Abstract
The segrosome is the nucleoprotein complex that mediates accurate plasmid segregation. In addition to its multifunctional role in segrosome assembly, the ParG protein of multiresistance plasmid TP228 is a transcriptional repressor of the parFG partition genes. ParG is a homodimeric DNA binding protein, with C-terminal regions that interlock into a ribbon-helix-helix fold. Antiparallel β-strands in this fold are presumed to insert into the OF operator major groove to exert transcriptional control as established for other ribbon-helix-helix factors. The OF locus comprises eight degenerate tetramer boxes arranged in a combination of direct and inverted orientation. Each tetramer motif likely recruits one ParG dimer, implying that the fully bound operator is cooperatively coated by up to eight dimers. OF was subdivided experimentally into four overlapping 20-bp sites (A to D), each of which comprises two tetramer boxes separated by AT-rich spacers. Extensive interaction studies demonstrated that sites A to D individually are bound with different affinities by ParG (C > A ≈ B ≫ D). Moreover, comprehensive scanning mutagenesis revealed the contribution of each position in the site core and flanking sequences to ParG binding. Natural variations in the tetramer box motifs and in the interbox spacers, as well as in flanking sequences, each influence ParG binding. The OF operator apparently has evolved with sites that bind ParG dissimilarly to produce a nucleoprotein complex fine-tuned for optimal interaction with the transcription machinery. The association of other ribbon-helix-helix proteins with complex recognition sites similarly may be modulated by natural sequence variations between subsites.
The accurate segregation of chromosomal and extrachromosomal DNA requires specialized molecular machines, both in procaryotes and in eucaryotes (8, 19, 38, 41). For bacterial low-copy-number plasmids, the segrosome is the nucleoprotein complex that ensures their precise segregation to daughter cells at cytokinesis (13, 19, 20, 38, 39). Segrosomes can be categorized into subtypes based on their molecular components (39), but the best-characterized complexes consist of either a Walker (ParA) or actin-type ATPase, a cis-acting centromere site, and a centromere binding factor. ATP-mediated assembly of the actin-like ATPase into a bipolar spindle elicits bidirectional polymer growth, forcing the attached plasmids in opposite directions (15). Evidence is accumulating that the widespread ParA-type plasmid segregation ATPases also polymerize in response to nucleotide binding and that this polymerization mediates plasmid segregation. However, the molecular mechanisms that underpin this behavior and how plasmid segregation is achieved remain to be fully unraveled (1, 3, 5, 12, 14, 26, 27).
The tripartite segrosome of multiresistance plasmid TP228 consists of the ParA homolog, ParF, and the ParG centromere binding protein, which assemble on the parH centromere (4, 18). ParF (22.0 kDa) is a member of the ParA superfamily of segregation proteins that are widely encoded by eubacterial and archaeal chromosomes and plasmids. Like its homologs, ParF is a weak ATPase whose nucleotide hydrolysis is enhanced by the partner protein, ParG (5). Strikingly, ATP binding promotes the polymerization of ParF into extensive multistranded filaments. ParG (8.6 kDa) enhances ParF polymerization independently of ATP but also superstimulates filamentation in the presence of ATP. In contrast, ParF polymerization is blocked by ADP (3, 5). This suggests that ParF action during partitioning may involve a cycle of polymerization and depolymerization in which, following segrosome formation at the centromere, the binding of ATP and ParG initially augments ParF filamentation. Stimulation of ParF nucleotide hydrolysis by ParG subsequently may induce the formation of ParF-ADP species within polymers, blocking further filament growth. Pushing of plasmids by ParF polymer extension or plasmid pulling by filament depolymerization may drive replicated plasmids to either side of the septal plane (5).
ParG is a homodimeric DNA binding protein (4) with C-terminal regions that interweave into a ribbon-helix-helix (RHH) fold and mobile N-terminal tails (16). Dimerization, DNA binding at the operator site, and interaction with ParF are mediated by the C-terminal regions (3, 7, 16). The flexible N termini are also multifunctional. First, the N-terminal tail of ParG includes an arginine finger-like motif that enhances ATP hydrolysis by ParF (3). The motif may be part of a semiflexible loop that intercalates into the ParF nucleotide binding pocket, analogous to arginine fingers in proteins such as human Ras-GAPs (2, 31). Arginine finger loops stabilize the transition state during nucleotide hydrolysis by their partner proteins (6), and the same may be the case with ParF-ParG. Stimulation of nucleotide hydrolysis by ParG may be an important step in the ParF polymerization-depolymerization cycle (3). Second, and separately from its role in ATPase enhancement, the ParG mobile tail is required for stimulation of ParF polymerization (3). ParG may either bundle filaments more extensively or stabilize ParF protomers within filaments (3, 5). The flexible tails within each ParG dimer potentially enwrap ParF monomers, either on the same or on adjacent protofilaments, or might act at points of polymer disassembly. Thus, ParG may be functionally analogous to microtubule-associated proteins that regulate tubulin kinetics or to formins and other factors that modulate growth and retraction of eucaryotic actin filaments (27). Third, ParG is a transcriptional repressor of the parFG genes (7), with antiparallel β-strands thought to insert into the major groove at the operator site as established for other RHH proteins (17, 34, 36, 42). The tail modulates the interaction of ParG with the operator site (here designated OF) during autoregulation. Specifically, a transient β-strand element in the tail that associates with the RHH domain is implicated in formation of the repressive nucleoprotein complex (7). Here, we probe further the interaction between ParG and the operator: the locus comprises a complex set of related motifs that ParG recognizes with different affinities, suggesting that the operator has evolved with sites that bind ParG dissimilarly to generate a nucleoprotein complex that is optimized for parFG regulation.
MATERIALS AND METHODS
Strains, growth medium, and protein purification.
Escherichia coli was grown at 37°C in Luria-Bertani medium with ampicillin (100 μg/ml) and/or kanamycin (50 μg/ml) when required for plasmid maintenance. Strain DH5α (43) was used for molecular cloning, and strain BL21(DE3) (Novagen) was used for ParG overproduction and transcriptional fusion assays. His-tagged ParG was purified by Ni2+ affinity chromatography as described elsewhere (4).
CDO reporter assays.
Transcriptional fusions of the wild-type parFG promoter-operator region (ABCD) and the same region in which site B was disrupted (AXCD, where X indicates the mutated site within OF) to a xylE reporter gene in plasmid pDM3.0 have been described previously (7). Equivalent fusions bearing sites XBCD, ABXD, and ABCX were constructed by annealing four overlapping, single-stranded oligonucleotides in each case (121 to 124; 125 to 128; and 125, 126, 129, and 130, respectively) (see Table S1 in the supplemental material) and cloning the assembled fragments in the BamHI site of pDM3.0. The sequences of the cloned inserts were verified. Catechol 2,3-dioxygenase (CDO) assays were performed essentially as outlined elsewhere (7). Briefly, E. coli BL21(DE3) was cotransformed with a pDM3.0 derivative bearing the xylE transcriptional fusion of interest and with pET22b or this vector expressing parG. Colonies were inoculated in Luria-Bertani broth under selective conditions and grown at 37°C until the A600 was ∼0.5. After growth for an additional 60 min, cells were pelleted, resuspended in 100 mM potassium phosphate buffer (pH 7.4) with 10% acetone, and sonicated. The cell lysate was clarified by centrifugation, and protein concentrations were estimated using the Bio-Rad protein assay. CDO activity was measured by monitoring the A375 change for 40 s at 24°C with 0.2 mM catechol. Measurements were performed at least in duplicate. One CDO unit is the amount of enzyme that oxidizes 1 μmol catechol/min at 24°C.
