Abstract
Clostridium difficile is a gram-positive, spore-forming enteric anaerobe which can infect humans and a wide variety of animal species. Recently, the incidence and severity of human C. difficile infection has markedly increased. In this study, we evaluated the genomic content of 73 C. difficile strains isolated from humans, horses, cattle, and pigs by comparative genomic hybridization with microarrays containing coding sequences from C. difficile strains 630 and QCD-32g58. The sequenced genome of C. difficile strain 630 was used as a reference to define a candidate core genome of C. difficile and to explore correlations between host origins and genetic diversity. Approximately 16% of the genes in strain 630 were highly conserved among all strains, representing the core complement of functional genes defining C. difficile. Absent or divergent genes in the tested strains were distributed across the entire C. difficile 630 genome and across all the predicted functional categories. Interestingly, certain genes were conserved among strains from a specific host species, but divergent in isolates with other host origins. This information provides insight into the genomic changes which might contribute to host adaptation. Due to a high degree of divergence among C. difficile strains, a core gene list from this study offers the first step toward the construction of diagnostic arrays for C. difficile.
Clostridium difficile is a gram-positive, spore-forming enteric anaerobic pathogen that infects or colonizes humans and multiple animal species. Clinical manifestations in humans range from asymptomatic colonization or mild diarrhea to pseudomembranous colitis and death (18). Antibiotic use among hospitalized patients is the primary risk factor for the development of C. difficile infection (CDI). It is believed that antibiotic therapy disrupts the normal colonic microflora, providing a niche in which C. difficile can multiply and produce toxins. Recently, there have been marked increases in the incidence and severity of CDI (29). Several recent outbreaks in North America and Europe have been caused by an emergent highly virulent strain, characterized as toxinotype III, restriction endonuclease type BI, PCR ribotype 027, and North American pulsed-field gel electrophoresis (PFGE) type 1 (NAP1) (28). This epidemic strain is characteristically resistant to fluoroquinolones (28).
Strains of C. difficile that cause colitis produce toxins A (TcdA, an enterotoxin) and B (TcdB, a cytotoxin). The corresponding genes, tcdA and tcdB, respectively, are located in a pathogenicity locus (PaLoc) together with a holin-like pore-forming protein (tcdE) (49) and genes encoding two transcriptional regulators (tcdC and tcdR) (13). Certain alleles of tcdC, which encodes a negative regulator of tcdA and tcdB, are characterized by the presence of single-nucleotide mutations, including Δ117 and C184T, that result in a truncated nonfunctional protein and corresponding in-frame deletions (18 bp or 39 bp) that can be used as markers (11, 14, 26). Variations within the PaLoc provide the basis for a commonly used classification scheme, toxinotyping, that assigns C. difficile strains into more than 20 types (34, 36, 43). Nontoxigenic strains do not cause colitis, although strains producing TcdB alone are virulent (1, 2). In addition to TcdA and TcdB, some strains including BI/027/NAP1 (28) produce a binary toxin (CdtA/CdtB) that may play an adjunct role in the pathogenesis of CDI (15).
Several techniques are used to understand the epidemiology and pathogenicity of C. difficile strains. Toxinotyping, mentioned above, is based on variations in the PaLoc. Other typing methods include multilocus variable number tandem-repeat analysis, amplified fragment length polymorphism, surface layer protein A gene sequencing, PCR-ribotyping, restriction endonuclease analysis, multilocus sequence typing, and PFGE (19). Recently, the genomic sequence of strain 630, a multidrug-resistant C. difficile strain isolated from a Swiss patient with severe CDI, was made available (40). Stabler et al. performed comparative phylogenomic studies of C. difficile strains using a microarray with PCR probes specific to the 630 genome (41). Their data revealed extensive variation in the genetic contents of each strain (41).
The genomic sequence of another human-associated C. difficile strain, QCD-32g58, has also been completely annotated and publicly available at an NIAID Bioinformatics Resource Center Pathema (http://pathema.jcvi.org/cgi-bin/Clostridium/pathemattomepage.cgi). The hypervirulent strain QCD-32g58 is responsible for a multi-institutional outbreak and is representative of the predominant NAP1/BI/027 strain in Quebec, Ontario, Canada (22). In the present study, we performed oligonucleotide-based comparative genomic hybridization (CGH) using the genomic sequences of strains 630 and QCD-32g58 to evaluate gene conservation and diversity among C. difficile strains. Compared to PCR-based arrays, microarrays made from specific oligonucleotide probes provide technical benefits such as less cross-hybridization, no phage contamination of the cDNA library, enhanced specificity, and finer control over probe concentration (24). Seventy-three C. difficile isolates of clinical origin recovered from diverse geographic regions and host species were tested. This report concentrates on the genes which are universally present in the genome of C. difficile and draws correlations between genetic diversity and host origin. We also compared the CGH data with PFGE patterns for the grouping of each isolate.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
All C. difficile isolates used in this study are listed in Table S1 in the supplemental material. Isolates were recovered from humans (n = 35), horses (n = 14), cattle (n = 17), and pigs (n = 8). The human isolates were from five countries, including 14 states in the United States. The equine isolates were isolated from horses with diarrhea admitted to the Ontario Veterinary College-Teaching Hospital, Guelph, Ontario, Canada, and 10 bovine isolates were recovered from calves with diarrhea in a veal farm operation in Ontario, Canada. Seven additional bovine isolates and all swine isolates were obtained from the University of Arizona, Tucson, AZ. Reference strain 630 is a multidrug-resistant isolate from a patient with severe pseudomembranous colitis (40). All strains were cultivated in prereduced anaerobically sterilized peptone yeast extract broth with glucose (Anaerobe Systems, Morgan Hill, CA) at 37°C for 48 h under anaerobic conditions. All isolates were confirmed to be C. difficile by colony morphology, growth on cycloserine-cefoxitin fructose agar, characteristic p-cresol odor, yellow-green fluorescence under long-wave UV light, negative indole reaction, and positive PRO reaction.
Microarray construction.
