated the csgD transcript level to approximately 60%, as determined by quantitative real-time RT-PCR. RNA was isolated from cells grown at 28°C for 20 h on LB agar plates without salt. Average values with standard deviations are displayed. **, P ≤ 0.01; *, P ≤ 0.05. The negative control was S. Typhimurium MAE46 ΔompR; the positive control was MAE52, showing csgD expression enhanced threefold compared to that in UMR1 (31).
Effect of STM1344 on intracellular c-di-GMP levels.
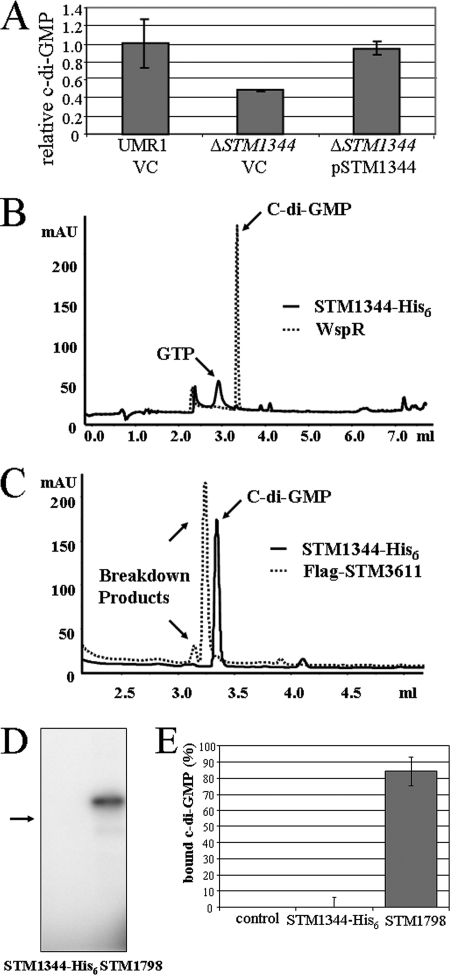
c-di-GMP positively affects the expression of CsgD (16). Since STM1344 changed the expression of CsgD, the change in c-di-GMP levels by STM1344 was investigated. The ΔSTM1344 mutant displayed a reduction in the c-di-GMP levels compared to those in wild-type UMR1, while the c-di-GMP levels in the ΔSTM1344 mutant complemented with the STM1344 gene on a plasmid reached wild-type levels (Fig. 6A). The elevation of the c-di-GMP concentration by STM1344 is consistent with the phenotypes observed for a ΔSTM1344 mutant, as described above. The reduction of c-di-GMP levels in a ΔSTM1344 mutant suggested that STM1344 has a direct or indirect role in the c-di-GMP signaling network.
FIG. 6.
Role of STM1344 in c-di-GMP metabolism in S. Typhimurium UMR1. (A) The total cellular c-di-GMP concentration is downregulated in the ΔSTM1344 mutant compared to those in wild-type S. Typhimurium UMR1 and the ΔSTM1344 mutant complemented with the STM1344-His6 sequence cloned into pBAD30. Cells were grown for 20 h on LB agar plates without salt at 28°C. Averages with standard deviations, which are based on two independent experiments with three technical replicates, are displayed. VC, pBAD30 vector control. (B to E) Purified STM1344 does not exhibit c-di-GMP-metabolizing and c-di-GMP binding activities. (B) Purified STM1344 does not exhibit diguanylate cyclase activity, in contrast to the positive control, constitutively active WspR* (18). Protein at 5 μM was incubated with 100 μM GTP for 1 h. The HPLC elution profiles of the reaction products are as follows: c-di-GMP elutes at 3.35 ml, whereas GTP elutes before 3 ml. The dashed-line curve represents the WspR* sample, whereas the solid-line curve represents the STM1344 sample. (C) Purified STM1344 does not exhibit c-di-GMP-specific phosphodiesterase activity, in contrast to the positive control, STM3611 (38). Protein at 1 μM was incubated with 100 μM c-di-GMP for 1 h at 37C. The HPLC elution profiles of the reaction products are as follows: c-di-GMP elutes at 3.35 ml; the first breakdown product, GpGp, elutes at 3.25 ml; and GMP elutes at 3.15 ml. The dashed-line curve represents the STM3611 sample, whereas the solid-line curve represents the STM1344 sample. mAU, milli-absorbance units. (D and E) Purified STM1344 does not show c-di-GMP binding, in contrast to the positive control, STM1798 (YcgR) (34), as determined by UV cross-linking (D) and equilibrium dialysis (E). The arrow in panel D indicates the position of STM1344-His6 UV cross-linking was performed with c-di-[32P]GMP. Protein at 10 μM was incubated with 100 μM c-di-GMP. For equilibrium dialysis, 200 pmol of c-di-GMP or other tested nucleotides was mixed with the purified proteins (200 pmol) in a dialysis cell surrounded by a membrane with an 8-kDa molecular mass cutoff. Equilibrium was reached after 16 h at room temperature with slow shaking. Average values with standard deviations are displayed.
