SUMMARY
The involvement of Toll-like receptor 4 (TLR4) in immunity against human herpesviruses has not been previously demonstrated. We show that infection of endothelial cells with Kaposi sarcoma herpesvirus (KSHV), a human oncogenic virus, leads to rapid suppression of TLR4 expression. This is a mechanism of immune escape as TLR4 mediates innate immunity against KSHV. In vitro, cells lacking TLR4 are more susceptible to KSHV infection, whereas activation of TLR4 protects cells from infection. In vivo, HIV-1-infected individuals carrying a mutant TLR4 allele appear more likely to have multicentric Castleman's disease, a lymphoproliferation associated with enhanced KSHV replication. ERK activation by KSHV structural proteins and the KSHV-encoded vGPCR plays a key role in the TLR4 downregulation, whereas the KSHV vIRF1 also contributes to this effect. Our findings reveal a role for TLR4 in innate immunity against herpesviruses and suggest the potential use of TLR4 agonists for the treatment of KSHV-related neoplasms.
INTRODUCTION
Toll-like receptors (TLR) are a family of membrane pattern recognition receptors recognizing highly conserved metabolic products of invading pathogens, termed pathogen-associated molecular patterns (PAMPs). TLRs are type I glycoproteins consisting of an extracellular PAMP recognition domain and a cytoplasmic Toll/interleukin-1 receptor homology signaling domain. Effective pathogen recognition can be achieved through a small number of TLRs (ten in humans). As PAMPs are indispensable for the invading pathogen, no escape mutants can evolve. Upon PAMP recognition, TLR activation leads to the initiation of an innate inflammatory response through activation of the nuclear factor κB (NF-κB), mitogen-activated protein kinase (MAPK), and interferon regulatory factor (IRF) pathways (Akira et al., 2006; Janeway and Medzhitov, 2002). In parallel, a complex regulatory network of TLR signaling exists to regulate this innate response (Liew et al., 2005).
The first characterized mammalian TLR was the human TLR4 (Medzhitov et al., 1997). It is expressed by cells of the immune system, such as dendritic cells, macrophages, and B lymphocytes, but also by endothelial cells. TLR4 acts in combination with serum or membrane CD14 and the adaptor protein MD2 as the receptor of bacterial lipopolysaccharide (LPS), playing a key role in innate responses against Gram-negative bacteria (Akira et al., 2006). TLR4 also recognizes other ligands such as the glycoprotein F of respiratory syncytia virus (RSV) (Kurt-Jones et al., 2000), as well as endogenous proteins including heat shock proteins, fibrinogen, and high-mobility-group box 1 (Apetoh et al., 2007; Marshak-Rothstein, 2006). Based on its exogenous ligands, TLR4 is generally associated with responses to Gram-negative bacteria and RSV. Notably, two single nucleotide polymorphisms (SNP) in the human TLR4 gene (Asp299Gly resulting from an A/G substitution, rs4986790, and Ile399Thr resulting from a C/T substitution, rs4986791) have been associated with severity of RSV-associated bronchiolitis and Gram-negative sepsis (Schroder and Schumann, 2005). Activation of TLR4 by a mouse RNA virus (mouse mammary tumor virus; MMTV) is thought to lead to subversion of the immune response (Burzyn et al., 2004). Furthermore, TLR4 has been implicated in responses against hepatitis C virus (Machida et al., 2006) and in mouse models of hepatitis B virus infection (Isogawa et al., 2005), whereas TLR4 activation by LPS in mouse macrophages inhibits replication of murine herpesvirus 68 (MHV68) by induction of an IRF3- and IRF7-mediated response (Doyle et al., 2002). However, a role for TLR4 in responses against human herpesviruses has not previously been demonstrated.
Kaposi sarcoma-associated herpesvirus (KSHV; also known as human herpesvirus 8) is an oncogenic human γ-herpesvirus associated with the development of Kaposi sarcoma (KS), a neoplasm of lymphatic endothelial-type cells (Wang et al., 2004), and certain lymphoproliferations, such as multicentric Castleman's disease (MCD) (Boshoff and Weiss, 2002). As a herpesvirus, KSHV establishes life-long infection in its host. In latently infected cells, only a fraction of viral genes are expressed, maintaining the viral episome, and providing a proliferative and immune escape advantage to infected cells. The viral genes expressed during latency include the latency-associated nuclear antigen 1 (LANA-1), viral cyclin, and viral FLICE inhibitory protein (vFLIP; reviewed in Moore and Chang [2003]). When infected cells enter a productive (lytic) program of gene expression, most of the viral genes are expressed, virions are formed, and new cells are recruited to be infected. Lytic KSHV genes include the viral G protein-coupled receptor (vGPCR), which is a multifunctional viral oncogene homologous to the human IL8 receptor, and the viral IRF1 (vIRF1), which inhibits interferon signaling (reviewed in Cannon [2007] and Moore and Chang [2003]). As herpesviruses have evolved to establish persistent infection (and therefore an immunological equilibrium with their host), most of the viral mechanisms of immune evasion occur during lytic and primary infection (Coscoy, 2007; Moore and Chang, 2003). This equilibrium is disturbed in immunosuppressed individuals, resulting in uncontrolled proliferation of infected cells and occasional development of cancer. Thus, the incidence of KSHV-related malignancies is dramatically increased among transplant recipients during iatrogenic immunosuppression and in HIV-1-infected individuals, where KS is the most common AIDS-related cancer (Boshoff and Weiss, 2002).
Although several aspects of the KSHV-specific immune responses and viral mechanisms of immune escape have been studied (Coscoy, 2007), the role and regulation of TLRs in innate immunity against KSHV remain unknown. Here we show that TLR4 mRNA levels and protein expression are significantly downregulated upon primary KSHV infection of human lymphatic endothelial cells (LEC). Using macrophages from TLR4-knockout mice, siRNA-mediated silencing of TLR4 in human LEC, and activation of endogenous TLR4 signaling, we demonstrate that TLR4 mediates an innate immune response against KSHV. The in vivo relevance of these findings is demonstrated by the higher incidence of the Asp299Gly TLR4 SNP among HIV-1-infected individuals with MCD, a KSHV-driven lymphoproliferation associated with high KSHV load. Consistently, primary LEC carrying this TLR4 allele are more susceptible to KSHV infection than LEC carrying the wild-type TLR4 gene. Furthermore, we demonstrate that KSHV suppresses TLR4 through activation of the extracellular signal-regulated kinase (ERK) MAPK pathway during viral binding (viral gene expression-independent mechanism), and by way of vIRF1 and ERK activation by vGPCR (viral gene expression-dependent mechanism).