Using a standalone implementation of the BLAST package, a BLAST-P genome comparison between strains 630 and QCD-32g58 (GenBank accession numbers AM180355 and AAML00000000, respectively) was performed with the E value of 1e5 and a minimum percentage identity threshold of 70% to identify coding sequences (CDS) common to both strains, as well as a list of CDS specific for each strain. CDS-specific oligonucleotides of 55 to 70 bases with a matched melting temperature of ∼60°C were designed using Operon Biotechnologies (Huntsville, AL) proprietary software and selected based upon several parameters, including the uniqueness in the genome, sequence complexity, lack of self-binding, GC content, binding energy, and proximity to the 3′ end of the gene. The array consisted of 13,824 spots corresponding to 3,309 CDS present in both strains, 365 CDS specific to strains 630, and 251 CDS specific to QCD-32g58; 126 CDS that lacked gene-specific unique regions were excluded from this study (see Table S2 in the supplemental material). Each gene was represented by three replicates of one sequence; some genes (chosen at random based on available array space) had six replicates. Probes were designed and synthesized by Operon Biotechnologies. Microarrays were printed in a single batch by the Cornell University Microarray Core Facility (http://cores.lifesciences.cornell.edu/brcinfo/) using probes (final concentration, 30 μM) resuspended in a final volume of 10 μl print buffer containing 1× SSC (0.15 M NaCl plus 0.015 M sodium citrate) and 0.005% sarcosyl onto Corning UltraGaps coated glass slides (Corning, Lowell, MA). Autoblanks were included as negative controls. The slides were processed according to the manufacturer's instructions, using hydration and UV irradiation, and stored in a dark, dust-free environment as previously described (32, 38).
Genomic DNA extraction and labeling.
Prior to proceeding with the genomic DNA extraction and array hybridization, all the strains were blind coded. Genomic DNA was isolated by using DNeasy blood and tissue mini kits (Qiagen, Valencia, CA) as described in the manufacturer's protocol for gram-positive bacteria. DNA was quantified and checked for purity using a DU800 spectrophotometer (Beckman Coulter, Fullerton, CA.). Genomic DNA (3 μg) from each strain was fragmented by DraI digestion at 37°C for 3 h and purified using a QIAquick PCR purification kit (Qiagen). Fragmented DNA was then labeled with 10 μg of exoresistant random primers (Fermentas, Glen Burnie, MD), 25 U of Klenow fragment (New England Biolabs, Ipswich, MA), deoxynucleoside triphosphate mix (0.12 mM each dATP, dCTP, and dGTP and 0.03 mM dTTP), and a 0.1 mM concentration of either Cy3- or Cy5-dUTP (Amersham Biosciences, Piscataway, NJ). The probes were purified from unincorporated dyes by use of the QIAquick PCR purification kit (Qiagen). Labeled DNA sample yields and dye incorporation efficiencies were determined by spectrometry as previously described (32, 38).
Hybridizations.
Genomic DNA from C. difficile 630 was used as a reference for all hybridizations. Dye swaps were performed for each comparison to rule out potential bias introduced by inherent differences in dye incorporation. The microarray slides were prehybridized with 25% formamide, 5× SSC, 0.2% (wt/vol) sarcosyl, and 10 mg/ml bovine serum albumin at 42°C for 1 h. The slides were then washed in MilliQ water and blow dried using compressed nitrogen gas. Equivalent amounts of labeled probes from reference and tested strains were pooled, lyophilized, and resuspended in 70 μl of hybridization buffer (25% formamide, 5× SSC, 0.2% sodium dodecyl sulfate, 1 mg/ml salmon sperm DNA, and 1 mg/ml yeast tRNA). The probes were then denatured at 99°C for 5 min, centrifuged briefly at 10,000 × g, and applied to the microarray under a 22-mm by 60-mm LifterSlip (Erie Scientific Co., Portsmouth, NH). The hybridizations were performed at 42°C for 16 h in a sealed humidified hybridization chamber (Corning), following by a 5-min wash in 2× SSC, 1% sodium dodecyl sulfate at 42°C and three 5-min washes in 0.1% SSC at room temperature. The slides were then dried and scanned immediately. All the experiments were performed by one person, and failed hybridizations were repeated based on a whisker plot of all arrays before data analysis.
Data analysis.
Arrays were scanned using a GenePix 4000B scanner (Molecular Devices, Sunnyvale, CA). The Cy3 and Cy5 signals and the local background intensities were quantified using GenePix Pro 6.1 software. The local background value was subtracted from the intensity of each spot. Spots were examined manually, and poor spots were flagged for elimination from the analysis. Data were then globally normalized using a Lowess algorithm. The mean normalized log2 value (ratio of tester signal/reference signal) and standard deviation were calculated from all replicates (at least two slides each with three spotted replicates). All subsequent data analyses were performed using Microsoft Excel and GACK (Genomotyping Analysis by Charlie Kim) software to determine present/divergent genes (20). A divergent CDS is one which is absent from the tested strain or whose sequence has diverged to a degree that hybridization cannot be detected. Prior to the experiments, we performed the quality control hybridizations, in which we hybridized (i) Cy3- and Cy5-labeled genomic DNA of the reference strain 630, (ii) the strain QCD-32g58, and (iii) the dye swap of the strains 630 and QCD-32g58 in order to validate the specificity of the probes. For stringent analyses of the present and absent/divergent genes, the estimated probability of presence cutoff values were set at 100% and 0%, respectively, to minimize any uncertainty in the present/divergent predictions. Core gene analysis was performed by applying the GACK program to the data set and analyzing the output in Excel to identify spots that were present in all strains. The percent presence analysis used to analyze the percentage of the C. difficile strain 630 genes shared by each test strain was calculated by determining the number of absent, missing, and present spots for each strain in the data set. The CGH data were clustered using various algorithms in Avadis Prophetic 3.3 software (Strand Genomics, Union City, CA) (32). CDS that were specific to QCD-32g58 (and not present in the reference strain 630) could not be analyzed using the GACK program, as this was a one-color analysis. A separate analysis was performed to determine the presence or absence of these genes using the Microbial Diagnostic Array Workstation (39). CDS from QCD-32g58 were considered present in the tested strains if the log2 ratio (signal median/background median) was more than the mean of the log2 values minus one standard deviation.
Confirmation of variable regions by amplification.
Five common CDS and five divergent CDS were randomly chosen for PCR analysis to verify the CGH results. The primer sequences are listed in Table S3 in the supplemental material. For each gene, a PCR was performed on the DNA from all tested C. difficile strains. The parameters for amplification were 94°C for 5 min, 30 cycles of 94°C for 1 min, 50°C for 1 min, and 72°C for 1 min, and a final extension cycle of 72°C for 10 min. The PCR products were separated by agarose gel electrophoresis to confirm the presence of a band of the expected size.
Strain typing assays.