DNase I footprinting.
A 231-bp fragment covering the parFG promoter region, including sites ABCD, and 30 bp downstream of the parF translational start was amplified from plasmid pDM-Oper (7) using oligonucleotides 17/18 (see Table S1 in the supplemental material). Oligonucleotides carrying sites A, B, C, D, or CD (19/20, 21/22, 23/24, 25/26, and 27/28, respectively) were cloned separately between the EcoRI and BamHI sites of pUC18 (32). The nucleotide sequences of the inserts were confirmed. The recombinant plasmids were used as templates in PCRs with primers 15/16 (see Table S1 in the supplemental material) to generate fragments (215 to 239 bp) in which either the top or bottom strand was 5′ biotinylated. PCR products were electrophoresed on 10% polyacrylamide gels in 0.5× Tris-borate-EDTA (TBE) buffer. Fragments were excised from gels and electroeluted in 0.5× TBE for 30 min at 100 V. The DNA was concentrated and purified with the QIAquick PCR purification kit (Qiagen) and resuspended in 50 μl sterile water. Footprinting reactions were performed essentially as described previously (22). Briefly, reaction mixtures containing biotinylated DNA (5 nM final concentration) and ParG (0.1 to 5 μM) were mixed in binding buffer [10 mM Tris-HCl, pH 7.5, 50 mM KCl, 1 mM dithiothreitol, 5 mM MgCl2, 0.05 mg/ml poly(dI-dC)] in final volumes of 20 μl and incubated for 20 min at 25°C. Samples were treated with 0.0075 units of DNase I (Roche) diluted in 20 mM Tris-HCl, pH 7.5, 50 mM NaCl, 7.5 mM MgCl2, 5 mM CaCl2 for 45 s at 25°C. Reactions were stopped by addition of 200 μl of 10 mM EDTA, pH 7.0, 300 mM sodium acetate, followed by extraction with an equal volume of phenol-chloroform (1:1). The upper phase was collected, and 1 μl of glycogen (20 mg/ml; Roche) and 500 μl of ethanol were added. The DNA was precipitated at −80°C for 30 min and harvested by centrifugation, and the pellets were washed with 70% ethanol. Pellets were dried, resuspended in 10 μl of loading buffer (95% formamide, 20 mM EDTA, 0.05% bromophenol blue, 0.05% xylene cyanol), and heated at 99°C for 10 min. Reaction mixtures were electrophoresed on equilibrated 6% sequencing gels (SequaGel [GeneFlow]) at 60 W in 1× TBE buffer for 2 h. DNA was transferred by capillary action to positively charged nylon membranes (Roche), and the transferred DNA fragments were immobilized by UV cross-linking. Detection of the biotin end-labeled DNA was performed using the LightShift chemiluminescent electrophoretic mobility shift assay (EMSA) kit (Pierce) (4).
EMSAs.
5′ biotinylated DNA substrates were obtained by PCR using labeled primers or by annealing complementary oligonucleotides (see Table S1 in the supplemental material). The resulting fragments were electrophoresed on 10% polyacrylamide gels in 0.5× TBE buffer followed by electroelution and further purification as described above. EMSAs with the intact operator DNA (amplified from plasmid pDM-Oper using oligonucleotides 13/14) or with variants possessing sites XBCD, AXCD, ABXD, and ABCX (prepared by annealing oligonucleotides 3/4, 7/8, 1/2, and 5/6, respectively) and XXCD or XXXD (amplified from pUC18 derivatives with primers 9/10 and 11/12, respectively) were performed with 98-bp substrates. In EMSA with single repeat motifs, synthetic 50-bp oligonucleotides carrying site A, B, C, or D (oligonucleotides 19/20, 21/22, 23/24, and 25/26, respectively) or site C in which the right half-site was inverted (Cinv) (oligonucleotides 29/30) were used. For scanning mutagenesis, a 48-bp synthetic, double-stranded fragment (oligonucleotides 33/34) corresponding to sites AB and a replica of this oligonucleotide in which site B was randomized (site AX; oligonucleotides 31/32) were employed as core sequences from which mutated sites, each carrying either a single substitution in one site or paired substitutions in two sites, were derived. Oligonucleotides 37/38 through 119/120 (see Table S1 in the supplemental material) carried the mutated sites. Biotinylated DNA (2 nM final concentration) was incubated at 25°C for 20 min in binding buffer [10 mM Tris-HCl, pH 7.5, 50 mM KCl, 1 mM dithiothreitol, 5 mM MgCl2, 0.05 mg/ml poly(dI-dC)] with ParG (concentrations given in figure legends). Reaction mixtures were electrophoresed on 10% polyacrylamide gels in 0.5× TBE buffer for 90 min at 80 V at 22°C. Gels were processed further as outlined above. Images were captured on FujiFilm RX NIF film and scanned into a digital format. The intensity of each band was determined using Gel-Pro Analyzer 3.1 (Media Cybernetics) software after subtracting background from a blank area adjacent to each sample band. ParG affinity for different DNA sites was determined as the apparent equilibrium dissociation constant Kapp (defined as the concentration of protein required to generate 50% occupancy), which was estimated from binding curves, similar to previous analysis of the MetJ RHH protein (21). The term Kapp is used because protein concentration and activity (the effective concentration) are different (29) due to the use of a competitor in binding reactions (24, 25). Results shown in figures are representative images of experiments performed at least in duplicate.
SPR.
Surface plasmon resonance (SPR) measurements were obtained with a Biacore 3000 instrument (Biacore AB) primed with immobilization buffer (10 mM Tris-HCl, pH 7.5, 200 mM NaCl, 1 mM EDTA). Streptavidin (SA) chips (Biacore) were conditioned with three consecutive 2-minute injections of activation buffer (1 M NaCl, 50 mM NaOH) prior to immobilization of biotinylated DNA (2 nM injected at 10 μl/min). Oligonucleotides (50 bp) possessing sites A to D were bound to the surface of the chip with similar response units (∼350) to allow comparison of results with different substrates. An oligonucleotide with an identical base composition but with an unrelated sequence was immobilized in one flow cell as a reference. The SA chips were primed with running buffer (10 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5 mM MgCl2, 1 mM dithiothreitol) prior to injection of proteins (100 nM) in running buffer at a flow rate of 50 μl/min at 25°C. The chip surface was regenerated by injection (15 μl) of 2 M guanidine-HCl (25 μl/min) after each protein sample. Data were reference subtracted using the oligonucleotide of unrelated sequence (oligonucleotides 35/36; see Table S1 in the supplemental material) and analyzed using BIAevaluation 3.1 software (Biacore AB). None of the kinetic models proposed by the software gave a satisfactory close curve-fitting for the interactions, thereby excluding them from derivation of binding constants for the ParG-DNA associations.
RESULTS
Refinement of the OF operator site boundaries.
