Abstract
Mycobacterium tuberculosis is one of the most successful pathogens of humankind. During infection, M. tuberculosis must cope with and survive against a variety of different environmental conditions. Sigma factors likely facilitate the modulation of the pathogen's gene expression in response to changes in its extracellular milieu during infection. σH, an alternate sigma factor encoded by the M. tuberculosis genome, is induced by thiol-oxidative stress, heat shock, and phagocytosis. In response to these conditions, σH induces the expression of σB, σE, and the thioredoxin regulon. In order to more effectively characterize the transcriptome controlled by σH, we studied the long-term effects of the induction of σH on global transcription in M. tuberculosis. The M. tuberculosis isogenic mutant of σH (Δ-σH) is more susceptible to diamide stress than wild-type M. tuberculosis. To study the long-term effects of σH induction, we exposed both strains to diamide, rapidly washed it away, and resumed culturing in diamide-free medium (post-diamide stress culturing). Analysis of the effects of σH induction in this experiment revealed a massive temporal programming of the M. tuberculosis transcriptome. Immediately after the induction of σH, genes belonging to the functional categories “virulence/detoxification” and “regulatory proteins” were induced in large numbers. Fewer genes belonging to the “lipid metabolism” category were induced, while a larger number of genes belonging to this category were downregulated. σH caused the induction of the ATP-dependent clp proteolysis regulon, likely mediated by a transcription factor encoded by Rv2745c, several members of the mce1 virulence regulon, and the sulfate acquisition/transport network.
Tuberculosis (TB) is responsible for over 2 million deaths annually (42). The failure of the BCG vaccine (1) and the emergence of drug-resistant strains of Mycobacterium tuberculosis have worsened this situation (34). To develop effective anti-TB drugs and vaccines, we need to define the mechanisms that help establish a long-term infection by M. tuberculosis.
Sigma (σ) factors bind to the RNA polymerase and influence its promoter specificity (27). Most eubacteria contain a principal σ factor, which regulates the transcription of housekeeping genes. A variable number of alternate σ factors control responses to specific environmental stimuli, adaptation to stress, and bacterial virulence (35). The pathogenesis of TB involves multiple phases and is believed to involve a carefully deployed series of adaptive bacterial virulence factors. It is conceivable that temporal expression of specific regulons controlled by the induction or availability of one or more sigma factors (10, 13) allows M. tuberculosis to survive in multiple phases of the disease process (16). One such factor, σH, is induced after heat, redox, nitrosative, and acid stress (25, 30, 41, 45) and phagocytosis (16). Its activity is regulated in a redox-dependent manner by the antisigma factor RshA (50) as well as by the protein kinase PknB (38). An isogenic Δ-σH mutant is attenuated in a mouse model and fails to induce typical pulmonary granulomatous pathology (25). σH directs the transcription of 31 genes, including those which encode extracytoplasmic function σ factors σE and σB and proteins involved in the maintenance of intrabacterial reducing capacity (25, 30). Expression of σH is induced after entry of M. tuberculosis into the host cells (19, 45), and this state of induced σH expression likely persists for a long period of time. Such continuing expression of σH must further affect gene expression in M. tuberculosis. The regulation of other transcription factors by σH creates a regulatory network with a potential for downstream modulation of gene expression. Induction of these proteins must in turn have further indirect impact on M. tuberculosis gene expression as a function of time. The study of these longer-term, enduring transcriptome changes will allow us to better understand the response of M. tuberculosis to phagocytosis and infection. We therefore sought to more clearly define these enduring effects of σH.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Wild-type M. tuberculosis and the Δ-σH mutant were grown at 37°C in Middlebrook 7H9-albumin-dextrose-catalase-Tween 80-glycerol broth or 7H10-Dubos oleic acid complex-glycerol agar.
Diamide stress.
We measured the effect of diamide on the viability of wild-type M. tuberculosis and Δ-σH by the CFU method. Aliquots of cultures 1, 2, 3, and 4 h after addition of diamide were serially diluted and plated. These results were compared to those for the experiment where diamide was withdrawn after 1 h. Cultures grown to mid-log phase were exposed to 10 mM diamide for 1 h. The cultures were centrifuged (1,455 × g, 5 min), washed in diamide-free medium, and further cultured in an equal amount of this medium. These cultures were designated post-diamide stress (PDS) cultures. Aliquots obtained before diamide addition and at 0, 30, 60, 90, 120, and 180 min PDS were serially diluted and plated.
Isolation of RNA.
RNA was isolated using the Trizol bead beater method (15), DNase treated, and purified (RNeasy; Qiagen) and its quantity assessed using a NanoDrop spectrophotometer.
Microarray studies.
Whole-genome oligonucleotide microarrays representing ∼4,750 M. tuberculosis open reading frames were obtained from the Pathogen Functional Genomic Resource Center (PFGRC) and used as recommended (ftp://ftp.jcvi.org/pub/data/PFGRC/pdf_files/protocols/). Biological replicate hybridizations were performed using PDS samples from wild-type M. tuberculosis and the mutant. Spots with imperfect morphologies, low signal-to-noise ratios, saturated intensities, and high degrees of variability between replicates were excluded from the analysis. Data were analyzed using S+ (Spotfire DecisionSite; TIBCO-Spotfire, Inc.) to determine statistically significant changes in expression, with adjustment for intensity bias by locally weighted scatter plot smoothing normalization (57). Genes were considered significantly and differentially expressed if they showed >1.75-fold changes with P values of <0.05.
Reverse transcription and relative quantification of mRNA by quantitative PCR (qPCR).
Quantitative real-time reverse transcription-PCR was performed with cDNA corresponding to 50 ng RNA by using a SYBR green Supermix kit (Applied Biosystems) to interrogate the expression of σH, σE, σB, trxB2, hsp, ctpG, bpoB, Rv2745c, clpP1, and mce1A. Expression levels were normalized using σA as an invariant transcript, and the average relative expression levels were determined for four replicate experiments by using the following formula: (PDS level for wild-type M. tuberculosis − pre-diamide stress level for wild-type M. tuberculosis) − (PDS level for Δ-σH − pre-diamide stress level for Δ-σH).
Generation of rabbit polyclonal anti-Rv2745c antibody.
A predicted highly antigenic peptide sequence specific to the C terminus of the Rv2745c-encoded protein (VREVV GDVLR GARMS QGRTL REV-C) was used to immunize rabbits. The animals were boosted 4 and 6 weeks after prime immunization. Antisera were collected 2, 3, and 4 weeks after the final boost, titrated, and affinity purified. The antibody specifically recognized the product of the M. tuberculosis Rv2745c gene.
Isolation of proteins, SDS-polyacrylamide gel electrophoresis, and Western blotting.
Total protein was isolated from 50 ml of each culture by using a B-PER (Pierce) kit. Sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis was performed on a 12% gel by using 20 μg protein. The membrane was incubated with anti-Rv2745c antibody (1:1,000) and then with a goat anti-rabbit antibody (1:10,000) conjugated to horseradish peroxidase (Bio-Rad).
Acetamide-induced expression of the M. tuberculosis Rv2745c gene.
The M. tuberculosis CDC1551 MT2816 (Rv2745c) gene was fused to the Mycobacterium smegmatis acetamidase promoter (Pace) on an integrative vector, pSCW38 (22). Following transformation of M. tuberculosis, the resulting strain (M. tuberculosis::pSM12) and its vector-only control (M. tuberculosis::pSCW38) were grown to log phase and induced with acetamide for 1 h. RNAs isolated from induced and uninduced cultures were used to perform qPCR as described earlier.
Microarray data accession number.
Microarray data can be accessed from Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/), using the provisional accession number GPL8382.
RESULTS
Wild-type M. tuberculosis and the Δ-σH mutant exhibit comparable inhibition levels PDS.
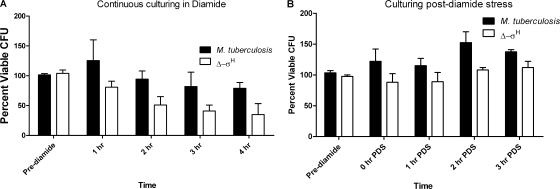
σH protects M. tuberculosis from heat, oxidative, nitrosative, and acid stress conditions. The Δ-σH mutant is more susceptible to diamide, a thiol-oxidative agent, which induces the expression of σH (30, 41). Prolonged culturing of the mutant in the presence of diamide would therefore lead to significant viability differences relative to wild-type M. tuberculosis, thus adversely affecting a global analysis of gene expression. When cultured continuously in the presence of diamide for 4 h, the viability of the Δ-σH mutant was reduced by more than 50% relative to that of wild-type M. tuberculosis (Fig. 1A). Therefore, we devised an alternate strategy (PDS culturing) to study the long-term global effects of σH induction on the M. tuberculosis transcriptome. After 1 h, diamide was rapidly washed away and culturing resumed in normal, diamide-free medium. However, the viabilities of the two cultures remained comparable during the PDS culturing (Fig. 1B).
FIG. 1.
Wild-type M. tuberculosis and the Δ-σH mutant exhibit comparable inhibition levels PDS. Both wild-type M. tuberculosis and the Δ-σH mutant were cultured in the presence of 10 mM diamide for 1, 2, 3, and 4 h (A) and 0 h (0 min), 1 h (60 min), 2 h (120 min), and 3 h (180 min) PDS (B). Data are means of results from three independent experiments, with plating for CFU performed in quadruplicates in each experiment, and the error bars represent standard deviations.
Transcriptome comparison of wild-type M. tuberculosis and the Δ-σH mutant PDS.
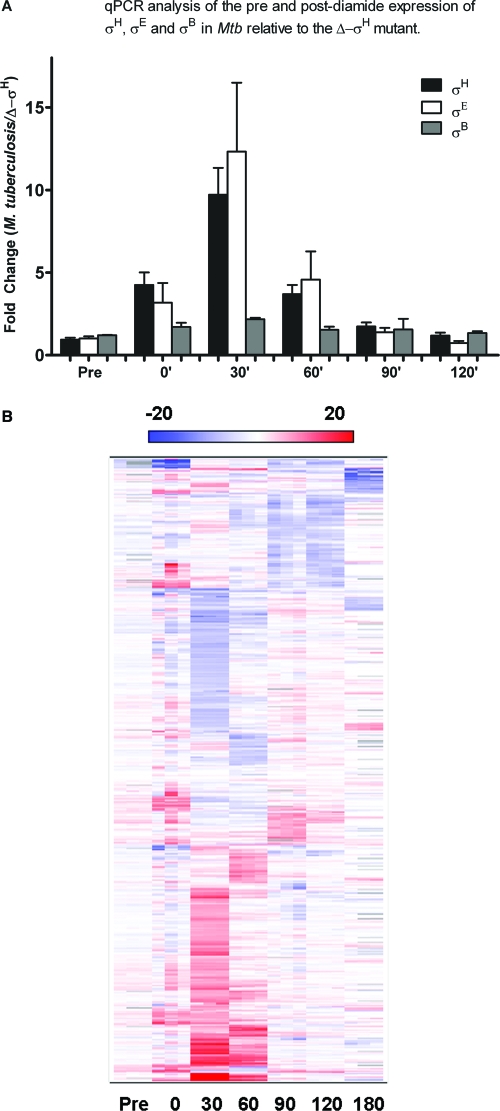
In order to verify that the expression of σH was induced during PDS culturing, we analyzed the presence of σH-specific transcripts at all PDS time points by using qPCR. As shown in Fig. 2A, expression of σH was induced at 0 min PDS in wild-type M. tuberculosis relative to that in the Δ-σH mutant, peaked at 30 min PDS, and thereafter declined, reaching background levels by 120 min. Since the expressions of both σE and σB are known to be induced in a σH-dependent manner following diamide stress, we also analyzed the expressions of these genes in PDS samples. The expressions of both σE and σB were induced at 0 min PDS and remained at significantly high levels until the 30-min and 60-min PDS time points, respectively (Fig. 2A).
FIG. 2.
Analysis of the comparative transcriptome levels of wild-type M. tuberculosis and the Δ-σH mutant PDS. (A) The expression levels of σH, σE, and σB in wild-type M. tuberculosis relative to the levels for the Δ-σH mutant in PDS samples were assessed by qPCR at different time points. Values represent mean differences (n-fold) ± standard deviations of results from quadruplicate experiments. (B) A two-dimensional clustering heat map shows M. tuberculosis genes expressed in a statistically significant manner for at least one PDS time point (0, 30, 60, 90, 120, or 180 min) relative to the pre-diamide induction time point. The intensity of the red color correlates with the degree of expression in wild-type M. tuberculosis relative to the level for the Δ-σH mutant, while the intensity of the blue color correlates with the degree of expression in the mutant relative to the level for wild-type M. tuberculosis. For each time point, changes are shown for two distinct microarrays, each of which was hybridized to a biologically independent RNA sample, along with an average value.