PFGE strain typing of all isolates was performed as previously described (21). PFGE patterns were assigned based on comparison to the CDC C. difficile database with BioNumerics 4.0 software (Applied Maths, Austin, TX). The isolates were assigned to a specific NAP type if PFGE patterns demonstrated >80% identity to established NAP types. A dendrogram of isolates in this study was constructed with BioNumerics 4.0 software by the unweighted-pair group method with arithmetic mean using Dice coefficients and 1.1% position tolerance and optimization, as previously published (19). Toxinotyping assays were performed according to the method of Rupnik et al. (35). The binary toxin gene, cdtB, was detected by PCR with primers 5′-CTTAATGCAAGTAAATACTGAG-3′ and 5′-AACGGATCTCTTGCTTCAGTC-3′. Deletions in tcdC were detected by PCR with primers 5′-GCACCTCATCACCATCTTC-3′ and 5′-TGGTTCAAAATGAAAGACGAC-3′, followed by electrophoresis in 2% MetaPhor agarose (Lonza, Allendale, NJ). The tcdC gene was amplified and sequenced using primers 5′-TTAATTAATTTTCTCTACAGCTATCC-3′ and 5′-TCTAATAAAAGGGAGATTGTATTATG-3′.
Microarray data accession number.
All datasets have been deposited in Gene Expression Omnibus (GEO) with platform accession number GPL6118 and series accession number GSE9693.
RESULTS AND DISCUSSION
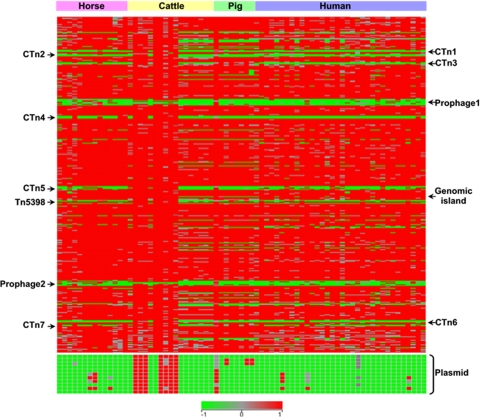
The genomic content of 73 strains from humans, horses, cattle, and pigs were analyzed by CGH to obtain broad information on the gene profiles of C. difficile using the genome of strain 630 as the reference. To assess the quality of our microarray data, five each of the conserved and divergent CDS were randomly chosen for confirmation tests by PCR amplification in all tested strains. Only 35 of 730 reactions from different targets and strains did not match the CGH results. The error rate for microarray predictions can therefore be estimated to be ∼4.8%. The pattern of presence or absence/divergence of CDS in the tested strains is shown in Fig. 1.
FIG. 1.
Comparative genomic analysis of 73 strains of C. difficile isolated from various hosts. Two arrays with dye swap for each strain were averaged and analyzed with GACK software. The CDS were arranged according to the numbering of the C. difficile 630 genes, with CD0001 at the top and CD3680 at the bottom, followed by CDS from the plasmid pCD630 (CDP01 to CDP11). Each column represents an isolate, and each row corresponds to a specific CDS. The status of each CDS is indicated by color as follows: red, present/conserved; green, absent; and gray, divergent. The positions of the putative conjugative transposon (CTn1 to CTn7), prophages, and the genomic island are indicated.
Hierarchical clustering by CGH and dendrogram from PFGE patterns.
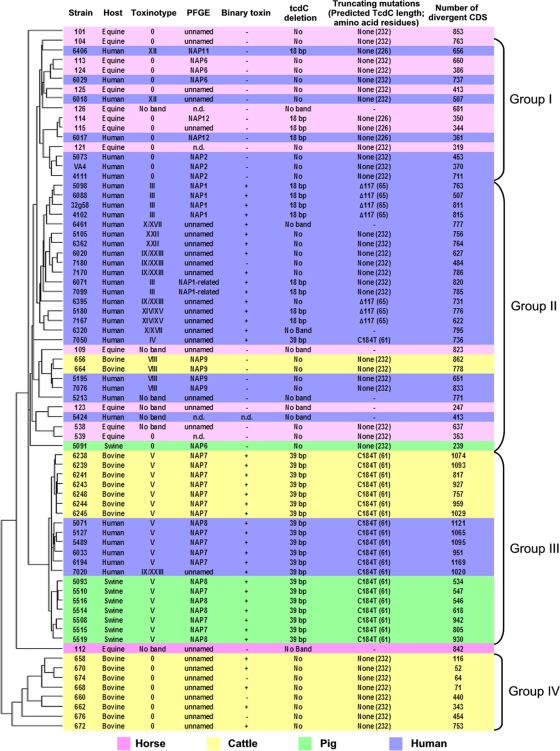
We performed hierarchical clustering analyses to evaluate the patterns of gene conservation and divergence among the tested strains. The application of various clustering methods, including the use of the Pearson absolute as a distance metric, did not substantially change the configuration of the tree (data not shown). The hierarchical clustering of 73 C. difficile isolates based on the overall variability in the CGH data revealed differences in genetic content among the tested strains (Fig. 2). Interestingly, most strains clustered into four groups (I to IV), leaving equine strains 101 and 112 as unrelated branches. Group I (n = 15) contained 8/14 equine isolates and 7/35 human isolates. Most (12/15; 80%) of the isolates in group I were of toxinotype 0, and none were positive for binary toxin. A single equine isolate in group I (strain 126) was nontoxigenic. Group II (n = 28) contained 21/35 human isolates, 4/14 equine isolates, two Canadian bovine isolates, and one swine isolate. The isolates in group II were of various toxinotypes, 16/28 were binary toxin positive, and 5 (4 equine, 1 human) were nontoxigenic. The isolates identified by PFGE as NAP1 (n = 6) made up 22% of this group and included strain QCD-32g58. Group III (n = 20) comprised 7/8 swine isolates, 7/7 bovine isolates from Arizona, and 6/35 human isolates. All isolates in this group were positive for binary toxin, and 20/21 were NAP7 or NAP8 and toxinotype V and carried a 39-bp deletion in tcdC. Group IV (n = 8) consisted entirely of bovine strains from Ontario that were toxinotype 0 and had no deletion in tcdC.
FIG. 2.
Hierarchical clustering of 73 strains of C. difficile based on the microarray data. The strain numbers, host origins, toxinotypes, PFGE types, presence of binary toxin, tcdC deletion observed, and tcdC stop codon detected by sequencing are indicated in addition to the number of divergent CDS compared to that of reference strain 630. n.d., not determined.
In general, the results from the microarray analyses were concordant with the PFGE results (Fig. 3). The group III and IV clusters are clearly evident in the PFGE dendrogram, whereas groups I and II are less distinct. The cluster of isolates in group IV were collected from the same veterinary hospital and veal farm operations in Ontario, whereas the bovine isolates from Arizona (group III) appear unrelated to the Ontario isolates by both CGH and PFGE.
FIG. 3.
Dendrogram of C. difficile isolates based on PFGE results. The group numbers based on CGH clustering and PFGE types are indicated.