To elucidate if STM1344 had a direct role in c-di-GMP metabolism, the STM1344 gene with a sequence encoding a C-terminal His tag was cloned into the expression vector pET23a(+), overexpressed, and purified as outlined in Materials and Methods. STM1344-His6 was obtained with over 99% purity as estimated by Coomassie blue staining of protein gels (data not shown). Functional analysis using relevant positive controls, however, showed that STM1344 was neither a c-di-GMP-dependent phosphodiesterase nor a diguanylate cyclase (Fig. 6B and C).
Surprisingly, no c-di-GMP binding by STM1344 was detected in two different assays using UV cross-linking and equilibrium dialysis (Fig. 6D and E), although the binding of c-di-GMP by the positive control YcgR, a PilZ domain-containing c-di-GMP binding protein (34), was readily detected. In addition, STM1344 did not bind related nucleotides such as cyclic GMP, cyclic AMP, or cyclic di-AMP (data not shown).
Role of STM1344 in the c-di-GMP signaling network.
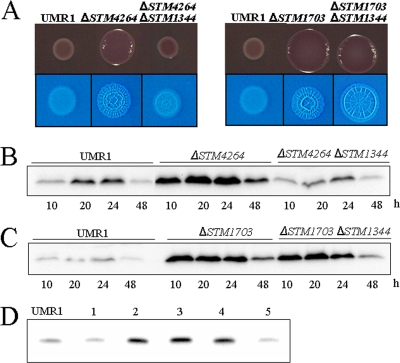
To analyze the position of STM1344 in the c-di-GMP signaling network, double mutants with the STM1344 knockout and mutations in genes coding for phosphodiesterases previously identified to affect rdar morphotype and CsgD expression, namely, STM1703, STM1827, STM3611, and STM4264, were constructed. While mutations in single genes upregulated rdar morphotype and CsgD expression as reported previously (37), the knockout of STM1344 had different effects in different strain backgrounds (Fig. 7; also data not shown). Upon the deletion of STM1344 in the STM4264 and STM1827 mutant backgrounds, the downregulation of rdar morphotype and CsgD expression to the level in the UMR1 wild type was observed, while in the STM1703 and STM3611 mutant backgrounds, CsgD expression remained at the levels in the corresponding single mutants (Fig. 7; also data not shown). On the other hand, swarming motility was partially restored by the deletion of STM1344 in the STM3611 mutant background, suggesting that STM1344 affects swarming motility through STM3611 and through another pathway (data not shown). Consequently, STM1344 acts upstream of STM1703 and STM3611 in the regulation of CsgD, whereas STM1344 is either located downstream of STM4264 and STM1827 or is part of an independent pathway overriding CsgD regulation by STM4264 and STM1827.
FIG. 7.
Position of STM1344 in the c-di-GMP signaling network regulating CsgD expression in S. Typhimurium UMR1. (A) Expression of the rdar morphotype (top) and the calcofluor white binding phenotype (bottom) in wild-type S. Typhimurium UMR1, the corresponding mutants with knockouts of the STM4264 and STM1703 phosphodiesterase genes, and the respective double mutants with STM1344 deleted. While the knockout of STM1344 downregulated rdar morphotype expression in the STM4264 mutant background, only slightly altered rdar morphotype expression in the STM1703 background was observed. Cells were grown for 20 h at 28°C on plates of LB agar without salt, supplemented with Congo red or calcofluor white. (B to D) Effects of STM1344 deletion on CsgD expression in the STM4264 (B), STM1703 (C), STM3611 (D), and STM1827 (D) mutant backgrounds. CsgD was detected by Western blot analysis. Cells were grown on LB agar plates without salt at 28°C. In the analysis presented in panel D, cells were grown for 20 h. Lanes: 1, ΔSTM1344 mutant; 2, ΔSTM3611 mutant; 3, ΔSTM1344 ΔSTM3611 double mutant; 4, ΔSTM1827 mutant; 5, ΔSTM1344 ΔSTM1827 double mutant.
