Abstract
The primary sigma factor of Staphylococcus aureus, σSA, regulates the transcription of many genes, including several essential genes, in this bacterium via specific recognition of exponential growth phase promoters. In this study, we report the existence of a novel staphylococcal phage G1-derived growth inhibitory polypeptide, referred to as G1ORF67, that interacts with σSA both in vivo and in vitro and regulates its activity. Delineation of the minimal domain of σSA that is required for its interaction with G1ORF67 as amino acids 294 to 360 near the carboxy terminus suggests that the G1 phage-encoded anti-σ factor may occlude the −35 element recognition domain of σSA. As would be predicted by this hypothesis, the G1ORF67 polypeptide abolished both RNA polymerase core-dependent binding of σSA to DNA and σSA-dependent transcription in vitro. While G1ORF67 profoundly inhibits transcription when expressed in S. aureus cells in mode of action studies, our finding that G1ORF67 was unable to inhibit transcription when expressed in Escherichia coli concurs with its inability to inhibit transcription by the E. coli holoenzyme in vitro. These features demonstrate the selectivity of G1ORF67 for S. aureus RNA polymerase. We predict that G1ORF67 is one of the central polypeptides in the phage G1 strategy to appropriate host RNA polymerase and redirect it to phage reproduction.
Transcription initiation is a critical regulatory step in cell metabolism, and components of the transcription machinery are validated targets for antibacterial drug discovery, as witnessed with the successful use of rifampin over the last 4 decades (5, 37, 44). In bacteria, the RNA polymerase (RNAP) core enzyme associates with one of several sigma factors to form the RNAP holoenzyme, thereby directing efficient transcription from specific promoters (4, 14, 26, 31). During exponential growth, σ70 of Escherichia coli and its orthologs from other bacteria are the primary σ factors, responsible for transcribing most growth-related and housekeeping genes. All primary σ factor orthologs share four distinct regions (σ1 to σ4) of highly conserved amino acid sequence with similar functions (4, 31). Regions σ2 and σ4 are involved in the direct contact with −10 and −35 promoter elements, respectively (4, 31).
Alternate σ factors recognize different promoter sequences and thereby direct the core enzyme to transcribe specific genes in response to changes in environmental conditions (14, 26, 41). The global gene expression profile is primarily exerted at the level of competition between various σ factors for the core enzyme (26). In Staphylococcus aureus, four different σ factors have been reported to date: the primary σ factor σSA that directs the transcription of housekeeping genes during exponential growth phase (6) and three alternate σ factors, namely SigB, which modulates the expression of stress response genes (7, 19); SigH, which is required for transcribing competence genes (30); and a novel extracytoplasmic function sigma factor named SigS that appears to be an important component of the stress and pathogenic responses (41).
The diversity of σ factors, their abundance in the cell, and their relative affinities for the RNAP core enzyme provide a sophisticated mechanism dictating the coordinated spatiotemporal expression of genes in response to specific environmental conditions (14, 26). Given this key role, σ factors are targeted by a wide range of transcriptional regulators, such as activators (43), repressors (28), competitors (26), small RNAs (45), and anti-σ factors (13). The binding of an anti-σ factor to its cognate σ factor leads to specific inhibition of transcription of the genes that are regulated by that particular σ factor (13). As an example, the bacteriophage T4-encoded anti-σ factor AsiA (32, 36) exerts a critical shift in the phage infective cycle by redirecting transcription by host RNAP to phage middle promoters. Recent structural studies reveal that AsiA binding to E. coli σ70 results in remodeling of domains that contact the −35 element of the promoter in conjunction with the T4 MotA protein while leaving the contacts between σ and the −10 region intact (4, 11, 22). Predicted orthologs of AsiA have been found in the genomes of other T4-like phages, as well as in genomes of some gram-negative bacteria, including E. coli and Pseudomonas aeruginosa (8, 15, 16, 18). This family of anti-σ70 factors shares key amino acid residues known to be crucial for the binding of AsiA to E. coli σ70 (36).
We recently reported the results from a phage genomics and functional genomics study based on protein-protein interaction between phage-carried growth inhibitory polypeptides and proteins of the bacterial host (23). In Staphylococcus aureus, such an approach identified a variety of host proteins that regulated key metabolic pathways, such as DNA replication and transcription. In most cases, these host proteins were shown to be essential for cell viability.
In the present study, we extend our studies to the staphylococcal bacteriophage G1, a member of the Myoviridae phage family, like T4 (21), and characterize a phage-encoded growth inhibitory polypeptide, designated G1ORF67, as an anti-σ factor that binds tightly to the primary sigma factor of S. aureus. The G1ORF67 binding domain on σSA is delineated, and the consequences of such interaction are assessed by functional assays both in vivo and in vitro.
MATERIALS AND METHODS
Reagents.
Restriction and modification enzymes were obtained from New England Biolabs. Lysostaphin, Na-arsenite, poly(dI-dC), streptomycin sulfate, phenylmethylsulfonyl fluoride, heart muscle kinase, silver nitrate, and glutathione agarose resin were purchased from Sigma. Ni++-nitrilotriacetate agarose resin was purchased from Qiagen. Affi-gel resin and protein assay kit were from Bio-Rad. Protease inhibitor cocktail was obtained from Roche Diagnostics. RNasin pancreatic RNase inhibitor was obtained from Promega. The E. coli RNAP holoenzyme and core enzyme were purchased from Epicentre Technologies. Radioactive nucleotides and precursors were from GE Healthcare. Europium (Eu)-conjugated anti-His6 antibody was purchased from CIS-Bio International; anti-influenza hemagglutinin antibody was from Babco. Allophycocyanin (APC)-anti-glutathione S-transferase (GST) antibody and -streptavidin conjugates were from Prozyme. Multiscreen GF/B plates were purchased from Millipore.
Growth inhibitory property of the phage polypeptide.
The growth inhibitory property of G1ORF67 was characterized in a time-kill broth assay essentially as described previously (23). Briefly, ORF67 was amplified by PCR from phage G1 (21) by using a sense primer, 5′-CGGGATCCATGAAATTAAAGATTTTAGA-3′, in conjunction with the antisense primer 5′-CCCAAGCTTCTATTTACTAATTTTTTTCA-3′. The PCR product was digested with BamHI/HindIII and cloned into the unique BamHI/HindIII sites of expression vector pTM under the control of Na-arsenite (23). S. aureus RN4220 was used as the host strain for monitoring the growth inhibitory property of G1ORF67 in the time-kill assay.
Identification of the bacterial target for G1ORF67 by affinity chromatography.
The G1ORF67 polypeptide (NCBI Entrez protein accession number YP_240941 [21]) was purified from E. coli BL21(DE3) as a His6 fusion using Ni++-chelate chromatography and cross-linked to Affi-gel 10 resin at protein/resin concentrations ranging from 0 to 7 mg/ml. Subsequent chromatographic steps with lysate from S. aureus RN4220 were performed as previously described (23).
Validation of G1ORF67-σSA interaction.