Wild-type M. tuberculosis transcripts made before diamide stress and at 0, 30, 60, 90, and 120 min PDS were then directly compared to the Δ-σH mutant transcripts by using microarrays. Figure 2B shows a hierarchical cluster of ∼800 genes differentially expressed significantly in the two strains for at least one of the PDS time points, vis-à-vis the pretreatment control. Forty-six genes were upregulated at 0 min PDS (Table 1), most of which were previously known to be dependent on σH for their expression (25, 30, 41). The expression of a much larger number of genes was perturbed (∼380 induced and ∼340 repressed) at the 30-min time point (see Tables 3 and 4). These substantial changes were perhaps due to the induction of σB, σE, and several other transcription factors by σH. Each transcription factor in turn influences a distinct group of genes, leading to such profound global effects in gene expression. In contrast, 111 genes were induced and 120 repressed in wild-type M. tuberculosis, relative to the levels for the mutant, at 60 min PDS. The number of perturbed genes continued to decline with the passage of time. At 90 min PDS, 74 genes were induced and 32 genes repressed in wild-type M. tuberculosis relative to the levels for the mutant. At 120 min PDS, only 23 genes were induced. Clearly, σH continues to exert its influence on specific M. tuberculosis regulons, long after its initial induction. A detailed analysis of these interactions will enhance our understanding of the M. tuberculosis stress biology.
TABLE 1.
Genes induced in wild-type M. tuberculosis, relative to the levels for the Δ-σH mutant, at 0 min PDSa
| Function | Locus tag | Gene | Fold expression
|
P | |
|---|---|---|---|---|---|
| Pre-diamide stress | 0 min PDS | ||||
| Heat shock | Rv0384c | clpB | 1.148 ± 0.043 | 3.760 ± 0.559 | 0.043 |
| Rv3417c | groEL1 | 1.051 ± 0.047 | 4.266 ± 0.768 | 0.049 | |
| Rv3418c | groES | 1.051 ± 0.047 | 2.562 ± 0.282 | 0.050 | |
| Rv0251c | hsp | 0.951 ± 0.048 | 2.141 ± 0.143 | 0.018 | |
| Rv0350 | dnaK | 1.068 ± 0.004 | 2.902 ± 0.416 | 0.050 | |
| Rv0469 | dnaJ1 | 1.003 ± 0.026 | 2.607 ± 0.321 | 0.040 | |
| Transcription | Rv2710 | sigB | 0.998 ± 0.017 | 1.998 ± 0.159 | 0.042 |
| Rv1221 | sigE | 0.874 ± 0.0004 | 2.602 ± 0.392 | 0.050 | |
| Rv3223c | sigH | 0.869 ± 0.029 | 3.471 ± 0.502 | 0.045 | |
| Rv0014c | pknB | 0.926 ± 0.120 | 1.902 ± 0.111 | 0.002 | |
| Rv1674c | 1.056 ± 0.125 | 5.065 ± 0.181 | 0.003 | ||
| Transport | Rv2397c | cysA1 | 0.944 ± 0.006 | 2.622 ± 0.324 | 0.043 |
| Rv2399c | cysT | 0.831 ± 0.035 | 2.146 ± 0.229 | 0.043 | |
| Rv2398c | cysW | 1.110 ± 0.012 | 2.085 ± 0.139 | 0.035 | |
| Rv1859 | modC | 1.208 ± 0.035 | 2.128 ± 0.239 | 0.049 | |
| Rv0924c | mntH | 0.909 ± 0.003 | 1.862 ± 0.196 | 0.046 | |
| Rv1473 | 0.876 ± 0.003 | 1.774 ± 0.087 | 0.016 | ||
| Detoxification | Rv2641 | cadI | 0.965 ± 0.016 | 4.139 ± 0.680 | 0.048 |
| Rv3913 | trxB2 | 0.979 ± 0.018 | 4.550 ± 0.780 | 0.047 | |
| Rv2899c | fdhD | 1.234 ± 0.010 | 3.036 ± 0.310 | 0.039 | |
| Molybdopterin biosynthesis | Rv2453c | mobA | 1.236 ± 0.025 | 3.301 ± 0.430 | 0.048 |
| Rv3206c | moeB1 | 1.095 ± 0.015 | 3.209 ± 0.018 | 0.003 | |
| Sulfate metabolism | Rv1336 | cysM | 0.914 ± 0.058 | 5.058 ± 0.173 | 0.006 |
| Rv1286 | cysN | 0.774 ± 0.246 | 3.511 ± 0.827 | 0.047 | |
The functional categorization of M. tuberculosis genes was based on their known functions as assigned by the TubercuList database (http://genolist.pasteur.fr/TubercuList/). For each gene, the changes in wild-type M. tuberculosis relative to the levels for the Δ-σH mutant in the pre-diamide stress samples and the 0-min-PDS samples are shown, along with standard deviations. Statistical significance was assigned using the ANOVA function within the S+ ArrayAnalyzer script.
TABLE 3.
Genes induced in wild-type M. tuberculosis, relative to the levels for the Δ-σH mutant, at 30 min PDSa
| Function | Locus tag | Gene | Fold expression
|
P | |
|---|---|---|---|---|---|
| Pre-diamide stress | 30 min PDS | ||||
| Heat shock | Rv0384c | clpB | 1.148 ± 0.043 | 21.896 ± 0.344 | 0.004 |
| Rv3417c | groEL1 | 1.051 ± 0.020 | 1.831 ± 0.037 | 0.003 | |
| Rv0440 | groEL2 | 1.075 ± 0.0003 | 1.799 ± 0.158 | 0.049 | |
| Rv3418c | groES | 1.342 ± 0.003 | 1.680 ± 0.100 | 0.067 | |
| Rv0351 | grpE | 1.057 ± 0.005 | 2.939 ± 0.074 | 0.008 | |
| Rv0251c | hsp | 0.951 ± 0.048 | 20.969 ± 0.201 | 0.002 | |
| Rv0353 | hspR | 0.977 ± 0.007 | 3.491 ± 0.088 | 0.007 | |
| Rv0563 | htpX | 0.915 ± 0.027 | 1.840 ± 0.005 | 0.005 | |
| Rv0469 | dnaJ1 | 1.003 ± 0.026 | 1.638 ± 0.030 | 0.002 | |
| Rv0250c | 1.352 ± 0.470 | 16.275 ± 1.063 | 0.008 | ||
| Virulence/detoxification | MT1966 | aceA2 | 1.366 ± 0.002 | 1.900 ± 0.065 | 0.025 |
| Rv1123c | bpoB | 1.017 ± 0.060 | 2.404 ± 0.100 | 0.003 | |
| Rv2641 | cadI | 0.965 ± 0.060 | 4.698 ± 0.464 | 0.026 | |
| Rv3648c | cspA | 0.954 ± 0.006 | 2.588 ± 0.092 | 0.011 | |
| Rv3874 | esxB | 0.993 ± 0.035 | 1.799 ± 0.159 | 0.051 | |
| Rv3890c | esxC | 1.045 ± 0.010 | 1.763 ± 0.002 | 0.002 | |
| Rv3891c | esxD | 0.907 ± 0.011 | 2.786 ± 0.083 | 0.008 | |
| Rv3017c | esxQ | 1.061 ± 0.035 | 2.804 ± 0.054 | 0.002 | |
| MT3105 | esxS | 0.855 ± 0.007 | 3.564 ± 0.159 | 0.013 | |
| Rv0169 | mce1A | 0.763 ± 0.013 | 2.924 ± 0.010 | 0.0003 | |
| Rv0170 | mce1B | 1.055 ± 0.015 | 1.766 ± 0.041 | 0.017 | |
| Rv0171 | mce1C | 1.016 ± 0.006 | 2.290 ± 0.052 | 0.010 | |
| Rv0172 | mce1D | 1.023 ± 0.009 | 1.780 ± 0.124 | 0.039 | |
| Rv0173 | mce1E (lprK) | 1.110 ± 0.009 | 2.505 ± 0.295 | 0.050 | |
| Rv0174 | mce1F | 1.005 ± 0.013 | 1.750 ± 0.020 | 0.010 | |
| Rv3846 | sodA | 1.344 ± 0.135 | 3.484 ± 0.338 | 0.049 | |
| Rv1471 | trxB1 | 0.986 ± 0.019 | 46.508 ± 1.171 | 0.005 | |
| Rv3913 | trxB2 | 0.979 ± 0.012 | 21.066 ± 0.647 | 0.007 | |
| Rv3914 | trxC | 1.214 ± 0.303 | 24.457 ± 0.204 | 0.004 | |
| Rv2525c | 1.167 ± 0.002 | 3.111 ± 0.172 | 0.020 | ||
| Rv1740 | 0.817 ± 0.019 | 2.590 ± 0.166 | 0.023 | ||
| Rv2595 | 0.795 ± 0.011 | 2.505 ± 0.128 | 0.015 | ||
| Rv2829c | 1.131 ± 0.026 | 2.342 ± 0.039 | 0.012 | ||
| Rv0660c | 0.757 ± 0.026 | 2.048 ± 0.078 | 0.018 | ||
| Rv2547 | 0.770 ± 0.015 | 1.908 ± 0.007 | 0.004 | ||
| Rv2526 | 0.772 ± 0.013 | 1.853 ± 0.038 | 0.032 | ||
| Rv2526 | 0.772 ± 0.013 | 1.853 ± 0.038 | 0.032 | ||
| Transcription | Rv2745c | clgR | 1.015 ± 0.061 | 5.495 ± 0.903 | 0.042 |
| Rv1994c | cmtR | 0.929 ± 0.021 | 6.574 ± 0.641 | 0.024 | |
| Rv1909c | furA | 1.289 ± 0.059 | 3.610 ± 0.051 | 0.010 | |
| Rv2711 | ideR | 1.026 ± 0.007 | 3.610 ± 0.114 | 0.009 | |
| Rv3291c | lrpA | 1.119 ± 0.016 | 3.072 ± 0.194 | 0.020 | |
| Rv0981 | mprA | 1.157 ± 0.017 | 2.360 ± 0.016 | 0.0002 | |
| Rv2710 | sigB | 1.255 ± 0.017 | 2.541 ± 0.030 | 0.008 | |
| Rv3414c | sigD | 1.174 ± 0.041 | 1.832 ± 0.015 | 0.019 | |
| Rv1221 | sigE | 0.874 ± 0.0004 | 19.133 ± 0.236 | 0.002 | |
| Rv3223c | sigH | 0.870 ± 0.029 | 5.883 ± 0.013 | 0.0007 | |
| Rv3911 | sigM | 0.994 ± 0.009 | 1.907 ± 0.102 | 0.027 | |
| Rv3232c | pvdS | 0.928 ± 0.020 | 2.163 ± 0.206 | 0.033 | |
| Rv2358 | zurR | 0.736 ± 0.044 | 1.977 ± 0.114 | 0.012 | |
| Rv1674c | 1.056 ± 0.125 | 6.603 ± 0.400 | 0.021 | ||
| Rv2884 | 0.660 ± 0.059 | 4.062 ± 0.131 | 0.012 | ||
| Rv3295 | 1.017 ± 0.037 | 3.167 ± 0.138 | 0.018 | ||
| Rv3291c | 1.119 ± 0.016 | 3.072 ± 0.194 | 0.020 | ||
| Cell wall associated | Rv0835 | lpqQ | 0.944 ± 0.097 | 2.707 ± 0.068 | 0.021 |
| Rv0847 | lpqS | 1.066 ± 0.088 | 5.901 ± 0.423 | 0.023 | |
| Rv1252c | lprE | 1.591 ± 0.020 | 2.607 ± 0.048 | 0.015 | |
| Rv1541c | lprI | 0.936 ± 0.011 | 2.433 ± 0.038 | 0.004 | |
| Rv1899c | lppD | 1.310 ± 0.063 | 2.219 ± 0.086 | 0.005 | |
| Rv2080 | lppJ | 1.175 ± 0.022 | 2.567 ± 0.057 | 0.012 | |
| Rv1338 | murI | 0.746 ± 0.050 | 6.006 ± 0.378 | 0.018 | |
| Rv3810 | pirG | 1.396 ± 0.004 | 2.750 ± 0.111 | 0.017 | |
| Transport | Rv1992c | ctpG | 0.916 ± 0.060 | 3.355 ± 0.134 | 0.006 |
| Rv2397c | cysA1 | 0.944 ± 0.006 | 2.962 ± 0.106 | 0.011 | |
| Rv2399c | cysT | 0.831 ± 0.035 | 2.893 ± 0.192 | 0.024 | |
| Rv0655 | mkl | 1.092 ± 0.035 | 2.021 ± 0.054 | 0.014 | |
| Rv2400c | subI | 1.386 ± 0.028 | 2.889 ± 0.235 | 0.039 | |
| MT0955 | pstS1 | 1.693 ± 0.007 | 2.485 ± 0.042 | 0.014 | |
| Rv1668c | 0.977 ± 0.035 | 2.171 ± 0.013 | 0.004 | ||
| Rv2477c | 1.032 ± 0.035 | 2.067 ± 0.035 | 0.0006 | ||
| Sulfate metabolism | Rv1286 | cysCN | 0.778 ± 0.246 | 2.924 ± 0.008 | 0.025 |
| Rv1285 | cysD | 1.115 ± 0.100 | 6.483 ± 0.162 | 0.001 | |
| Rv2335 | cysE | 0.854 ± 0.124 | 1.817 ± 0.019 | 0.024 | |
| Rv0848 | cysK2 | 0.842 ± 0.072 | 11.028 ± 1.308 | 0.027 | |
| Rv1336 | cysM | 0.914 ± 0.058 | 11.442 ± 0.094 | 0.0007 | |
| Rv1335 | cysO | 0.925 ± 0.003 | 6.606 ± 0.183 | 0.007 | |
| Hypothetical proteins | Rv2466c | 1.161 ± 0.004 | 39.813 ± 0.476 | 0.002 | |
| Rv0140 | 1.362 ± 0.006 | 30.307 ± 6.028 | 0.046 | ||
| Rv3463 | 0.996 ± 0.016 | 26.869 ± 0.082 | 0.0005 | ||
| MT3140 | 1.151 ± 0.032 | 24.694 ± 1.428 | 0.013 | ||
The functional categorization of M. tuberculosis genes was based on their known functions as assigned by the TubercuList database (http://genolist.pasteur.fr/TubercuList/). For each gene, the changes in wild-type M. tuberculosis relative to the levels for the Δ-σH mutant in the pre-diamide stress samples and the 30-min-PDS samples are shown, along with standard deviations. Statistical significance was assigned using the ANOVA function within the S+ ArrayAnalyzer script.