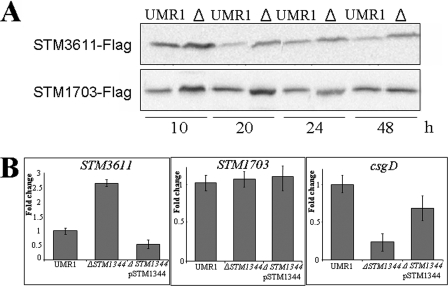
As STM1344 exerts its effect on CsgD expression through STM1703 and STM3611, we tested whether STM1344 has an effect on the expression of STM1703 and STM3611. To monitor the expression of the STM1703 and STM3611 proteins, sequences encoding C-terminally FLAG-tagged fusion proteins were constructed on the chromosome. Subsequent Western blot analysis revealed that, indeed, the expression of STM1703 and STM3611 was elevated in the ΔSTM1344 mutant (Fig. 8A). Quantitative real-time RT-PCR analysis revealed that STM1344 downregulated STM3611 transcript levels, but not STM1703 transcript levels (Fig. 8B). Therefore, STM1344, directly or indirectly, represses STM1703 and STM3611 expression, yet on different levels.
FIG. 8.
Effect of STM1344 knockout in S. Typhimurium UMR1 on the expression of the phosphodiesterases STM3611 and STM1703. (A) STM1344 downregulates the expression of STM3611 and STM1703 throughout the growth phase, as upregulated expression of STM3611 and STM1703 in the ΔSTM1344 mutant is observed. The STM3611 and STM1703 genes on the chromosome were linked with a FLAG tag sequence, and the proteins were detected with a FLAG antibody by Western blot analysis. Cells were grown on LB agar plates without salt at 28°C for the indicated times. Δ indicates the STM1344 knockout mutant of S. Typhimurium UMR1. (B) STM1344 affects the STM3611 gene transcript level but does not affect the STM1703 gene transcript level as determined by quantitative real-time RT-PCR. The csgD transcript was monitored as a positive control. RNA was isolated from cells grown for 16 h on LB agar plates without salt at 28°C. Average values with standard deviations are displayed.
DISCUSSION
In this study, a function for the EAL-like protein STM1344 in the transition between sessility and motility in S. Typhimurium was assigned. STM1344 positively regulated the expression of the multicellular rdar morphotype behavior and its major regulator CsgD and suppressed motility. STM1344, however, does not seem to play a direct role in c-di-GMP metabolism but regulated the rdar morphotype and motility indirectly by affecting the expression of the c-di-GMP-dependent phosphodiesterases STM1703 and STM3611 (Fig. 9).
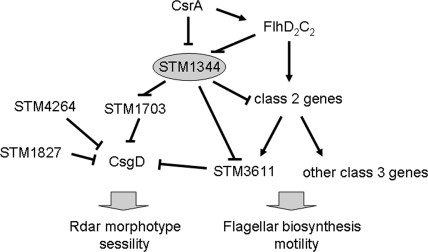
FIG. 9.
Model illustrating the position of STM1344 in the c-di-GMP signaling network regulating the transition between sessility and motility. STM1344 positively affects the expression of the major regulator of the multicellular rdar morphotype behavior, CsgD, through the downregulation of the expression of the phosphodiesterases STM1703 and STM3611 (37). However, the regulation of STM3611 by STM1344 is probably indirect, as STM1344 represses not only STM3611, but also transcription from class 2 and class 3 promoters of the flagellar regulon (45). On the other hand, the expression of CsrA and the FlhD2C2 activator complex (expressed by the class 1 promoters) at the top of the flagellar regulon hierarchy downregulates STM1344 expression (45; Jonas et al., unpublished). In addition, CsrA positively affects FlhD2C2 (44). STM4264 and STM1827 are two other phosphodiesterases that downregulate the expression of CsgD independently of STM1344 (37).