The interaction between the phage polypeptide and the bacterial protein was validated essentially as described previously for S. aureus DnaI and the 77ORF104 polypeptide (NCBI Entrez protein accession number NP_958646 [23]). For far-Western analysis, S. aureus σSA was purified from E. coli BL21(DE3) as a fusion protein tagged at its N terminus with the heart muscle kinase phosphorylation site (17) and a His6 tag. The recombinant protein was radiolabeled with [γ-32P]ATP and heart muscle kinase and used as a probe with immobilized phage polypeptides. In the time-resolved fluorescence resonance energy transfer (TR-FRET) (27) assay, G1ORF67 was purified from E. coli BL21(DE3) as a GST fusion; its interaction with purified His6-tagged σSA was detected by using antitag antibodies conjugated to APC and Europium (Eu), respectively, as described previously (23). Yeast-two-hybrid analysis was performed using a Matchmaker two-hybrid system 3 according to the manufacturer's instructions (CLONTECH Laboratories).
Overexpression and purification of S. aureus RNAP core enzyme.
The S. aureus rpoA gene encoding the α subunit of RNAP was PCR amplified from genomic DNA of S. aureus strain RN4220 using the sense oligonucleotide 5′-CGGGATCCATGATAGAAATCGAAAAACCTAGA-3′ and the antisense oligonucleotide 5′-ACGCGTCGACACTATCTTCTTTTCTTAATCCTAA-3′. The PCR product was digested with BamHI/SalI and cloned into pTM (23) as a C-terminal fusion with tandem affinity purification tags consisting of His6 and the biotin acceptor domain (2) and used to transform S. aureus RN4220. Cells were grown in tryptic soy broth (TSB) with 30 μg/ml kanamycin to an optical density at 540 nm of 0.5 and induced with 10 μM Na-arsenite for 2 h at 37°C. Bacteria were harvested by centrifugation. The bacterial pellet from a 30-liter culture was resuspended in 400 ml HNG-1000 buffer (20 mM HEPES-KOH, pH 8.0, 1 M NaCl, and 10% glycerol) supplemented with 10 mM imidazole, protease inhibitor cocktail, 1 mM phenylmethylsulfonyl fluoride, and approximately 30,000 U of lysostaphin. The cell suspension was incubated at 37°C for 30 min, and cells were lysed by sonication. Nucleic acids were precipitated with 3% streptomycin sulfate for 20 min at 4°C. The RNAP core enzyme was purified essentially free of σSA by Ni++-nitrilotriacetic acid chromatography using wash buffers HNG-1000 and TGEN (10 mM Tris-HCl, pH 8.0, 5% glycerol, 150 mM NaCl, and 0.1 mM EDTA), each supplemented with 10 mM imidazole, and using as elution buffer TGEN with 200 mM imidazole. The identities of the purified subunits (α, β, and β′) were confirmed by mass spectrometry of tryptic digests (23; data not shown).
In vitro transcription assays.
In vitro transcription assay reactions were similar to those described previously (6) and were performed with increasing concentrations (range, 0 to 500 nM) of σSA and 25 nM of E. coli core enzyme in a total volume of 25 μl containing 40 mM Tris-acetate, pH 7.9; 100 mM NaCl; 5 mM MgCl2; 1 mM dithiothreitol; 0.1 mg/ml bovine serum albumin (BSA); 0.5 mM of ATP, GTP, and CTP; 0.25 mM UTP; 5 μCi [α-32P]UTP (3,000 Ci/mmol); 1 U RNasin; and 40 ng pB6 template DNA. Plasmid pB6 is a derivative of the previously described pZE21 vector harboring a cassette driven by the λPL promoter, the kanamycin gene, and the ColE1 RNA1 gene (25). To investigate the effect of G1ORF67 on σSA-dependent transcription, the σSA-specific phage polypeptide G1ORF67 and the negative control polypeptide 77ORF104 (23) were purified as GST fusions and preincubated (final concentration, 2 μM) with σSA for 10 min on ice prior to the addition of the other reagents. Reaction mixtures were incubated at 37°C for 15 min, stopped with formamide loading buffer, and electrophoresed on a denaturing gel.
Alternatively, the gel-based assay was converted to a miniaturized high-throughput trichloroacetic acid (TCA)-based assay as follows: σSA (100 nM) was mixed with S. aureus core enzyme (50 nM) in a total volume of 25 μl containing 40 mM Tris-acetate, pH 7.9; 100 mM NaCl; 5 mM MgCl2; 1 mM dithiothreitol; 0.1 mg/ml BSA; 150 μM each ATP, GTP, and CTP; 30 μM UTP; 100,000 cpm [α-32P]UTP (3,000 Ci/mmol); 1 U RNasin; and 40 ng pB6 template DNA. The effect of G1ORF67 was monitored by including the purified phage polypeptide or a negative-control polypeptide (each at a final concentration of 10 μM) in the reaction mixture, followed by an incubation of 1 h at 37°C in a 96-well PCR plate. Samples were transferred to 96-well multiscreen plates and subjected to a 10% TCA precipitation step in the presence of 10 μg salmon sperm carrier DNA. The radiolabeled RNA product was counted by using a liquid scintillation counter (Trilux 1450 Microbeta; PerkinElmer).
In vitro DNA-binding studies.
A TR-FRET assay for formation of RNAP-promoter oligonucleotide nucleoprotein complexes was developed as follows. The 5′ end of the sense strand of the −41 to −12 sequence of the λPR promoter oligonucleotide (10) was biotinylated and annealed to its complementary strand for use as a probe. The assay was performed in a 24-μl volume containing 20 mM HEPES, pH 8.0, 100 mM KCl, 1 mM EDTA, 400 mM KF, 200 nM BSA, 3% glycerol, 50 nM biotin-tagged oligonucleotide probe, 32 nM His-tagged σSA, and 10 nM of E. coli core enzyme. The reaction mixture was incubated for 15 min at room temperature, and 6 μl of a mixture of Eu-conjugated anti-His6 and APC-conjugated streptavidin was added to final concentrations of 3 and 15 nM, respectively. Samples were mixed, and 25 μl of the mixture was transferred to a black 96-well plate (Molecular Devices). After 45 min of incubation at room temperature, the fluorescence signals (excitation, 340 nm; Eu emission, 612 nm; and APC emission, 665 nm) were measured using an Ultra plate reader (Tecan). The specificity of the interaction was monitored by including one of three nonbiotinylated oligonucleotides (−35 sequence is underlined and in boldface): (i) parental λPR, 5′-ATGATATTGACTTATTGAATAAAATTGGGT-3′; (ii) λPL, 5′-GATAGAGATTGACATCCCTATCAGTGATAGAGATACTGAGCACAT CAGC-3′; or (iii) Mut λPR, 5′-ATGATAACTTTGTATTGAATAAAATTGGGT-3′ (as control, with scrambled −35 element). In competition analyses, an excess of untagged oligonucleotide was added to the reaction mixture prior to the addition of the Eu and APC conjugates. The effect of G1ORF67 on the DNA-binding activity of σSA was monitored by including increasing amounts of the phage polypeptide (0 to 4 μM) in the reaction mixture prior to the addition of Eu and APC conjugates.
RESULTS
Characterization of phage G1ORF67 as an inhibitor of S. aureus growth.