The CDS (n = 251) present in strain QCD-32g58 but not in strain 630 were also included in our microarrays. As expected, strain 630 clustered outside groups I to IV compared only against the QCD-32g58-specific CDS (Fig. 4). The clustering patterns of all the isolates based on CDS present in strain QCD-32g58 are consistent with the results obtained using strain 630 as a reference. Similarly to our findings, Stabler et al. (41) reported four major clades, including the hypervirulent (HY), toxin-defective (A− B+), and two human/animal (HA1 and HA2) clades. The subcluster in group II containing toxinotype VIII, NAP9, isolates appears to correspond to Stabler's A− B+ clade (41).
FIG. 4.
Hierarchical clustering of C. difficile isolates based on the microarray data with CDS from strain QCD-32g58. Each column represents a specific CDS, and each row corresponds to a strain. The clustering method is based on the Pearson absolute with both rows and columns. The status of each CDS is indicated by color as follows: red, present/conserved; green, absent; and gray, divergent.
Patients with CDI acquire the organism from the environment (31) and interspecies transmission of C. difficile may be possible (4). Because the spores of C. difficile are heat resistant, a role for food (especially meats) in the transmission of C. difficile may be possible. Our group III contained isolates from humans, cattle, and pigs, suggesting that animals may be a source for C. difficile transmission to humans or vice versa.
Functional core of the C. difficile genome.
It is apparent from Fig. 1 that C. difficile, as a species, exhibits a high level of genomic variability. Of the 3,674 CDS spotted on the microarray slides, we found that 586 (16% of the genes in the strain 630 genome) were highly conserved in all strains, representing the core of the functional genes defining the species C. difficile. These genes are found outside the regions that appear to contain mobile or exogenously acquired DNA. The common genes in this study were classified and grouped with respect to their functional categories following the genome annotation by Bioinformatics Resource Center Pathema (http://pathema.jcvi.org/) (Table 1), and the list of common CDS in each functional category is shown in Table S4 in the supplemental material. Approximately, 20% (118/586) of the core CDS are classified as genes encoding hypothetical proteins, and the remainder are homologous to genes involved in housekeeping functions such as metabolism, biosynthesis, DNA replication, transcription, translation, transport, and cell division. We found 251 CDS specific to strain QCD-32g58 that are not present in the reference strain 630, although 14 of these were present in all of the other tested strains (see Table S5 in the supplemental material).
TABLE 1.
Expected functional categories of core components for all C. difficile strains and divergent genes according to host origin
| Role | Total no. of CDS | No. (%) of core CDS | No. (%) of divergent CDS from indicated host
|
|||
|---|---|---|---|---|---|---|
| Equine | Bovine | Swine | Human | |||
| Amino acid biosynthesis | 85 | 21 (24.7) | 31 (36.5) | 39 (45.9) | 22 (25.9) | 43 (50.6) |
| Biosynthesis of cofactors, prosthetic groups, and carriers | 88 | 17 (19.3) | 42 (47.7) | 44 (50.0) | 18 (20.5) | 59 (67.0) |
| Cell envelope | 206 | 43 (20.9) | 101 (49.0) | 117 (56.8) | 56 (27.2) | 126 (61.2) |
| Cellular processes | 178 | 30 (16.9) | 101 (56.7) | 112 (62.9) | 57 (32.0) | 118 (66.3) |
| Central intermediary metabolism | 98 | 17 (17.3) | 44 (44.9) | 58 (59.2) | 22 (22.4) | 65 (66.3) |
| DNA metabolism | 139 | 24 (17.3) | 86 (61.9) | 86 (61.9) | 40 (28.8) | 99 (71.2) |
| Energy metabolism | 259 | 55 (21.2) | 124 (47.9) | 130 (50.2) | 51 (19.7) | 164 (63.3) |
| Hypothetical protein | 920 | 118 (12.8) | 513 (55.8) | 632 (68.7) | 345 (37.5) | 665 (72.3) |
| Lipid metabolism | 37 | 7 (18.9) | 19 (51.4) | 20 (54.1) | 7 (18.9) | 22 (59.5) |
| Mobile and extrachromosomal element functions | 164 | 3 (1.8) | 152 (92.7) | 151 (92.1) | 143 (87.2) | 156 (95.1) |
| Purines, pyrimidines, nucleosides, and nucleotides | 59 | 6 (10.2) | 37 (62.7) | 36 (61.0) | 6 (10.2) | 43 (72.9) |
| Protein fate | 121 | 20 (16.5) | 64 (52.9) | 68 (56.2) | 25 (20.7) | 85 (70.2) |
| Protein synthesis | 124 | 26 (21.0) | 77 (62.1) | 60 (48.4) | 9 (7.3) | 81 (65.3) |
| Regulatory functions | 330 | 48 (14.5) | 164 (49.7) | 205 (62.1) | 96 (29.1) | 237 (71.8) |
| Signal transduction | 151 | 24 (15.9) | 70 (46.4) | 99 (65.6) | 43 (28.5) | 105 (69.5) |
| Transcription | 35 | 8 (22.9) | 17 (48.6) | 18 (51.4) | 7 (20.0) | 22 (62.9) |
| Transport and binding proteins | 392 | 76 (19.4) | 172 (43.9) | 230 (58.7) | 102 (26.0) | 250 (63.8) |
| Unknown function | 288 | 43 (14.9) | 141 (49.0) | 180 (62.5) | 80 (27.8) | 186 (64.6) |
| Total | 3,674 | 586 (15.9) | 1,955 (53.2) | 2,285 (62.2) | 1,129 (30.7) | 2,526 (68.8) |
A similarly low core gene content has previously been observed in C. difficile in a study by Stabler et al. (41) in which a similar number of strains and diversity of host origins was evaluated. Interestingly, only 153 genes were found to be conserved in both studies. A comparison of the log2-transformed signals from the single strain that was assayed on both platforms showed a pairwise correlation of 0.82, and the Cronbach's standardized α item reliability (3, 10) was calculated to be 0.9 (data not shown). This is remarkable, considering the differences between the platforms, and provides a measure of confidence that results obtained from the two different platforms should be comparable. It has also been reported that there is little gene conservation between C. difficile and other closely related clostridial species (38). This is in contrast to a high degree of genome conservation in species like Escherichia coli and Salmonella spp. To further confirm this, we performed a BLAST-P comparison of the C. difficile 630 genome with 40 other available clostridial genomes in RefSeq v29 (http://www.ncbi.nlm.nih.gov/RefSeq/). An E value threshold of 1e5 and 50% identities revealed only 57 CDS present across all clostridial spp. Most of these conserved genes were ribosomal proteins and genes coding essential metabolic functions. Because the lowest number of core genes reported so far is for Helicobacter pylori with an estimated core genome of 70% (16), the fact that C. difficile exhibited only 16% of the genome conservation is remarkable.