The closest homologue of STM1344 is the EAL domain protein STM3611. Not only do these two proteins have the same domain structure (only an EAL domain), but also, of all EAL domain proteins in S. Typhimurium, STM3611 has the amino acid sequence most similar to that of STM1344 (26). However, there are variations in the details. Many amino acids conserved in STM1344 and STM3611 are located outside of the motifs conserved in functional phosphodiesterases and known to be required for maximal activity against c-di-GMP (Fig. 1) (25). STM1344 is also lacking several amino acids highly conserved in functional phosphodiesterases, some of which have been shown previously to be required for c-di-GMP-dependent phosphodiesterase activity (Fig. 1) (25, 36). If present, these conserved amino acids, such as N85 (Fig. 1), do not have an influence on STM1344 function. Only the F168A substitution gave a protein which had lost its regulatory function. Functionally, STM1344 and STM3611 are also the only two EAL domain proteins which influence both CsgD expression and motility in the S. Typhimurium UMR1 strain background, albeit in opposite manners (Fig. 2).
CsgD expression and motility are not the only phenotypes regulated by STM1344 in S. Typhimurium. STM1344 was originally identified as a protein required for virulence in a mouse model of systemic infection (13). In particular, STM1344 was shown to mediate resistance to hydrogen peroxide and to restrict fast killing of macrophages.
In this previous work, a sevenfold-higher c-di-GMP concentration in the ΔSTM1344 mutant than in the wild type was found (13). We, however, observed the downregulation of the total cellular c-di-GMP levels in the ΔSTM1344 mutant. The reason for the inconsistency in the effect of STM1344 on the c-di-GMP concentration between previous work and our study is unknown but may be methodology related. Only c-di-GMP present in the soluble fraction of a bacterial lysate was measured previously, while we extracted c-di-GMP from whole cells.
The effect of STM1344 on c-di-GMP concentrations as determined in this study is, however, in agreement with the effect of STM1344 on the multicellular and motility phenotypes, e.g., the expression of STM1344 leads to higher c-di-GMP levels, the upregulation of CsgD (as CsgD expression is activated by c-di-GMP [16]), and the inhibition of motility (as motility is inhibited by c-di-GMP [38]). The previously suggested correlation between STM1344 expression and c-di-GMP concentration contributed to the hypothesis that high c-di-GMP levels inhibit the acute infection phenotype (41). While low c-di-GMP levels are indeed required for specific virulence phenotypes (42), systemic infection of mice with S. Typhimurium is complex. Therefore, it is more likely that dynamic regulation of c-di-GMP concentrations is required for successful infection.
How does STM1344 exert its function? A recent study has shown that an EAL domain with noncanonical motifs can interact with a transcriptional regulator and thus influence gene expression (43). This study shows that STM1344, directly or indirectly, represses the expression of the phosphodiesterases STM1703 and STM3611. Although direct biochemical evidence strongly suggests that STM1344 lacks c-di-GMP-metabolizing activity, STM1344 may still interact with functional GGDEF, EAL, or c-di-GMP binding domains and thus influence c-di-GMP metabolism, with subsequent expression of EAL domain proteins. Equally likely, however, STM1344 may have acquired an alternative function and interact with, e.g., a transcriptional regulator.
EAL domain proteins, like GGDEF domain proteins, form a large superfamily of bacterial proteins. However, not all the proteins have retained their core functions, namely, c-di-GMP-degrading and c-di-GMP-synthesizing activities; some have evolved alternative functions. Those functions may be tightly associated with the c-di-GMP network components, as GGDEF domains with nonconsensus GGDEF motifs located in the N-terminal direction from EAL domains in the same protein function as GTP sensors and signal transducers (3, 6). A more distant function independent of c-di-GMP network components is performed by the GGDEF-EAL domain protein CsrD, as it aids in the degradation of small RNAs by RNAse E (40). As CsrD, however, regulates the activity of the carbon storage regulator A (CsrA), which affects the expression of a number of GGDEF and EAL domain proteins (15), even CsrD is still part of a c-di-GMP signaling network.