We applied a functional genomics approach (23) to characterize staphylococcal bacteriophage G1 (21). Accordingly, a total of 214 open reading frames (ORFs) of at least 33 amino acids were predicted to be encoded by the phage genome (21). In dot screen-based assays (23), seven ORFs were shown to abolish bacterial growth when their expression was induced in S. aureus strain RN4220 (data not shown). One of these ORFs, termed G1ORF67, with a predicted molecular mass of 25 kDa, is the subject of the current study. In order to establish the kinetics of the growth inhibition mediated by G1ORF67, we induced its expression and monitored its effect on S. aureus viability, measured as CFU, over time. The results indicate that the phage polypeptide exerted a bacteriostatic effect upon its induction since, unlike the uninduced growth control, bacterial CFU remained constant from 40 min to 240 min following induction of G1ORF67 synthesis (Fig. 1). As a negative control, another phage polypeptide whose expression did not show inhibition of bacterial growth in a dot screen-based assay was also examined in an identical manner (Fig. 1). The observed growth inhibitory effect of G1ORF67 appeared to be specific to S. aureus, since its expression in Streptococcus pneumoniae and Pseudomonas aeruginosa did not yield any growth inhibitory effect (data not shown).
FIG. 1.
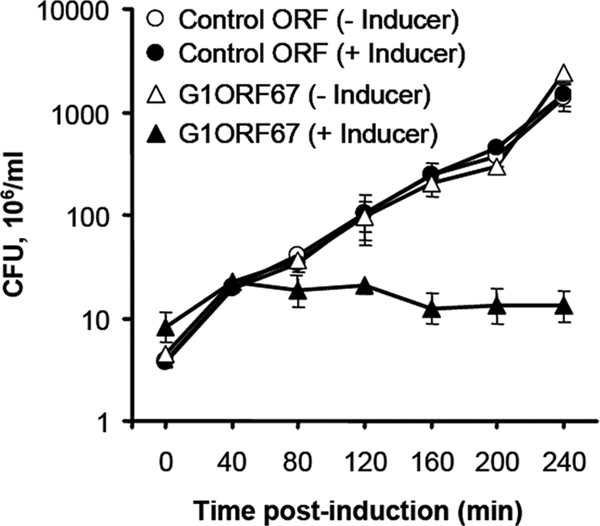
Growth inhibition kinetics of S. aureus expressing G1ORF67. Clones of S. aureus RN4220 harboring either G1ORF67 or a control ORF (which has no impact on S. aureus growth) under the regulation of an arsenite-regulatable promoter were grown in tryptic soy broth supplemented with 30 μg/ml kanamycin with or without 5 μM NaAsO2. At different time intervals, aliquots of the cultures were plated onto tryptic soy agar plates supplemented with kanamycin in order to determine the number of CFU. Results are expressed as the means ± standard deviations of the results for three independent clones.
G1ORF67 binds directly to the primary sigma factor of S. aureus, σSA.
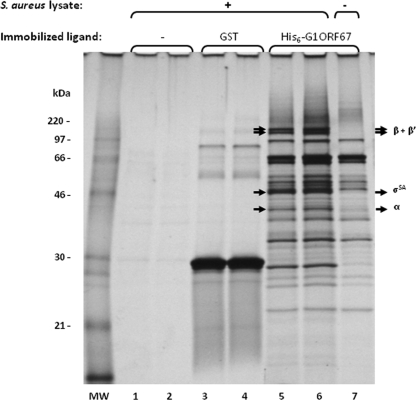
In an attempt to identify cellular proteins that mediated the growth inhibitory effect of G1ORF67, we expressed and purified G1ORF67 as a His6 fusion protein from E. coli and used it as a column ligand in affinity chromatography studies. Whole-cell lysates from S. aureus were then loaded onto a His6-G1ORF67 column and a GST control column. After extensive washes, bound proteins were eluted with 1% sodium dodecyl sulfate (SDS), resolved by SDS-polyacrylamide gel electrophoresis (PAGE), and stained with silver nitrate (Fig. 2). The bands of interest were excised from the gel and subjected to trypsin digestion, and their identities were revealed by mass spectrometry as described previously (23). Accordingly, four polypeptides with apparent masses of 35 kDa, 45 kDa, and >100 kDa bound selectively to the His6-G1ORF67 column (Fig. 2, lanes 5 and 6) and not to the GST column (Fig. 2, lane 3 and 4). As a further control, the four polypeptides were absent in eluates from a His6-G1ORF67 column that had not received S. aureus lysate (Fig. 2, lane 7). Hence, the recovery of these polypeptides was dependent on both His6-G1ORF67 and S. aureus lysate (Fig. 2). Mass spectrometry of tryptic digests of the polypeptides indicated that they are components of S. aureus RNAP holoenzyme, consisting of the α subunit (35-kDa band); the primary sigma factor, σSA (45-kDa band); and the β and β′ subunits (>100-kDa bands). Under the experimental conditions described here and for three independent G1ORF67 affinity chromatography experiments, no targets other than the RNAP holoenzyme were reproducibly identified.
FIG. 2.
G1ORF67 interacts with the RNAP holoenzyme of S. aureus. Results of SDS-PAGE of 1% SDS eluates from His-tagged G1ORF67 (lanes 5 to 7) or GST (lanes 3 and 4) in affinity chromatography. Experiments were performed in duplicate with a mock-immobilized resin and S. aureus lysate (lanes 1 and 2), with a resin containing immobilized GST with S. aureus lysate (lanes 3 and 4), and with a resin containing immobilized His-tagged G1ORF67 and S. aureus lysate (lanes 5 and 6). A resin containing immobilized His-tagged G1ORF67 with no input lysate (lane 7) served as a further control. The positions of migration of the β, β′, σSA, and α polypeptides are indicated to the right of the gel image. The masses, in kDa, of protein standards are indicated to the left of lane “MW.” Eluates were separated by SDS-PAGE and visualized with silver nitrate. Specific polypeptide bands were excised from the gel and subjected to tryptic peptide mass determination by liquid chromatography and electrospray tandem mass spectrometry. +, present; −, absent.
To validate the interaction between G1ORF67 and subunit(s) of the S. aureus RNAP, we performed a series of in vitro and cell-based interaction assays. We focused initially on σSA as a potential interacting protein, since the corresponding primary sigma factor of E. coli, σ70, is known to be the target of the T4 phage-carried transcriptional regulator AsiA (32, 36). In protein affinity (far-Western) blotting, a concentration-dependent hybridization signal was detected between the immobilized G1ORF67 polypeptide and a radiolabeled σSA probe (Fig. 3A), thereby confirming that the interaction between G1ORF67 and σSA, as initially detected by affinity chromatography, was direct. No interaction between the σSA probe and immobilized control polypeptide 77ORF104 was detected (Fig. 3A). Like G1ORF67, 77ORF104 inhibits the growth of S. aureus when expressed intracellularly (23); however, 77ORF104 binds to a different S. aureus protein (DnaI) (23), and its sequence is unrelated to that of G1ORF67 (11% similarity at the amino acid level).
FIG. 3.