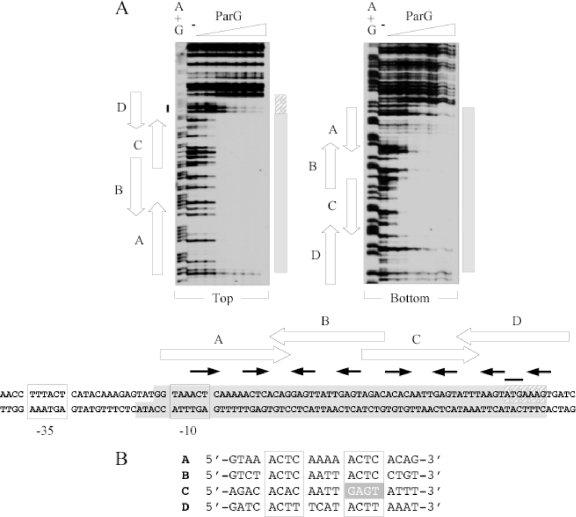
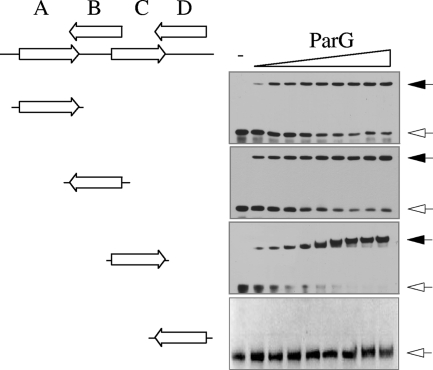
Based on in silico analysis, we recently proposed that the operator site (here designated OF) for negative autoregulation of parFG expression comprised an imperfect inverted repeat (IR) that overlaps the putative −10 promoter box (7). Disruption of the IR abolished ParG-mediated repression, establishing the repeat as a regulatory site. To determine the boundaries of the region contacted by ParG, a 231-bp fragment that spans the parFG regulatory region was subjected to DNase I footprinting in the presence of ParG (Fig. 1A). The protein protected a region on both strands from digestion that was almost twice the length of the IR previously identified. More specifically, protection included the IR, as well as the entire region between the repeat and the parF translation start codon. Examination of the nucleotide sequence of the protected region revealed eight 5′-ACTC-3′ (or variant) tetramer motifs separated by 4-bp AT-rich spacers. Three of the tetramers are in direct orientation, whereas five of the motifs are inverted. A single tetramer box is insufficient for ParG binding in EMSA (see Fig. 8), but two appropriately spaced boxes produce a single nucleoprotein complex with the protein (7). This correlates with observations with other RHH proteins which assemble on adjoining sites as dimers of dimers (36). Thus, the tetramer boxes in OF can be arranged functionally as sites A to D: sites A and B correspond approximately to the IR previously identified (7), each consisting of a pair of 5′-ACTC-3′ tetramer motifs spaced by 4-bp AT-rich tracts and flanked by 4 bp on either side. In the case of site D, the motifs are 5′-ACTT-3′, whereas in site C the motifs are 5′-ACAC-3′ and its imperfect inverted counterpart 5′-GAGT-3′ (Fig. 1B).
FIG. 1.
DNase I footprinting of the OF locus. (A) Footprinting reactions were performed as outlined in Materials and Methods using PCR fragments biotinylated at the 5′ ends of either top or bottom strands. ParG concentrations (μM monomer, left to right): 0, 0.05, 0.1, 0.2, 0.4, 0.6, 0.8, and 1.0. The locations of sites A to D are marked by open arrows. Tetramer boxes within the sites are indicated by filled arrows. Shaded boxes denote the regions protected from DNase I digestion by ParG. The adjoining hatched box indicates the segment of site D that is only partially protected from DNase I digestion by ParG. The black line denotes the position of the parF translation start codon. A+G, Maxam-Gilbert sequencing reactions. The relative dispositions on the top and bottom strands of the parFG promoter-operator region that are protected from DNase I digestion are illustrated in the bottom panel. Putative −10 and −35 promoter motifs are boxed. (B) Alignment (5′ to 3′) of the top strands of sites A and C and the bottom strands of sites B and D. The degenerate 5′-ACTC-3′ tetramer motifs are boxed. The inverted motif in site C is shaded.
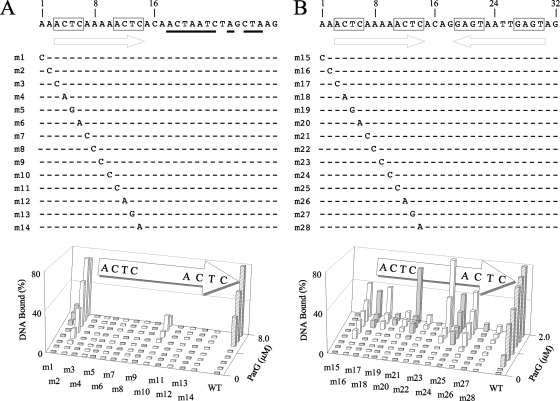
FIG. 8.
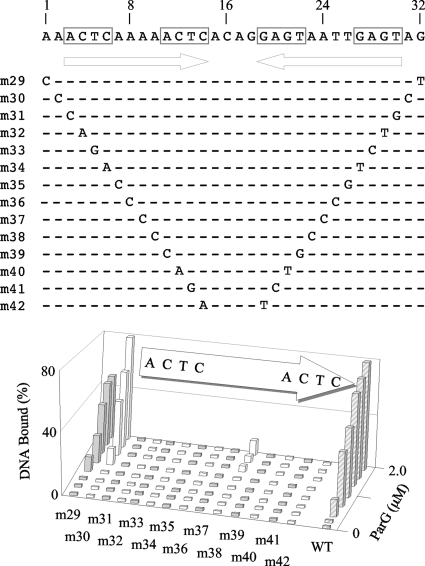
Scanning mutagenesis of the A site flanked either by randomized sequences (A) or by site B (B). Randomized nucleotides are underlined. Purine-to-pyrimidine and pyrimidine-to-purine substitution mutations at each position within the 12-bp site A, as well as at two flanking base pairs, were studied by EMSA using 48-bp oligonucleotides in both contexts. ParG concentrations up to 8 and 2 μM were used in panels A and B, respectively. Data quantitation using Gel-Pro Analyzer 3.1 (Media Cybernetics) software is shown. The results shown are representative of experiments performed at least in duplicate. WT, wild type.
Close inspection of the DNase I footprinting patterns with OF suggests that site C was protected from digestion at slightly lower ParG concentrations than were the other sites, most obviously on the bottom DNA strand (Fig. 1A). Conversely, site D required larger amounts of ParG for protection and was not fully protected on the top strand even at the highest protein concentration that was tested.
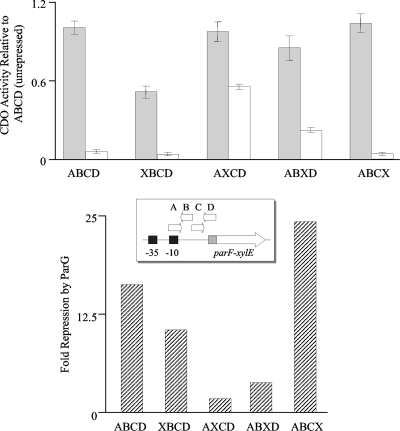
In view of the extension of the OF boundaries described above, multiple mutations were introduced separately into each of the sites A to D, and the effects of the alterations on transcriptional repression by ParG were assessed by fusing the mutated promoter-operator regions and the 5′ end of parF to a promoterless xylE cassette in plasmid pDM3.0 as described previously (7). The unrepressed promoter activities were similar for transcriptional fusions bearing the wild-type and each of the mutated operator regions, except for the fusion possessing the XBCD operator, which exhibited ∼50% of the CDO activity of the unmutated site (Fig. 2). It is likely that the parFG promoter sequences overlap site A within the operator (Fig. 1A), causing a reduction in basal promoter levels when the latter is mutated. Transcriptional fusions bearing the wild-type and AXCD operators were repressed ∼16- and ∼2-fold, respectively, by ParG provided in trans (Fig. 2), which correlates well with previous data (7). Disruption of sites A (XBCD) and C (ABXD) in OF also decreased transcriptional repression by ParG to ∼10- and 3.8-fold, respectively. In contrast, a transcriptional fusion containing multiple mutations in site D was repressed ∼24-fold by ParG. The reason for this apparent increase in repression remains to be investigated. In summary, transcriptional fusion studies revealed that sites A, B, and C are required for full repression by ParG at the OF operator. Moreover, disruption of the different sites elicited different effects on repression. The role of site D in vivo remains unclear, although, as described below, it is bound specifically by ParG in vitro.