TABLE 4.
Genes repressed in wild-type M. tuberculosis, relative to the levels for the Δ-σH mutant, at 30 min PDSa
| Function | Locus tag | Gene | Fold expression
|
P | |
|---|---|---|---|---|---|
| Pre-diamide stress | 30 min PDS | ||||
| Lipid metabolism | Rv0973c | accA2 | 1.206 ± 0.105 | 0.265 ± 0.061 | 0.039 |
| Rv0904c | accD3 | 1.059 ± 0.029 | 0.590 ± 0.058 | 0.041 | |
| Rv3252c | alkB | 1.427 ± 0.029 | 0.213 ± 0.041 | 0.013 | |
| Rv2243 | fabD | 0.784 ± 0.062 | 0.357 ± 0.056 | 0.002 | |
| Rv0859 | fadA | 1.548 ± 0.045 | 0.428 ± 0.193 | 0.047 | |
| Rv2590 | fadD9 | 0.985 ± 0.008 | 0.498 ± 0.111 | 0.047 | |
| Rv3089 | fadD13 | 0.965 ± 0.022 | 0.098 ± 0.005 | 0.004 | |
| Rv0244c | fadE5 | 0.968 ± 0.036 | 0.229 ± 0.009 | 0.014 | |
| Rv2724c | fadE20 | 1.261 ± 0.024 | 0.475 ± 0.102 | 0.036 | |
| Rv0469 | umaA | 1.214 ± 0.008 | 0.567 ± 0.095 | 0.030 | |
| Intermediary metabolism | Rv3086 | adhD | 1.421 ± 0.187 | 0.049 ± 0.018 | 0.027 |
| Rv3841 | bfrB | 0.762 ± 0.002 | 0.182 ± 0.132 | 0.050 | |
| Rv3617 | ephA | 0.961 ± 0.038 | 0.431 ± 0.063 | 0.043 | |
| Rv3854c | ethA | 0.844 ± 0.019 | 0.097 ± 0.016 | 0.00008 | |
| Rv3251c | rubA | 1.003 ± 0.051 | 0.161 ± 0.132 | 0.048 | |
| Rv3250c | rubB | 1.220 ± 0.059 | 0.175 ± 0.085 | 0.031 | |
| Cell wall associated | Rv0341 | iniB | 0.301 ± 0.163 | 0.294 ± 0.134 | 0.050 |
| Rv3084 | lipR | 1.412 ± 0.009 | 0.049 ± 0.028 | 0.006 | |
| Rv1217c | 1.120 ± 0.009 | 0.273 ± 0.054 | 0.011 | ||
| Rv2846c | 0.908 ± 0.085 | 0.293 ± 0.083 | 0.035 | ||
The functional categorization of M. tuberculosis genes was based on their known functions as assigned by the TubercuList database (http://genolist.pasteur.fr/TubercuList/). For each gene, the changes in wild-type M. tuberculosis relative to the levels for the Δ-σH mutant in the pre-diamide stress samples and the 30-min-PDS samples are shown, along with standard deviations. Statistical significance was assigned using the ANOVA function within the S+ ArrayAnalyzer script.
Many genes induced at 0 h PDS were known to be dependent on σH for their expression, e.g., the σH, σB, σE, trxB2, trxC, groEL1, clpB, hsp, cysA1, cysT, cysW, and Rv2466c genes (Table 1). These genes play important roles in maintaining cellular redox homeostasis, as peptidases or molecular chaperones, and in sulfate acquisition (52). Genes involved in molybdenum cofactor biosynthesis and transport and modulation of cell division were also induced. Genes with lower levels of expression in wild-type M. tuberculosis than in the mutant encode ABC transporters and DNA recombination or fatty acid biosynthesis proteins (Table 2). The expression levels of a majority of genes induced at 0 min PDS continued to be elevated at 30 min PDS, with the expression levels of σH, σE, σB, trxB2, clpB, and hsp peaking at this time point (Table 3). Other highly induced genes at this time point included cadI, CYP132 to CYP135A1, lipL, lpqS, nuoF to nuoK, rubA, rubB, Rv0303, Rv0331, Rv0692, Rv0790c, Rv1674c, Rv2348c, Rv2884, and Rv3753c. Many of these genes are known to be expressed under nonproliferating, microaerophilic conditions (55); starvation (5); antibiotic treatment (7); or phagocytosis (19, 45), indicating an overlap in the transcriptional programs of the pathogen in response to different stress conditions that mimic intraphagosomal stress. Genes induced in M. tuberculosis at this time were related to detoxification; stress; virulence; biosynthesis of molybdenum; transport of metal cations, RNA, sulfate/thiosulfate, and phosphate; and respiration (Table 3). The genes repressed in M. tuberculosis at this time were involved in biosynthesis of fatty acids, lipid virulence factors phiocerol mycocerosate and sulfolipid-1, polyketides, mycobactin siderophores, peptidoglycan, heme-porphyrin, and amino acids (Table 4) (56). The expression levels of the mce1 operon, heat shock proteins, sulfate transporters, the thioredoxin regulon, and Rv2745c genes continued to be elevated in M. tuberculosis at 60 min PDS (Table 5). The expression of σH itself continued to be elevated (2.88-fold) but declined from its peak value at 30 min. Other genes induced in M. tuberculosis were involved in mycothiol-dependent detoxification; stress; fatty acid degradation; biosynthesis of phospholipids, cobalamin, and amino acids; immune-response/virulence; protein processing; energy metabolism; and macromolecule salvage (Table 5). Genes repressed in M. tuberculosis were involved in biosynthesis of fatty acids, pthiocerol dimycocerosates, polyketides, glycoproteins, polysaccharides, proto-heme, and pantothenate and transport of phosphate, drugs, glycine betaine, sugar, and Mg2+ (Table 6). The genes induced at 90 min PDS (Tables 7 and 8) were involved in biosynthesis of fatty acids, polyketides, the cell wall, molybdenum, polysaccharides, and thiosulfates; detoxification; transport of asparagine, cations, sulfate, phosphate, and drugs; DNA repair; protein processing; sensory regulation; and immune response. At 120 min PDS, global changes in gene expression had, to a large extent, subsided (Tables 9 and 10). These results should be interpreted in light of the fact that diamide-dependent expression of σH results in induction of σE, σB, and several other transcription factors in wild-type M. tuberculosis relative to the level for the Δ-σH mutant.
TABLE 2.
Genes repressed in wild-type M. tuberculosis, relative to the levels for the Δ-σH mutant, at 0 min PDSa
| Function | Locus tag | Gene | Fold expression
|
P | |
|---|---|---|---|---|---|
| Pre-diamide stress | 0 min PDS | ||||
| Lipid metabolism | Rv0973c | accA2 | 1.206 ± 0.105 | 0.265 ± 0.061 | 0.039 |
| Rv0904c | accD3 | 1.059 ± 0.029 | 0.590 ± 0.058 | 0.041 | |
| Rv3252c | alkB | 1.427 ± 0.029 | 0.213 ± 0.041 | 0.013 | |
| Rv2243 | fabD | 0.784 ± 0.062 | 0.357 ± 0.056 | 0.002 | |
| Rv0859 | fadA | 1.548 ± 0.045 | 0.428 ± 0.193 | 0.047 | |
| Rv2590 | fadD9 | 0.985 ± 0.008 | 0.498 ± 0.111 | 0.047 | |
| Rv3089 | fadD13 | 0.965 ± 0.022 | 0.098 ± 0.005 | 0.004 | |
| Rv0244c | fadE5 | 0.968 ± 0.036 | 0.229 ± 0.009 | 0.014 | |
| Rv2724c | fadE20 | 1.261 ± 0.024 | 0.475 ± 0.102 | 0.036 | |
| Rv0469 | umaA | 1.214 ± 0.008 | 0.567 ± 0.095 | 0.030 | |
| Intermediary metabolism | Rv3086 | adhD | 1.421 ± 0.187 | 0.049 ± 0.018 | 0.027 |
| Rv3841 | bfrB | 0.762 ± 0.002 | 0.182 ± 0.132 | 0.050 | |
| Rv3617 | ephA | 0.961 ± 0.038 | 0.431 ± 0.063 | 0.043 | |
| Rv3854c | ethA | 0.844 ± 0.019 | 0.097 ± 0.016 | 0.00008 | |
| Rv3251c | rubA | 1.003 ± 0.051 | 0.161 ± 0.132 | 0.048 | |
| Rv3250c | rubB | 1.220 ± 0.059 | 0.175 ± 0.085 | 0.031 | |
| Cell wall associated | Rv0341 | iniB | 0.301 ± 0.163 | 0.294 ± 0.134 | 0.050 |
| Rv3084 | lipR | 1.412 ± 0.009 | 0.049 ± 0.028 | 0.006 | |
| Rv1217c | 1.120 ± 0.009 | 0.273 ± 0.054 | 0.011 | ||
| Rv2846c | 0.908 ± 0.085 | 0.293 ± 0.083 | 0.035 | ||
The functional categorization of M. tuberculosis genes was based on their known functions as assigned by the TubercuList database (http://genolist.pasteur.fr/TubercuList/). For each gene, the changes in wild-type M. tuberculosis relative to the levels for the Δ-σH mutant in the pre-diamide stress samples and the 0-min-PDS samples are shown, along with standard deviations. Statistical significance was assigned using the ANOVA function within the S+ ArrayAnalyzer script.
TABLE 5.