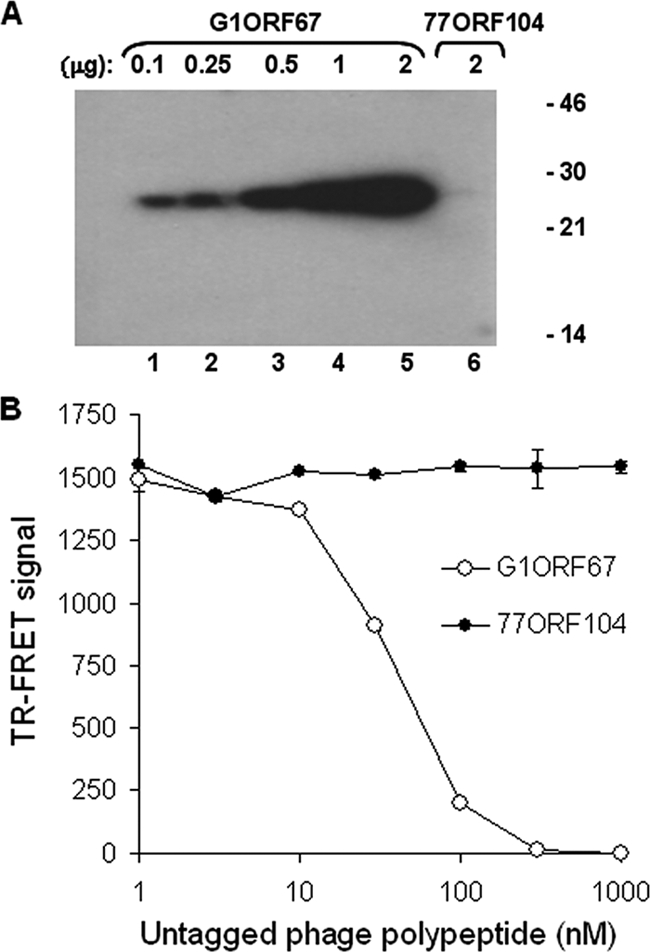
Confirmation of the direct interaction between G1ORF67 and S. aureus σSA. (A) Far-Western analysis demonstrates the direct interaction of σSA with G1ORF67. Increasing amounts of purified His-tagged G1ORF67 (100 ng to 2 μg, lanes 1 to 5) or 2 μg of 77ORF104 (lane 6) were separated by SDS-PAGE, immobilized onto a nitrocellulose membrane, and probed with 32P-labeled σSA. Protein standards (masses in kDa) are indicated on the right. (B) Dose-response study of the interaction between G1ORF67 and σSA by TR-FRET as described in Materials and Methods. Untagged G1ORF67 or 77ORF104 polypeptides were used as competitors. Error bars show standard deviations.
The interaction between G1ORF67 and σSA was also confirmed by the TR-FRET assay (27), a solution-phase assay for monitoring protein-protein interactions (23). Accordingly, the 50% inhibitory concentration, at which 50% of the signal was inhibited, for the interaction between the phage polypeptide and σSA, as determined by competition with untagged G1ORF67, was estimated to be approximately 30 nM (Fig. 3B).
The G1ORF67 polypeptide binds to a region of σSA comprising conserved regions 4.1 and 4.2.
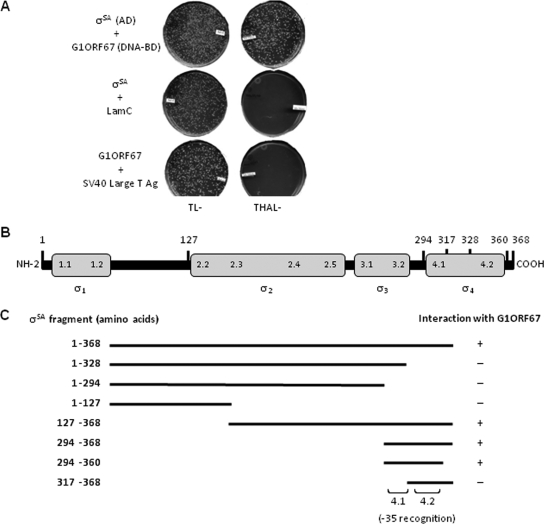
To delineate the minimal region of σSA capable of binding to G1ORF67, we employed the yeast two-hybrid assay. Coexpression of G1ORF67 and full-length σSA allowed growth of the recombinant yeast strain under selective conditions (Fig. 4A), regardless of whether the phage polypeptide or the bacterial protein was expressed as a fusion with the GAL4 transactivation or DNA-binding domain (Fig. 4A and data not shown). This finding indicated that under the conditions of the yeast two-hybrid assay, G1ORF67 and σSA interact, in concurrence with the results from far-Western blotting and the TR-FRET assay presented above. Next, a series of σSA truncation mutants were tested with full-length G1ORF67 as combinatorial pairs for their ability to confer growth on yeast on selective medium. A fragment of 67 amino acids (residues 294 to 360) near the C terminus of σSA was sufficient for its binding to G1ORF67 (Fig. 4C). Interestingly, this region contains conserved regions 4.1 and 4.2, of which region 4.2 is involved in the direct contact with the −35 consensus element (9). Truncation of the N-terminal portion of this 67-amino-acid fragment, resulting in loss of amino acids 294 to 316 (and thereby most of region 4.1, which comprises amino acids 300 to 321 in σSA), led to the loss of G1ORF67 binding (Fig. 4B and C). Similarly, truncation of the C terminus of full-length σSA, resulting in the loss of amino acids 329 to 368 (and thereby practically all of region 4.2, which comprises amino acids 328 to 354 in σSA), resulted in an inability to support growth of the recombinant strain on selective medium (Fig. 4B and C). Hence, the two-hybrid system delineated residues 294 to 360 of σSA, containing regions 4.1 and 4.2, as the minimal region required for binding to G1ORF67.
FIG. 4.
In vivo interaction of G1ORF67 with S. aureus σSA. (A) Yeast two-hybrid analysis. Constructs encoding full-length σSA (amino acids 1 to 368) as a fusion with GAL4 activation domain (AD) and G1ORF67 as a fusion with GAL4 DNA-binding domain (DNA BD) were generated and used to transform the yeast strain. Cotransformants were plated on selective medium lacking tryptophan and leucine (TL−) or lacking tryptophan, histidine, adenine, and leucine (THAL−). (B) Schematic representation of domain organization of S. aureus σSA. (C) Mapping of the minimal domain of S. aureus σSA that interacts with G1ORF67. Constructs encoding full-length σSA (amino acids 1 to 368) or its truncated derivatives were cloned as fusions with the GAL4 activation domain and used in combination with full-length G1ORF67/GAL4 DNA-binding domain in transformations. Cotransformants were plated on selective medium, and the growth or absence of growth of the yeast strain was interpreted as an interaction (+) or absence of an interaction (−), respectively, between σSA and G1ORF67.
The RNAP core-dependent DNA-binding ability of σSA is abolished by G1ORF67.