FIG. 2.
Repression analysis of the wild-type parFG promoter-operator region (ABCD) and the same region bearing disruptions of one of the sites within the OF operator (XBCD, AXCD, ABXD, and ABCX) assessed by CDO activity of parF-xylE transcriptional fusions. The top panel shows CDO activities relative to the levels of the unrepressed wild-type region for each fusion in the absence (filled bars) and presence (open bars) of ParG provided in trans. The inset shows a schematic representation of the parF-xylE constructs. The bottom panel illustrates the repression by ParG of each transcriptional fusion relative to its unrepressed levels.
Effect of site replacement on OF binding by ParG.
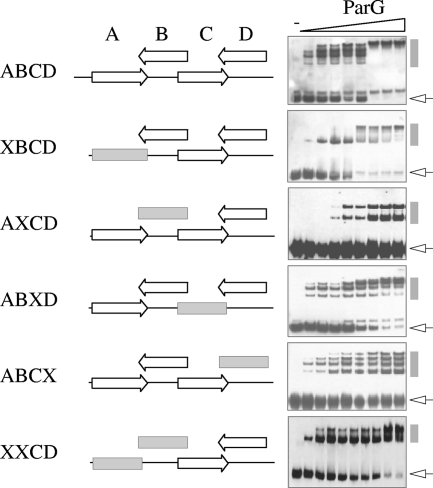
ParG loads on a substrate bearing sites AB initially as a tetramer (dimer of dimers) and, at elevated protein concentrations, as a pair of tetramers. This behavior is reflected in the formation of two nucleoprotein complexes with distinct migrations in EMSA (7). Analogously, ParG initially formed one major complex with sites CD, with a second complex produced at higher ParG concentrations (XXCD in Fig. 3). A fragment containing the complete OF site (repeats ABCD) formed four principal species, and some minor complexes, with ParG (Fig. 3). The DNA was entirely assembled into the slowest-migrating complex at the highest protein concentrations. It is tempting to speculate that the principal nucleoprotein complexes arise from increasing occupation of the four sites by a ParG dimer of dimers, each species formed by the sequential addition of a pair of dimers to a preexisting complex. OF fragments in which any one of the four sites was replaced by a random sequence showed altered binding patterns with ParG (Fig. 3). Although removal of site A (XBCD) did not completely prevent binding by the protein, only a single intermediate complex was evident. Similarly, fragment ABXD was almost fully bound at the highest ParG concentrations tested, although multiple intermediate complexes were apparent. Moreover, the pattern of complex formation differed from that observed with the intact OF site, with faster-migrating complexes persisting at ParG concentrations at which OF was fully bound. Two nucleoprotein complexes with approximately equimolar concentrations were evident when site B was replaced (AXCD), but binding was incomplete, with a significant fraction of the DNA unbound even at the highest ParG concentration tested. Finally, the protein generated a ladder of complexes of similar intensities with fragment ABCX. In summary, the variable and complex ParG binding patterns observed with the OF operator lacking any single site demonstrate that all four of the sites are required for proper ParG binding in vitro and that removal of any one site perturbs the normal pattern of complex formation observed with the intact OF locus.
FIG. 3.
EMSA of the full-length operator OF (ABCD) and variants in which one site was replaced by a random sequence (designated X and marked by a gray horizontal box). Biotinylated oligonucleotides (98 bp) (see Table S1 in the supplemental material) were tested as described in Materials and Methods using ParG concentrations (μM monomer, left to right): 0, 0.2, 0.4, 0.6, 0.8, 1.0, 1.2, 1.4, and 1.6. Unbound DNAs and nucleoprotein complexes are indicated by open arrows and gray vertical bars, respectively. A second species that migrates close to the unbound DNA in some blots in this and other figures is likely to be the same fragment with an atypical secondary structure.
ParG binds the OF sites with different avidities.
The analyses presented in Fig. 2 and 3 suggest that the four OF sites and the tetramer boxes within them may be recognized with different affinities by ParG, leading to the diverse patterns observed in EMSA with DNA fragments possessing any three of the sites. Sites A to D were tested further individually by EMSA, DNase I footprinting, and SPR. A double-stranded oligonucleotide bearing site C was bound entirely into a single nucleoprotein complex by ParG (Fig. 4). Similarly, repeats A and B each formed a single complex with ParG, although <75% of the DNA was assembled into a complex at the highest ParG concentration that was tested. Site D failed to form a complex with ParG in repeated experiments, suggesting either that this repeat is bound poorly by the protein or that the complex is unstable during electrophoresis. Kapp values revealed that the affinity of ParG for these sites was in the order C > A ≈ B ≫ D.
FIG. 4.
EMSA of isolated OF sites. Biotinylated oligonucleotides (50 bp) (see Table S1 in the supplemental material) were tested as described in Materials and Methods using ParG concentrations (μM monomer, left to right): 0, 0.5, 0.75, 1.0, 1.5, 2.0, 2.5, 3.0, 3.5, and 4.0, except for site D from which the 0.75 μM concentration was omitted The oligonucleotides include the 12-bp site cores flanked on either side by 4 bp from the site's normal context (Fig. 1). Unbound DNAs and nucleoprotein complexes are indicated by open and filled arrows, respectively.
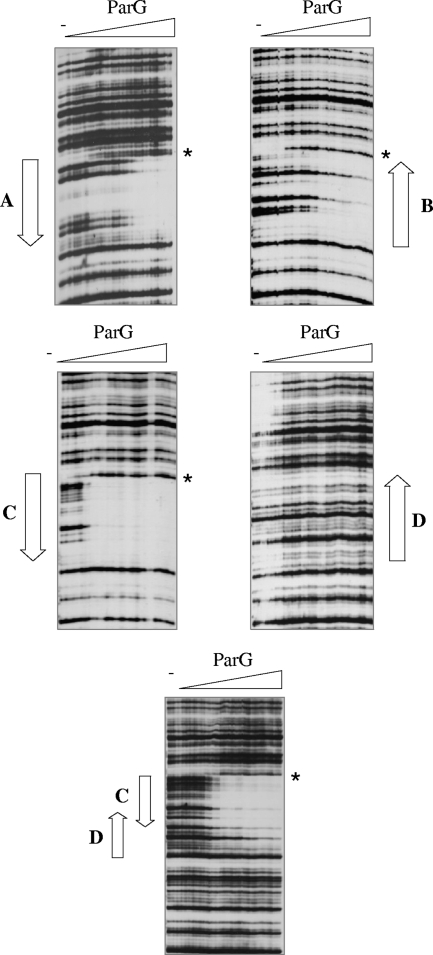
DNase I footprinting was employed to investigate further the interaction between ParG and the OF sites. Each of the repeats was cloned individually in plasmid pUC18. The recombinant plasmids were used in PCRs, in which one of the primers was 5′ biotinylated, to produce labeled fragments that were used in footprinting studies with ParG titrations (Fig. 5). Repeat C was the only site that was fully protected from DNase I digestion at 0.3 to 0.4 μM ParG. This result correlates with the observation that site C is also preferentially protected within the intact OF site, most noticeably on the bottom strand (Fig. 1A). Sites A and B individually required up to 1.4 to 1.6 μM ParG for protection, although the latter remained partially unprotected even at the highest ParG concentration that was used (Fig. 5). The regions protected from DNase I digestion with separate sites A, B, and C fully spanned the sites and extended 1 or 2 bp into flanking sequences. Interestingly, identical DNase I-hypersensitive sites were located at the junctions between the protected and unprotected zones for reactions with sites A, B, and C, suggesting that binding of ParG perturbs the DNA at these positions.