Genes induced in wild-type M. tuberculosis, relative to the levels for the Δ-σH mutant, at 60 min PDSa
| Function | Locus tag | Gene | Fold expression
|
P | |
|---|---|---|---|---|---|
| Pre-diamide stress | 60 min PDS | ||||
| Heat shock | Rv0384c | clpB | 1.148 ± 0.043 | 5.072 ± 0.851 | 0.050 |
| Rv0351 | grpE | 1.0571 ± 0.005 | 3.721 ± 0.363 | 0.031 | |
| Rv0353 | hspR | 0.977 ± 0.007 | 4.653 ± 0.521 | 0.031 | |
| Rv0352 | dnaJ1 | 1.003 ± 0.026 | 10.429 ± 1.595 | 0.038 | |
| Rv2373c | dnaJ2 | 0.851 ± 0.011 | 1.992 ± 0.186 | 0.034 | |
| Rv3417c | groEL1 | 1.051 ± 0.047 | 6.590 ± 1.161 | 0.048 | |
| Rv0563 | htpX | 0.915 ± 0.027 | 1.987 ± 0.151 | 0.037 | |
| Rv0350 | dnaK | 1.068 ± 0.020 | 4.734 ± 0.078 | 0.006 | |
| Virulence/detoxification | Rv0170 | mce1B | 1.055 ± 0.015 | 2.300 ± 0.233 | 0.039 |
| Rv0172 | mce1D | 1.023 ± 0.009 | 1.856 ± 0.164 | 0.050 | |
| Rv0174 | mce1F | 1.005 ± 0.013 | 2.368 ± 0.168 | 0.025 | |
| Rv3757c | proW | 0.946 ± 0.021 | 1.814 ± 0.022 | 0.0002 | |
| Rv3891c | esxD | 0.907 ± 0.011 | 2.208 ± 0.311 | 0.050 | |
| Rv3890c | esxC | 1.045 ± 0.010 | 1.976 ± 0.071 | 0.014 | |
| Rv3913 | trxB2 | 0.979 ± 0.018 | 5.060 ± 0.319 | 0.005 | |
| Rv2094c | tatA | 0.979 ± 0.018 | 1.750 ± 0.100 | 0.020 | |
| Transcription | Rv0182c | sigG | 1.066 ± 0.018 | 1.674 ± 0.018 | 0.003 |
| Rv3286c | sigF | 0.287 ± 0.002 | 1.963 ± 0.194 | 0.025 | |
| Rv1221 | sigE | 0.875 ± 0.0004 | 2.944 ± 0.317 | 0.034 | |
| Rv3223c | sigH | 0.869 ± 0.029 | 2.745 ± 0.371 | 0.047 | |
| Rv2711 | ideR | 1.026 ± 0.007 | 1.957 ± 0.186 | 0.046 | |
| Rv1963c | mce3R | 1.078 ± 0.004 | 2.148 ± 0.057 | 0.011 | |
| Rv3295 | 1.017 ± 0.037 | 1.887 ± 0.002 | 0.009 | ||
| Rv2745c | 1.015 ± 0.061 | 3.903 ± 0.496 | 0.033 | ||
| Rv2884 | 0.659 ± 0.059 | 2.237 ± 0.407 | 0.048 | ||
| Rv1674c | 1.056 ± 0.125 | 3.504 ± 0.412 | 0.048 | ||
| Transport | Rv2397c | cysA1 | 0.944 ± 0.006 | 6.604 ± 1.143 | 0.0005 |
| Rv2399c | cysT | 0.831 ± 0.035 | 4.595 ± 0.462 | 0.003 | |
| Rv2398c | cysW | 1.110 ± 0.012 | 3.995 ± 0.408 | 0.031 | |
| Rv0655 | mkl | 1.092 ± 0.029 | 1.935 ± 0.036 | 0.038 | |
| Rv1992c | ctpG | 0.916 ± 0.060 | 3.354 ± 0.134 | 0.001 | |
| Rv3270 | ctpC | 1.081 ± 0.022 | 2.987 ± 0.400 | 0.049 | |
| Rv2400c | subI | 1.386 ± 0.028 | 5.680 ± 0.459 | 0.022 | |
| Sulfate metabolism | Rv0848 | cysK2 | 0.842 ± 0.072 | 2.452 ± 0.395 | 0.044 |
| Rv1336 | cysM | 0.914 ± 0.058 | 7.884 ± 1.464 | 0.044 | |
| Cell wall associated | Rv1338 | murI | 0.746 ± 0.050 | 2.777 ± 0.393 | 0.048 |
| Rv0847 | lpqS | 1.066 ± 0.088 | 5.469 ± 0.135 | 0.002 | |
| Rv2349c | plcC | 0.991 ± 0.011 | 1.726 ± 0.180 | 0.050 | |
| Rv2051c | ppm1 | 1.370 ± 0.181 | 1.827 ± 0.274 | 0.045 | |
| MT0056 | ponA | 0.988 ± 0.013 | 1.722 ± 0.172 | 0.047 | |
| Protein turnover | Rv0782 | ptrBb | 1.122 ± 0.008 | 1.770 ± 0.296 | 0.009 |
| Rv2110c | prcB | 0.814 ± 0.034 | 1.624 ± 0.087 | 0.036 | |
| Rv0983 | pepD | 1.080 ± 0.026 | 1.692 ± 0.416 | 0.029 | |
| Rv3596c | clpC1 | 1.077 ± 0.015 | 2.553 ± 0.137 | 0.009 | |
| Rv2461c | clpP1 | 1.118 ± 0.011 | 2.258 ± 0.010 | 0.004 | |
| Rv2460c | clpP2 | 1.103 ± 0.014 | 2.835 ± 0.125 | 0.018 | |
| Rv2457c | clpX | 1.306 ± 0.016 | 1.901 ± 0.140 | 0.046 | |
| Energy metabolism | Rv3053c | nrdH | 1.126 ± 0.003 | 1.896 ± 0.170 | 0.046 |
| Rv3146 | nuoB | 0.917 ± 0.013 | 1.998 ± 0.170 | 0.032 | |
| Rv3147 | nuoC | 0.673 ± 0.012 | 2.095 ± 0.286 | 0.046 | |
| Rv3150 | nuoF | 1.015 ± 0.031 | 2.011 ± 0.260 | 0.050 | |
| Rv3153 | nuoI | 1.008 ± 0.021 | 2.410 ± 0.330 | 0.048 | |
| Rv3154 | nuoJ | 0.920 ± 0.00008 | 1.881 ± 0.212 | 0.049 | |
| Rv3155 | nuoK | 0.916 ± 0.005 | 1.860 ± 0.223 | 0.039 | |
| Rv3158 | nuoN | 0.848 ± 0.013 | 1.667 ± 0.155 | 0.045 | |
The functional categorization of M. tuberculosis genes was based on their known functions as assigned by the TubercuList database (http://genolist.pasteur.fr/TubercuList/). For each gene, the changes in wild-type M. tuberculosis relative to the levels for the Δ-σH mutant in the pre-diamide stress samples and the 60-min-PDS samples are shown, along with standard deviations. Statistical significance was assigned using the ANOVA function within the S+ ArrayAnalyzer script.
TABLE 6.
Genes repressed in wild-type M. tuberculosis, relative to the levels for the Δ-σH mutant, at 60 min PDSa
| Function | Locus tag | Gene | Fold expression
|
P | |
|---|---|---|---|---|---|
| Pre-diamide stress | 60 min PDS | ||||
| Lipid metabolism | Rv0973c | accA2 | 1.203 ± 0.105 | 0.488 ± 0.023 | 0.040 |
| Rv2247 | accD6 | 0.840 ± 0.009 | 0.600 ± 0.027 | 0.035 | |
| Rv3252c | alkB | 1.427 ± 0.029 | 0.250 ± 0.025 | 0.010 | |
| Rv1094 | desA2 | 0.655 ± 0.006 | 0.394 ± 0.062 | 0.048 | |
| Rv1185c | fadD21 | 1.061 ± 0.057 | 0.504 ± 0.051 | 0.002 | |
| Rv3089 | fadD13 | 0.965 ± 0.002 | 0.220 ± 0.053 | 0.009 | |
| Rv0404 | fadD30 | 1.135 ± 0.045 | 0.555 ± 0.037 | 0.002 | |
| Rv2950c | fadD29 | 1.051 ± 0.003 | 0.396 ± 0.033 | 0.012 | |
| Rv2930 | fadD26 | 1.066 ± 0.022 | 0.121 ± 0.043 | 0.012 | |
| Rv1483 | fabG1 | 1.788 ± 0.025 | 0.421 ± 0.133 | 0.017 | |
| Rv2524c | fas | 0.870 ± 0.047 | 0.419 ± 0.055 | 0.050 | |
| Rv2384 | mbtA | 1.097 ± 0.080 | 0.545 ± 0.074 | 0.002 | |
| Rv2382c | mbtC | 0.681 ± 0.043 | 0.424 ± 0.007 | 0.042 | |
| Rv2381c | mbtD | 0.801 ± 0.040 | 0.375 ± 0.054 | 0.050 | |
| Rv2379c | mbtF | 0.854 ± 0.003 | 0.488 ± 0.064 | 0.037 | |
| Rv2931 | ppsA | 0.991 ± 0.016 | 0.256 ± 0.100 | 0.036 | |
| Rv2932 | ppsB | 0.808 ± 0.067 | 0.407 ± 0.059 | 0.004 | |
| Rv2933 | ppsC | 1.293 ± 0.033 | 0.337 ± 0.072 | 0.024 | |
| Rv2934 | ppsD | 1.303 ± 0.107 | 0.527 ± 0.205 | 0.029 | |
| Rv2935 | ppsE | 0.852 ± 0.001 | 0.498 ± 0.076 | 0.047 | |
| Rv0405 | pks6 | 1.068 ± 0.020 | 0.390 ± 0.010 | 0.007 | |
| Information pathways | Rv0651 | rplJ | 1.022 ± 0.037 | 0.566 ± 0.095 | 0.028 |
| Rv2442c | rplU | 1.051 ± 0.023 | 0.589 ± 0.022 | 0.022 | |
| Rv1015c | rplY | 0.875 ± 0.008 | 0.531 ± 0.008 | 0.011 | |
| Rv2057c | rpmG1 | 1.032 ± 0.085 | 0.609 ± 0.016 | 0.036 | |
| Rv0718 | rpsH | 1.117 ± 0.121 | 0.500 ± 0.006 | 0.041 | |
| Rv2055c | rpsR2 | 0.814 ± 0.013 | 0.552 ± 0.013 | 0.017 | |
| Rv0710 | rpsQ | 1.115 ± 0.030 | 0.609 ± 0.003 | 0.013 | |
| Transport | Rv0917 | betP | 0.890 ± 0.022 | 0.541 ± 0.083 | 0.039 |
| Rv2937 | drrB | 0.781 ± 0.019 | 0.496 ± 0.002 | 0.017 | |
| Rv2938 | drrC | 1.059 ± 0.019 | 0.527 ± 0.028 | 0.054 | |
| Rv2846c | efpA | 0.906 ± 0.085 | 0.417 ± 0.085 | 0.0001 | |
| Rv1811 | mgtC | 0.941 ± 0.031 | 0.635 ± 0.008 | 0.016 | |
| Rv2281 | pitB | 1.112 ± 0.001 | 0.490 ± 0.057 | 0.020 | |
| MT0955 | pstS1 | 1.693 ± 0.007 | 0.596 ± 0.029 | 0.007 | |
| Rv1217c | 1.120 ± 0.009 | 0.608 ± 0.064 | 0.032 | ||
| Rv0986 | 0.789 ± 0.034 | 0.452 ± 0.109 | 0.050 | ||
| Rv0987 | 0.996 ± 0.052 | 0.477 ± 0.049 | 0.044 | ||
| Rv1349 | 0.904 ± 0.014 | 0.566 ± 0.060 | 0.050 | ||
The functional categorization of M. tuberculosis genes was based on their known functions as assigned by the TubercuList database (http://genolist.pasteur.fr/TubercuList/). For each gene, the changes in wild-type M. tuberculosis relative to the levels for the Δ-σH mutant in the pre-diamide stress samples and the 60-min-PDS samples are shown, along with standard deviations. Statistical significance was assigned using the ANOVA function within the S+ ArrayAnalyzer script.
TABLE 7.