The finding that the −35 consensus element-binding region of σSA was necessary and sufficient for interaction with G1ORF67 prompted us to test the hypothesis that core-dependent DNA-binding activity of σSA would be impaired in the presence of G1ORF67. As a prelude to this experiment, we developed a solution-phase TR-FRET assay to monitor RNAP core-dependent DNA-binding activity of σSA. This assay used as a probe a biotinylated duplex oligonucleotide representing a truncated version of λPR consisting of the sequence from −41 to −12 (10). This probe thus contains the −35 consensus element but lacks the −10 sequence. The biotinylated, truncated λPR oligonucleotide was incubated with His-tagged σSA and RNAP core enzyme; Eu-conjugated anti-His6 antibodies and APC-conjugated streptavidin were then added. In the absence of G1ORF67, a strong TR-FRET signal from the APC fluorescence acceptor was detected upon excitation of the Eu fluorescence donor (Fig. 5A, 0 nM competitor). Since in this assay RNAP core was strictly required for binding of σSA to the λPR oligonucleotide (data not shown), this result indicated that σSA possesses core-dependent DNA-binding activity on the λPR oligonucleotide containing the consensus −35 element.
FIG. 5.
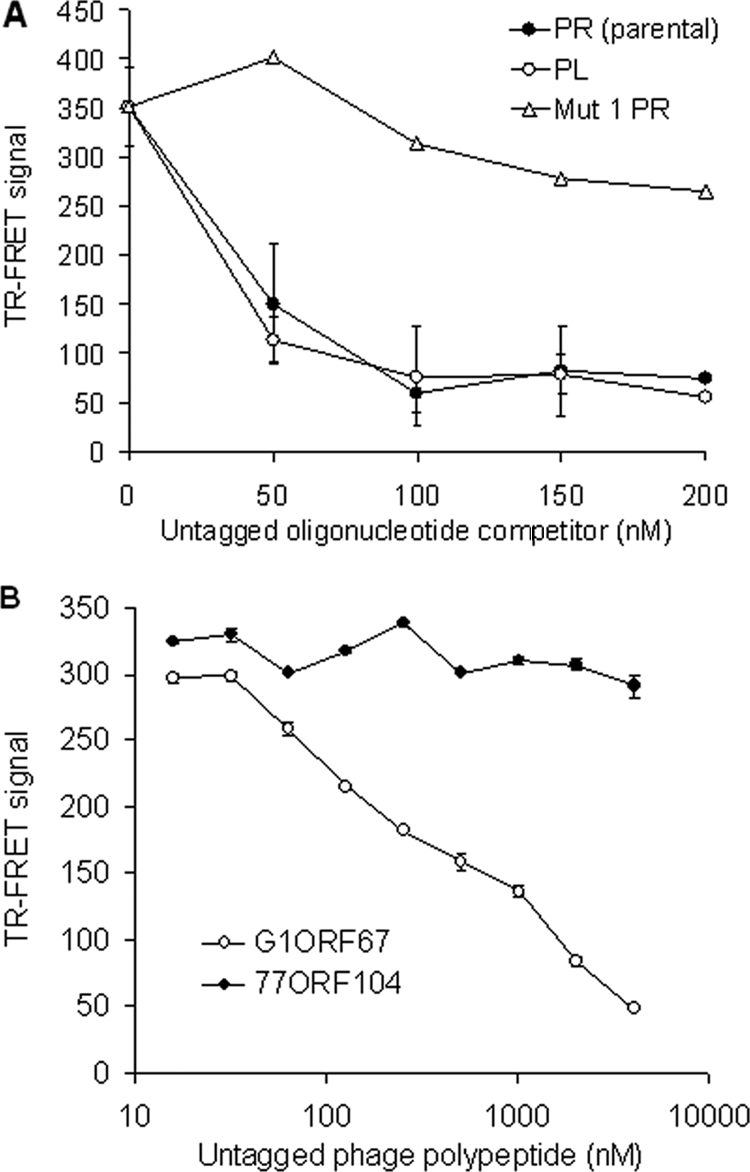
Core-dependent DNA binding of σSA and inhibition by G1ORF67. (A) Competition for core-σSA-λPR oligonucleotide binding as monitored by TR-FRET. Nonbiotinylated λPR and λPL oligonucleotides and a nonbiotinylated mutant version of the λPR oligonucleotide containing a scrambled −35 regulatory sequence element (Mut 1 PR) were used as competitors. (B) Dose-dependent inhibition of the binding of core-σSA to λPR by G1ORF67 as determined by TR-FRET. An unrelated growth-inhibitory phage polypeptide, 77ORF104, was used as negative control. Error bars show standard deviations.
In competition analyses, an excess of nonbiotinylated oligonucleotide was added to the reaction mixture prior to the addition of the Eu and APC conjugates. The parental λPR and λPL oligonucleotides were found to compete efficiently in a concentration-dependent manner for the interaction between RNAP holoenzyme and biotinylated λPR promoter fragment (Fig. 5A). As would be predicted for a specific interaction between the RNAP holoenzyme and a duplex DNA bearing both −35 and −10 consensus sequences, a mutated version of the λPR oligonucleotide containing a scrambled −35 consensus sequence (A-35CTTTG in place of T-35TGACT) failed to compete for binding (Fig. 5A).
The effect of G1ORF67 on the core-dependent DNA-binding activity of σSA was monitored by including increasing amounts of the purified phage polypeptide in the RNAP holoenzyme-promoter oligonucleotide mixture prior to the addition of Eu and APC conjugates. G1ORF67 specifically inhibited the ability of core-σSA to bind to its cognate promoter DNA in a dose-dependent manner (Fig. 5B), whereas the purified control polypeptide 77ORF104 had no impact on core-dependent DNA binding by σSA.
The G1ORF67 polypeptide inhibits σSA-dependent transcription in vitro.
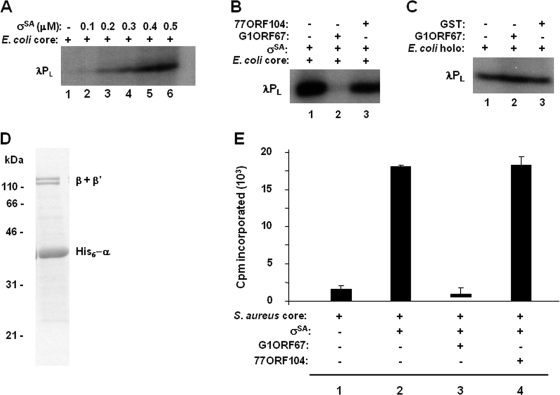
The findings that G1ORF67 both interacted with σSA, as shown above, and specifically inhibited transcription in S. aureus cells (23) prompted us to develop a σSA-dependent in vitro functional assay to directly test the effect of purified G1ORF67 polypeptide on transcription. Initially, in vitro transcription assays comprised the E. coli RNAP core enzyme complemented with purified σSA and a DNA template containing σSA-regulated promoters; radiolabeled transcription products were monitored by gel electrophoresis and autoradiography. Transcription from the λPL promoter was stimulated by σSA in a dose-dependent manner (Fig. 6A). The RNA1 gene from ColE1 was also transcribed by σSA in a dose-dependent manner (data not shown), a finding consistent with previous work of Deora and Misra (6). The addition of purified G1ORF67 polypeptide to the reaction mixture abolished σSA-dependent transcription (Fig. 6B; compare lane 2 to lane 1). To determine whether the inhibition observed was specific to G1ORF67, we tested the effect of purified phage polypeptide 77ORF104 on transcription and found that transcription was unaffected in the presence of the unrelated phage polypeptide (Fig. 6B, lane 3). Interestingly, G1ORF67 lacked inhibitory activity in transcription assays with the E. coli RNAP holoenzyme (Fig. 6C). This outcome is consistent with the inability of G1ORF67 to interact with E. coli σ70 in the yeast two-hybrid system (data not shown). Taken together, these results demonstrate selectivity of G1ORF67 for the primary sigma factor of S. aureus.