RESULTS
KSHV Infection of Lymphatic Endothelial Cells Leads to Suppression of TLR4 Expression
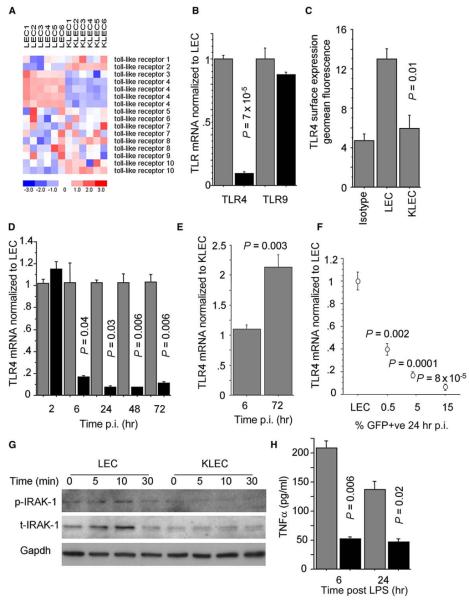
We have previously determined the immune transcriptional signature of KSHV-infected LEC (KLEC) using gene expression microarray (GEM) profiling (Lagos et al., 2007). We analyzed these data focusing on TLRs and observed a significant downregulation of TLR4 levels (false discovery rate threshold q < 0.005; Figure 1A). The only other significant effects of infection on TLR expression were an upregulation of TLR2 and a downregulation of TLR3. However, the normalized signal for basal TLR3 mRNA levels in LEC was low (Figure S1A), suggesting that TLR3 is not significantly expressed in these cells. Thus, we considered only TLR4 as significantly downregulated after infection. TLR4 acts as the functional LPS receptor, in the presence of soluble CD14 (sCD14), on dermal microvasculature, umbilical vein, and lymphatic endothelial cells (Faure et al., 2000; Pegu et al., 2008; Pugin et al., 1993). In our system, we confirmed that TLR4 is expressed and functional in response to LPS (through upregulation of ICAM-1) in primary LEC (Figures S1B and S1C). Following this, we validated our GEM data by quantitative reverse transcriptase polymerase chain reaction (qRTPCR) and confirmed that TLR4 mRNA levels are downregulated after KSHV infection, whereas mRNA levels of TLR9, the TLR typically associated with responses to DNA viruses (Guggemoos et al., 2008; Muller et al., 2008), were not affected by KSHV (Figure 1B). The TLR4 mRNA decrease led to significant downregulation of TLR4 surface expression (reducing TLR4 geomean fluorescence to levels similar to isotype control; Figures 1C and S1D). Notably, TLR4 mRNA levels were downregulated as early as 6 hr postinfection (p.i.) and the effect persisted up to 72 hr p.i. (Figure 1D). Early TLR4 downregulation (6 hr p.i.) also occurred when LEC were exposed to UV-inactivated KSHV (Figure 1E) inferring that structural KSHV proteins contribute to this effect. However, at later time points (72 hr p.i.), TLR4 levels significantly increased in LEC exposed to UV-inactivated KSHV compared to KLEC (Figure 1E). Furthermore, TLR4 downregulation correlated with number of KSHV-infected cells (Figure 1F). These findings indicated that KSHV employed mechanisms independent of and driven by viral gene expression to downregulate TLR4. To exclude any confounding effects of contaminating traces of LPS on TLR4 expression, we showed that LPS did not affect basal TLR4 expression in LEC (Figure S1E) and that the TLR4 downregulation still occurred when the infection was performed in the presence of polymyxin-B (an inhibitor of the LPS-TLR4 interaction; Figure S1F).
Figure 1. KSHV Suppresses TLR4 Expression.
(A) Heatmap of TLR expression in LEC and KSHV-infected LEC (KLEC; 72 hr p.i.), based on GEM profiling. Rows correspond to TLR probe sets. Red and blue denote high and low expression respectively, the color scale indicating units of standard deviation from the mean.
(B) TLR4 and TLR9 mRNA levels in LEC (gray bars) and KLEC (black bars) 72 hr p.i.
(C) Surface TLR4 expression in LEC and GFP-expressing KLEC (72 hr p.i.).
(D) Time course of TLR4 mRNA levels in LEC (gray bars) and KLEC (black bars). For each time point, mRNA levels are normalized to LEC TLR4 mRNA levels.
(E) TLR4 mRNA levels in LEC infected with UV-inactivated KSHV normalized to KLEC TLR4 mRNA at 6 hr and 72 hr p.i. P value between UV-inactivated samples and KLEC is shown.
(F) TLR4 mRNA levels in relation to percentage of GFP-expressing KLEC (24 hr p.i.). Values normalized to LEC and P values shown.
(G) Levels of phosphorylated and total IRAK1 (p-IRAK1 and t-IRAK1, respectively) following stimulation of LEC and KLEC (72 hr p.i.) with 100ng/ml LPS/1 μg/ml sCD14 for the indicated time. GAPDH levels are shown as loading controls. Note that the increase of the t-IRAK1 signal in LEC following LPS stimulation represents an increase in the activated form of the protein rather than an increase in the total protein levels (Hatao et al., 2004).
(H) Levels of secreted TNF-α following stimulation of LEC (gray bars) and KLEC (black bars; 72 hr p.i.) with LPS/sCD14 for 6 hr and 24 hr. For (B)–(D) and (H), P values between KLEC and LEC, samples are shown. In all panels, error bars represent standard deviation.
Next we assessed the functional relevance of the TLR4 downregulation by KSHV. One of the first events upon TLR4 activation involves the phosphorylation, ubiquitination, and degradation of IRAK1 (Gottipati et al., 2008). In LEC, we observed transient phosphorylation of IRAK1 within 5–10 min of stimulation with LPS followed by protein degradation. IRAK1 activation after LPS stimulation was significantly impaired in KLEC (72 hr p.i.; Figure 1G). Similarly, transcriptional induction and secretion of TNF-α after stimulation with LPS (one of the endpoints of the TLR4 signaling cascade) was significantly impaired in KLEC (72 hr p.i.; Figure 1H).
TLR4 Mediates an Innate Immune Response Against KSHV
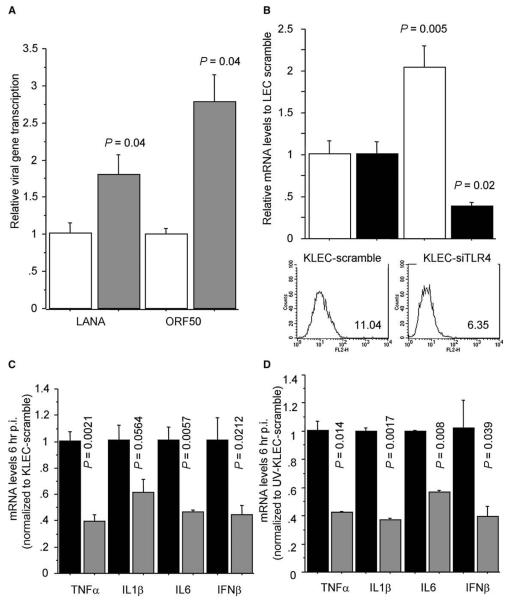
The rapid TLR4 downregulation during primary KSHV infection of LEC suggested that TLR4 might play a role in the innate response against this virus. Although KSHV does not infect mice with functional T lymphocytes, it has been shown using NOD/SCID mice that macrophages are one of the viral cellular targets in vivo (Parsons et al., 2006). Therefore, we first tested the role of TLR4 in the response against KSHV using peritoneal macrophages from C57BL10/ScNJ mice, which lack the Tlr4 gene, and wild-type controls (C57BL10/ScSnJ). KSHV infection of macrophages from the Tlr4 knockout mice led to a 2- to 3-fold higher viral gene expression compared to wild-type controls (Figure 2A).
Figure 2. Increased Susceptibility of Cells Lacking TLR4 to KSHV Infection.
(A) mRNA levels of KSHV-encoded LANA-1 and ORF50 transcripts after KSHV infection (48 hr p.i.) of peritoneal macrophages from C57BL10/ScSnJ mice (wild-type, white bars) and C57BL10/ScNJ mice (Tlr4−/−, gray bars). Levels normalized to average gene expression in KSHV-infected C57BL10/ScSnJ macrophages. P values indicate statistical significance of changes between C57BL10/ScNJ and C57BL10/ScSnJ macrophages.
(B) LANA-1 mRNA levels (white bars) in KLEC (48 hr p.i.). LEC were transfected with nontargeting siRNA (KLEC scramble) or TLR4-targeting siRNA (KLEC-siTLR4) 48 hr before KSHV infection. TLR4 surface expression (histograms; TLR4 geometrical mean fluorescence shown) and mRNA levels (black bars) were determined 48 hr post siRNA transfection. mRNA levels are normalized to KLEC-scramble levels.
(C) TNF-α, IL1-β, IL-6, and IFN-β mRNA levels 6 hr after KSHV infection of KLEC scramble (black bars) or KLEC-siTLR4 (gray bars). Levels are normalized to KLEC scramble.
(D) As in (C), but after exposure of LEC to UV-inactivated KSHV. In (B)–(D), P values indicate statistical significance of changes between mRNA levels in KLEC scramble and KLEC-siTLR4. In all panels, error bars represent standard deviation.