FIG. 5.
DNase I footprinting of isolated OF sites. Footprinting reactions were performed as outlined in Materials and Methods using PCR fragments biotinylated at the 5′ ends of the bottom strands. ParG concentrations (μM monomer, left to right): 0.0, 0.2, 0.3, 0.4, 0.8, 1.0, 1.2, 1.4, 1.6, 1.8, 2.0, 2.5, 3.5, 4.0, and 5.0. The locations of sites A to D are marked by arrows. Positions that are hypersensitive to DNase I cleavage in the presence of ParG are highlighted by the stars.
In contrast with sites A, B, and C, an isolated site D was not protected from DNase I digestion at any ParG concentration (Fig. 5). This result agrees with footprinting studies of the entire OF site in which site D was incompletely protected by ParG, most obviously on the top strand, at protein concentrations that were sufficient for full protection of the other sites (Fig. 1A). To assess whether the partial protection of site D in OF necessitated the presence of sites A, B, and C or whether the adjacent site C was sufficient, a fragment bearing sites CD was analyzed by DNase I footprinting (Fig. 5). Following full protection of repeat C, incomplete protection of site D was apparent at elevated ParG concentrations. This correlates with the appearance of a second, slower-migrating species in EMSA analysis with the XXCD fragment at high ParG concentrations (Fig. 3). Thus, site D alone is bound poorly, if at all, by ParG but can be at least partially occupied when site C is also present. More generally, the DNase I footprinting results support EMSA data that the OF sites are recognized with different affinities by the protein and that cooperative interactions between ParG dimers mediate assembly of the nucleoprotein complex at the operator.
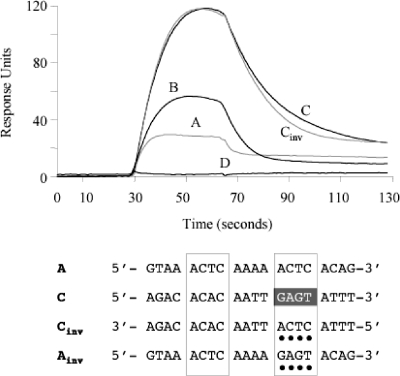
SPR experiments were performed to examine the ParG-site interactions in real time. Using a Biacore 3000 instrument, SA sensor chips (Biacore) were derivatized with biotinylated 50-bp oligonucleotides bearing site A, B, C, or D and with an unrelated DNA of the same length and base composition as a reference in one flow cell. Optimal conditions for the experiments were first identified using a range of ParG concentrations and flow rates. Subsequently, ParG was passed over the immobilized DNAs at 0.1 μM (monomer equivalents) for 36 s (association) and allowed to wash off subsequently for 70 s (dissociation) (Fig. 6). The protein bound well to site C under these conditions, reproducibly generating ∼120 response units. In contrast, ParG did not bind repeat D more appreciably than the unrelated DNA. Repeats A and B both produced intermediate response curves with ParG. The SPR results correlate well with the combined data from EMSA and DNase I footprinting, which reveal that ParG recognizes the OF sites preferentially in the order C > A ≈ B ≫ D.
FIG. 6.
SPR analysis of isolated OF sites. Biotinylated oligonucleotides (50 bp) (see Table S1 in the supplemental material) were tested as described in Materials and Methods. The oligonucleotides include the 12-bp site cores flanked on either side by 4 bp from the site's normal context (Fig. 1). Representative sensorgrams of ParG binding (100 nM monomer) to the sites are shown. The bottom panel shows an alignment of sites A and C and variant sites. The degenerate 5′-ACTC-3′ motifs are boxed. Nucleotide differences between wild-type and variant sites are highlighted with dots.
The most obvious feature of site C that may lead to it being recognized most avidly by ParG is the presence of a 5′-ACAC-3′ tetramer box spaced by 4 bp from its imperfect inverted counterpart 5′-GAGT-3′. In contrast, sites A, B, and D, which are bound less well by ParG, instead contain two directly repeated tetramer motifs (Fig. 1B). To assess whether converting repeat C to a site bearing directly repeated tetramer boxes affected the interaction of ParG with the site, the synthetic site Cinv was tested in SPR. The 5′-GAGT-3′ motif in C is inverted to 5′-ACTC-3′ in Cinv. However, the sensorgram patterns of C and Cinv were very similar (Fig. 6). Conversely, inversion of one of the 5′-ACTC-3′ motifs in site A to generate a synthetic site (Ainv) that carried symmetrical 5′-ACTC-3′ and 5′-GAGT-3′ boxes, like site C (Fig. 6), did not improve ParG binding (data not shown). These results indicate that the inverted arrangement of tetramer boxes in site C is not the major factor that makes it a preferred binding site for ParG.
Positions flanking the 12-bp core site are crucial for ParG binding.
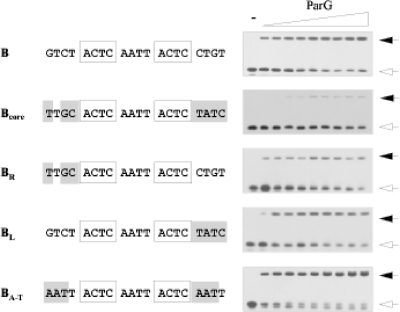
The 4 bp that separate the tetramer boxes in the 12-bp core sites are AT rich and elicit reduced ParG binding when mutated (see below). In contrast, the 4 bp between sites A and B and between sites B and C have a higher GC content (Fig. 1B). Fragments comprising the 12-bp core of site B flanked by wild-type or mutated 4-bp flanking regions were individually tested by EMSA. Mutations that altered three or four nucleotides in both of these regions greatly reduced ParG binding (Bcore; Fig. 7). Thus, a 12-bp core is capable only of weak ParG binding. Moreover, disruption of either the left (BR) or, to a lesser extent, the right (BL) flanking sequences independently reduced binding by ParG. However, not all mutations in the regions that flank the 12-bp core region are deleterious: conversion of these sequences to AT-only base pairs (BA-T) noticeably improved ParG binding to site B (Fig. 7). Thus, although the tetramer boxes likely harbor critical information for ParG interactions, both the 4-bp AT-rich sequences in the centers of the 12-bp sites (see below) and the nucleotides that flank the sites (Fig. 7) are also important determinants for proper ParG binding. In particular, the AT-rich centers of sites A to D may allow greater DNA flexibility, thereby promoting interactions between ParG dimers bound to adjacent tetramers.
FIG. 7.
Mutational analysis of regions flanking the 12-bp core in site B. The sequence of the bottom strand of site B with 4 bp of flanking nucleotides on either side is shown at the top. The 5′-ACTC-3′ motifs are boxed. Mutated sites are illustrated underneath with mutated positions shaded. Oligonucleotides (50 bp) bearing these sites were tested in EMSA as described in Materials and Methods using ParG concentrations (μM monomer, left to right): 0, 0.5, 0.75, 1.0, 1.5, 2.0, 2.5, 3.0, 3.5, and 4.0. Unbound DNAs and nucleoprotein complexes are indicated by open and filled arrows, respectively. The ParG titration shown for site B is the same as that shown in Fig. 4.