FIG. 6.
Inhibition of σSA-dependent transcription in vitro by G1ORF67. (A) Dose-dependent stimulation of the transcriptional activity of E. coli core enzyme from λPL promoter. The amount of σSA is indicated above each lane. The core enzyme was used at 25 nM. Transcription products were separated on a denaturing polyacrylamide gel and visualized by autoradiography. (B) Specific inhibition of σSA-dependent transcription from λPL promoter by G1ORF67. E. coli core enzyme and σSA were used at 25 nM and 500 nM, respectively. Where present, G1ORF67 and the negative-control phage polypeptide (77ORF104) were added to 2 μM (final concentration). (C) G1ORF67 does not inhibit in vitro transcription from the E. coli RNAP holoenzyme (E. coli holo). E. coli holoenzyme was used at 25 nM. Where present, G1ORF67 and the negative-control polypeptide (GST) were added to 2 μM (final concentration). (D) SDS-PAGE analysis of the endogenous RNAP core enzyme purified from S. aureus RN4220. Proteins were resolved by SDS-PAGE and stained with Coomassie blue. Protein standards (masses in kDa) are indicated to the left of the gel image. His6-α and β + β′ indicate the migration positions of the histidine-tagged α subunit and of untagged β and β′ subunits of S. aureus RNAP as determined by tryptic fingerprinting and mass spectrometry. (E) Inhibition of σSA-dependent transcriptional activity of S. aureus core enzyme by G1ORF67 as monitored by TCA precipitation and liquid scintillation counting. The concentrations of S. aureus core enzyme and σSA were 50 nM and 100 nM, respectively. Where present, G1ORF67 and the negative-control phage polypeptide (77ORF104) were added to 10 μM (final concentration). Error bars show standard deviations. +, present; −, absent.
We sought to determine whether G1ORF67 also had an inhibitory effect on σSA-dependent transcription in the context of S. aureus RNAP. To this end, the core enzyme of S. aureus was copurified to near homogeneity (Fig. 6D). Transcription products from the S. aureus RNAP core, from the holoenzyme, and from the holoenzyme in the presence of phage polypeptides were subjected to TCA precipitation and quantitated by liquid scintillation counting. The S. aureus RNAP core enzyme, which had been purified under conditions of high stringency to remove σSA, had only weak activity (Fig. 6E, bar 1). In contrast, when purified σSA was added back to the core enzyme, transcription was stimulated 10-fold (Fig. 6E, bar 2). As was seen above with σSA-dependent transcription by the E. coli RNAP core, the ability of σSA to stimulate transcription by the S. aureus core was abolished with G1ORF67 (Fig. 6E, bar 3) but not with 77ORF104 (Fig. 6E, bar 4). Under these conditions, the 50% inhibitory concentration of G1ORF67 for the in vitro transcription reaction was approximately 0.2 μM (data not shown). These results clearly indicated that G1ORF67 inhibits σSA-dependent transcription regardless of whether σSA is in association with the RNAP core enzyme of S. aureus or E. coli. Together, the findings illustrate that G1ORF67 interacts with σSA and acts as a transcription inhibitor at σSA-regulated promoters.
DISCUSSION
In the present study, we characterized the mechanism by which G1ORF67, a staphylococcal bacteriophage polypeptide, inhibits the growth of S. aureus: it binds to the primary sigma factor of S. aureus and inhibits transcription at σSA-dependent promoters. These results extend our previous observation of specific inhibition of transcription in S. aureus cells upon expression of G1ORF67 in that they suggest a mechanism by which the phage polypeptide may inhibit S. aureus transcription at −35 consensus promoters. This activity may be central to host shutoff early in the G1 phage infective cycle. Given both its direct interaction with σSA and its ability to inhibit σSA function, we conclude that G1ORF67 is a phage-encoded anti-σ factor directed against σSA of S. aureus.
AsiA of phage T4 is a prototypic anti-σ factor with specificity for σ70, the primary sigma factor of E. coli (32). Recent models propose that AsiA first binds to free σ70 and that this complex then binds to the RNAP core to form AsiA-bound holoenzyme (1, 12, 22, 24, 38, 42). The AsiA binding determinants of σ70 have been mapped to the C-terminal region; they consist of conserved regions 4.1 and 4.2 (29, 39, 40), of which region 4.2 is primarily responsible for the binding of σ70 to the canonical −35 promoter element (4, 24, 29). Binding of AsiA to σ70 has been demonstrated to preclude the binding of σ70 to the canonical −35 promoter element, thereby inhibiting transcription of cellular genes that require an authentic −35 element (12, 36, 42). Similarly to AsiA, Rsd was shown to bind to region 4.2 and to inhibit transcription from σ70 promoters (15, 40); however, in contrast to AsiA, Rsd prevents the binding of σ70 to the core (35, 46). Likewise, region 4 of the flagellar-specific sigma factor σ28 of Salmonella enterica serovar Typhimurium is required for binding the anti-sigma factor FlgM (20).
G1ORF67 is unrelated to AsiA (21% similarity at the amino acid level) and only marginally similar to Rsd (20% identity and 33% similarity), suggesting that these anti-sigma factors may inhibit transcription at −35 consensus promoters by different mechanisms. Alternatively, despite the significant level of amino acid sequence homology between region 4 of σ70 and σSA (43% identity and 62% similarity), anti-sigma factors may require different interactions for transcription inhibition or sigma appropriation (11). Using the yeast two-hybrid system under experimental conditions that validated the interaction between G1ORF67 and σSA, we were unable to detect AsiA-σSA and G1ORF67-σ70 interactions (data not shown). There are at least two possible mechanisms that could explain the observed inhibitory effect of G1ORF67 on σSA, as demonstrated in functional assays. The first possibility is that the phage polypeptide interacts with both free and RNAP core-bound σSA, thereby preventing its binding to the −35 promoter sequence and ultimately inhibiting transcription of genes requiring an authentic −35 consensus element. Alternatively, G1ORF67 may prevent the binding of σSA to the RNAP core enzyme, thereby inactivating both σSA and the core. However, our finding that in affinity chromatography, the α, β, and β′ subunits were coeluted along with σSA (Fig. 2) supports the formation of a ternary complex consisting of G1ORF67-σSA-RNAP core, as was found for AsiA-σ70-core (38). This finding argues against a mechanism by which G1ORF67 prevents σSA from binding to the core enzyme. That the G1ORF67 binding site was mapped to a region containing regions 4.1 and 4.2 of σSA supports this possibility and predicts that in the putative ternary complex, the surface on σSA required for binding to the −35 recognition element would be occluded by G1ORF67.