Interestingly, many potential regulatory genes, including 5/31 transcriptional antiterminators, 9/45 two-component system genes, and 13 phosphotransferase system genes, were found in all isolates tested. These proteins may play an important role in the monitoring of external environments and appropriate adaptation to corresponding conditions. Several gene clusters were common to all strains; for example, the CD1550 to CD1552 cluster is homologous to the hisBHA genes, which are responsible for the interconnection of histidine biosynthesis to nitrogen metabolism and the de novo biosynthesis of purines (8). This gene cluster is also conserved in a wide range of bacteria including other clostridia, such as C. perfringens and C. acetobutylicum (8).
CDS encoding potential transport and binding proteins were also conserved in all isolates tested (Table 1). An example is the gene cluster CD1591 to CD1593, which is homologous to the kdpABC operon encoding three cytoplasmic membrane proteins that form a potassium-transporting P-type ATPase system for the regulation of cytoplasmic potassium in response to osmotic stress in C. acetobutylicum (48). Homologs to the ferrous iron transport system feoAB (CD1478 to CD1479 and CD3274) were also found in all isolates examined. Iron is involved in a wide variety of biochemical processes in many microorganisms, and its limitation plays a pivotal role in host defense against infection by restricting bacterial replication (33). The impaired function of these transporters leads to decreased ferrous iron uptake and gut colonization by Escherichia coli in mice (42). These proteins may play a role in C. difficile virulence by facilitating colonization.
Genomic diversity in C. difficile.
The number of CDS divergent from strain 630 varied among the isolates, ranging from 52 (1.4%) in an Ontario bovine isolate (strain 670) to 1,169 (31.8%) in a human isolate (strain 6194) (Fig. 2). Taken together, ∼84.1% of the total number of CDS was absent or divergent. Our results showed a comparable degree of strain-to-strain variability compared to that of data previously reported (41). It is noteworthy that absent/divergent genes in the tested strains seemed to be distributed across the entire C. difficile 630 sequence and across all the predicted functional categories. The number of divergent CDS in C. difficile from different host origins in each functional category, as well as the percentage of genes in each category observed to be divergent, is shown in Table 1. As C. difficile possesses a large number of mobile genetic elements, including seven conjugative transposons and prophages (40), it is not surprising to find a high number of divergent CDS in the functional group comprising mobile and extrachromosomal elements.
Genes with host association.
In addition to the previously characterized regions, there were multiple regions throughout the genome of the reference strain 630 that contain uncharacterized putative proteins. These clusters are of interest because some of them represent genes which may have a host association. For example, the region including the CD1871 to CD1878B genes, which encodes hypothetical proteins (CD1871, CD1871A, CD1878, CD1878A, and CD1878B), putative membrane transporters (CD1872 to CD1875), and putative two-component system regulators (CD1876 and CD1877), is conserved among all bovine isolates from Canada but divergent in other isolates. The importance of two-component signal transduction systems in the response of C. difficile and other clostridia to environmental stimuli has been reviewed (25). They are involved in sensing cell envelope stress and regulate genes important for cell envelope integrity, detoxification, and virulence. This information may provide clues for host adaptation.
Variations in carbohydrate utilization patterns are traditionally used for the identification of bacteria (7). C. difficile is a heterotrophic anaerobe able to metabolize a wide range of carbohydrate substrates such as oligosaccharides and sugar alcohols. A region from CD0762 to CD0768 was present in all swine isolates, but was divergent among those with other origins. This locus is homologous to a gene system involved with glucitol metabolism in various bacteria including Clostridium spp. (44). Previous reports document the usefulness of glucitol fermentation patterns in the serotyping of C. difficile (12, 30). It may be possible that the ability to use glucitol provides advantages in the adaptation and survival of C. difficile in swine colonic environments.
Genes associated with virulence.
C. difficile colonizes the mucosal surfaces of the colon and is able to evade the early components of the host immune response to cause antibiotic-associated diarrhea, potentially leading to life-threatening disease (17). The genome sequence of strain 630 revealed several groups of genes that may be associated with virulence (40). Variations in pathogenicity may arise as a result of the uptake of genetic materials (horizontal gene transfer) that confer antibiotic resistance, toxin production, or adhesion to host cells or through gene loss during adaptation to a given environment (27). Some conserved CDS that may serve as potential virulence factors include six cell surface proteins (CD1469, CD1751, CD1987, CD2767, CD2784, and CD2799), one of the two fibronectin binding protein fbpA homologs that Stabler et al. (41) described as conserved (CD2592) (Fig. 3), and tellurium resistance protein homologs (CD1634, CD1652, and CD1799) (Fig. 4). The distribution of the CDS for known or potential virulent factors among C. difficile strains is shown in Fig. 5.
FIG. 5.
Distribution of the known or putative virulence-related CDS among C. difficile strains. Each row corresponds to a CDS, and each column in each panel represents a test strain. The status of each CDS is indicated by color as follows: red, present/conserved; green, absent; and gray, divergent. The designations of these CDS are indicated on the right. Intergenic regions are not shown.
(i) Flagella-related genes.
Flagella-associated proteins may play a role in intestinal colonization (45, 46). All bovine isolates from Ontario, but none from Arizona, retained most flagella-associated CDS. The first locus (CD0226 to CD0240) was present but divergent across all strains, but 7/7 bovine isolates from Arizona and 6/35 human isolates had a high level of divergence in the second locus (CD0245 to CD0271). Less than 80% of the complement of flagella-associated genes in human strains 5071, 5127, 5489, 6033, 6194, and 7020 were divergent. Taken together, these results support the hypothesis of Stabler et al. (41) that motility might not be required for the virulence of C. difficile in humans.
(ii) Toxin-related genes.
We compared the results of the PaLoc analysis by CGH with toxinotype. In this study, the tested strains were classified into 10 toxinotypes (0, III, IV, V, VIII, IX/XXIII, X/XVII, XII, XIV/XV, and XXII) or were nontoxigenic (Fig. 2). Microarray results suggested that both tcdA and tcdB were absent or highly divergent from four human isolates (5213, 5424, 6320, and 6461) and six equine isolates (109, 112, 123, 126, 538, and 539). However, only 2/4 human and 5/6 equine isolates appeared to be truly nontoxigenic. These human isolates were of toxinotypes X/XVII, both of which should exhibit a TcdA− TcdB+ phenotype (34). This anomaly might be due to divergence in tcdB in these strains, leading to reduced hybridization and failed signal detection. Furthermore, consistent with the microarray data, toxinotype VIII isolates, which contain a deletion in tcdA (34), included the bovine isolates 656 and 664 and human strains 5125 and 7076. Interestingly, all of the nontoxigenic isolates were obtained from humans or animals with diarrhea attributed to CDI. These findings suggest that symptomatic animals and humans may be simultaneously infected with both toxigenic and nontoxigenic C. difficile strains.