Genes induced in wild-type M. tuberculosis, relative to the levels for the Δ-σH mutant, at 90 min PDSa
| Function | Locus tag | Gene | Fold expression
|
P | |
|---|---|---|---|---|---|
| Pre-diamide stress | 90 min PDS | ||||
| Lipid metabolism | Rv3667 | acs | 0.921 ± 0.057 | 1.722 ± 0.154 | 0.050 |
| Rv0468 | fadB2 | 1.355 ± 0.033 | 2.628 ± 0.040 | 0.007 | |
| Rv1185c | fadD21 | 1.106 ± 0.057 | 2.284 ± 0.167 | 0.042 | |
| Rv0231 | fadE4 | 1.166 ± 0.016 | 1.755 ± 0.087 | 0.026 | |
| Rv0242c | fabG4 | 1.091 ± 0.023 | 2.196 ± 0.076 | 0.010 | |
| Rv1493 | mutB | 1.106 ± 0.016 | 1.727 ± 0.172 | 0.055 | |
| Rv1182 | papA3 | 1.265 ± 0.091 | 2.071 ± 0.148 | 0.052 | |
| Rv2946c | pks1 | 0.850 ± 0.038 | 1.660 ± 0.225 | 0.051 | |
| Rv2935 | ppsE | 0.852 ± 0.001 | 1.791 ± 0.153 | 0.036 | |
| Transport | Rv2920c | amt | 1.029 ± 0.053 | 1.667 ± 0.197 | 0.049 |
| Rv2397c | cysA1 | 0.944 ± 0.006 | 2.194 ± 0.056 | 0.011 | |
| Rv3117 | cysA3 | 1.141 ± 0.014 | 1.702 ± 0.051 | 0.014 | |
| Rv2398c | cysW | 1.110 ± 0.012 | 1.778 ± 0.043 | 0.010 | |
| Rv0908 | ctpE | 1.030 ± 0.033 | 1.978 ± 0.262 | 0.051 | |
| Rv1811 | mgtC | 0.941 ± 0.031 | 1.564 ± 0.153 | 0.001 | |
| Rv2281 | pitB | 1.112 ± 0.001 | 2.018 ± 0.211 | 0.046 | |
| Rv1273c | 0.872 ± 0.034 | 3.093 ± 0.576 | 0.055 | ||
| Rv1458c | 1.119 ± 0.024 | 2.166 ± 0.150 | 0.027 | ||
| Cell wall associated | Rv3793 | embC | 0.953 ± 0.004 | 1.982 ± 0.211 | 0.046 |
| Rv0399c | lpqK | 0.974 ± 0.113 | 4.103 ± 0.553 | 0.047 | |
| Rv1235 | lpqY | 1.080 ± 0.035 | 2.113 ± 0.014 | 0.004 | |
| Rv1921c | lppF | 1.244 ± 0.266 | 3.160 ± 0.567 | 0.035 | |
| Rv2945c | lppX | 0.783 ± 0.029 | 2.214 ± 0.272 | 0.045 | |
| Rv0451c | mmpS4 | 0.893 ± 0.013 | 2.082 ± 0.165 | 0.033 | |
| Rv2350c | plcB | 0.974 ± 0.010 | 2.247 ± 0.272 | 0.045 | |
| Rv3682 | ponA2 | 1.135 ± 0.019 | 1.771 ± 0.152 | 0.046 | |
| Rv0284 | 0.846 ± 0.015 | 2.213 ± 0.370 | 0.050 | ||
| Rv1184c | 1.486 ± 0.037 | 3.460 ± 0.010 | 0.003 | ||
| PE/PPE family | Rv2768c | PPE43 | 0.878 ± 0.049 | 1.850 ± 0.082 | 0.030 |
| Rv3478 | PPE60 | 1.150 ± 0.024 | 1.652 ± 0.140 | 0.050 | |
| Rv1172c | PE12 | 0.910 ± 0.0003 | 1.720 ± 0.174 | 0.047 | |
| Rv1787 | PPE25 | 1.128 ± 0.003 | 2.219 ± 0.230 | 0.047 | |
| Rv2519 | PE26 | 1.047 ± 0.052 | 1.643 ± 0.189 | 0.050 | |
| Rv1789 | PPE26 | 0.871 ± 0.037 | 1.680 ± 0.144 | 0.049 | |
| Rv1430 | PE16 | 1.013 ± 0.046 | 1.746 ± 0.209 | 0.049 | |
| Rv3533c | PPE62 | 1.117 ± 0.033 | 2.557 ± 0.390 | 0.050 | |
| 0.890 ± 0.022 | 0.541 ± 0.083 | 0.039 | |||
| Protein processing | Rv3596c | clpC1 | 1.077 ± 0.015 | 1.651 ± 0.026 | 0.016 |
| Rv2460c | clpP2 | 1.103 ± 0.014 | 1.838 ± 0.191 | 0.051 | |
| Rv2461c | clpP1 | 1.118 ± 0.011 | 1.624 ± 0.049 | 0.016 | |
| Transcription | Rv3132c | devS | 1.037 ± 0.069 | 2.556 ± 0.432 | 0.050 |
| Rv1479 | moxR1 | 0.875 ± 0.0008 | 1.965 ± 0.013 | 0.003 | |
| Rv2088 | pknJ | 0.984 ± 0.159 | 1.771 ± 0.326 | 0.047 | |
| Rv1032c | trcS | 1.012 ± 0.080 | 1.949 ± 0.137 | 0.044 | |
The functional categorization of M. tuberculosis genes was based on their known functions as assigned by the TubercuList database (http://genolist.pasteur.fr/TubercuList/). For each gene, the changes in wild-type M. tuberculosis relative to the levels for the Δ-σH mutant in the pre-diamide stress samples and the 90-min-PDS samples are shown, along with standard deviations. Statistical significance was assigned using the ANOVA function within the S+ ArrayAnalyzer script.
TABLE 8.
Genes repressed in wild-type M. tuberculosis, relative to the levels for the Δ-σH mutant, at 90 min PDSa
| Function | Locus tag | Gene | Fold expression
|
P | |
|---|---|---|---|---|---|
| Pre-diamide stress | 90 min PDS | ||||
| Virulence | Rv3667 | hbhA | 1.203 ± 0.010 | 0.502 ± 0.185 | 0.051 |
| Lipid metabolism | Rv3825c | pks2 | 2.535 ± 0.003 | 0.550 ± 0.320 | 0.037 |
| Hypothetical proteins | MT1775 | 1.283 ± 0.003 | 0.339 ± 0.117 | 0.031 | |
The functional categorization of M. tuberculosis genes was based on their known functions as assigned by the TubercuList database (http://genolist.pasteur.fr/TubercuList/). For each gene, the changes in wild-type M. tuberculosis relative to the levels for the Δ-σH mutant in the pre-diamide stress samples and the 90-min-PDS samples are shown, along with standard deviations. Statistical significance was assigned using the ANOVA function within the S+ ArrayAnalyzer script.
TABLE 9.
Genes induced in wild-type M. tuberculosis, relative to the levels for the Δ-σH mutant, at 120 min PDSa
| Function | Locus tag | Gene | Fold expression
|
P | |
|---|---|---|---|---|---|
| Pre-diamide stress | 120 min PDS | ||||
| Transport | Rv1183 | mmpL10 | 0.911 ± 0.221 | 1.666 ± 0.074 | 0.044 |
| Rv0929 | pstC2 | 1.086 ± 0.034 | 1.756 ± 0.059 | 0.031 | |
| Rv2281 | pitB | 1.112 ± 0.001 | 1.883 ± 0.164 | 0.047 | |
| Rv2397c | cysA1 | 0.944 ± 0.006 | 1.610 ± 0.148 | 0.050 | |
| Intermediary metabolism | Rv1318c | adc | 1.130 ± 0.295 | 1.996 ± 0.384 | 0.023 |
| Rv3303c | lpdA | 1.309 ± 0.026 | 1.809 ± 0.032 | 0.034 | |
| Rv0557 | pimB | 1.385 ± 0.021 | 3.026 ± 0.348 | 0.044 | |
| Rv0886 | fprB | 1.022 ± 0.077 | 1.841 ± 0.113 | 0.051 | |
| Rv0373c | coxM | 1.336 ± 0.029 | 3.246 ± 0.336 | 0.042 | |
The functional categorization of M. tuberculosis genes was based on their known functions as assigned by the TubercuList database (http://genolist.pasteur.fr/TubercuList/). For each gene, the changes in wild-type M. tuberculosis relative to the levels for the Δ-σH mutant in the pre-diamide stress samples and the 120-min-PDS samples are shown, along with standard deviations. Statistical significance was assigned using the ANOVA function within the S+ ArrayAnalyzer script.
TABLE 10.
Genes repressed in wild-type M. tuberculosis, relative to the levels for the Δ-σH mutant, at 120 min PDSa
| Function | Locus tag | Gene | Fold expression
|
P | |
|---|---|---|---|---|---|
| Pre-diamide stress | 120 min PDS | ||||
| Heat shock | Rv3418c | groES | 1.342 ± 0.003 | 0.580 ± 0.023 | 0.007 |
| Rv0440 | groEL2 | 1.075 ± 0.0003 | 0.580 ± 0.0003 | 0.010 | |
| Rv0251c | hsp | 0.951 ± 0.048 | 0.613 ± 0.050 | 0.032 | |
| Virulence | Rv3875 | esxA | 1.083 ± 0.0005 | 0.590 ± 0.037 | 0.017 |
| Rv3874 | esxB | 0.993 ± 0.035 | 0.580 ± 0.023 | 0.006 | |
| Rv0287 | esxG | 0.941 ± 0.018 | 0.580 ± 0.023 | 0.003 | |
| Rv0288 | esxH | 1.176 ± 0.021 | 0.580 ± 0.021 | 0.016 | |
| Rv1793 | esxN | 1.068 ± 0.036 | 0.570 ± 0.009 | 0.020 | |
| Rv3019c | esxR | 1.215 ± 0.018 | 0.586 ± 0.032 | 0.004 | |
| Molybdenum biosynthesis | MT3193 | moaB1 | 1.798 ± 0.118 | 0.539 ± 0.111 | 0.001 |
| Rv3111 | moaC1 | ND | 0.622 ± 0.026 | 0.093 | |
| Rv3112 | moaD1 | ND | 0.345 ± 0.067 | 0.023 | |
| Rv0868c | moaD2 | 1.233 ± 0.024 | 0.565 ± 0.082 | 0.019 | |
The functional categorization of M. tuberculosis genes was based on their known functions as assigned by the TubercuList database (http://genolist.pasteur.fr/TubercuList/). For each gene, the changes in wild-type M. tuberculosis relative to the levels for the Δ-σH mutant in the pre-diamide stress samples and the 120-min-PDS samples are shown, along with standard deviations. Statistical significance was assigned using the ANOVA function within the S+ ArrayAnalyzer script. ND, not determined.
Verification of microarray data.
To verify our transcriptome results, we performed SYBR green qPCR assays (Fig. 3A). The expression of trxB2 was induced in wild-type M. tuberculosis relative to the level for the Δ-σH mutant at 0 min PDS and remained at significantly high levels at all time points. The expression of hsp was induced at 0 min PDS and peaked at 30 min PDS. The expression of ctpG was induced only at the 30-min-PDS time point. The expression of bpoB was induced at both the 30 (peak)- and the 60-min-PDS time points, though its expression had been induced only at the earlier time point in microarray studies. The Rv2745c gene was induced between 30 and 90 min PDS, peaking at the 60-min time point. The expression of clpP1 was induced as early as 0 min PDS and peaked at 60 min PDS. The expression of mce1A was induced at and peaked at 30 min PDS (Fig. 3A).
FIG. 3.
Confirmation of the PDS microarray data. (A) qPCR-based interrogation of the PDS expression levels of certain genes with perturbed expression in wild-type M. tuberculosis relative to the level for the Δ-σH mutant at different time points. Results are shown for trxB2, hsp, ctpG, bpoB, Rv2745c, clpP1, and mce1A. Values represent mean differences (n-fold) ± standard deviations of results from quadruplicate experiments. Statistically significant changes in expression are denoted by * (P < 0.05), ** (P < 0.01), or *** (P < 0.005). (B) Western blot of M. tuberculosis wild-type and Δ-σH mutant lysates obtained from cultures treated pre-diamide stress and PDS shows a σH-dependent expression of Rv2745c. Lysates were probed with an affinity-purified polyclonal antibody raised against an epitope on the protein encoded by the M. tuberculosis Rv2745c gene. Lane A, prestained protein ladder (Invitrogen); lane B, lysate from log-phase pre-diamide stress M. tuberculosis culture; lane C, lysate from log-phase pre-diamide stress Δ-σH culture; lanes D, F, H, and J, lysates from 0-, 30-, 60-, and 90-min-PDS M. tuberculosis cultures; lanes E, G, I, and K, lysates from 0-, 30-, 60-, and 90-min-PDS Δ-σH cultures.
A polyclonal antibody against the Rv2745c-encoded transcription factor specifically detected the induced levels of this protein at 30, 60, and 90 min PDS only in wild-type M. tuberculosis (Fig. 3B). The corresponding band was not detected in comparable samples obtained from the Δ-σH mutant. It has previously been reported that the expression of Rv2745c is controlled by σE (14, 51). The inability of the Δ-σH mutant to induce the expression of σE likely results in the lack of induction of the Rv2745c gene.