STM1344 is a novel component that contributes to the fine-tuned regulation of the transition between sessility and motility (Fig. 1 and 9). Swarming motility, a form of multicellular movement on surfaces (12), but not swimming motility is highly affected by STM1344. Indeed, STM1344 does not influence the swimming speed but does influence the onset of swimming motility. Collectively, these observations lead to the conclusion that STM1344 is specifically required for the sessility-motility transition on surfaces. STM1344 favors sessility by repressing the expression of the two phosphodiesterases, STM1703 and STM3611, required for the downregulation of CsgD expression (37). Whether the repression of phosphodiesterase expression by STM1344 is direct remains to be shown. In any case, STM1344 connects sessility with motility, as the STM3611 gene, a class 3 flagellar gene, is part of the flagellar regulon (17). The repression of STM3611 expression and motility by STM1344 is in agreement with the recently reported inhibition of flagellar class 2 and class 3 promoters by STM1344 (45). STM1344 is an integral part of the sessility-motility switch but not the most upstream component, as the expression of STM1344 is directly repressed by the binding of the carbon storage regulator CsrA, a sessility-motility switcher, to the STM1344 mRNA (K. Jonas, A. N. Edwards, I. Ahmad, A. Lamprokostopoulou, T. Romeo, U. Römling, and Ö. Melefors, unpublished results) and by the action of the master regulator of the flagellar regulon, FlhD2C2, when the flagellar biosynthesis machinery is switched on (45).
Acknowledgments
We are grateful to Stephen Lory for providing constitutively active WspR* and to Stephanie Kamann for experimental contributions during project work.
This work was supported by grants from the Karolinska Institutet (elitforskartjänst to U.R.), the Swedish Research Council (621-2004-3979 and 621-2007-6509), and the Carl Trygger Foundation (CTS-07:306).
Footnotes
Published ahead of print on 17 April 2009.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1994. Current protocols in molecular biology. John Wiley & Sons, New York, NY.
- 2.Brombacher, E., A. Baratto, C. Dorel, and P. Landini. 2006. Gene expression regulation by the curli activator CsgD protein: modulation of cellulose biosynthesis and control of negative determinants for microbial adhesion. J. Bacteriol. 1882027-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Christen, M., B. Christen, M. Folcher, A. Schauerte, and U. Jenal. 2005. Identification and characterization of a cyclic di-GMP-specific phosphodiesterase and its allosteric control by GTP. J. Biol. Chem. 28030829-30837. [DOI] [PubMed] [Google Scholar]
- 4.Cotter, P. A., and S. Stibitz. 2007. c-di-GMP-mediated regulation of virulence and biofilm formation. Curr. Opin. Microbiol. 1017-23. [DOI] [PubMed] [Google Scholar]
- 5.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 976640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duerig, A., S. Abel, M. Folcher, M. Nicollier, T. Schwede, N. Amiot, B. Giese, and U. Jenal. 2009. Second messenger-mediated spatiotemporal control of protein degradation regulates bacterial cell cycle progression. Genes Dev. 2393-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galperin, M. Y. 2005. A census of membrane-bound and intracellular signal transduction proteins in bacteria: bacterial IQ, extroverts and introverts. BMC Microbiol. 535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerstel, U., C. Park, and U. Römling. 2003. Complex regulation of csgD promoter activity by global regulatory proteins. Mol. Microbiol. 49639-654. [DOI] [PubMed] [Google Scholar]
- 9.Gerstel, U., and U. Römling. 2003. The csgD promoter, a control unit for biofilm formation in Salmonella typhimurium. Res. Microbiol. 154659-667. [DOI] [PubMed] [Google Scholar]
- 10.Gibson, D. L., A. P. White, S. D. Snyder, S. Martin, C. Heiss, P. Azadi, M. Surette, and W. W. Kay. 2006. Salmonella produces an O-antigen capsule regulated by AgfD and important for environmental persistence. J. Bacteriol. 1887722-7730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1774121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harshey, R. M. 1994. Bees aren't the only ones: swarming in gram-negative bacteria. Mol. Microbiol. 13389-394. [DOI] [PubMed] [Google Scholar]