As differences in the activation of mouse and human TLR4 genes have been reported (Kawasaki et al., 2001) and KSHV is considered to be species restricted, we next tested the role of TLR4 in the innate response during primary infection of human LEC. Similar to the data we obtained using macrophages from Tlr4 knockout mice, infection of human LEC after siRNA-mediated TLR4 silencing resulted in higher viral gene expression (Figure 2B). This correlated with an impaired innate response as siRNA-mediated TLR4 silencing also caused impaired induction of TNF-α, IL1-β, IL-6, and IFN-β after KSHV infection of LEC (Figure 2C). Together, these results demonstrated that TLR4 contributes to the innate response during primary KSHV infection. This antiviral activity is likely to be the result of TLR4 activation by a viral envelope glycoprotein, as exposure of LEC to UV-inactivated KSHV also led to a TLR4-dependent induction of TNF-α, IL1-β, IL-6, and IFN-β (Figure 2D).
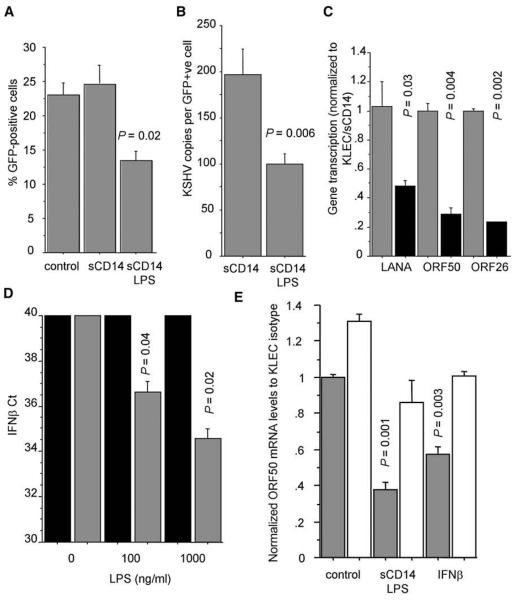
Next, we investigated whether it was possible to inhibit KSHV infection by activating TLR4 before exposure to the virus. Activation of TLR4 signaling with LPS resulted in decreased number of infected cells (Figure 3A), a reduction in the number of viral genome copies per infected cell (Figure 3B), and lower latent, early lytic, and late lytic viral gene expression (Figure 3C). To exclude nonspecific effects of LPS on KSHV infection, we showed that LPS treatment did not affect KSHV infection of macrophages from C57BL10/ScNJ Tlr4−/− mice or human 293T cells (Figure S2), which do not express TLR4 and do not respond to LPS. Furthermore, we showed that TLR4 activation by LPS resulted in IFN-β induction (Figure 3D), and that inhibition of type I interferon activity reversed the protective effect of TLR4 activation against KSHV infection (Figure 3E).
Figure 3. TLR4 Activation by LPS Protects LEC from KSHV Infection.
(A) Percentage of GFP-expressing cells in KLEC, KLEC pretreated with sCD14 (1 μg/ml), and KLEC pretreated with sCD14 (1 μg/ml) and LPS (100 ng/ml).
(B) Average number of KSHV genomes per GFP-expressing cell. Cells were pretreated as described in (A).
(C) mRNA levels of KSHV latent, immediate lytic, and late lytic genes (LANA, ORF50, and ORF26, respectively) in KLEC pretreated with sCD14 (gray bars) or sCD14 and LPS (black). Levels are normalized to sCD14 pretreated KLEC. In (A)–(C), P values between KLEC pretreated with sDC14 and sCD14/LPS are shown, and experiments are performed 72 hr p.i.
(D) IFN-β expression in LEC treated with PBS (black bars) or 1 μg/ml sCD14 (gray bars) and varying amounts of LPS (x axis) for 6 hr. Ct values (y axis) indicate cycle number when PCR products become detectable. Samples are normalized for GAPDH Ct. P values between PBS/LPS- and sCD14/LPS-treated LEC for each LPS concentration are shown.
(E) ORF50 mRNA levels in KLEC and in KLEC pre-treated with sCD14 and LPS or IFN-β (1000U/ml) in the presence of an IFNaRII neutralizing antibody (white bars) or an isotype control (gray bars). IFN-β treatment was used to assess the neutralizing efficiency of the antibody. P values indicate significant changes compared with to KLEC control. In all panels, error bars represent standard deviation.
Increased Incidence of the Asp299Gly TLR4 SNP among HIV-1-Infected Individuals with MCD
Based on the above results we investigated whether SNPs associated with the deregulation of TLR4 activity (Ferwerda et al., 2007; Tulic et al., 2007) result in higher KSHV load in vitro and in vivo and therefore development of KSHV-associated neoplasms. First, we compared primary human LEC carrying a heterozygous Asp299Gly mutant allele with wild-type TLR4 LEC. In agreement with previous observations with transfected wild-type and mutant TLR4 constructs (Tulic et al., 2007), we observed that LEC-TLR4A/A (wild-type) express higher levels of surface TLR4 than LEC-TLR4A/G (mutant), whereas TLR4 mRNA levels did not differ between the two cell types. The two cell types were also matched for TLR9 mRNA levels (Figure 4A). To determine the TLR4 activity in the wild-type and mutant cells, we examined their response to LPS stimulation. In agreement with previous findings, we observed that stimulation of LEC-TLR4A/G with LPS resulted in significantly higher induction of proinflammatory cytokine transcription compared to wild-type LEC. Notably, in contrast to the inflammatory gene profile, our data also suggested that LEC-TLR4A/G produced less IFN-β upon TLR4 stimulation with LPS (Figure 4B). As we showed that KSHV activates TLR4 (Figure 2B), we speculated that the combination of an impaired interferon and an enhanced inflammatory TLR4-mediated response could make the mutant cells more susceptible to KSHV infection. Indeed, we showed that KSHV infection of LEC-TLRA/G was significantly more efficient compared to LEC-TLR4A/A (Figures 4C and 4D).
Figure 4. Increased Susceptibility of LEC-TLR4A/G to KSHV Infection.
(A) TLR4 expression in LEC-TLR4A/A (wild-type TLR4) and LEC-TLR4A/G (heterozygous mutation in TLR4 gene). Inserts show the corresponding pyrograms showing a 50% loss of the fourth peak (corresponding to A) and 50% increase of the third peak (corresponding to a G) in the mutant cells. Bar graph shows mRNA levels of TLR4 and TLR9 in LEC-TLR4A/A (gray bars) and LEC-TLR4A/G (black bars).
(B) mRNA levels of inflammatory cytokines and IFN-β in LEC-TLR4A/A (gray bars) and LEC-TLR4A/G (black bars) 6 hr poststimulation with LPS/sCD14. P values indicate significant changes compared to LEC-TLR4A/A. mRNA levels are corrected for background transcription (in nonstimulated LEC).
(C) Representative dot plot of KSHV infected LEC-TLR4A/A and LEC-TLR4A/G. Numbers correspond to percentage of infected cells.
(D) KSHV gene expression in KLEC-TLR4A/A (gray bars) and KLEC-TLR4A/G (black bars) 72 hr p.i. P values indicate significant changes compared to KLEC-TLR4A/A. In all panels, error bars represent standard deviation.
The above results indicated that cells carrying TLR4 SNP may be more susceptible to KSHV infection. To assess this hypothesis in vivo, we determined the frequency of two TLR4 SNPs in a cohort of 109 HIV-1-infected individuals. In this cohort, 43 individuals were diagnosed with KSHV-associated cancers (23 KS, 9 KS and MCD, 11 MCD), and 66 HIV-1-infected controls had non-KSHV-associated cancers or no cancer (Table S1). The frequencies of the Asp299Gly and Ile399Thr polymorphisms in the entire cohort were 12.8% and 4.6%, respectively. All individuals were heterozygous for the polymorphisms and the Ile399Thr SNP cosegregated with the Asp299Gly mutation (Figure 5A). We observed a statistically significant higher frequency of the Asp299Gly SNP among individuals with MCD in comparison with non-KSHV cancer controls (30% versus 10.6%, p = 0.044; Figures 5A and 5B) and in comparison to individuals with only KS (30% versus 4.3%, p = 0.026). Furthermore, our data showed that HIV-1-infected individuals with the Asp299Gly SNP were more likely to have MCD when compared to individuals carrying the wild-type TLR4 allele (odds ratio 4.34, 95% confidence interval 1.3–14.4, p = 0.021). We also confirmed in this cohort that MCD (with or without KS) is associated with higher plasma KSHV load compared to KS (Figure 5C; KSHV load is shown at time of diagnosis). These findings infer that HIV-1/KSHV-co-infected individuals with the Asp299Gly TLR4 SNP are more likely to have MCD and are consistent with our in vitro results demonstrating the role of TLR4 in immunity against KSHV.