Scanning mutagenesis of the OF locus.
The contribution of each base pair in the OF locus to the interaction with ParG was probed by assessing the binding properties of synthetic double-stranded oligonucleotides bearing substitution mutations. As the intact OF region displays a complex pattern of ParG binding in EMSA (Fig. 3), mutations were analyzed in more informative contexts. Site A located within a 48-bp fragment forms a single retarded complex with ParG in EMSA (7) (Fig. 4). Mutations in site A in this context (here renamed AX for consistency with the analyses described above) (Fig. 8A) provide a useful comparison with equivalent substitution mutations in other contexts that were analyzed (Fig. 7 and 8). Purine-to-pyrimidine and pyrimidine-to-purine substitution mutations at each position within the 12-bp core of site A, as well as at two flanking base pairs, were studied. Protein concentrations up to 8 μM were tested, at which point ∼80% of the wild-type DNA is assembled into a single nucleoprotein complex. All single base pair mutations in substrate AX entirely abolished ParG binding, except at a single position between the 5′-ACTC-3′ tetramer boxes, which retained weak binding at high ParG concentrations (mutant m10; Fig. 8A). Mutations in the two base pairs immediately 5′ of the core site were tolerated well (m2) or poorly (m1).
In contrast with oligonucleotide AX, a fragment bearing sites AB produces two well-separated complexes with ParG in EMSA: a faster-migrating species containing a ParG dimer of dimers and, at higher protein concentrations, a slower-migrating complex that includes a pair of ParG tetramers (7). These complexes form efficiently at ParG concentrations of 1 to 2 μM and can include >80% of the target oligonucleotide. Therefore, the set of mutations that was analyzed in the AX context (Fig. 8A) was also examined as pairs of symmetrical mutations in the AB substrate in EMSA with ParG (Fig. 9). Only one position between the tetramer boxes was weakly tolerant of simultaneous substitution in the two sites (mutant m38). Detectable binding was eliminated by all other double mutations. The substitutions in mutant m38 are at the position equivalent to that of the mutation in mutant m10, which is the only alteration that was weakly tolerated in the AX context (Fig. 8A). As noted in binding studies with the AX oligonucleotide, mutations in positions immediately flanking the sites reduced (mutant m29) or had no major effect (mutant 30) in the AB context (Fig. 9).
FIG. 9.
Symmetrical scanning mutagenesis of the AB sites. Complementary purine-to-pyrimidine and pyrimidine-to-purine substitution mutations at each position within the two sites, as well as at flanking base pairs, were studied by EMSA using 48-bp oligonucleotides. ParG concentrations up to 2 μM were used. Data quantitation using Gel-Pro Analyzer 3.1 (Media Cybernetics) software is shown. The results shown are representative of experiments performed at least in duplicate. WT, wild type.
In view of the strong effects on ParG binding noted with most of the mutations examined as single changes in the AX context and as double mutations in the AB substrate, the mutations were also tested as single substitutions in the AB context (Fig. 8B). In contrast with the previous analyses, more diverse effects on ParG binding were noted, which may reflect that the variant oligonucleotides all possess an intact B site in addition to the mutated A site: ParG assembled on the intact site may stabilize protein bound more weakly to the mutated site. Mutations at the third or fourth position in either of the 5′-ACTC-3′ boxes in the A site greatly reduced, but did not entirely abolish, ParG binding in EMSA (mutants m19, m20, m27, and m28). Mutations at other positions in the tetramer boxes exerted detectable, but less pronounced, effects (mutants m17, m18, m25, and m26). Analogously, mutations in the poly(A) tract that separates the tetramer boxes had either strong (mutants m22 and m23) or less severe (mutants m21 and m24) effects on ParG binding. The result with mutant m24 correlates with the residual binding observed with the equivalent single mutation (m10) in the AX context (Fig. 8A) and with double mutations (m38) in the AB context (Fig. 9). In summary, ParG interaction with variant OF sites was examined in different contexts. This analysis revealed that ParG binding is highly sensitive to purine-to-pyrimidine and pyrimidine-to-purine substitution mutations. Moreover, mutations in either the 5′-ACTC-3′ boxes or the 4-bp AT-rich intervening sequences (Fig. 1B) reduce ParG binding, with the third and fourth positions in the tetramer boxes and the central dinucleotides in the spacers specially sensitive to mutation.
DISCUSSION
RHH proteins are a widely disseminated class of transcriptional repressors found in diverse eubacteria and archaea (36). The OF operator site bound by the ParG RHH protein consists of eight variant 5′-ACTC-3′ tetramer motifs, three arranged in direct orientation and five in inverted orientation. The motifs are separated precisely by 4-bp spacers, thereby providing a center-to-center distance of 8 bp between the motifs (Fig. 1). This is similar to the center-to-center separation of binding sites for RHH homologs such as MetJ but is shorter than the 11 bp between binding sites for Arc, for example. Differences in spacing between adjacent binding sites are dictated by protein-protein interactions between RHH dimers bound to these sites. An oligonucleotide substrate bearing four 5′-ACTC-3′ boxes, two in direct orientation and two in inverted orientation, first loads a pair of ParG dimers. This is followed by the recruitment of two additional dimers at higher protein concentrations (7). In view of the latter 1:1 ratio between protein dimer and tetramer motif, it is plausible that each motif is bound by a single ParG dimer, implying that the entire OF site is coated by up to eight dimers. This interpretation correlates with observations with other RHH factors that bind as a dimer to a single DNA site and cooperatively oligomerize on adjoining sites (36). A fragment bearing a single 5′-ACTC-3′ box is not detectably bound by ParG in vitro (7). Instead, at least two adjacent boxes are required for complex formation (Fig. 4). Cooperative interactions between ParG dimers bound to neighboring boxes likely generate a nucleoprotein complex that is sufficiently stable to be detected experimentally, as has been noted with other RHH repressors (21, 42). Cooperative interactions in the assembly of ParG on OF are most obvious in the case of site D. Instead of the canonical 5′-ACTC-3′ motifs, this site consists of two 5′-ACTT-3′ motifs separated by a 4-bp spacer that, unlike the AT-rich spacer in other sites, includes a C residue (Fig. 1B). This combination of substitutions abolishes ParG binding to the isolated site in vitro (Fig. 3 to 5). However, in the presence of the adjacent site C, site D is partially protected from DNase I digestion by ParG (Fig. 5) and also forms a second complex in EMSA with the protein (Fig. 3) that is not evident when site C alone is tested. However, disruption of site D did not reduce repression by ParG of a transcriptional fusion of the parFG promoter-operator region to a reporter gene (Fig. 2), leaving the contribution of this site to ParG repression at OF uncertain.