- 13.Hisert, K. B., M. MacCoss, M. U. Shiloh, K. H. Darwin, S. Singh, R. A. Jones, S. Ehrt, Z. Zhang, B. L. Gaffney, S. Gandotra, D. W. Holden, D. Murray, and C. Nathan. 2005. A glutamate-alanine-leucine (EAL) domain protein of Salmonella controls bacterial survival in mice, antioxidant defence and killing of macrophages: role of cyclic diGMP. Mol. Microbiol. 561234-1245. [DOI] [PubMed] [Google Scholar]
- 14.Jaroszewski, L., L. Rychlewski, Z. Li, W. Li, and A. Godzik. 2005. FFAS03: a server for profile-profile sequence alignments. Nucleic Acids Res. 33(Web server issue)W284-W288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jonas, K., A. N. Edwards, R. Simm, T. Romeo, U. Römling, and Ö. Melefors. 2008. The RNA binding protein CsrA controls cyclic di-GMP metabolism by directly regulating the expression of GGDEF proteins. Mol. Microbiol. 70236-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kader, A., R. Simm, U. Gerstel, M. Morr, and U. Römling. 2006. Hierarchical involvement of various GGDEF domain proteins in rdar morphotype development of Salmonella enterica serovar Typhimurium. Mol. Microbiol. 60602-616. [DOI] [PubMed] [Google Scholar]
- 17.Ko, M., and C. Park. 2000. Two novel flagellar components and H-NS are involved in the motor function of Escherichia coli. J. Mol. Biol. 303371-382. [DOI] [PubMed] [Google Scholar]
- 18.Kulesekara, H., V. Lee, A. Brencic, N. Liberati, J. Urbach, S. Miyata, D. G. Lee, A. N. Neely, M. Hyodo, Y. Hayakawa, F. M. Ausubel, and S. Lory. 2006. Analysis of Pseudomonas aeruginosa diguanylate cyclases and phosphodiesterases reveals a role for bis-(3′-5′)-cyclic-GMP in virulence. Proc. Natl. Acad. Sci. USA 1032839-2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Latasa, C., A. Roux, A. Toledo-Arana, J. M. Ghigo, C. Gamazo, J. R. Penades, and I. Lasa. 2005. BapA, a large secreted protein required for biofilm formation and host colonization of Salmonella enterica serovar Enteritidis. Mol. Microbiol. 581322-1339. [DOI] [PubMed] [Google Scholar]
- 20.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25402-408. [DOI] [PubMed] [Google Scholar]
- 21.Merighi, M., V. T. Lee, M. Hyodo, Y. Hayakawa, and S. Lory. 2007. The second messenger bis-(3′-5′)-cyclic-GMP and its PilZ domain-containing receptor Alg44 are required for alginate biosynthesis in Pseudomonas aeruginosa. Mol. Microbiol. 65876-895. [DOI] [PubMed] [Google Scholar]
- 22.Newell, P. D., R. D. Monds, and G. A. O'Toole. 2009. LapD is a bis-(3′,5′)-cyclic dimeric GMP-binding protein that regulates surface attachment by Pseudomonas fluorescens Pf0-1. Proc. Natl. Acad. Sci. USA 1063461-3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paul, R., S. Weiser, N. C. Amiot, C. Chan, T. Schirmer, B. Giese, and U. Jenal. 2004. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev. 18715-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pei, J., and N. V. Grishin. 2001. GGDEF domain is homologous to adenylyl cyclase. Proteins 42210-216. [DOI] [PubMed] [Google Scholar]
- 25.Rao, F., Y. Yang, Y. Qi, and Z. X. Liang. 2008. Catalytic mechanism of cyclic di-GMP-specific phosphodiesterase: a study of the EAL domain-containing RocR from Pseudomonas aeruginosa. J. Bacteriol. 1903622-3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Römling, U. 2005. Characterization of the rdar morphotype, a multicellular behaviour in Enterobacteriaceae. Cell. Mol. Life Sci. 621234-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Römling, U., and D. Amikam. 2006. Cyclic di-GMP as a second messenger. Curr. Opin. Microbiol. 9218-228. [DOI] [PubMed] [Google Scholar]
- 28.Römling, U., Z. Bian, M. Hammar, W. D. Sierralta, and S. Normark. 1998. Curli fibers are highly conserved between Salmonella typhimurium and Escherichia coli with respect to operon structure and regulation. J. Bacteriol. 180722-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Römling, U., W. Bokranz, W. Rabsch, X. Zogaj, M. Nimtz, and H. Tschäpe. 2003. Occurrence and regulation of the multicellular morphotype in Salmonella serovars important in human disease. Int. J. Med. Microbiol. 293273-285. [DOI] [PubMed] [Google Scholar]
- 30.Römling, U., M. Rohde, A. Olsen, S. Normark, and J. Reinköster. 2000. AgfD, the checkpoint of multicellular and aggregative behaviour in Salmonella typhimurium regulates at least two independent pathways. Mol. Microbiol. 3610-23. [DOI] [PubMed] [Google Scholar]