Conditional expression of the Rv2745c gene in M. tuberculosis induces the expression of the clp regulon.
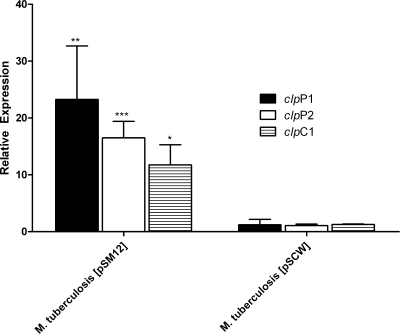
The Rv2745c gene was cloned downstream of the M. smegmatis Pace in an integrative vector and used to transform M. tuberculosis. The conditional expression of Rv2745c gene was induced in this strain by adding acetamide at log phase. As measured by qPCR, the Pace-dependent expression of Rv2745c for 1 as well as 2 h led to high levels of induction of clpP1, clpP2, and clpC1 (Fig. 4). Such induction was not observed in an M. tuberculosis strain transformed with a control vector, conclusively showing that the expression of the M. tuberculosis clp proteolysis regulon is controlled by the Rv2745c-encoded protein ClgR.
FIG. 4.
qPCR-based analysis of the expression of clp regulon genes following acetamide-dependent induction of the Rv2745c gene in M. tuberculosis. qPCR was used to interrogate the expression levels of clpP1, clpP2, and clpC1 following induction of the Rv2745c gene. Results are shown for M. tuberculosis(pSM12) (carrying an integrative vector with Rv2745c in a transcriptional fusion with the M. smegmatis acetamidase promoter) and M. tuberculosis(pSCW38) (carrying an integration-proficient vector with the acetamidase promoter without any gene, as a control). Values represent mean differences (n-fold) ± standard deviations of results from quadruplicate experiments. Statistically significant changes in expression are denoted by * (P < 0.05), ** (P < 0.01), or *** (P < 0.005).
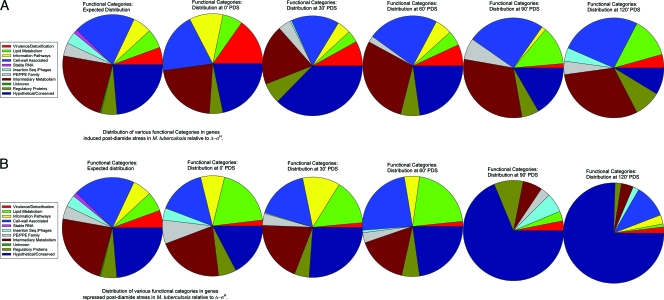
Functional categorization of the wild-type M. tuberculosis PDS transcriptome relative to that for the Δ-σH mutant.
The results obtained in this study were analyzed based on the M. tuberculosis genome functional categories. The expected number of genes in a given category was calculated for a given data set, based on the total number of genes assigned to each category in the genome. This was compared to the actual number of genes of each functional category induced or repressed at a given time point (Fig. 5A and B).
FIG. 5.
Functional characterization of the PDS microarray data. Microarray data for each time point (0, 30, 60, 90, and 120 min PDS) were analyzed based on the distribution of various functional categories as defined by the TubercuList database (http://genolist.pasteur.fr/TubercuList/) for genes that were induced (A) or repressed (B) in wild-type M. tuberculosis, relative to the level for the Δ-σH mutant. These results were compared to the expected distribution of different functional categories (pie on the left). The pie sections are arranged in an anticlockwise manner, beginning with a red section for category 0, followed by green and yellow sections for categories 1 and 2, respectively.
DISCUSSION
The ability of M. tuberculosis to survive within the host requires resistance to various physiological and environmental stresses. Sigma factors encode a family of transcriptional regulators necessary for such adaptation processes. Expression of sigma factors σB, σE, and σH has been shown to be required for the resistance of the pathogen to various environmental stresses likely to be encountered in vivo (25, 26, 29-31, 41). A subset of these sigma factors is induced during the growth of M. tuberculosis within macrophages (19) and required for virulence in animal models (2, 25, 30, 32). In this report, we shed more light on the complex transcriptional network controlled by σH. We interpreted these results in a global context by studying the relative abundances of genes belonging to a specific functional category. Initially (at 0 and 30 min PDS), and not unexpectedly, the expression levels of a larger-than-expected number of genes belonging to the virulence/detoxification category were upregulated in M. tuberculosis. Diamide is a thiol-oxidative agent. Detoxifying proteins Hsp, GroEL (which helps properly fold proteins) (52), ClpB and HtpX (peptidases) (52), SodA (which destroys free radicals toxic to M. tuberculosis) (11), and BpoB (which controls damaging reactive oxygen species during phagocytosis) are induced to likely help the pathogen effectively deal with the consequences of such stress. Initial oxidative damage by the host macrophages may serve as an environmental cue for M. tuberculosis to turn on the expression of key virulence genes, which may in turn allow the pathogen to invade secondary cells and adapt to the host. Virulence genes induced by M. tuberculosis immediately following diamide stress include members of the mce1 operon (49); Rv2525c, a substrate of the twin-arginine translocation system, which exports folded proteins (47); Rv1740, Rv2526, Rv2547, Rv2595, and Rv2829c, PIN domain antitoxins (3); CspA, an RNA chaperone that interacts with TA networks (58); and invasin, involved in interaction with phagocytes. However, fewer than the expected number of “lipid metabolism” genes were induced at 0 and 30 min PDS (Fig. 5A). This represents a marked shift in the metabolic profile of the pathogen immediately after phagocytosis. By silencing expensive functions at a time when it is faced with a high degree of potentially destructive stress, M. tuberculosis appears to conserve cellular energy and protect its lipids and mycolic acids from oxidative damage. Further reinforcing this point, the expression levels of more than the expected number of “lipid metabolism” genes, including those involved in mycolic acid synthesis, biogenesis of mycobactin siderophores, and lipid virulence factors, were repressed at 0 and 30 min PDS (Fig. 5B). Beyond 60 min PDS, the expression levels of more than the expected number of “intermediary metabolism” genes were upregulated in M. tuberculosis. This represents an adaptation from the initial “stress defense” mode to one where the pathogen prepares metabolites for the subsequent replication and proliferation. These genes are involved in arginine biosynthesis, macromolecular salvage, and protein processing. It is energetically more expensive for M. tuberculosis to biosynthesize l-arginine than for this organism to acquire it from the external environment (17, 54). That M. tuberculosis still synthesizes arginine reflects its inability to sequester it following oxidative stress. An increased need for protein processing and turnover may be necessitated due to damage to protein structure by oxidative stress and a subsequent inability of the heat shock network to completely restore the structures of damaged proteins. Such proteins may need to be completely degraded via the ATP-dependent clp proteolytic pathway. At 60 min PDS, the “lipid metabolism” category continued to be repressed in M. tuberculosis (Fig. 5B). The 90-min-PDS time point marked a significant shift in the pattern of wild-type M. tuberculosis gene expression relative to the level for the Δ-σH mutant. At both 90 and 120 min PDS, the expression levels of a large number of “lipid metabolism” and “cell wall-associated” genes were elevated. This included many genes whose expression levels were initially repressed. A larger-than-expected number of “lipid metabolism” genes were no longer repressed. This demonstrates the temporal plasticity of gene expression in M. tuberculosis in response to stress. The pathogen is able to effectively silence expensive, energy-consuming cellular processes, like fatty acid biosynthesis, immediately following oxidative stress, under conditions where lipids can also be oxidized and permanently damaged. With the passage of time, the deleterious effects of oxidative stress are nullified and M. tuberculosis resumes fatty acid biosynthesis.
Our results show that σH induces the expression of a large number of M. tuberculosis transcriptional regulators. This in turn reshapes global transcription in M. tuberculosis, silencing lipid and virulence factor biosynthesis until the stress has been effectively managed, while inducing the expression of key regulons involved in virulence, detoxification, and protein processing. Here, we focus our attention on a few key transcription networks controlled by σH.
The ATP-dependent Clp proteolysis regulon is upregulated PDS.
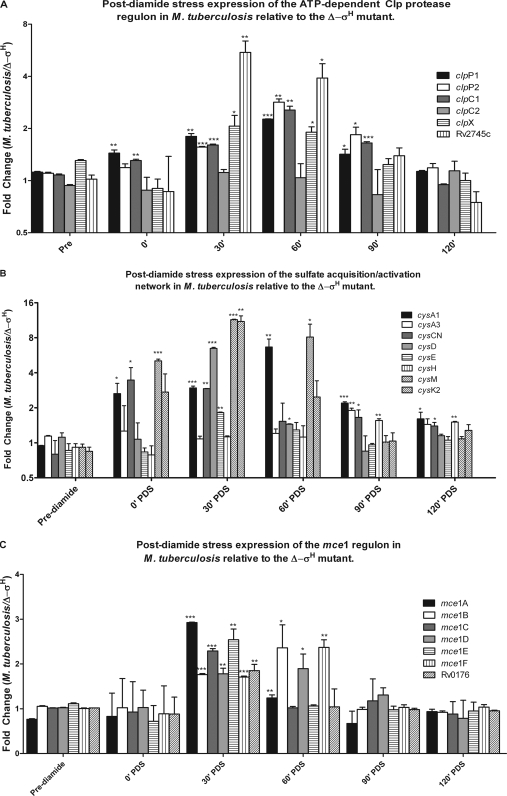
Clp proteases perform regulatory functions by controlling the availability of key enzymes or regulators (18). The M. tuberculosis genome encodes two proteolytic (clpP1 and clpP2) and three catalytic (clpC1, clpC2, and clpX) Clp complex subunits (10, 13). In Corynebacterium glutamicum and Streptomyces coelicolor, expression of the clp operon is regulated by a transcription factor, ClgR (clp gene regulator), a homolog of Rv2745c (4, 12). The expression of the Rv2745c gene was induced beginning at 30 min PDS (Fig. 6A). The expressions of clpP1, clpP2, clpC1 (but not clpC2), clpX, and clpS were also induced at 30 min PDS and peaked at the 60-min-PDS time point. When the M. tuberculosis CDC1551 MT2816 gene (Rv2745c) was conditionally induced, the expression levels of the clp regulon genes clpP1, clpP2, and clpC1 were highly upregulated (Fig. 4). We conclude that the Rv2745c gene product positively regulates the expression of the M. tuberculosis ATP-dependent clp proteolysis regulon. The expression of Rv2745c is dependent on σE (14, 51), which is itself induced in a σH-dependent manner upon diamide stress (25, 30) as well as in a σH-dependent manner under yet other stress conditions (21). At this time, we are not certain if σH indirectly induces Rv2745c expression via σE or if it can also directly initiate transcription from the Rv2745c promoter. The promoter sequences recognized by M. tuberculosis σH and σE are remarkably similar, with the exception of the 3′ position of the −35 element (51). It is conceivable that the expression of Rv2745c is induced by σE under certain conditions where σH is not induced, e.g., upon cell wall damage by SDS (28), or under conditions where neither σH nor σE is induced, e.g., upon treatment with thioridizine (N. K. Dutta, S. Mehra, and D. Kaushal, submitted for publication), and by both σH and σE during oxidative and heat stress.
FIG. 6.
Genes belonging to the ATP-dependent clp regulon, the mammalian cell entry 1 (mce1) operon, and the sulfate acquisition/activation network are expressed in a σH-dependent manner PDS. The expression levels of clp regulon genes clpP1, clpP2, clpC1, clpC2, clpX, and Rv2745c are presented as means of results from two independent microarray experiments (A). The expression levels of the sulfate transport/activation genes cysA1, cysA3, cysCN, cysD, cysE, cysH, cysM, and cysK2 are presented as means of results from two independent array experiments (B). The expression levels of mce1 operon genes mce1A, mce1B, mce1C, mce1D, mce1E, mce1F, and Rv0176 are presented as means of results from two independent experiments (C). Statistically significant changes in expression are denoted by * (P < 0.05), ** (P < 0.01), or *** (P < 0.005).
Regulation of sulfate transport following PDS.