Figure 5. Association of TLR4 SNP with MCD.
(A) TLR4 SNP map of 109 HIV-1-infected individuals. PN, patient number, DS, disease status (black, MCD and KS; dark gray, MCD; light gray, KS; white, other), A/G, Asp299Gly, and C/T, Ile399Thr. Red and green boxes indicate presence of the corresponding SNP.
(B) Clinical characteristics and SNP incidence in the studied cohort. P values are calculated with Fischer Exact Probability Test.
(C) Box plot of plasma KSHV load at time of diagnosis in individuals with MCD (with or without KS) and KS only (available for 16 participants with KS only). Boxes indicate the lower and upper quartiles, and the line indicates the median viral load for each group. The ends of the lines outside the boxes indicate minimum and maximum loads, whereas circles represent outliers. Median and range of each group are also shown on top of each box. P value between the two groups is also shown.
vGPCR and vIRF1 Contribute to the TLR4 Downregulation by KSHV
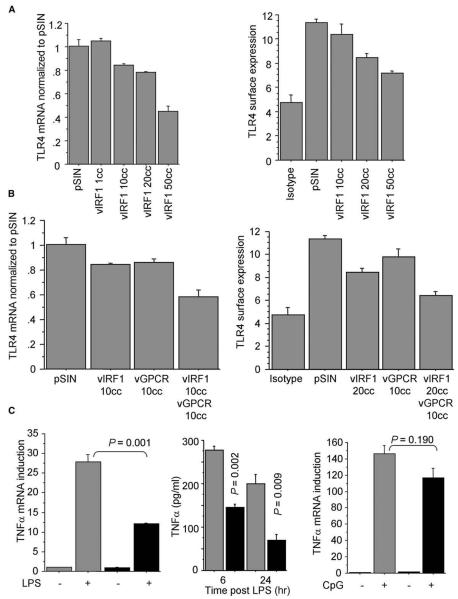
We have shown here that TLR4 mediates an antiviral response and is significantly downregulated upon KSHV infection. We therefore investigated the mechanism employed by KSHV to suppress TLR4. We determined TLR4 expression in LEC expressing 25 individual KSHV genes using a lentiviral gene delivery vector as previously reported (Lagos et al., 2007; Vart et al., 2007) to achieve efficient and stable viral gene expression in LEC. An initial screen revealed that expression of the KSHV vGPCR or vIRF1 caused a decrease in TLR4 mRNA levels, whereas no viral genes resulted in significant upregulation of TLR4 (Table S2). These findings were first confirmed for vIRF1 at the mRNA and protein levels (Figures 6A and S3), showing a decrease of TLR4 expression by vIRF1 in a dose-dependent manner. Furthermore, we showed that vIRF1 is primarily responsible for the TLR4 downregulation, but also that vGPCR contributes to suppression of TLR4 expression as coexpression of vIRF1 and vGPCR in LEC led to an additive effect on TLR4 expression (Figure 6B). In these experiments, the viral genes were expressed at levels where no cytopathic effects were observed (a maximum of 10 copies/cell for vGPCR, 50 copies/cell for vIRF1 when expressed alone, and 20 copies/cell when expressed with vGPCR). Notably, at these expression levels, vIRF1 and vGPCR resulted in a decrease in TLR4 protein expression comparable to that observed in KLEC (72 hr p.i.), although the TLR4 mRNA downregulation was more pronounced in KLEC (Figures 1B, 1C, and 6B). We have previously shown that both vIRF1 and vGPCR are expressed in KLEC at 72 hr p.i. (Lagos et al., 2007; Vart et al., 2007).
Figure 6. vIRF1 and vGPCR Contribute to the Downregulation of TLR4.
(A) TLR4 mRNA and surface expression (geometrical mean fluorescence) levels in LEC expressing increasing amounts of vIRF1 (shown as average number of lentiviral copies per cell, cc) and empty lentiviral vector (pSIN). Levels normalized to LEC-pSIN.
(B) TLR4 mRNA and surface expression levels in LEC expressing pSIN, vIRF1, vGPCR, and vIRF1 and vGPCR.
(C) Fold mRNA TNF-α induction and TNF-α secreted levels in LEC-expressing vIRF1 and vGPCR (black bars) or pSIN (gray bars) after stimulation with LPS/sCD14 and fold mRNA TNF-α induction after stimulation with CpG DNA. P values between stimulated LEC-pSIN and LEC-vIRF1/vGPCR are shown. For all panels, experiments were performed 72 hr p.i. In all panels, error bars represent standard deviation.
To confirm that the TLR4 downregulation by vIRF1 and vGPCR was functionally relevant, we tested whether these genes affected response to LPS stimulation in LEC. We found that similar to KLEC, LEC expressing vIRF1 and vGPCR displayed impaired TNF-α induction after TLR4 activation, whereas response to TLR9 stimulation was similar between these cells and controls (Figure 6C), excluding that the observed hyporesponsiveness to LPS was due to inhibition of TLR signaling by these viral genes at a different level downstream of TLR4.
The Role of the ERK/MAPK Pathway in TLR4 Regulation by KSHV in Endothelial Cells
The KSHV-encoded vIRF1 has been shown to inhibit IRF-driven transcription (Coscoy, 2007; Moore and Chang, 2003), whereas basal human TLR4 expression is regulated by interferon consensus-binding protein (ICSBP) (Rehli et al., 2000), which is the IRF with the highest sequence similarity to vIRF1. On the other hand, vGPCR activates several signaling pathways (reviewed in Cannon [2007]), none of which are directly associated with negative regulation of TLR4. We therefore investigated the mechanism by which vGPCR causes suppression of TLR4 expression. This was addressed by determining TLR4 expression after treating vGPCR-expressing LEC with chemical inhibitors of pathways that are activated by this gene, such as the NF-κB, phosphatidylinositol-3-kinase (PI3K), ERK and Jun N-terminal kinase (JNK) MAPK pathways, or those involved in TLR4 expression in other cell types, such as the p38/MAPK pathway. We found that only inhibitors of ERK phosphorylation were able to completely reverse the observed TLR4 downregulation (Figure 7A) in a dose-dependent manner (Figure 7B). To exclude any nonspecific effects of the synthetic inhibitors we also showed that siRNA-mediated silencing of ERK1 and ERK2 reversed the vGPCR-mediated TLR4 suppression (Figure 7C), without affecting expression of vGPCR (Figure S4A).
Figure 7. ERK Activation Contributes to TLR4 Downregulation in KLEC.
(A) TLR4 mRNA levels in LEC-pSIN (black bar) and LEC-expressing vGPCR (gray bars) after treatment with the chemical inhibitors LY294002 (LY), BAY11-7082 (BAY), SB202190 (SB), JNK inhibitor II (JNK), PD98059 (PD), and UO126 (UO). Controls were treated with 0.1% DMSO.
(B) TLR4 mRNA levels in LEC-pSIN (black bar) and LEC-vGPCR (gray bars) after treatment with DMSO, 1 μM and 10 μM of UO126. Inset shows protein levels of phosphorylated and total ERK in LEC-pSIN and LEC-vGPCR. For (A) and (B), levels are normalized to LEC-pSIN/0.1% DMSO.
(C) TLR4 expression in LEC-pSIN (black bars) and LEC-vGPCR (72 hr p.i., gray bars). LEC were transfected with nontargeting siRNA (scramble) or siRNAs targeting ERK1 and ERK2 (siERK) 48 hr before infection with pSIN or vGPCR lentivirus. Inset shows levels of ERK1 (top band) and ERK2 (bottom band) in scramble and siERK LEC (48 hr posttransfection). Significant P values between LEC-pSIN and LEC-vGPCR are shown. Bar graph shows levels of ERK1 and ERK2 in LEC-siERK compared to LEC-scramble 48 hr posttransfection.