- 31.Römling, U., W. D. Sierralta, K. Eriksson, and S. Normark. 1998. Multicellular and aggregative behaviour of Salmonella typhimurium strains is controlled by mutations in the agfD promoter. Mol. Microbiol. 28249-264. [DOI] [PubMed] [Google Scholar]
- 32.Ryan, R. P., Y. Fouhy, J. F. Lucey, L. C. Crossman, S. Spiro, Y. W. He, L. H. Zhang, S. Heeb, M. Camara, P. Williams, and J. M. Dow. 2006. Cell-cell signaling in Xanthomonas campestris involves an HD-GYP domain protein that functions in cyclic di-GMP turnover. Proc. Natl. Acad. Sci. USA 1036712-6717. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33.Ryan, R. P., Y. Fouhy, J. F. Lucey, and J. M. Dow. 2006. Cyclic di-GMP signaling in bacteria: recent advances and new puzzles. J. Bacteriol. 1888327-8334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ryjenkov, D. A., R. Simm, U. Römling, and M. Gomelsky. 2006. The PilZ domain is a receptor for the second messenger c-di-GMP. The PilZ domain protein YcgR controls motility in Enterobacteria. J. Biol. Chem. 28130310-30314. [DOI] [PubMed] [Google Scholar]
- 35.Ryjenkov, D. A., M. Tarutina, O. V. Moskvin, and M. Gomelsky. 2005. Cyclic diguanylate is a ubiquitous signaling molecule in bacteria: insights into biochemistry of the GGDEF protein domain. J. Bacteriol. 1871792-1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmidt, A. J., D. A. Ryjenkov, and M. Gomelsky. 2005. The ubiquitous protein domain EAL is a cyclic diguanylate-specific phosphodiesterase: enzymatically active and inactive EAL domains. J. Bacteriol. 1874774-4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simm, R., A. Lusch, A. Kader, M. Andersson, and U. Römling. 2007. Role of EAL-containing proteins in multicellular behavior of Salmonella enterica serovar Typhimurium. J. Bacteriol. 1893613-3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simm, R., M. Morr, A. Kader, M. Nimtz, and U. Römling. 2004. GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol. Microbiol. 531123-1134. [DOI] [PubMed] [Google Scholar]
- 39.Simm, R., M. Morr, U. Remminghorst, M. Andersson, and U. Römling. 2009. Quantitative determination of cyclic diguanosine monophosphate concentrations in nucleotide extracts of bacteria by matrix-assisted laser desorption/ionization-time-of-flight mass spectrometry. Anal. Biochem. 38653-58. [DOI] [PubMed] [Google Scholar]
- 40.Suzuki, K., P. Babitzke, S. R. Kushner, and T. Romeo. 2006. Identification of a novel regulatory protein (CsrD) that targets the global regulatory RNAs CsrB and CsrC for degradation by RNase E. Genes Dev. 202605-2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tamayo, R., J. T. Pratt, and A. Camilli. 2007. Roles of cyclic diguanylate in the regulation of bacterial pathogenesis. Annu. Rev. Microbiol. 61131-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tischler, A. D., and A. Camilli. 2005. Cyclic diguanylate regulates Vibrio cholerae virulence gene expression. Infect. Immun. 735873-5882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tschowri, N., S. Busse, and R. Hengge. 2009. The BLUF-EAL protein YcgF acts as a direct anti-repressor in a blue-light response of Escherichia coli. Genes Dev. 23522-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wei, B. L., A. M. Brun-Zinkernagel, J. W. Simecka, B. M. Pruss, P. Babitzke, and T. Romeo. 2001. Positive regulation of motility and flhDC expression by the RNA-binding protein CsrA of Escherichia coli. Mol. Microbiol. 40245-256. [DOI] [PubMed] [Google Scholar]
- 45.Wozniak, C. E., C. Lee, and K. T. Hughes. 2008. T-POP array identifies EcnR and PefI-SrgD as novel regulators of flagellar gene expression. J. Bacteriol. 1911498-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zeghouf, M., J. Li, G. Butland, A. Borkowska, V. Canadien, D. Richards, B. Beattie, A. Emili, and J. F. Greenblatt. 2004. Sequential peptide affinity (SPA) system for the identification of mammalian and bacterial protein complexes. J. Proteome Res. 3463-468. [DOI] [PubMed] [Google Scholar]
- 47.Zogaj, X., M. Nimtz, M. Rohde, W. Bokranz, and U. Römling. 2001. The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol. Microbiol. 391452-1463. [DOI] [PubMed] [Google Scholar]