(iii) Cell surface proteins.
Most CDS in the capsule-related cluster (CD2769 to -CD2780) and the type IV pilus-associated loci (CD3294 to CD3297 and CD3503 to CD3513) were present across all strains with a low level of divergence. It was previously reported that both loci responsible for pilus biosynthesis are core components of the C. difficile genome (41). Certain virulence determinants, such as a putative collagen protease (CD1228) and a fibronectin binding protein, Fbp68 (CD2592), were also conserved across all strains, but others exhibited differing degrees of divergence. For instance, >80% of the strains possessed cysteine protease Cwp84 (CD2787), a putative S-layer protein precursor (CD2791), and a putative collagen-binding protein (CD2831), whereas only ∼30% possessed heat-shock-inducible adhesin Cwp66 (CD2789), the S-layer protein SlpA (CD2793), and a putative collagen-binding surface protein (CD3392). This is consistent with the known two-domain structure of SlpA and Cwp66; the extracellular domain is highly variable (9, 37) and in fact, the probes for these CDS were specific for the hypervariable regions of their respective proteins. In this instance, it is possible that the lack of signal indicates substantial divergence from the 630 sequence, rather than the absence of these conserved genes.
(iv) Antibiotic resistance genes.
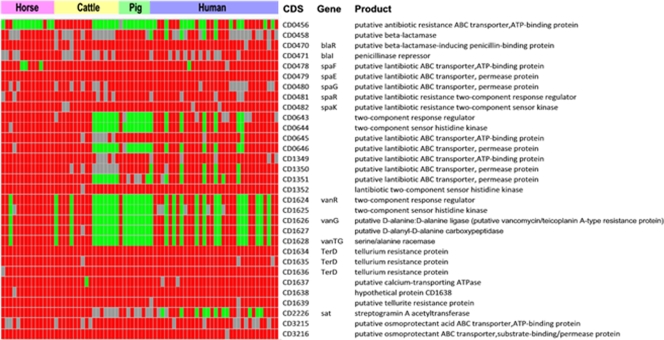
One contributing cause to the emergence of the NAP1/BI/027 strain is its increased resistance to antibiotics, including fluoroquinolones (6). A large repertoire of CDS potentially involved in antibiotic resistance has been identified (40). In our study, CDS potentially responsible for tellurium resistance showed a low level of divergence, and some (CD1634, CD1652, and CD1799) were conserved in most of the strains we tested (Fig. 6). Tellurium compounds are relatively rare in the environment, but many pathogenic bacteria possess tellurium resistance genes (47). These genes may encode the enzymes capable of utilizing tellurate and other metalloids as electron acceptors in anaerobic respiration (5). Three lantibiotic resistance homologs were identified (CD0478 to CD0482, CD0643 to CD0646, and CD1349 to CD1352). The first locus was divergent to a lower degree than the latter two loci, with a high level of divergence in swine and human isolates. Other CDS, including a daunomycin resistance homolog (CD0456), beta-lactam resistance homologs (CD0458, CD0470, and CD0471), and a streptogramin A acetyltransferase homolog (CD2226), exhibited different levels of divergence across the strains tested in this collection (Fig. 6). Thirty-one of 235 ATP-binding cassette (ABC) transporters were conserved. Most conserved ABC proteins in C. difficile are homologous to those responsible for the transport of essential nutrients such as amino acids and phosphate. Divergent or absent ABC transporters might account for the different antimicrobial resistance characteristics of this bacterium, since many ABC transporters are associated with multidrug resistance traits (23).
FIG. 6.
Patterns of CDS potentially associated with antibiotic resistance among C. difficile strains. Each row corresponds to a CDS, and each column in each panel represents a test strain. The status of each CDS is indicated by color as follows: red, present/conserved; green, absent; and gray, divergent. The designations of these CDS are indicated on the right. Intergenic regions are not shown.
Conclusion.
The use of CGH microarrays allowed us to gain a substantial amount of data that support epidemiologic evidence. The information obtained by CGH contributes important and novel information to the understanding of bacterial pathogenesis because gene content information derived from this study cannot be obtained with traditional typing techniques. However, the pitfall of CGH microarrays is that it generates information of a one-way character. Genes divergent in the tested genome that are present in the control strain can easily be detected, but genes that are unique to the tested strains cannot be monitored. In our analyses, the cutoff point was set at 100% estimated probability of presence, which ensured absolute confidence in the present CDS but could introduce some false negatives. Our CGH analyses revealed large genome plasticity and diversity among C. difficile isolates from various sources and certain genes with host-specific association. Due to a low level of gene conservation among C. difficile strains, a list of the C. difficile core genes is therefore valuable for future investigation strategies for C. difficile pathogenesis. The clustering of isolates based on microarray analyses, as well as PFGE, sheds light on host-specific adaptation and the possible interspecies transmission of C. difficile. We hope that this study will serve as the first step toward understanding the complex mechanisms underlying host adaptation and pathogenesis. The sequencing of additional C. difficile isolates from different host origins is warranted to further explore the genetic variability among them.
Supplementary Material
Acknowledgments
This project was supported with federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under contract N01-AI-30054, project no. ZC005-06.
We thank Dale Gerding and Andre Dascal for providing us with C. difficile strains 630 and 32g58, respectively. We thank L. Clifford McDonald and Michael J. Stanhope for helpful discussion.
Footnotes
Published ahead of print on 17 April 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.al-Barrak, A., J. Embil, B. Dyck, K. Olekson, D. Nicoll, M. Alfa, and A. Kabani. 1999. An outbreak of toxin A negative, toxin B positive Clostridium difficile-associated diarrhea in a Canadian tertiary-care hospital. Can. Commun. Dis. Rep. 2565-69. [PubMed] [Google Scholar]
- 2.Alfa, M. J., A. Kabani, D. Lyerly, S. Moncrief, L. M. Neville, A. Al-Barrak, G. K. Harding, B. Dyck, K. Olekson, and J. M. Embil. 2000. Characterization of a toxin A-negative, toxin B-positive strain of Clostridium difficile responsible for a nosocomial outbreak of Clostridium difficile-associated diarrhea. J. Clin. Microbiol. 382706-2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allen, M. J., and W. M. Yen (ed.). 2002. Introduction to measurement theory. Waveland Press, Long Grove, IL.