(D) TLR4 expression in LEC (black bars) and KLEC (72 hr p.i., gray bars) after treatment of cells with DMSO, SB202190, JNK inhibitor II, UO126, PD98059, or LY294002. P values indicate statistical significance of changes between KLEC-DMSO and KLEC-UO or KLEC-PD. Inset shows protein levels of phosphorylated and total ERK in LEC-DMSO, KLEC-DMSO, and KLEC treated with UO126 MEK-inhibitor. P values indicate statistically significant changes between LEC-DMSO and KLEC samples.
(E) TLR4 expression in LEC (black bars) and KLEC (6 hr p.i., gray bars) after treatment with DMSO (control for chemical inhibitors) or media (control for antioxidants), N-acetyl-cysteine and ascorbic acid (anti-oxidants), LY294002, and UO126. TLR9 mRNA levels in LEC-DMSO, KLEC-DMSO, and KLEC treated with UO126 (6 hr p.i.).
(F) TLR4 expression in LEC (black bars) and KLEC (6 hr p.i., gray bars). Cells were transfected with nontargeting siRNA (scramble) or siRNAs targeting ERK1 and ERK2 (siERK) 48 hr before infection with KSHV. P values indicate significant changes between LEC and KLEC. In all panels, error bars represent standard deviation.
Next, we confirmed that ERK was phosphorylated in KLEC and showed that inhibition of ERK activation partly restored TLR4 levels in KSHV-infected LEC (72 hr p.i.; Figure 7D). Although it has been shown that inhibition of ERK activation before KSHV infection inhibits viral gene expression (Sharma-Walia et al., 2005), treatment of KLEC (72 hr p.i.) with ERK-phosphorylation inhibitors did not affect viral gene expression (Figure S4B). Inhibitors of PI3K, JNK, and p38 activation did not affect the KSHV-induced TLR4 downregulation, showing that the effect of ERK-phosphorylation inhibitors was not due to nonspecific inhibition of other kinases. These findings were consistent with a contribution of vGPCR to the observed TLR4 downregulation. The contribution of the NF-κB pathway to TLR4 downregulation in KLEC was excluded, as NF-κB inhibitors did not affect the vGPCR-mediated TLR4 downregulation, and vFLIP, the other major viral activator of NF-κB, did not affect TLR4 expression (Table S2).
Following this, we tested the effect of ERK activation inhibitors in the early TLR4 regulation by KSHV (6 hr p.i.), as it has been shown that ERK activation is also caused by viral binding on the surface of endothelial cells (Sharma-Walia et al., 2005). ERK phosphorylation inhibitors and siRNA-mediated silencing of ERK1 and ERK2 prior to infection reversed early TLR4 suppression in KLEC (6 hr p.i.; Figure 7D). Another possible mechanism of TLR4 downregulation in endothelial cells during early stages of infection, mediated by induction of reactive oxygen species (Ishida et al., 2002), did not affect TLR4 expression in KLEC. Inhibitors of ERK activation did not affect TLR9 mRNA levels in KLEC (Figure S4C). This apparent specific effect of ERK activation on TLR4 in LEC could be explained by the fact that conserved ETS binding sites are present in the TLR4 promoter, whereas the human TLR9 promoter does not include any ETS (conserved or nonconserved) sites (Figure S4D). Together, these data showed that ERK phosphorylation due to viral binding mediates TLR4 suppression during the early stages of KSHV infection, whereas ERK phosphorylation by vGPCR also contributes to TLR4 downregulation at 72 hr p.i.
DISCUSSION
Understanding the mechanisms of the host immune response against viral infections and the strategies employed by viruses to escape this response is crucial for designing new therapies for infectious diseases. In the case of cancers caused by oncogenic viruses, such as KSHV, current treatment regimes rely on systemic cytotoxic therapy and immune system reconstitution through antiretroviral therapy (ART), but take little advantage of our current knowledge of KSHV immunobiology (Hansen et al., 2007). In this respect, revealing the role and mechanisms of regulation of the TLR-mediated response to KSHV is essential, as clinically approved agonists and antagonists of several TLRs are available (Kanzler et al., 2007). In this study, we define a particular TLR (TLR4), which mediates an innate response against KSHV. To our knowledge, this is the first report that implicates TLR4 in immune responses against human herpesviruses.
TLR4 mediates antiviral immunity through induction of an interferon antiviral gene expression program and production of inflammatory cytokines. Its antiviral activity has been demonstrated mainly against RNA viruses such as RSV and hepatitis C virus (Kurt-Jones et al., 2000; Machida et al., 2006). We extend the antiviral role of TLR4 to the family of human herpesviruses by demonstrating that cells with reduced (siRNA silencing) or no (knockout mice) TLR4 expression display an impaired immune response during primary KSHV infection and increased viral gene expression (Figure 2). Our findings also suggest that this antiviral activity is due to direct binding between TLR4 and a KSHV envelope glycoprotein, such as glycoprotein B or K8.1 (Figure 2). Importantly, although our data were obtained using endothelial cells, it is likely that TLR4 also plays a role in innate immunity during KSHV infection of B lymphocytes as these cells express functional TLR4, and they are responsive to TLR4 ligands (Peng, 2005).
The only other virus, which has been shown to induce innate immunity by direct interaction with TLR4 is RSV (Kurt-Jones et al., 2000). In the context of RSV infection, the Asp299Gly and Ile399Thr TLR4 mutations have been associated with severity of RSV-induced bronchiolitis (Tal et al., 2004). It has been suggested that these mutations affect TLR4 function by impairing transport of the receptor to the cell membrane (Tulic et al., 2007) and that the Asp299Gly SNP is associated with a proinflammatory phenotype (Ferwerda et al., 2007). In our study we observed that LEC heterozygous for the Asp299Gly mutation are more susceptible to KSHV infection (Figure 4), and we show an association of the Asp299Gly SNP among HIV-1-infected individuals with MCD (Figure 5), a lymphoproliferation associated with high KSHV load (Boivin et al., 2002; Oksenhendler et al., 2000). Interestingly, our data suggest that this association with MCD may be due to a combination of the impaired interferon response and a proinflammatory phenotype of cells carrying the Asp299Gly allele. Such a proinflammatory environment is strongly associated with increased KSHV gene expression during primary KSHV infection in vitro (Sadagopan et al., 2007) and specifically MCD development in vivo (Oksenhendler et al., 2000). Further larger prospective studies should evaluate these findings and the association of these SNPs with KSHV-associated malignancies in other geographical and/or HIV-1-negative cohorts. However, the results presented here provide the first crucial insight into the role of TLR4 in controlling KSHV infection in humans and identify the first genetic factor that could be associated with MCD development.
Our results reveal KSHV as the first virus, which targets TLR4 expression as an immune evasion mechanism. Interestingly, it has been reported that TLR4 is not expressed in the KS biopsy-derived SLK endothelial cell line (Livengood et al., 2007). However, SLK cells have no detectable KSHV episome or viral gene expression; therefore, the lack of TLR4 expression in this cell line is unlikely to be associated with KSHV. Moreover, TLR4 is a mediator of an innate response, and its regulation by the virus should be rapid and occur during primary infection and lytic replication when new cells are about to be infected. Importantly, the maintenance of KSHV latency relies on this de novo infection of cells (Grundhoff and Ganem, 2004). Consistent with the above, in our primary infection model we observe a rapid and dramatic drop of TLR4 expression in infected endothelial cells (Figure 1). Of note immune evasion mechanisms employed by herpesviruses have evolved to delay immune responses allowing establishment of latency, rather than to completely escape immune surveillance. Thus, it is not surprising that cells lacking TLR4 are more susceptible to infection by KSHV (Figure 2) despite the fact that KSHV has evolved mechanisms to downregulate TLR4. Furthermore, we excluded the possibility that this early downregulation is a result of the previously reported mRNA shutoff caused during the early stages of infection (Glaunsinger and Ganem, 2004) by showing that TLR9 mRNA levels are not affected by infection (Figure 1). We used TLR9 as control because it is the TLR typically associated with responses to DNA viruses (Akira et al., 2006), including the related murine gammaherpesvirus-68 (Guggemoos et al., 2008). Our data do not exclude that TLR9 could be regulated by KSHV at the posttranscriptional level.