Although our results from σSA-DNA-binding assays strongly support the notion that inhibition of σSA-DNA binding by G1ORF67 is due to interference with the required −35 contact region, they did not address whether this interference is direct (amino acid residues of σSA required for its direct contact with the −35 consensus element are also involved in the interaction with G1ORF67) or indirect (through conformational change rendering regions 4.1 and 4.2 of σSA inaccessible for binding to the −35 consensus element).
While the expression of G1ORF67 in growing S. aureus cells results in rapid and profound inhibition of host transcription (23), one of the outstanding questions raised by this study is the functional implication of G1ORF67-σSA interaction for phage biology during the infection. That is, how is the genome of phage G1 transcribed if σSA activity is impaired? Additionally, which phage G1 genes govern the expression of early versus middle or late functions and does G1ORF67 play a role in this process? In the context of phage T4, AsiA protein cooperates with T4 MotA to coactivate transcription from T4 middle promoters (3, 33, 34). Based on amino acid sequence homology, we did not find a MotA homologue in the genome of phage G1. This is perhaps not surprising given the absence of sequence homology between G1ORF67 and T4 AsiA. We predict that the binding of G1ORF67 to σSA and the resulting inhibition of host σSA-dependent RNAP activity trigger a shift in the transcriptional profile of phage genes during infection. The importance of such a shift in gene expression would be to promote the expression of genes involved in phage maturation and release. G1ORF67 would thus appear to be one of the central polypeptides in phage G1's strategy to appropriate host RNAP and redirect it to phage reproduction.
Acknowledgments
We thank Tom Parr, Adel Rafai Far, and Geoff McKay for their advice and critical reading of the manuscript. We are also indebted to Marko Virta for his generous gift of the pTOO21 vector. We thank all the members of Targanta Therapeutics for their support during the course of this project.
We thank the National Research Council of Canada for financial support. M.D., J.L., and G.M. are recipients of an Industrial Research Fellowship from the National Sciences and Engineering Research Council of Canada (NSERC).
Footnotes
Published ahead of print on 17 April 2009.
REFERENCES
- 1.Adelman, K., G. Orsini, A. Kolb, L. Graziani, and E. N. Brody. 1997. The interaction between the AsiA protein of bacteriophage T4 and the sigma70 subunit of Escherichia coli RNA polymerase. J. Biol. Chem. 27227435-27443. [DOI] [PubMed] [Google Scholar]
- 2.Beckett, D., E. Kovaleva, and P. J. Schatz. 1999. A minimal peptide substrate in biotin holoenzyme synthetase-catalyzed biotinylation. Protein Sci. 8921-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonocora, R. P., G. Caignan, C. Woodrell, M. H. Werner, and A. M. Hinton. 2008. A basic/hydrophobic cleft of the T4 activator MotA interacts with the C-terminus of E. coli sigma70 to activate middle gene transcription. Mol. Microbiol. 69331-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burgess, R. R., and L. Anthony. 2001. How sigma docks to RNA polymerase and what sigma does. Curr. Opin. Microbiol. 4126-131. [DOI] [PubMed] [Google Scholar]
- 5.Campbell, E. A., N. Korzheva, A. Mustaev, K. Murakami, S. Nair, A. Goldfarb, and S. A. Darst. 2001. Structural mechanism for rifampicin inhibition of bacterial RNA polymerase. Cell 104901-912. [DOI] [PubMed] [Google Scholar]
- 6.Deora, R., and T. K. Misra. 1996. Characterization of the primary sigma factor of Staphylococcus aureus. J. Biol. Chem. 27121828-21834. [DOI] [PubMed] [Google Scholar]
- 7.Deora, R., T. Tseng, and T. K. Misra. 1997. Alternative transcription factor σSB of Staphylococcus aureus: characterization and role in transcription of the global regulatory locus sar. J. Bacteriol. 1796355-6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deretic, V., and W. M. Konyecsni. 1989. Control of mucoidy in Pseudomonas aeruginosa: transcriptional regulation of algR and identification of the second regulatory gene, algQ. J. Bacteriol. 1713680-3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dove, S. L., S. A. Darst, and A. Hochschild. 2003. Region 4 of sigma as a target for transcription regulation. Mol. Microbiol. 48863-874. [DOI] [PubMed] [Google Scholar]
- 10.Fenton, M. S., S. J. Lee, and J. D. Gralla. 2000. Escherichia coli promoter opening and −10 recognition: mutational analysis of σ70. EMBO J. 191130-1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hinton, D. M., S. Pande, N. Wais, X. B. Johnson, M. Vuthoori, A. Makela, and I. Hook-Barnard. 2005. Transcriptional takeover by sigma appropriation: remodelling of the sigma70 subunit of Escherichia coli RNA polymerase by the bacteriophage T4 activator MotA and co-activator AsiA. Microbiology 1511729-1740. [DOI] [PubMed] [Google Scholar]
- 12.Hinton, D. M., and S. Vuthoori. 2000. Efficient inhibition of Escherichia coli RNA polymerase by the bacteriophage T4 AsiA protein requires that AsiA binds first to free sigma70. J. Mol. Biol. 304731-739. [DOI] [PubMed] [Google Scholar]
- 13.Hughes, K. T., and K. Mathee. 1998. The anti-sigma factors. Annu. Rev. Microbiol. 52231-286. [DOI] [PubMed] [Google Scholar]
- 14.Ishihama, A. 2000. Functional modulation of Escherichia coli RNA polymerase. Annu. Rev. Microbiol. 54499-518. [DOI] [PubMed] [Google Scholar]
- 15.Jishage, M., D. Dasgupta, and A. Ishihama. 2001. Mapping of the Rsd contact site on the sigma 70 subunit of Escherichia coli RNA polymerase. J. Bacteriol. 1832952-2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jishage, M., and A. Ishihama. 1998. A stationary phase protein in Escherichia coli with binding activity to the major sigma subunit of RNA polymerase. Proc. Natl. Acad. Sci. USA 954953-4958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaelin, W. G., Jr., W. Krek, W. R. Sellers, J. A. DeCaprio, F. Ajchenbaum, C. S. Fuchs, T. Chittenden, Y. Li, P. J. Farnham, M. A. Blanar, et al. 1992. Expression cloning of a cDNA encoding a retinoblastoma-binding protein with E2F-like properties. Cell 70351-364. [DOI] [PubMed] [Google Scholar]
- 18.Kato, J., L. Chu, K. Kitano, J. D. DeVault, K. Kimbara, A. M. Chakrabarty, and T. K. Misra. 1989. Nucleotide sequence of a regulatory region controlling alginate synthesis in Pseudomonas aeruginosa: characterization of the algR2 gene. Gene 8431-38. [DOI] [PubMed] [Google Scholar]