- 4.Arroyo, L. G., S. A. Kruth, B. M. Willey, H. R. Staempfli, D. E. Low, and J. S. Weese. 2005. PCR ribotyping of Clostridium difficile isolates originating from human and animal sources. J. Med. Microbiol. 54163-166. [DOI] [PubMed] [Google Scholar]
- 5.Baesman, S. M., T. D. Bullen, J. Dewald, D. Zhang, S. Curran, F. S. Islam, T. J. Beveridge, and R. S. Oremland. 2007. Formation of tellurium nanocrystals during anaerobic growth of bacteria that use Te oxyanions as respiratory electron acceptors. Appl. Environ. Microbiol. 732135-2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blossom, D. B., and L. C. McDonald. 2007. The challenges posed by reemerging Clostridium difficile infection. Clin. Infect. Dis. 45222-227. [DOI] [PubMed] [Google Scholar]
- 7.Bochner, B. R. 1989. Sleuthing out bacterial identities. Nature 339157-158. [DOI] [PubMed] [Google Scholar]
- 8.Brilli, M., and R. Fani. 2004. Molecular evolution of hisB genes. J. Mol. Evol. 58225-237. [DOI] [PubMed] [Google Scholar]
- 9.Calabi, E., S. Ward, B. Wren, T. Paxton, M. Panico, H. Morris, A. Dell, G. Dougan, and N. Fairweather. 2001. Molecular characterization of the surface layer proteins from Clostridium difficile. Mol. Microbiol. 401187-1199. [DOI] [PubMed] [Google Scholar]
- 10.Cronbach, L. J. 1951. Coefficient alpha and the internal structure of tests. Psychometrika 16297-334. [Google Scholar]
- 11.Curry, S. R., J. W. Marsh, C. A. Muto, M. M. O'Leary, A. W. Pasculle, and L. H. Harrison. 2007. tcdC genotypes associated with severe TcdC truncation in an epidemic clone and other strains of Clostridium difficile. J. Clin. Microbiol. 45215-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delmee, M., M. Homel, and G. Wauters. 1985. Serogrouping of Clostridium difficile strains by slide agglutination. J. Clin. Microbiol. 21323-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dineen, S. S., A. C. Villapakkam, J. T. Nordman, and A. L. Sonenshein. 2007. Repression of Clostridium difficile toxin gene expression by CodY. Mol. Microbiol. 66206-219. [DOI] [PubMed] [Google Scholar]
- 14.Dupuy, B., R. Govind, A. Antunes, and S. Matamouros. 2008. Clostridium difficile toxin synthesis is negatively regulated by TcdC. J. Med. Microbiol. 57685-689. [DOI] [PubMed] [Google Scholar]
- 15.Geric, B., R. J. Carman, M. Rupnik, C. W. Genheimer, S. P. Sambol, D. M. Lyerly, D. N. Gerding, and S. Johnson. 2006. Binary toxin-producing, large clostridial toxin-negative Clostridium difficile strains are enterotoxic but do not cause disease in hamsters. J. Infect. Dis. 1931143-1150. [DOI] [PubMed] [Google Scholar]
- 16.Gressmann, H., B. Linz, R. Ghai, K. P. Pleissner, R. Schlapbach, Y. Yamaoka, C. Kraft, S. Suerbaum, T. F. Meyer, and M. Achtman. 2005. Gain and loss of multiple genes during the evolution of Helicobacter pylori. PLoS Genet. 1e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hurley, B. W., and C. C. Nguyen. 2002. The spectrum of pseudomembranous enterocolitis and antibiotic-associated diarrhea. Arch. Intern. Med. 1622177-2184. [DOI] [PubMed] [Google Scholar]
- 18.Kelly, C. P., and J. T. LaMont. 1998. Clostridium difficile infection. Annu. Rev. Med. 49375-390. [DOI] [PubMed] [Google Scholar]
- 19.Killgore, G., A. Thompson, S. Johnson, J. Brazier, E. Kuijper, J. Pepin, E. H. Frost, P. Savelkoul, B. Nicholson, R. J. van den Berg, H. Kato, S. P. Sambol, W. Zukowski, C. Woods, B. Limbago, D. N. Gerding, and L. C. McDonald. 2008. Comparison of seven techniques for typing international epidemic strains of Clostridium difficile: restriction endonuclease analysis, pulsed-field gel electrophoresis, PCR-ribotyping, multilocus sequence typing, multilocus variable-number tandem-repeat analysis, amplified fragment length polymorphism, and surface layer protein A gene sequence typing. J. Clin. Microbiol. 46431-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim, C. C., E. A. Joyce, K. Chan, and S. Falkow. 2002. Improved analytical methods for microarray-based genome-composition analysis. Genome Biol. 3RESEARCH0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klaassen, C. H., H. A. van Haren, and A. M. Horrevorts. 2002. Molecular fingerprinting of Clostridium difficile isolates: pulsed-field gel electrophoresis versus amplified fragment length polymorphism. J. Clin. Microbiol. 40101-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loo, V. G., L. Poirier, M. A. Miller, M. Oughton, M. D. Libman, S. Michaud, A. M. Bourgault, T. Nguyen, C. Frenette, M. Kelly, A. Vibien, P. Brassard, S. Fenn, K. Dewar, T. J. Hudson, R. Horn, P. Rene, Y. Monczak, and A. Dascal. 2005. A predominantly clonal multi-institutional outbreak of Clostridium difficile-associated diarrhea with high morbidity and mortality. N. Engl. J. Med. 3532442-2449. [DOI] [PubMed] [Google Scholar]
- 23.Lubelski, J., W. N. Konings, and A. J. Driessen. 2007. Distribution and physiology of ABC-type transporters contributing to multidrug resistance in bacteria. Microbiol. Mol. Biol. Rev. 71463-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lyons, P. 2003. Advances in spotted microarray resources for expression profiling. Brief. Funct. Genomic. Proteomic. 221-30. [DOI] [PubMed] [Google Scholar]
- 25.Mascher, T. 2006. Intramembrane-sensing histidine kinases: a new family of cell envelope stress sensors in Firmicutes bacteria. FEMS Microbiol. Lett. 264133-144. [DOI] [PubMed] [Google Scholar]
- 26.Matamouros, S., P. England, and B. Dupuy. 2007. Clostridium difficile toxin expression is inhibited by the novel regulator TcdC. Mol. Microbiol. 641274-1288. [DOI] [PubMed] [Google Scholar]