We show that ERK activation due to the activity of vGPCR or KSHV structural proteins, such as K8.1 or glycoprotein B (Sharma-Walia et al., 2005), plays a key role in the TLR4 downregulation in endothelial cells. Interestingly, it has been reported that in murine dendritic cells ERK activation leads to TLR4 upregulation (An et al., 2002), which suggests that TLR4 regulation is cell-type specific. Moreover, we speculate that TLR4 regulation through ERK activation in KLEC may be part of a more complex signaling network involving the simultaneous activation of other signaling cascades including crosstalk between different arms of the MAPK signaling.
We identify two lytic KSHV genes, vIRF1 and vGPCR, that contribute to TLR4 transcriptional repression in endothelial cells. Our results demonstrate that vIRF1 and vGPCR lead to TLR4 downregulation through two independent mechanisms. vIRF1 is a known viral inhibitor of IRF-mediated transcription (Coscoy, 2007; Moore and Chang, 2003), with high homology to the cellular ICSBP, a transcription factor necessary for basal TLR4 expression (Rehli et al., 2000). We have previously shown that there is background IRF-mediated transcription in LEC even in the absence of interferon (Lagos et al., 2007), which could explain the effect of vIRF1 on TLR4 transcription. On the other hand, we show that vGPCR-mediated TLR4 suppression occurs by way of ERK activation (Figure 6). Of note, among the three main MAPK pathways, p38 is mainly and positively associated with TLR4 transcription, as the TLR4 gene has a PU.1 element in its promoter.
The driving force in the evolution of herpesviruses is the establishment of latency in the host. This strongly depends on efficient escape from the immune system, mainly during primary infection and lytic replication. Our data reveal that evasion of TLR4-mediated response is one such target for KSHV, and we show that one of the mechanisms that the virus employs to escape this response involves suppression of TLR4 expression. As we show that TLR4 simulation impedes KSHV gene expression (Figure 3), our findings suggest the potential of TLR4 agonists in KSHV-related neoplasm prevention or treatment. Activation of TLR4 could boost antigen presentation (Kanzler et al., 2007) and potentiate cytotoxic therapy (Apetoh et al., 2007), but also exert a direct antiviral effect. A future clinical study could be to investigate whether activation of TLR4 signaling with synthetic TLR4 agonists leads to reduced circulating KSHV load and therefore could be used for the treatment of KSHV-related malignancies.
EXPERIMENTAL PROCEDURES
Animals and Cells
C57BL10/ScNJ (Jax mice number 003752) and C57BL10/ScSnJ mice (wild-type for the Il12rβ gene; Jax mice number 000476) were purchased from Jackson Laboratory. Thioglycollate-induced peritoneal macrophages were grown in Dulbecco's modified Eagle's medium (GIBCO), supplemented with 10% FCS. LEC were grown as described (Lagos et al., 2007). All experiments were performed before passage 5. TLR4 signaling in LEC was activated with 1 μg/ml sCD14 and 100 ng/ml LPS for 6 hr. Under these conditions, LPS treatment did not cause any apoptosis in LEC (tested with the MTT assay; not shown). TLR9 signaling was activated by incubation of LEC with 5 μg/ml of CpG type A, B, and C (ODN2336, ODN10103, ODN2395; Coley Pharma) for 16 hr. BCBL1-1C and 293T cells were grown as previously described (Lagos et al., 2007).
KSHV Production and Infection of LEC
KSHV was produced and LEC were infected as previously described (Lagos et al., 2007). KSHV was inactivated by UV irradiation for 30 min in a Stratalinker 2400 (Stratagene). LANA mRNA (by qRTPCR) was undetectable in LEC treated with UV-inactivated KSHV 48 hr p.i. GFP expression in KLEC was used as a surrogate marker of infection and infections typically resulted in 25%–40% GFP-expressing KLEC (72 hr p.i.). KSHV infection of mouse macrophages was detected by qRTPCR.
GEM Analysis
GEM profiles of six pairs of LEC and KLEC (3 days p.i.) were obtained and analyzed as described (Lagos et al., 2007), using Affymetrix hg-u133+2 GeneChips. The GEM data are available in the ArrayExpress database (accession number E-MEXP-561).
Antibodies
Surface expression of proteins was assessed by flow cytometry (FACSCalibur, Becton Dickinson). We used RPE labeled antibodies against human TLR4 (Santa Cruz), ICAM1 (PharMingen), and the corresponding isotype controls. Analyses were done after gating live cells. The levels of ERK activation were determined by western blot analysis using antibodies specific for phoshorylated ERK1/2 (Cell Signaling Technology, Inc.) and total ERK1/2 (Upstate) as described (Vart et al., 2007). Levels of phosphorylated and total IRAK1 were determined using antibodies from Cell Signaling Technology, Inc. A neutralizing antibody against the human IFNαRII was obtained from Merck.
Participants and TLR4 SNP Analysis
Blood samples were collected from HIV-1-infected individuals attending the St. Stephen's Centre at the Chelsea and Westminster Hospital. Sample collection, preparation, and DNA extraction was carried out under ethical approval of the Riverside Research Ethics Committee, and all participants gave informed consent. Genomic DNA was extracted from 5×106 peripheral blood mononuclear cells using the Nucleon Genomic DNA Extraction Kit (Tepnel Life Sciences) according to the manufacturer's instructions. DNA was resuspended in RNase/DNase free water (Sigma) to a final concentration of 50 ng/μl and stored at −20°C until required. TLR4 SNPs were assessed by pyrosequencing (Biotage, UK). Primer sequences were designed using Pyrosequencing Assay Design Software (Biotage) and were as follows: Forward PCR primer GACGATTAGCATACTTAGACTACTACCTC, reverse biotinylated PCR primer AGCCCAAGAAGTTTGAACTCAT, Asp299Gly sequencing primer ATACTTAGACTACTACCTCG, and Ile399Thr sequencing primer CAAAGTGATTTTGGGAC. The biotin-labeled PCR products were purified with Steptavidin TM Sepharose Beads (GE Healthcare) using the Pyrosequencing Preparation Station (Biotage) and annealed to the sequencing primer at 80°C for 2 min. The samples were then run according to the instrument (PyroMarkTMMD) parameters.
Lentiviral Expression of KSHV Genes
KSHV genes were cloned from the BC3 and BC1 PEL cell lines and were expressed using a modified pSIN-MCS lentiviral vector as described (Vart et al., 2007). The number of lentiviral copies per cell was determined by qPCR and viral gene expression was confirmed by RT-PCR. All experiments shown were performed 3 days postlentivirus infection.
qRTPCR and qPCR
Genomic DNA and total RNA were extracted using the QIAamp DNA mini and RNEasy mini kits, respectively (both from QIAGEN). TLR4, TLR9, TNF-α, IL1-β, IL-6, LANA, ORF50, vIRF-1, vGPCR, and ORF26 mRNA levels were quantified by qRTPCR using the SYBR Green Master Mix (Applied Biosystems, ABI Prism 7700). Optimized forward and reverse primers for these genes and GAPDH (house-keeping reference gene) were used at final concentration of 300 nM. For ERK1, ERK2, and IFN-β qRTPCR, commercially available primers were used (Applied Biosytems). Cell-associated and plasma KSHV loads were determined by qPCR as described (Bourboulia et al., 2004).
TNF-α ELISA
TNF-α levels in supernatants from LPS stimulated LEC were determined using a commercially available ELISA (R&D Systems). TNF-α was not detectable in supernatants of nonstimulated cells.
RNA Interference
LEC were seeded in 6-well plates 1 day prior to transfection with ERK1-, ERK2-, or TLR4-targeting or nontargeting (scramble) siRNA (OnTargetPlus SmartPool from Dharmacon). The concentration of siRNA was 100 nM, and cells were transfected using oligofectamine reagent (Invitrogen). Cells were infected with KSHV 48 hr post siRNA transfection.