Sulfate assimilation is crucial for M. tuberculosis. It is an essential bionutrient with a key role in biosynthesis of cysteine, mycothiol, and coenzyme A. Moreover, extracellular presentation of sulfated metabolites may regulate host-pathogen and cell-cell communication (6). The CysTWA SubI ABC transporter complex is responsible for the active transport of inorganic sulfate across the mycobacterial cell membrane. Once imported, sulfate is phosphorylated to adenosine-5-phosphosulfate (APS) and then to 3′-phosphoadenosine 5′-phosphosulfate (PAPS), a universal sulfate donor, in a two-step reaction catalyzed by the bifunctional enzyme encoded by cysNC. GTP hydrolysis performed by the cysD product provides energy for this energetically unfavorable reaction. Together, the cysNC- and cysD-encoded proteins form a sulfate-activating complex. The sulfate moiety in APS can be reduced to sulfite by the cysH product in a reaction catalyzed by thioredoxin. Sulfite can be further reduced to sulfide by the nirA-encoded sulfite reductase (ferredoxin-dependent nitrite reductase). Sulfide is used to biosynthesize cysteine, methionine, coenzyme A, and mycothiol. De novo synthesis of cysteine involves the generation of O-acetyl-l-serine either by the cysE product, in which case it is condensed with a reduced sulfide by-product of cysK2 or cysM, or by the cysO and moeZ gene products. The expressions of cysD and cysNC were induced in wild-type M. tuberculosis, relative to the level for Δ-σH, at 30 min PDS. These genes have also been induced during stationary-phase growth (19) and upon reactive nitrogen (36) or oxidative (40) stress. The expressions of cysH and nirA (Rv2391) were induced at 90 and 120 min PDS in M. tuberculosis, although the level of cysH induction was below the stringent statistical-significance levels employed by us. The cysE, cysM, cysO, and moeZ genes were also induced in M. tuberculosis following stress. These results show that an entire network of genes involved in sulfate acquisition from the environment and its reduction to biologically utilizable forms for biosynthetic processes was activated. This underscores the crucial role of sulfate metabolism in the constitution of mycobacterial proteins, enzymes, and other biologically active compounds.
Regulation of the mce1 operon.
The mce1 operon is important for the virulence of M. tuberculosis (49). The six Mce1 proteins (Mce1A to Mce1F) interact with host components during infection and influence the immune response. The M. tuberculosis mce1 mutants have aberrant inflammatory cell migration and poor granuloma formation, resulting in uncontrolled bacterial growth and rapid death of mutant-infected mice (49). Members of the mce1 operon have been shown to be coexpressed with σH and σE upon phagocytosis by human macrophages (19). In this report, we show that the expressions of several members of the mce1 regulon are induced as a result of oxidative stress. These include mce1A, mce1C, and mce1E (30 min) and mce1B and mce1D (30 and 60 min). While we do not know the identity of the factor that directly mediates the expression of the mce1 regulon following oxidative stress, it is likely that σH is not directly involved. The induction of σH following oxidative stress perhaps serves as a cue for the pathogen to induce its key virulence functions in order to disseminate to secondary cells.
Transcriptional networks in M. tuberculosis following σH induction.
Modulation of the gene expression of numerous transcription factors helps σH mediate its long-term impact. The function of the Rv2745c-encoded transcription factor has already been discussed. Other transcription factors whose expression levels were perturbed in wild-type M. tuberculosis relative to the level for the mutant include Rv0273c, Rv0324, Rv0348, Rv0465c, Rv1019, Rv1049, Rv1129c, Rv1674c, Rv1956, Rv3291c, and Rv3692. Rv3291c encodes LrpA, a feast/famine family member that is induced during anaerobic conditions (55) and nutritional starvation (5) and is critical for persistence (39). Its Escherichia coli homolog regulates metabolic pathways in response to the availability of amino acids in the external environment (8). While we do not know which M. tuberculosis genes are regulated by lrpA, a large number of genes related to amino acid metabolism were induced at 60 min PDS, following the induction of lrpA at 30 min PDS.
The Rv0324-Rv0325-Rv0326 operon is induced in enduring hypoxia, along with the σH/σE regulon (48). The Rv0347-Rv0348-Rv0349 operon is induced in vivo at the late stage of chronic TB (53). Rv0348 potentiates the expression of σF, which orchestrates entry into the chronic stage of TB (15). The expression of Rv1049 is induced by heat shock (52), peroxide stress (48), and starvation (5). Rv1049 is required for survival in murine macrophages (43). Rv1956 is induced by starvation (5) and encodes a possible antitoxin. Rv0465c is induced following nutritional stress (20) in a σE- and σF-dependent manner. The expressions of Rv1129c and its adjoining genes Rv1130 and Rv1131 are induced in a σE (29)- and mprA-dependent manner (37).
The Rv1994c (cmtR)-encoded repressor CmtR senses the critical thresholds of Cd(II) and Pb(II) (9), is induced during starvation (5) and oxidative stress (48), and is required for in vivo survival (24). We propose that diamide treatment oxidizes the numerous cysteine thiols that CmtR senses metal ions through. This derepresses the cmtR regulon, leading to the induction of transporters Rv1993 and ctpG. This allows M. tuberculosis to transport out the high levels of Cd(II) and Pb(II). CmtR is able to sense metal ions upon the restoration of redox homeostasis in M. tuberculosis, eventually repressing the Rv1993-ctpG operon. We also observed the induction of the cadI gene by diamide treatment in M. tuberculosis, in contrast to an earlier report (23). However, it must be considered that the earlier report involved the use of the H37Rv strain, not the CDC1551 strain, of M. tuberculosis. Alternatively, it is possible that a duration of diamide treatment other than 1 h or a concentration of diamide other than 10 mM was used in the previous report. The function of M. tuberculosis CadI appears to be similar to that of metallothioneins, which are low-molecular-weight, cysteine-rich proteins that protect against metal toxicity. Since diamide oxidizes cysteine residues on proteins, an increased induction of cadI may be an attempt by M. tuberculosis to overcome the decreased activity of this protein.
furA encodes a global repressor that uses Fe2+ as a cofactor to bind the katG operator. katG is transcribed from two promoters, one generating a bicistronic furA-katG message, expressed initially during infection, and another generating a monocistronic katG mRNA, which peaks at a later stage (33). The expression of furA was induced at 30 min PDS and then declined. The expression pattern of katG was the mirror opposite; this gene was repressed at 30 min PDS, presumably due to repression by furA, and then induced as the furA levels declined, peaking at 90 min PDS. The iron-binding repressor IdeR reduces the expression of genes involved in siderophore biosynthesis, iron storage, and transport (44), including mbtABCDEFGI, fadE14, Rv3402c, and Rv3403c. All these genes exhibited reduced expression in wild-type M. tuberculosis, relative to the level for the mutant, at 30 min PDS. The expression of ideR is induced by σB, which is itself induced in a σH-dependent manner following oxidative stress. Rv2358 encodes Zur, which regulates the homeostasis and uptake of zinc, an essential element that is included in numerous enzymes and proteins as a cofactor or a structural scaffold but can be toxic at higher concentrations (28). Rv0981 (mprA) encodes a two-component response transcriptional regulatory protein, MprA (mycobacterial persistence regulator), which is induced upon starvation and in a σE-dependent manner.
To summarize, we have shown that σH not only directs the transcription of a subset of genes involved in the maintenance of redox homeostasis and protein integrity but also induces a network of transcription factors, which further induce crucial bacterial functions, such as ATP-dependent proteolysis of specific targets, acquisition and utilization of environmental sulfate toward the biosynthesis of key mycobacterial metabolites, specific virulence- and pathogenesis-related operons that help M. tuberculosis survive and disseminate in vivo, and transcription factors that help M. tuberculosis sense and respond to the levels of various metal and trace metal ions. We demonstrate the dynamic nature of the pathogen's response to adverse environmental conditions, with the expression of genes involved in lipid biosynthesis being repressed following oxidative stress and resuming upon the removal of stress. We hope to extend these findings by interrogating their intraphagosomal and in vivo relevance.
Acknowledgments
This work was supported by NIH grants P20RR020159 and P51RR164, with additional support from the Tulane Research Enhancement Fund and the Louisiana Vaccine Center.
We acknowledge excellent technical, secretarial, and graphic design assistance from Sondra LeBreton, Avery MacLean, and Robin Rodriguez, respectively. We are grateful to the PFGRC for DNA microarrays and to Colorado State University for genomic DNA.
Footnotes
Published ahead of print on 17 April 2009.
REFERENCES
- 1.Andersen, P., and T. M. Doherty. 2005. The success and failure of BCG—implications for a novel tuberculosis vaccine. Nat. Rev. Microbiol. 3656-662. [DOI] [PubMed] [Google Scholar]
- 2.Ando, M., T. Yoshimatsu, C. Ko, P. J. Converse, and W. R. Bishai. 2003. Deletion of Mycobacterium tuberculosis sigma factor E results in delayed time to death with bacterial persistence in the lungs of aerosol-infected mice. Infect. Immun. 717170-7172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arcus, V. L., P. B. Rainey, and S. J. Turner. 2005. The PIN-domain toxin-antitoxin array in mycobacteria. Trends Microbiol. 13360-365. [DOI] [PubMed] [Google Scholar]
- 4.Bellier, A., and P. Mazodier. 2004. ClgR, a novel regulator of clp and lon expression in Streptomyces. J. Bacteriol. 1863238-3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Betts, J. C., P. T. Lukey, L. C. Robb, R. A. McAdam, and K. Duncan. 2002. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol. Microbiol. 43717-731. [DOI] [PubMed] [Google Scholar]
- 6.Bhave, D. P., W. B. Muse III, and K. S. Caroll. 2007. Drug targets in mycobacterial sulfur metabolism. Infect. Disord. Drug Targets 7140-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boshoff, H. I., T. G. Myers, B. R. Copp, M. R. McNeil, M. A. Wilson, and C. E. Barry III. 2004. The transcriptional responses of Mycobacterium tuberculosis to inhibitors of metabolism: novel insights into drug mechanisms of action. J. Biol. Chem. 27940174-40184. [DOI] [PubMed] [Google Scholar]
- 8.Calvo, J. M., and R. G. Mathews. 1994. Leucine responsive regulatory protein, a global regulator of metabolism in Escherichia coli. Microbiol. Rev. 58466-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cavet, J. S., A. I. Graham, W. Meng, and N. J. Robinson. 2003. A cadmium-lead-sensing ArsR-SmtB repressor with novel sensory sites. J. Biol. Chem. 27844560-44566. [DOI] [PubMed] [Google Scholar]
- 10.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, A. Krogh, J. McLean, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. A. Quail, M. A. Rajandream, J. Rogers, S. Rutter, K. Seeger, J. Skelton, R. Squares, S. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393537-544. [DOI] [PubMed] [Google Scholar]
- 11.Edwards, K. M., M. H. Cynamon, R. K. Voladri, C. C. Hager, M. S. DeStefano, K. T. Tham, D. L. Lakey, M. R. Bochan, and D. S. Kernodle. 2001. Iron-cofactored superoxide dismutase inhibits host responses to Mycobacterium tuberculosis. Am. J. Respir. Crit. Care Med. 642213-2219. [DOI] [PubMed] [Google Scholar]
- 12.Engels, S., J. E. Schweitzer, C. Ludwig, M. Bott, and S. Schaffer. 2004. clpC and clpP1P2 gene expression in Corynebacterium glutamicum is controlled by a regulatory network involving the transcriptional regulators ClgR and HspR as well as the ECF sigma factor sigmaH. Mol. Microbiol. 52285-302. [DOI] [PubMed] [Google Scholar]