Like other RHH proteins, the positively charged antiparallel β-strands in the ParG dimer are thought to insert in the DNA major groove and make nucleotide base-specific contacts (16). In particular, we speculate that alternating residues at three positions (Arg-36, Asn-38, and Asn-40) within the β-strands in ParG may make critical hydrogen bonding contacts with bases in the 5′-ACTC-3′ boxes. However, positions external to the tetramer boxes also play a vital role in ParG binding, and these positions may also be contacted by residues in the β-strands, as is the case with other RHH proteins (36). Site-specific substitutions in the 4-bp spacers within the 12-bp sites always reduced, and in most cases abolished, ParG binding (Fig. 7 and 8). Oligo(dA:dT)n tracts, where n = 3, impart significant intrinsic curvature on DNA (11, 23). Thus, the AT-rich spacers within the sites may contribute to formation of a nucleoprotein complex in which the curved topology of the spacer DNA promotes interactions between ParG dimers bound to adjoining 5′-ACTC-3′ boxes. DNA within the nucleoprotein complexes of the Arc, MetJ, and CopG RHH proteins is bent by up to 60°, with the bend centered either at the binding site or between adjacent sites (17, 34, 36). Less pronounced DNA curvature is evident in cocrystal structures of NikR and FitA RHH proteins with their cognate DNA sites, with a smooth bending of the DNA between better-separated binding sites (28, 37). Although intrinsic curvature has yet to be assessed rigorously in any site bound by an RHH protein, inherent curvature may contribute to the formation of the bend observed in protein-DNA structures.
Improved ParG binding to site B occurs when the AT content of the 4-bp sequences that flank the site is increased (compare sites B and BA-T in Fig. 7). In contrast with the AT-rich 4-bp spacers that separate the 5′-ACTC-3′ motifs, it is difficult to envisage how enhanced binding to site BA-T could be due to stronger ParG dimer-dimer interactions. Instead, in addition to interactions with the 5′-ACTC-3′ boxes, ParG may make contacts with the flanking regions that are improved by the higher AT content of site BA-T. In addition to residues in the β-strands, two amide nitrogens in the second α-helix of RHH factors make nonspecific contacts with the DNA phosphate backbone. In the case of ParG, additional interactions might also be achieved by amino acids in the flexible tails that extend from the folded RHH domain. The tail is known to modulate the binding of ParG to DNA, although the mechanism involved has yet to be elucidated fully (7). In summary, the ParG binding motifs in the OF operator comprise an array of 5′-ACTC-3′ boxes separated by 4-bp spacers. The tetramer boxes likely harbor the core sequences recognized by the β-strands in the RHH structure of ParG. The intervening AT-rich spacers may contribute to the intrinsic conformation of the OF site, as well as harboring crucial information for additional ParG contacts. Sequence variations among the tetramer boxes, the combination of inverted and directly repeated boxes, and the different AT contents of the interbox regions likely generate a nucleoprotein complex whose features are fine-tuned for optimal interaction with the transcription machinery and for mediating parFG repression and derepression. Elucidation of the ParG-DNA structure(s), combined with functional studies in vivo, will clarify these interactions further.
Among RHH proteins whose tertiary structures have been elucidated, the ω protein of plasmid pSM19305 and the ParR proteins specified by plasmids pSK41 and pB171 are functional analogs of ParG in plasmid segregation. All four dimeric proteins both autorepress transcription of the corresponding genes and bind to their cognate centromeres during segrosome assembly (4, 7, 9, 30, 33, 35, 40). However, the sequences and organization of the motifs recognized by the proteins differ. The ω protein binds arrays of highly conserved 7-bp repeats with the consensus 5′-(A/T)ATCAC(A/T)-3′. The arrays comprise 7 to 10 repeats depending on their location (10, 42). ParR of plasmid pSK41 binds eight 10-bp repeats that possess core 5′-TATA-3′ motifs (40). ParR of plasmid pB171 binds two 10-bp direct repeats with the sequence 5′-AATACTCAAT-3′ (35). By comparison, the multiple 5′-ACTC-3′ tetramer motifs recognized by ParG and the flanking sequences in which they are embedded are more degenerate. Like ParG, ω binds to a single repeat poorly, preferentially binding cooperatively to ≥2 heptad repeats. The heptad repeats bound by ω can be arranged in a variety of configurations, again reminiscent of the tetramer motifs recognized by ParG. In contrast, the 10-bp repeats bound by ParR of plasmid pSK41 are arranged exclusively in direct orientation. The pSK41 protein wraps its binding site about its positive convex surface to form an extended complex with an open, solenoid-shaped topology (40). Analogously, ParR of pB171 has been reported to form ring-like structures on its binding site (30). Further analysis of ParG and its interactions with DNA will generate key insights into DNA segregation and its control in bacteria.
Supplementary Material
Acknowledgments
This work was supported by grants from the Biotechnology and Biological Sciences Research Council (grant number G19857) and The Wellcome Trust (grant number 072661/Z/03/Z) to F.H.
Footnotes
Published ahead of print on 17 April 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Adachi, S., K. Hori, and S. Hiraga. 2006. Subcellular positioning of F plasmid mediated by dynamic localization of SopA and SopB. J. Mol. Biol. 356850-863. [DOI] [PubMed] [Google Scholar]
- 2.Ahmadian, M. R., P. Stege, K. Scheffzek, and A. Wittinghofer. 1997. Confirmation of the arginine-finger hypothesis for the GAP-stimulated GTP-hydrolysis reaction of Ras. Nat. Struct. Biol. 4686-689. [DOI] [PubMed] [Google Scholar]
- 3.Barillà, D., E. Carmelo, and F. Hayes. 2007. The tail of the ParG DNA segregation protein remodels ParF polymers and enhances ATP hydrolysis via an arginine finger-like motif. Proc. Natl. Acad. Sci. USA 1041811-1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barillà, D., and F. Hayes. 2003. Architecture of the ParF-ParG protein complex involved in procaryotic DNA segregation. Mol. Microbiol. 49487-499. [DOI] [PubMed] [Google Scholar]
- 5.Barillà, D., M. F. Rosenberg, U. Nobbmann, and F. Hayes. 2005. Bacterial DNA segregation dynamics mediated by the polymerizing protein ParF. EMBO J. 241453-1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bos, J. L., H. Rehmann, and A. Wittinghofer. 2007. GEFs and GAPs: critical elements in the control of small G proteins. Cell 129865-877. [DOI] [PubMed] [Google Scholar]
- 7.Carmelo, E., D. Barillà, A. P. Golovanov, L. Y. Lian, A. Derome, and F. Hayes. 2005. The unstructured N-terminal tail of ParG modulates assembly of a quaternary nucleoprotein complex in transcription repression. J. Biol. Chem. 28028683-28691. [DOI] [PubMed] [Google Scholar]
- 8.Cheeseman, I. M., and A. Desai. 2008. Molecular architecture of the kinetochore-microtubule interface. Nat. Rev. Mol. Cell Biol. 933-46. [DOI] [PubMed] [Google Scholar]