- 27.Maurelli, A. T., R. E. Fernandez, C. A. Bloch, C. K. Rode, and A. Fasano. 1998. “Black holes” and bacterial pathogenicity: a large genomic deletion that enhances the virulence of Shigella spp. and enteroinvasive Escherichia coli. Proc. Natl. Acad. Sci. USA 953943-3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McDonald, L. C., G. E. Killgore, A. Thompson, R. C. Owens, Jr., S. V. Kazakova, S. P. Sambol, S. Johnson, and D. N. Gerding. 2005. An epidemic, toxin gene-variant strain of Clostridium difficile. N. Engl. J. Med. 3532433-2441. [DOI] [PubMed] [Google Scholar]
- 29.McDonald, L. C., M. Owings, and D. B. Jernigan. 2006. Clostridium difficile infection in patients discharged from US short-stay hospitals, 1996-2003. Emerg. Infect. Dis. 12409-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakamura, S., M. Mikawa, S. Nakashio, M. Takabatake, I. Okado, K. Yamakawa, T. Serikawa, S. Okumura, and S. Nishida. 1981. Isolation of Clostridium difficile from the feces and the antibody in sera of young and elderly adults. Microbiol. Immunol. 25345-351. [DOI] [PubMed] [Google Scholar]
- 31.O'Neill, G., J. E. Adams, R. A. Bowman, and T. V. Riley. 1993. A molecular characterization of Clostridium difficile isolates from humans, animals and their environments. Epidemiol. Infect. 111257-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Palaniappan, R. U., Y. Zhang, D. Chiu, A. Torres, C. Debroy, T. S. Whittam, and Y. F. Chang. 2006. Differentiation of Escherichia coli pathotypes by oligonucleotide spotted array. J. Clin. Microbiol. 441495-1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ratledge, C., and L. G. Dover. 2000. Iron metabolism in pathogenic bacteria. Annu. Rev. Microbiol. 54881-941. [DOI] [PubMed] [Google Scholar]
- 34.Rupnik, M. 2008. Heterogeneity of large clostridial toxins: importance of Clostridium difficile toxinotypes. FEMS Microbiol. Rev. 32541-555. [DOI] [PubMed] [Google Scholar]
- 35.Rupnik, M., V. Avesani, M. Janc, C. von Eichel-Streiber, and M. Delmee. 1998. A novel toxinotyping scheme and correlation of toxinotypes with serogroups of Clostridium difficile isolates. J. Clin. Microbiol. 362240-2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rupnik, M., V. Braun, F. Soehn, M. Janc, M. Hofstetter, R. Laufenberg-Feldmann, and C. von Eichel-Streiber. 1997. Characterization of polymorphisms in the toxin A and B genes of Clostridium difficile. FEMS Microbiol. Lett. 148197-202. [DOI] [PubMed] [Google Scholar]
- 37.Savariau-Lacomme, M. P., C. Lebarbier, T. Karjalainen, A. Collignon, and C. Janoir. 2003. Transcription and analysis of polymorphism in a cluster of genes encoding surface-associated proteins of Clostridium difficile. J. Bacteriol. 1854461-4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scaria, J., R. U. Palaniappan, D. Chiu, J. A. Phan, L. Ponnala, P. McDonough, Y. T. Grohn, S. Porwollik, M. McClelland, C. S. Chiou, C. Chu, and Y. F. Chang. 2008. Microarray for molecular typing of Salmonella enterica serovars. Mol. Cell. Probes 22238-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scaria, J., A. Sreedharan, and Y. F. Chang. 2008. Microbial Diagnostic Array Workstation (MDAW): a web server for diagnostic array data storage, sharing and analysis. Source Code Biol. Med. 314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sebaihia, M., B. W. Wren, P. Mullany, N. F. Fairweather, N. Minton, R. Stabler, N. R. Thomson, A. P. Roberts, A. M. Cerdeno-Tarraga, H. Wang, M. T. Holden, A. Wright, C. Churcher, M. A. Quail, S. Baker, N. Bason, K. Brooks, T. Chillingworth, A. Cronin, P. Davis, L. Dowd, A. Fraser, T. Feltwell, Z. Hance, S. Holroyd, K. Jagels, S. Moule, K. Mungall, C. Price, E. Rabbinowitsch, S. Sharp, M. Simmonds, K. Stevens, L. Unwin, S. Whithead, B. Dupuy, G. Dougan, B. Barrell, and J. Parkhill. 2006. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat. Genet. 38779-786. [DOI] [PubMed] [Google Scholar]
- 41.Stabler, R. A., D. N. Gerding, J. G. Songer, D. Drudy, J. S. Brazier, H. T. Trinh, A. A. Witney, J. Hinds, and B. W. Wren. 2006. Comparative phylogenomics of Clostridium difficile reveals clade specificity and microevolution of hypervirulent strains. J. Bacteriol. 1887297-7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stojiljkovic, I., M. Cobeljic, and K. Hantke. 1993. Escherichia coli K-12 ferrous iron uptake mutants are impaired in their ability to colonize the mouse intestine. FEMS Microbiol. Lett. 108111-115. [DOI] [PubMed] [Google Scholar]
- 43.Stubbs, S., M. Rupnik, M. Gibert, J. Brazier, B. Duerden, and M. Popoff. 2000. Production of actin-specific ADP-ribosyltransferase (binary toxin) by strains of Clostridium difficile. FEMS Microbiol. Lett. 186307-312. [DOI] [PubMed] [Google Scholar]
- 44.Tangney, M., J. K. Brehm, N. P. Minton, and W. J. Mitchell. 1998. A gene system for glucitol transport and metabolism in Clostridium beijerinckii NCIMB 8052. Appl. Environ. Microbiol. 641612-1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tasteyre, A., M. C. Barc, T. Karjalainen, P. Dodson, S. Hyde, P. Bourlioux, and P. Borriello. 2000. A Clostridium difficile gene encoding flagellin. Microbiology 146957-966. [DOI] [PubMed] [Google Scholar]
- 46.Tasteyre, A., T. Karjalainen, V. Avesani, M. Delmee, A. Collignon, P. Bourlioux, and M. C. Barc. 2001. Molecular characterization of fliD gene encoding flagellar cap and its expression among Clostridium difficile isolates from different serogroups. J. Clin. Microbiol. 391178-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taylor, D. E. 1999. Bacterial tellurite resistance. Trends Microbiol. 7111-115. [DOI] [PubMed] [Google Scholar]
- 48.Treuner-Lange, A., A. Kuhn, and P. Durre. 1997. The kdp system of Clostridium acetobutylicum: cloning, sequencing, and transcriptional regulation in response to potassium concentration. J. Bacteriol. 1794501-4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Voth, D. E., and J. D. Ballard. 2005. Clostridium difficile toxins: mechanism of action and role in disease. Clin. Microbiol. Rev. 18247-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.