- 13.Fleischmann, R. D., D. Alland, J. A. Eisen, L. Carpenter, O. White, J. Peterson, R. DeBoy, R. Dodson, M. Gwinn, D. Haft, E. Hickey, J. F. Kolonay, W. C. Nelson, L. A. Umayam, M. Ermolaeva, S. L. Salzberg, A. Delcher, T. Utterback, J. Weidman, H. Khouri, J. Gill, A. Mikula, W. Bishai, W. R. Jacobs, Jr., J. C. Venter, and C. M. Fraser. 2002. Whole-genome comparison of Mycobacterium tuberculosis clinical and laboratory strains. J. Bacteriol. 1845479-5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fontán, P. A., V. Aris, M. E. Alvarez, S. Ghanny, J. Cheng, P. Soteropoulos, A. Trevani, R. Pine, and I. Smith. 2008. Mycobacterium tuberculosis sigma factor E regulon modulates the host inflammatory response. J. Infect. Dis. 198877-885. [DOI] [PubMed] [Google Scholar]
- 15.Geiman, D. E., D. Kaushal, C. Ko, S. Tyagi, Y. C. Manabe, B. G. Schroeder, R. D. Fleischmann, N. E. Morrison, P. J. Converse, P. Chen, and W. R. Bishai. 2004. Attenuation of late-stage disease in mice infected by the Mycobacterium tuberculosis mutant lacking the SigF alternate sigma factor and identification of SigF-dependent genes by microarray analysis. Infect. Immun. 721733-1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gomez, J. E., J. M. Chen, and W. R. Bishai. 1997. Sigma factors of Mycobacterium tuberculosis. Tuber. Lung Dis. 78175-183. [DOI] [PubMed] [Google Scholar]
- 17.Gordhan, B. G., D. A. Smith, H. Alderton, R. A. McAdam, G. J. Bancroft, and V. Mizrahi. 2002. Construction and phenotypic characterization of an auxotrophic mutant of Mycobacterium tuberculosis defective in l-arginine biosynthesis. Infect. Immun. 703080-3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gottesman, S. 1996. Proteases and their targets in Escherichia coli. Annu. Rev. Genet. 30465-506. [DOI] [PubMed] [Google Scholar]
- 19.Graham, J. E., and J. E. Clark-Curtiss. 1999. Identification of Mycobacterium tuberculosis RNAs synthesized in response to phagocytosis by human macrophages by selective capture of transcribed sequences (SCOTS). Proc. Natl. Acad. Sci. USA 9611554-11559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hampshire, T., S. Soneji, J. Bacon, B. W. James, J. Hinds, R. A. Stabler, P. D. Marsh, and P. D. Butcher. 2004. Stationary phase gene expression of Mycobacterium tuberculosis following a progressive nutrient depletion: a model for persistent organisms? Tuberculosis 84228-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He, H., R. Hovey, J. Kane, V. Singh, and T. C. Zahrt. 2006. MprAB is a stress-responsive two-component system that directly regulates expression of sigma factors SigB and SigE in Mycobacterium tuberculosis. J. Bacteriol. 1882134-2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hernandez-Abanto, S. M., S. C. Woolwine, S. K. Jain, and W. R. Bishai. 2006. Tetracycline-inducible gene expression in mycobacteria within an animal host using modified Streptomyces tcp830 regulatory elements. Arch. Microbiol. 186459-464. [DOI] [PubMed] [Google Scholar]
- 23.Hotter, G. S., T. Wilson, and D. M. Collins. 2001. Identification of a cadmium induced gene in Mycobacterium bovis and Mycobacterium tuberculosis. FEMS Microbiol. Lett. 200151-155. [DOI] [PubMed] [Google Scholar]
- 24.Jain, S. K., S. M. Hernandez-Abanto, Q.-J. Cheng, P. Singh, L. H. Ly, L. G. Klinkenberg, N. E. Morrison, P. J. Converse, E. Nuermberger, J. Grosset, D. N. McMurray, P. C. Karakousis, G. Lamichhane, and W. R. Bishai. 2007. Accelerated detection of Mycobacterium tuberculosis genes essential for bacterial survival in guinea pigs, compared with mice. J. Infect. Dis. 1951634-1642. [DOI] [PubMed] [Google Scholar]
- 25.Kaushal, D., B. G. Schroeder, S. Tyagi, T. Yoshimatsu, C. Scott, C. Ko, L. Carpenter, J. Mehrotra, Y. C. Manabe, R. D. Fleischmann, and W. R. Bishai. 2002. Reduced immunopathology and mortality despite tissue persistence in a Mycobacterium tuberculosis mutant lacking alternative sigma factor, SigH. Proc. Natl. Acad. Sci. USA 998330-8335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee, J. H., P. C. Karakousis, and W. R. Bishai. 2008. Roles of SigB and SigF in the Mycobacterium tuberculosis sigma factor network. J. Bacteriol. 190699-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lonetto, M., M. Gribskov, and C. A. Gross. 1992. The sigma 70 family: sequence conservation and evolutionary relationships. J. Bacteriol. 1743843-3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maciag, A., E. Dainese, G. M. Rodriguez, A. Milano, R. Provvedi, M. R. Pasca, I. Smith, G. Palu, G. Riccardi, and R. Manganelli. 2007. Global analysis of the Mycobacterium tuberculosis Zur (FurB) regulon. J. Bacteriol. 189730-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manganelli, R., M. I. Voskuil, G. K. Schoolnik, and I. Smith. 2001. The Mycobacterium tuberculosis ECF sigma factor sigmaE: role in global gene expression and survival in macrophages. Mol. Microbiol. 41423-437. [DOI] [PubMed] [Google Scholar]
- 30.Manganelli, R., M. I. Voskuil, G. K. Schoolnik, E. Dubnau, M. Gomez, and I. Smith. 2002. Role of the extracytoplasmic-function sigma factor sigma(H) in Mycobacterium tuberculosis global gene expression. Mol. Microbiol. 45365-374. [DOI] [PubMed] [Google Scholar]
- 31.Manganelli, R., R. Provvedi, S. Rodrigue, J. Beaucher, L. Gaudreau, and I. Smith. 2004. Sigma factors and global gene regulation in Mycobacterium tuberculosis. J. Bacteriol. 186895-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manganelli, R., L. Fattorini, D. Tan, E. Iona, G. Orefici, G. Altavilla, P. Cusatelli, and I. Smith. 2004. The extra cytoplasmic function sigma factor σE is essential for Mycobacterium tuberculosis virulence in mice. Infect. Immun. 723038-3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Master, S., T. C. Zahrt, J. Song, and V. Deretic. 2001. Mapping of Mycobacterium tuberculosis katG promoters and their differential expression in infected macrophages. J. Bacteriol. 1834033-4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matteelli, A., G. B. Migliori, D. Cirillo, R. Centis, E. Girard, and M. Raviglion. 2007. Multidrug-resistant and extensively drug-resistant Mycobacterium tuberculosis: epidemiology and control. Expert Rev. Anti Infect. Ther. 5857-871. [DOI] [PubMed] [Google Scholar]
- 35.Missiakas, D., and S. Raina. 1998. The extracytoplasmic function sigma factors: role and regulation. Mol. Microbiol. 281059-1066. [DOI] [PubMed] [Google Scholar]
- 36.Ohno, H., G. Zhu, V. P. Mohan, D. Chu, S. Kohno, W. R. Jacobs, Jr., and J. Chan. 2003. The effects of reactive nitrogen intermediates on gene expression in Mycobacterium tuberculosis. Cell. Microbiol. 5637-648. [DOI] [PubMed] [Google Scholar]
- 37.Pang, X., P. Vu, T. F. Byrd, S. Ghanny, P. Soteropoulos, G. V. Mukamolova, S. Wu, B. Samten, and S. T. Howard. 2007. Evidence for complex interactions of stress-associated regulons in an mprAB deletion mutant of Mycobacterium tuberculosis. Microbiology 1531229-1242. [DOI] [PubMed] [Google Scholar]
- 38.Park, S. T., C.-M. Kang, and R. N. Husson. 2008. Regulation of the SigH stress response regulon by an essential protein kinase in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 10513105-13110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parti, R. P., R. Shrivastava, S. Srivastava, A. R. Subramanian, R. Roy, B. S. Srivastava, and R. Srivastava. 2008. A transposon insertion mutant of Mycobacterium fortuitum attenuated in virulence and persistence in a murine infection model that is complemented by Rv3291c of Mycobacterium tuberculosis. Microb. Pathog. 45370-376. [DOI] [PubMed] [Google Scholar]
- 40.Pinto, R., Q. X. Tang, W. J. Britton, T. S., Leyh, and J. A. Triccas. 2004. The Mycobacterium tuberculosis cysD and cysNC genes form a stress-induced operon that encodes a tri-functional sulfate-activating complex. Microbiology 1501681-1686. [DOI] [PubMed] [Google Scholar]
- 41.Raman, S., T. Song, X. Puyang, S. Bardarov, W. R. Jacobs, Jr., and R. N. Husson. 2001. The alternative sigma factor SigH regulates major components of oxidative and heat stress responses in Mycobacterium tuberculosis. J. Bacteriol. 1836119-6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raviglione, M. C. 2003. The TB epidemic from 1992 to 2002. Tuberculosis (Edinburgh) 834-14. [DOI] [PubMed] [Google Scholar]
- 43.Rengarajan, J., B. R. Bloom, and E. J. Rubin. 2005. Genome-wide requirements for Mycobacterium tuberculosis adaptation and survival in macrophages. Proc. Natl. Acad. Sci. USA 1028327-8332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rodriguez, G. M., M. I. Voskuil, B. Gold, G. K. Schoolnik, and I. Smith. 2002. ideR, An essential gene in Mycobacterium tuberculosis: role of IdeR in iron-dependent gene expression, iron metabolism, and oxidative stress response. Infect. Immun. 703371-3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rohde, K. H., R. B. Abramovitch, and D. G. Russell. 2007. Mycobacterium tuberculosis invasion of macrophages: linking bacterial gene expression to environmental cues. Cell Host Microbe 2352-364. [DOI] [PubMed] [Google Scholar]
- 46.Rustad, T. R., M. I. Harrell, R. Liao, and D. R. Sherman. 2008. The enduring hypoxic response of Mycobacterium tuberculosis. PLoS ONE 3e1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saint-Joanis, B., C. Demangel, M. Jackson, P. Brodin, L. Marsollier, H. Boshoff, and S. T. Cole. 2006. Inactivation of Rv2525c, a substrate of the twin arginine translocation (Tat) system of Mycobacterium tuberculosis, increases ß-lactam susceptibility and virulence. J. Bacteriol. 1886669-6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schnappinger, D., S. Ehrt, M. I. Voskuil, Y. Liu, J. A. Mangan, I. M. Monahan, G. Dolganov, B. Efron, P. D. Butcher, C. Nathan, and G. K. Schoolnik. 2003. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal environment. J. Exp. Med. 198693-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shimono, N., L. Morici, N. Casali, S. Cantrell, B. Sidders, S. Ehrt, and L. W. Riley. 2003. Hypervirulent mutant of Mycobacterium tuberculosis resulting from disruption of the mce1 operon. Proc. Natl. Acad. Sci. USA 10015918-15923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Song, T., S. L. Dove, K. H. Lee, and R. N. Husson. 2003. RshA, an anti-sigma factor that regulates the activity of the mycobacterial stress response sigma factor SigH. Mol. Microbiol. 50949-959. [DOI] [PubMed] [Google Scholar]
- 51.Song, T., S. E. Song, S. Raman, M. Anaya, and R. N. Husson. 2008. Critical role of a single position in the −35 element for promoter recognition by Mycobacterium tuberculosis SigE and SigH. J. Bacteriol. 1902227-2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stewart, G. R., L. Wernisch, R. Stabler, J. A. Mangan, J. Hinds, K. G. Laing, D. B. Young, and P. D. Butcher. 2002. Dissection of the heat-shock response in Mycobacterium tuberculosis using mutants and microarrays. Microbiology 1483129-3138. [DOI] [PubMed] [Google Scholar]
- 53.Talaat, A. M., S. K. Ward, C. W. Wu, E. Rondon, C. Tavano, J. P. Bannantine, R. Lyons, and S. A. Johnston. 2007. Mycobacterial bacilli are metabolically active during chronic tuberculosis in murine lungs: insights from genome-wide transcriptional profiling. J. Bacteriol. 1894265-4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Talaue, M. T., V. Venketaraman, M. H. Hazbón, M. Peteroy-Kelly, A. Seth, R. Colangeli, D. Alland, and N. D. Connell. 2006. Arginine homeostasis in J774.1 macrophages in the context of Mycobacterium bovis BCG infection. J. Bacteriol. 1884830-4840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Voskuil, M. I., K. C. Visconti, and G. K. Schoolnik. 2004. Mycobacterium tuberculosis gene expression during adaptation to stationary phase and low-oxygen dormancy. Tuberculosis (Edinburgh) 84218-227. [DOI] [PubMed] [Google Scholar]
- 56.Walters, S. B., E. Dubnau, I. Kolesnikova, F. Laval, M. Daffe, and I. Smith. 2006. The Mycobacterium tuberculosis PhoPR two-component system regulates genes essential for virulence and complex lipid biosynthesis. Mol. Microbiol. 60312-330. [DOI] [PubMed] [Google Scholar]
- 57.Yang, Y. H., S. Dudoit, P. Luu, D. M. Lin, V. Peng, J. Ngai, and T. P. Speed. 2002. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 30e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhu, L., S. Phadtare, H. Nariya, M. Ouyang, R. N. Husson, and M. Inouye. 2008. The mRNA interferases, MazF-mt3 and MazF-mt7 from Mycobacterium tuberculosis target unique pentad sequences in single-stranded RNA. Mol. Microbiol. 69559-569. [DOI] [PMC free article] [PubMed] [Google Scholar]