- 9.de la Hoz, A. B., S. Ayora, I. Sitkiewicz, S. Fernández, R. Pankiewicz, J. C. Alonso, and P. Ceglowski. 2000. Plasmid copy-number control and better-than-random segregation genes of pSM19035 share a common regulator. Proc. Natl. Acad. Sci. USA 97728-733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de la Hoz, A. B., F. Pratto, R. Misselwitz, C. Speck, W. Weihofen, K. Welfle, W. Saenger, H. Welfle, and J. C. Alonso. 2004. Recognition of DNA by ω protein from the broad-host range Streptococcus pyogenes plasmid pSM19035: analysis of binding to operator DNA with one to four heptad repeats. Nucleic Acids Res. 323136-3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diekmann, S. 1992. Analyzing DNA curvature in polyacrylamide gels. Methods Enzymol. 21230-46. [DOI] [PubMed] [Google Scholar]
- 12.Ebersbach, G., and K. Gerdes. 2004. Bacterial mitosis: partitioning protein ParA oscillates in spiral-shaped structures and positions plasmids at mid-cell. Mol. Microbiol. 52385-398. [DOI] [PubMed] [Google Scholar]
- 13.Ebersbach, G., and K. Gerdes. 2005. Plasmid segregation mechanisms. Annu. Rev. Genet. 39453-479. [DOI] [PubMed] [Google Scholar]
- 14.Fogel, M. A., and M. K. Waldor. 2006. A dynamic, mitotic-like mechanism for bacterial chromosome segregation. Genes Dev. 203269-3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garner, E. C., C. S. Campbell, D. B. Weibel, and R. D. Mullins. 2007. Reconstitution of DNA segregation driven by assembly of a prokaryotic actin homolog. Science 3151270-1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Golovanov, A. P., D. Barillà, M. Golovanova, F. Hayes, and L. Y. Lian. 2003. ParG, a protein required for active partition of bacterial plasmids, has a dimeric ribbon-helix-helix structure. Mol. Microbiol. 501141-1153. [DOI] [PubMed] [Google Scholar]
- 17.Gomis-Rüth, F. X., M. Solá, P. Acebo, A. Párraga, A. Guasch, R. Eritja, A. González, M. Espinosa, G. del Solar, and M. Coll. 1998. The structure of plasmid-encoded transcriptional repressor CopG unliganded and bound to its operator. EMBO J. 177404-7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hayes, F. 2000. The partition system of multidrug resistance plasmid TP228 includes a novel protein that epitomizes an evolutionarily-distinct subgroup of the ParA superfamily. Mol. Microbiol. 37528-541. [DOI] [PubMed] [Google Scholar]
- 19.Hayes, F., and D. Barillà. 2006. The bacterial segrosome: a dynamic nucleoprotein machine for DNA trafficking and segregation. Nat. Rev. Microbiol. 4133-143. [DOI] [PubMed] [Google Scholar]
- 20.Hayes, F., and D. Barillà. 2006. Assembling the bacterial segrosome. Trends Biochem. Sci. 31247-250. [DOI] [PubMed] [Google Scholar]
- 21.He, Y., C. W. Garvie, S. Elworthy, I. W. Manfield, T. McNally, I. D. Lawrenson, S. E. V. Phillips, and P. G. Stockley. 2002. Structural and functional studies of an intermediate on the pathway to operator binding by Escherichia coli MetJ. J. Mol. Biol. 32039-53. [DOI] [PubMed] [Google Scholar]
- 22.Kędzierska, B., L. Y. Lian, and F. Hayes. 2007. Toxin-antitoxin regulation: bimodal interaction of YefM-YoeB with paired DNA palindromes exerts transcriptional autorepression. Nucleic Acids Res. 35325-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koo, H. S., H. M. Wu, and D. M. Crothers. 1986. DNA bending at adenine-thymine tracts. Nature 320501-506. [DOI] [PubMed] [Google Scholar]
- 24.Lane, D., P. Prentki, and M. Chandler. 1992. Use of gel retardation to analyze protein-nucleic acid interactions. Microbiol. Rev. 56509-528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Larouche, K., M. J. Bergeron, S. Leclerc, and S. L. Guérin. 1996. Optimization of competitor poly(dI-dC)·poly(dI-dC) levels is advised in DNA-protein interaction studies involving enriched nuclear proteins. BioTechniques 20439-444. [DOI] [PubMed] [Google Scholar]
- 26.Lim, G. E., A. I. Derman, and J. Pogliano. 2005. Bacterial DNA segregation by dynamic SopA polymers. Proc. Natl. Acad. Sci. USA 10217658-17663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Machón, C., T. J. G. Fothergill, D. Barillà, and F. Hayes. 2007. Promiscuous stimulation of ParF protein polymerization by heterogeneous centromere binding factors. J. Mol. Biol. 3741-8. [DOI] [PubMed] [Google Scholar]
- 28.Mattison, K., J. S. Wilbur, M. So, and R. G. Brennan. 2006. Structure of FitAB from Neisseria gonorrhoeae bound to DNA reveals a tetramer of toxin-antitoxin heterodimers containing pin domains and ribbon-helix-helix motifs. J. Biol. Chem. 28137942-37951. [DOI] [PubMed] [Google Scholar]
- 29.Michael, C., and T. S. Stephen. 2001. Transcriptional regulation in eukaryotes: concepts, strategies, and techniques. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 30.Moller-Jensen, J., S. Ringgard, C. P. Mercogliano, K. Gerdes, and J. Lowe. 2007. Structural analysis of the ParR/parC plasmid partition complex. EMBO J. 264413-4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nadanaciva, S., J. Weber, S. Wilke-Mounts, and A. E. Senior. 1999. Importance of F1-ATPase residue α-Arg-376 for catalytic transition state stabilization. Biochemistry 3815493-15499. [DOI] [PubMed] [Google Scholar]
- 32.Norrander, J., T. Kempe, and J. Messing. 1983. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene 26101-106. [DOI] [PubMed] [Google Scholar]
- 33.Pratto, F., A. Cicek, W. A. Weihofen, R. Lurz, W. Saenger, and J. G. Alonso. 2008. Streptococcus pyogenes pSM19035 requires dynamic assembly of ATP-bound ParA and ParB on parS DNA during plasmid segregation. Nucleic Acids Res. 363676-3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raumann, B. E., M. A. Rould, C. O. Pabo, and R. T. Sauer. 1994. DNA recognition by β-sheets in the Arc repressor-operator crystal structure. Nature 367754-757. [DOI] [PubMed] [Google Scholar]
- 35.Ringgaard, S., G. Ebersbach, J. Borch, and K. Gerdes. 2007. Regulatory cross-talk in the double par locus of plasmid pB171. J. Biol. Chem. 2823134-3145. [DOI] [PubMed] [Google Scholar]
- 36.Schreiter, E. R., and C. L. Drennan. 2007. Ribbon-helix-helix transcription factors: variations on a theme. Nat. Rev. Microbiol. 5710-720. [DOI] [PubMed] [Google Scholar]
- 37.Schreiter, E. R., S. C. Wang, D. B. Zamble., and C. L. Drennan. 2006. NikR-operator complex structure and the mechanism of repressor activation by metal ions. Proc. Natl. Acad. Sci. USA 10313676-13681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schumacher, M. A. 2007. Structural biology of plasmid segregation proteins. Curr. Opin. Struct. Biol. 17103-109. [DOI] [PubMed] [Google Scholar]
- 39.Schumacher, M. A. 2008. Structural biology of plasmid partition: uncovering the molecular mechanisms of DNA segregation. Biochem. J. 4121-18. [DOI] [PubMed] [Google Scholar]
- 40.Schumacher, M. A., T. C. Glover, A. J. Brzoska, S. O. Jensen, T. D. Dunham, R. A. Skurray, and N. Firth. 2007. Segrosome structure revealed by a complex of ParR with centromere DNA. Nature 4501268-1271. [DOI] [PubMed] [Google Scholar]
- 41.Thanbichler, M., and L. Shapiro. 2006. Chromosome organization and segregation in bacteria. J. Struct. Biol. 156292-303. [DOI] [PubMed] [Google Scholar]
- 42.Weihofen, W. A., A. Cicek, F. Pratto, J. C. Alonso, and W. Saenger. 2006. Structures of ω repressors bound to direct and inverted DNA repeats explain modulation of transcription. Nucleic Acids Res. 341450-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Woodcock, D. M., P. J. Crowther, J. Doherty, S. Jefferson, E. DeCruz, M. Noyer-Weidner, S. S. Smith, M. Z. Michael, and M. W. Graham. 1989. Quantitative evaluation of Escherichia coli host strains for tolerance to cytosine methylation in plasmid and phage recombinants. Nucleic Acids Res. 173469-3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.