Pharmacologic Inhibitions
We used the chemical inhibitors BAY11-7082 (NF-κB inhibitor, 5 μM), JNK inhibitor II (25 μM), SB202190 (p38/MAPK inhibitor, 10 μM), PD98059 (MEK inhibitor, 10 μM), UO126 (MEK inhibitor, 10 μM), and LY294002 (PI3K inhibitor, 5 μM), all from Calbiochem. For LEC and KLEC at 72 hr p.i., and LEC infected with lentivirus, the inhibitors were added to the cells for 6 hr, apart from BAY11-7082 and LY294002, which were added for 2 hr and 4 hr, respectively. For LEC and KLEC at 6 hr p.i., all inhibitors were added to the cells for 2 hr prior to infection and removed during infection. Polymyxin B, N-acetylcysteine, and ascorbic acid (all from Sigma) were used at 10 mg/ml, 2.5 mM, and 1 mM concentrations, respectively. Cell viability was not affected by any of the inhibitors in the above conditions (not shown).
TLR Promoter Analysis
The promoters (1 kB upstream the transcription start site) of human TLR4 and TLR9 were analyzed using DiAlign TF with incorporated MatInspector software.
Statistical Analysis
All experiments were performed in four to six independent replicates, and error bars correspond to standard deviation from the mean. Statistical significance (P values) was determined with a two-sided unpaired Student's t test. Statistical analysis of GEM was performed as described using a moderated t statistic and a false discovery rate correction (Lagos et al., 2007; Wang et al., 2004). Statistical significance of the frequency of the TLR4 SNPs among HIV-1-infected individuals was determined using the Fischer Exact Probability Test.
Supplementary Material
ACKNOWLEDGMENTS
We thank Drs. Jeffrey Vieira, Rolf Renne, and Benny Chain for providing reagents, and the Medical Research Council (MRC), Cancer Research UK, and Biotechnology and Biological Sciences Research Council (BBSRC) for funding.
Footnotes
The authors have no conflicting financial interests.
REFERENCES
- Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- An H, Yu Y, Zhang M, Xu H, Qi R, Yan X, Liu S, Wang W, Guo Z, Guo J, et al. Involvement of ERK, p38 and NF-kappaB signal transduction in regulation of TLR2, TLR4 and TLR9 gene expression induced by lipopolysaccharide in mouse dendritic cells. Immunology. 2002;106:38–45. doi: 10.1046/j.1365-2567.2002.01401.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apetoh L, Ghiringhelli F, Tesniere A, Obeid M, Ortiz C, Criollo A, Mignot G, Maiuri MC, Ullrich E, Saulnier P, et al. Toll-like receptor 4-dependent contribution of the immune system to anticancer chemotherapy and radiotherapy. Nat. Med. 2007;13:1050–1059. doi: 10.1038/nm1622. [DOI] [PubMed] [Google Scholar]
- Boivin G, Cote S, Cloutier N, Abed Y, Maguigad M, Routy JP. Quantification of human herpesvirus 8 by real-time PCR in blood fractions of AIDS patients with Kaposi's sarcoma and multicentric Castleman's disease. J. Med. Virol. 2002;68:399–403. doi: 10.1002/jmv.10217. [DOI] [PubMed] [Google Scholar]
- Boshoff C, Weiss R. AIDS-related malignancies. Nat. Rev. Cancer. 2002;2:373–382. doi: 10.1038/nrc797. [DOI] [PubMed] [Google Scholar]
- Bourboulia D, Aldam D, Lagos D, Allen E, Williams I, Cornforth D, Copas A, Boshoff C. Short- and long-term effects of highly active antiretroviral therapy on Kaposi sarcoma-associated herpesvirus immune responses and viraemia. AIDS. 2004;18:485–493. doi: 10.1097/00002030-200402200-00015. [DOI] [PubMed] [Google Scholar]
- Burzyn D, Rassa JC, Kim D, Nepomnaschy I, Ross SR, Piazzon I. Toll-like receptor 4-dependent activation of dendritic cells by a retrovirus. J. Virol. 2004;78:576–584. doi: 10.1128/JVI.78.2.576-584.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon M. The KSHV and other human herpesviral G protein-coupled receptors. Curr. Top. Microbiol. Immunol. 2007;312:137–156. doi: 10.1007/978-3-540-34344-8_5. [DOI] [PubMed] [Google Scholar]
- Coscoy L. Immune evasion by Kaposi's sarcoma-associated herpesvirus. Nat. Rev. Immunol. 2007;7:391–401. doi: 10.1038/nri2076. [DOI] [PubMed] [Google Scholar]
- Doyle S, Vaidya S, O'Connell R, Dadgostar H, Dempsey P, Wu T, Rao G, Sun R, Haberland M, Modlin R, Cheng G. IRF3 mediates a TLR3/TLR4-specific antiviral gene program. Immunity. 2002;17:251–263. doi: 10.1016/s1074-7613(02)00390-4. [DOI] [PubMed] [Google Scholar]
- Faure E, Equils O, Sieling PA, Thomas L, Zhang FX, Kirschning CJ, Polentarutti N, Muzio M, Arditi M. Bacterial lipopolysaccharide activates NF-kappaB through toll-like receptor 4 (TLR-4) in cultured human dermal endothelial cells. Differential expression of TLR-4 and TLR-2 in endothelial cells. J. Biol. Chem. 2000;275:11058–11063. doi: 10.1074/jbc.275.15.11058. [DOI] [PubMed] [Google Scholar]
- Ferwerda B, McCall MB, Alonso S, Giamarellos-Bourboulis EJ, Mouktaroudi M, Izagirre N, Syafruddin D, Kibiki G, Cristea T, Hijmans A, et al. TLR4 polymorphisms, infectious diseases, and evolutionary pressure during migration of modern humans. Proc. Natl. Acad. Sci. USA. 2007;104:16645–16650. doi: 10.1073/pnas.0704828104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaunsinger B, Ganem D. Lytic KSHV infection inhibits host gene expression by accelerating global mRNA turnover. Mol. Cell. 2004;13:713–723. doi: 10.1016/s1097-2765(04)00091-7. [DOI] [PubMed] [Google Scholar]
- Gottipati S, Rao NL, Fung-Leung WP. IRAK1: a critical signaling mediator of innate immunity. Cell. Signal. 2008;20:269–276. doi: 10.1016/j.cellsig.2007.08.009. [DOI] [PubMed] [Google Scholar]
- Grundhoff A, Ganem D. Inefficient establishment of KSHV latency suggests an additional role for continued lytic replication in Kaposi sarcoma pathogenesis. J. Clin. Invest. 2004;113:124–136. doi: 10.1172/JCI200417803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guggemoos S, Hangel D, Hamm S, Heit A, Bauer S, Adler H. TLR9 contributes to antiviral immunity during gammaherpesvirus infection. J. Immunol. 2008;180:438–443. doi: 10.4049/jimmunol.180.1.438. [DOI] [PubMed] [Google Scholar]
- Hansen A, Boshoff C, Lagos D. Kaposi sarcoma as a model of oncogenesis and cancer treatment. Expert Rev. Anticancer Ther. 2007;7:211–220. doi: 10.1586/14737140.7.2.211. [DOI] [PubMed] [Google Scholar]
- Hatao F, Muroi M, Hiki N, Ogawa T, Mimura Y, Kaminishi M, Tanamoto K. Prolonged Toll-like receptor stimulation leads to downregulation of IRAK-4 protein. J. Leukoc. Biol. 2004;76:904–908. doi: 10.1189/jlb.0504277. [DOI] [PubMed] [Google Scholar]
- Ishida I, Kubo H, Suzuki S, Suzuki T, Akashi S, Inoue K, Maeda S, Kikuchi H, Sasaki H, Kondo T. Hypoxia diminishes toll-like receptor 4 expression through reactive oxygen species generated by mitochondria in endothelial cells. J. Immunol. 2002;169:2069–2075. doi: 10.4049/jimmunol.169.4.2069. [DOI] [PubMed] [Google Scholar]