- 19.Kullik, I. I., and P. Giachino. 1997. The alternative sigma factor sigmaB in Staphylococcus aureus: regulation of the sigB operon in response to growth phase and heat shock. Arch. Microbiol. 167151-159. [DOI] [PubMed] [Google Scholar]
- 20.Kutsukake, K., S. Iyoda, K. Ohnishi, and T. Iino. 1994. Genetic and molecular analyses of the interaction between the flagellum-specific sigma and anti-sigma factors in Salmonella typhimurium. EMBO J. 134568-4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kwan, T., J. Liu, M. DuBow, P. Gros, and J. Pelletier. 2005. The complete genomes and proteomes of 27 Staphylococcus aureus bacteriophages. Proc. Natl. Acad. Sci. USA 1025174-5179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lambert, L. J., Y. Wei, V. Schirf, B. Demeler, and M. H. Werner. 2004. T4 AsiA blocks DNA recognition by remodeling sigma70 region 4. EMBO J. 232952-2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu, J., M. Dehbi, G. Moeck, F. Arhin, P. Bauda, D. Bergeron, M. Callejo, V. Ferretti, N. Ha, T. Kwan, J. McCarty, R. Srikumar, D. Williams, J. J. Wu, P. Gros, J. Pelletier, and M. DuBow. 2004. Antimicrobial drug discovery through bacteriophage genomics. Nat. Biotechnol. 22185-191. [DOI] [PubMed] [Google Scholar]
- 24.Lonetto, M. A., V. Rhodius, K. Lamberg, P. Kiley, S. Busby, and C. Gross. 1998. Identification of a contact site for different transcription activators in region 4 of the Escherichia coli RNA polymerase sigma70 subunit. J. Mol. Biol. 2841353-1365. [DOI] [PubMed] [Google Scholar]
- 25.Lutz, R., and H. Bujard. 1997. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 251203-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maeda, H., N. Fujita, and A. Ishihama. 2000. Competition among seven Escherichia coli sigma subunits: relative binding affinities to the core RNA polymerase. Nucleic Acids Res. 283497-3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mathis, G. 1995. Probing molecular interactions with homogeneous techniques based on rare earth cryptates and fluorescence energy transfer. Clin. Chem. 411391-1397. [PubMed] [Google Scholar]
- 28.Mika, F., and R. Hengge. 2005. A two-component phosphotransfer network involving ArcB, ArcA, and RssB coordinates synthesis and proteolysis of sigmaS (RpoS) in E. coli. Genes Dev. 192770-2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Minakhin, L., J. A. Camarero, M. Holford, C. Parker, T. W. Muir, and K. Severinov. 2001. Mapping the molecular interface between the sigma(70) subunit of E. coli RNA polymerase and T4 AsiA. J. Mol. Biol. 306631-642. [DOI] [PubMed] [Google Scholar]
- 30.Morikawa, K., Y. Inose, H. Okamura, A. Maruyama, H. Hayashi, K. Takeyasu, and T. Ohta. 2003. A new staphylococcal sigma factor in the conserved gene cassette: functional significance and implication for the evolutionary processes. Genes Cells 8699-712. [DOI] [PubMed] [Google Scholar]
- 31.Murakami, K. S., and S. A. Darst. 2003. Bacterial RNA polymerases: the wholo story. Curr. Opin. Struct. Biol. 1331-39. [DOI] [PubMed] [Google Scholar]
- 32.Orsini, G., M. Ouhammouch, J. P. Le Caer, and E. N. Brody. 1993. The asiA gene of bacteriophage T4 codes for the anti-σ70 protein. J. Bacteriol. 17585-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ouhammouch, M., K. Adelman, S. R. Harvey, G. Orsini, and E. N. Brody. 1995. Bacteriophage T4 MotA and AsiA proteins suffice to direct Escherichia coli RNA polymerase to initiate transcription at T4 middle promoters. Proc. Natl. Acad. Sci. USA 921451-1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ouhammouch, M., G. Orsini, and E. N. Brody. 1994. The asiA gene product of bacteriophage T4 is required for middle mode RNA synthesis. J. Bacteriol. 1763956-3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Patikoglou, G. A., L. F. Westblade, E. A. Campbell, V. Lamour, W. J. Lane, and S. A. Darst. 2007. Crystal structure of the Escherichia coli regulator of sigma70, Rsd, in complex with sigma70 domain 4. J. Mol. Biol. 372649-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pineda, M., B. D. Gregory, B. Szczypinski, K. R. Baxter, A. Hochschild, E. S. Miller, and D. M. Hinton. 2004. A family of anti-sigma70 proteins in T4-type phages and bacteria that are similar to AsiA, a transcription inhibitor and co-activator of bacteriophage T4. J. Mol. Biol. 3441183-1197. [DOI] [PubMed] [Google Scholar]
- 37.Sensi, P. 1983. History of the development of rifampin. Rev. Infect. Dis. 5(Suppl. 3)S402-S406. [DOI] [PubMed] [Google Scholar]
- 38.Severinova, E., K. Severinov, and S. A. Darst. 1998. Inhibition of Escherichia coli RNA polymerase by bacteriophage T4 AsiA. J. Mol. Biol. 2799-18. [DOI] [PubMed] [Google Scholar]
- 39.Sharma, U. K., and D. Chatterji. 2006. Both regions 4.1 and 4.2 of E. coli sigma(70) are together required for binding to bacteriophage T4 AsiA in vivo. Gene 376133-143. [DOI] [PubMed] [Google Scholar]
- 40.Sharma, U. K., and D. Chatterji. 2008. Differential mechanisms of binding of anti-sigma factors Escherichia coli Rsd and bacteriophage T4 AsiA to E. coli RNA polymerase lead to diverse physiological consequences. J. Bacteriol. 1903434-3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shaw, L. N., C. Lindholm, T. K. Prajsnar, H. K. Miller, M. C. Brown, E. Golonka, G. C. Stewart, A. Tarkowski, and J. Potempa. 2008. Identification and characterization of sigma, a novel component of the Staphylococcus aureus stress and virulence responses. PLoS ONE 3e3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Urbauer, J. L., K. Adelman, R. J. Urbauer, M. F. Simeonov, J. M. Gilmore, M. Zolkiewski, and E. N. Brody. 2001. Conserved regions 4.1 and 4.2 of sigma(70) constitute the recognition sites for the anti-sigma factor AsiA, and AsiA is a dimer free in solution. J. Biol. Chem. 27641128-41132. [DOI] [PubMed] [Google Scholar]
- 43.Vicente, M., K. F. Chater, and V. De Lorenzo. 1999. Bacterial transcription factors involved in global regulation. Mol. Microbiol. 338-17. [DOI] [PubMed] [Google Scholar]
- 44.Villain-Guillot, P., L. Bastide, M. Gualtieri, and J. P. Leonetti. 2007. Progress in targeting bacterial transcription. Drug Discov. Today 12200-208. [DOI] [PubMed] [Google Scholar]
- 45.Wassarman, K. M., and G. Storz. 2000. 6S RNA regulates E. coli RNA polymerase activity. Cell 101613-623. [DOI] [PubMed] [Google Scholar]
- 46.Westblade, L. F., L. L. Ilag, A. K. Powell, A. Kolb, C. V. Robinson, and S. J. Busby. 2004. Studies of the Escherichia coli Rsd-sigma70 complex. J. Mol. Biol. 335685-692. [DOI] [PubMed] [Google Scholar]