- Isogawa M, Robek MD, Furuichi Y, Chisari FV. Toll-like receptor signaling inhibits hepatitis B virus replication in vivo. J. Virol. 2005;79:7269–7272. doi: 10.1128/JVI.79.11.7269-7272.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janeway CA, Jr., Medzhitov R. Innate immune recognition. Annu. Rev. Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- Kanzler H, Barrat FJ, Hessel EM, Coffman RL. Therapeutic targeting of innate immunity with Toll-like receptor agonists and antagonists. Nat. Med. 2007;13:552–559. doi: 10.1038/nm1589. [DOI] [PubMed] [Google Scholar]
- Kawasaki K, Gomi K, Nishijima M. Gln22 of mouse MD-2 is essential for species-specific lipopolysaccharide mimetic action of taxol. J. Immunol. 2001;166:11–14. doi: 10.4049/jimmunol.166.1.11. [DOI] [PubMed] [Google Scholar]
- Kurt-Jones EA, Popova L, Kwinn L, Haynes LM, Jones LP, Tripp RA, Walsh EE, Freeman MW, Golenbock DT, Anderson LJ, Finberg RW. Pattern recognition receptors TLR4 and CD14 mediate response to respiratory syncytial virus. Nat. Immunol. 2000;1:398–401. doi: 10.1038/80833. [DOI] [PubMed] [Google Scholar]
- Lagos D, Trotter MW, Vart RJ, Wang HW, Matthews NC, Hansen A, Flore O, Gotch F, Boshoff C. Kaposi sarcoma herpesvirus-encoded vFLIP and vIRF1 regulate antigen presentation in lymphatic endothelial cells. Blood. 2007;109:1550–1558. doi: 10.1182/blood-2006-05-024034. [DOI] [PubMed] [Google Scholar]
- Liew FY, Xu D, Brint EK, O'Neill LA. Negative regulation of toll-like receptor-mediated immune responses. Nat. Rev. Immunol. 2005;5:446–458. doi: 10.1038/nri1630. [DOI] [PubMed] [Google Scholar]
- Livengood AJ, Wu CC, Carson DA. Opposing roles of RNA receptors TLR3 and RIG-I in the inflammatory response to double-stranded RNA in a Kaposi's sarcoma cell line. Cell. Immunol. 2007;249:55–62. doi: 10.1016/j.cellimm.2007.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida K, Cheng KT, Sung VM, Levine AM, Foung S, Lai MM. Hepatitis C virus induces toll-like receptor 4 expression, leading to enhanced production of beta interferon and interleukin-6. J. Virol. 2006;80:866–874. doi: 10.1128/JVI.80.2.866-874.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshak-Rothstein A. Toll-like receptors in systemic autoimmune disease. Nat. Rev. Immunol. 2006;6:823–835. doi: 10.1038/nri1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov R, Preston-Hurlburt P, Janeway CA., Jr. A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature. 1997;388:394–397. doi: 10.1038/41131. [DOI] [PubMed] [Google Scholar]
- Moore PS, Chang Y. Kaposi's sarcoma-associated herpesvirus immunoevasion and tumorigenesis: two sides of the same coin? Annu. Rev. Microbiol. 2003;57:609–639. doi: 10.1146/annurev.micro.57.030502.090824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller T, Hamm S, Bauer S. TLR9-mediated recognition of DNA. Handb. Exp. Pharmacol. 2008;183:51–70. doi: 10.1007/978-3-540-72167-3_3. [DOI] [PubMed] [Google Scholar]
- Oksenhendler E, Carcelain G, Aoki Y, Boulanger E, Maillard A, Clauvel JP, Agbalika F. High levels of human herpesvirus 8 viral load, human interleukin-6, interleukin-10, and C reactive protein correlate with exacerbation of multicentric castleman disease in HIV-infected patients. Blood. 2000;96:2069–2073. [PubMed] [Google Scholar]
- Parsons CH, Adang LA, Overdevest J, O'Connor CM, Taylor JR, Jr., Camerini D, Kedes DH. KSHV targets multiple leukocyte lineages during long-term productive infection in NOD/SCID mice. J. Clin. Invest. 2006;116:1963–1973. doi: 10.1172/JCI27249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegu A, Qin S, Fallert Junecko BA, Nisato RE, Pepper MS, Reinhart TA. Human Lymphatic Endothelial Cells Express Multiple Functional TLRs. J. Immunol. 2008;180:3399–3405. doi: 10.4049/jimmunol.180.5.3399. [DOI] [PubMed] [Google Scholar]
- Peng SL. Signaling in B cells via Toll-like receptors. Curr. Opin. Immunol. 2005;17:230–236. doi: 10.1016/j.coi.2005.03.003. [DOI] [PubMed] [Google Scholar]
- Pugin J, Ulevitch RJ, Tobias PS. A critical role for monocytes and CD14 in endotoxin-induced endothelial cell activation. J. Exp. Med. 1993;178:2193–2200. doi: 10.1084/jem.178.6.2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehli M, Poltorak A, Schwarzfischer L, Krause SW, Andreesen R, Beutler B. PU.1 and interferon consensus sequence-binding protein regulate the myeloid expression of the human Toll-like receptor 4 gene. J. Biol. Chem. 2000;275:9773–9781. doi: 10.1074/jbc.275.13.9773. [DOI] [PubMed] [Google Scholar]
- Sadagopan S, Sharma-Walia N, Veettil MV, Raghu H, Sivakumar R, Bottero V, Chandran B. Kaposi's sarcoma-associated herpesvirus induces sustained NF-kappaB activation during de novo infection of primary human dermal microvascular endothelial cells that is essential for viral gene expression. J. Virol. 2007;81:3949–3968. doi: 10.1128/JVI.02333-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder NW, Schumann RR. Single nucleotide polymorphisms of Toll-like receptors and susceptibility to infectious disease. Lancet Infect. Dis. 2005;5:156–164. doi: 10.1016/S1473-3099(05)01308-3. [DOI] [PubMed] [Google Scholar]
- Sharma-Walia N, Krishnan HH, Naranatt PP, Zeng L, Smith MS, Chandran B. ERK1/2 and MEK1/2 induced by Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) early during infection of target cells are essential for expression of viral genes and for establishment of infection. J. Virol. 2005;79:10308–10329. doi: 10.1128/JVI.79.16.10308-10329.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tal G, Mandelberg A, Dalal I, Cesar K, Somekh E, Tal A, Oron A, Itskovich S, Ballin A, Houri S, et al. Association between common Toll-like receptor 4 mutations and severe respiratory syncytial virus disease. J. Infect. Dis. 2004;189:2057–2063. doi: 10.1086/420830. [DOI] [PubMed] [Google Scholar]
- Tulic MK, Hurrelbrink RJ, Prele CM, Laing IA, Upham JW, Le Souef P, Sly PD, Holt PG. TLR4 polymorphisms mediate impaired responses to respiratory syncytial virus and lipopolysaccharide. J. Immunol. 2007;179:132–140. doi: 10.4049/jimmunol.179.1.132. [DOI] [PubMed] [Google Scholar]
- Vart RJ, Nikitenko LL, Lagos D, Trotter MW, Cannon M, Bourboulia D, Gratrix F, Takeuchi Y, Boshoff C. Kaposi's sarcoma-associated herpesvirus-encoded interleukin-6 and G-protein-coupled receptor regulate angiopoietin-2 expression in lymphatic endothelial cells. Cancer Res. 2007;67:4042–4051. doi: 10.1158/0008-5472.CAN-06-3321. [DOI] [PubMed] [Google Scholar]
- Wang HW, Trotter MW, Lagos D, Bourboulia D, Henderson S, Makinen T, Elliman S, Flanagan AM, Alitalo K, Boshoff C. Kaposi sarcoma herpesvirus-induced cellular reprogramming contributes to the lymphatic endothelial gene expression in Kaposi sarcoma. Nat. Genet. 2004;36:687–693. doi: 10.1038/ng1384. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.