Abstract
To elucidate the pathogenesis of hepatocellular carcinoma (HCC) and develop useful prognosis predictors, it is necessary to identify biologically relevant genomic alterations in HCC. In our study, we defined recurrently altered regions (RARs) common to many cases of HCCs, which may contain tumor-related genes, using whole-genome array-CGH and explored their associations with the clinicopathologic features. Gene set enrichment analysis was performed to investigate functional implication of RARs. On an average, 23.1% of the total probes were altered per case. Mean numbers of altered probes are significantly higher in high-grade, bigger and microvascular invasion (MVI) positive tumors. In total, 32 RARs (14 gains and 18 losses) were defined and 4 most frequent RARs are gains in 1q21.1-q32.1 (64.5%), 1q32.1-q44 (59.2%), 8q11.21-q24.3 (48.7%) and a loss in 17p13.3-p12 (51.3%). Through focusing on RARs, we identified genes and functional pathways likely to be involved in hepatocarcinogenesis. Among genes in the recurrently gained regions on 1q, expression of KIF14 and TPM3 was significantly increased, suggesting their oncogenic potential in HCC. Some RARs showed the significant associations with the clinical features. Especially, the recurrent loss in 9p24.2-p21.1 and gain in 8q11.21-q24.3 are associated with the high tumor grade and MVI, respectively. Functional analysis showed that cytokine receptor binding and defense response to virus pathways are significantly enriched in high grade-related RARs. Taken together, our results and the strategy of analysis will help to elucidate pathogenesis of HCC and to develop biomarkers for predicting behaviors of HCC.
Keywords: hepatocellular carcinoma, recurrently altered regions, array comparative genomic hybridization, KIF14, TPM3
Hepatocellular carcinoma (HCC) is one of the most common human malignancies and responsible for ~5% of all cancer-related deaths in the world.1 Given that the overall HCC incidence is still rising and prognosis of the disease remains poor, it is important to develop effective diagnostic and therapeutic modalities based on sound biological insights into hepatocarcinogenesis.2,3
The copy number alterations observed in human solid tumors are known to contribute to the tumorigenesis by affecting the activities of cancer-related genes in the altered chromosomal regions.4 Thus, genome-wide mapping of copy number alterations in cancer can facilitate the identification of cancer-related genes, which will improve the understanding of tumorigenesis. Using conventional cytogenetic tools such as comparative genomic hybridization (CGH), copy number gains on 1q, 8q and 20q, along with losses on 1p, 4q, 8p, 13q, 16q and 17p have been previously identified in HCC.5-7 However, the resolution of conventional cytogenetic analysis is insufficient to precisely identify submicroscopic changes. Recently introduced array-CGH, the combination of conventional CGH and microarray technology, enabled high-resolution screening of genome-wide copy number alterations containing potential cancer-related genes.8,9 Through array-CGH analysis, novel oncogenes such as JAB1 or differentiation-specific regions have been identified in HCC.5,10 Also etiology-dependent copy number alterations and genes relevant to hepatocarcinogenesis were suggested in HCC.11 But, it is still difficult to identify the biologically relevant changes and their functional significance in a systematic manner due to the extensive and complex nature of chromosomal alterations.
We hypothesized that recurrent copy number changes common to many HCC cases may contain essential genes for hepatocarcinogenesis. Using this strategy, recurrently altered regions (RARs) were defined in 76 primary HCCs using whole-genome array CGH analysis, and the associations between RARs and clinicopathologic features were examined. Also, we functionally categorized the genes located in the RARs.
Material and methods
Study materials
Frozen tissues (tumor and adjacent normal tissue pairs) were obtained from 76 primary HCC patients (65 males and 11 females) who underwent surgical resection. This study was performed under the approval of the Institutional Review Board of the Catholic University Medical College of Korea. Tumor stage was determined according to the standard tumor-node-metastasis classification of AJCC guidelines (6th edition). Clinicopathologic information about the 76 cases is available in Supporting Table 1. Ten-micrometer-thick frozen sections were prepared, and tumor cell-rich areas (tumor cells comprise more than 80–90% of the selected area) without tumor cell necrosis were microdissected, from which genomic DNA was extracted as described previously.12,13 Normal human genomic DNA was purchased from Promega (Madison, WI) and used as sex-matched reference DNA for array-CGH.
Array comparative genomic hybridization
A large insert clone array covering the entire human genome at 1 Mb resolution was used for profiling genomic alterations.14 Array-CGH was performed as described elsewhere using MAUI hybridization station (BioMicro Systems, Salt Lake city, UT).12,13 Data processing, normalization and realigning of raw array-CGH data were performed using web-based array-CGH analysis software ArrayCyGHt (http://genomics.catholic.ac.kr/arrayCGH/).15 We used print-tip loess normalization method for analysis. Large insert clones (n = 2,958) and genomic coordinates such as cytogenetic bands or gene positions were mapped according to the UCSC genome browser (http://genome.ucsc.edu/; May 2004 freeze).
Identification of copy number alteration
To set the cutoff value for chromosomal alterations of individual clones based on standard deviation (SD), we performed 6 independent hybridizations using DNA from normal individuals (4 sex-matched and 2 male-to-female), from which control SD values of individual clones were obtained. Chromosomal gain or loss was assigned when the normalized log2 intensity ratio of each data point exceeded or fell below ±3 SD derived from normal control hybridizations. Regional copy number change was defined as DNA copy number alteration stretching over 2 or more consecutive large insert clones, but not across an entire chromosomal arm. High-level amplification of clones was defined when their intensity ratios were higher than 1.0 in log2 scale and vice versa for homozygous deletion. RAR was defined as regional copy number changes observed in 15 (20%) or more samples out of 76 HCC samples. The array-CGH data described in our study are available at website (http://www.systemsbiology.co.kr/micro/CGH/hepato.htm).
Real-time quantitative PCR assay
The first-strand cDNA was synthesized from total RNA of 20 HCCs using M-MLV reverse transcriptase (Invitrogen, Carlsbad, CA). cDNA from the nonneoplastic part of liver, which showed no septal fibrosis or liver cirrhosis, was used as normal control. Real-time quantitative PCR was performed using Mx3000P qPCR system and MxPro Version 3.00 software (Stratagene, CA). Twenty microliter of real-time qPCR mixture contained 10 ng of cDNA, 1×SYBR® Green Tbr polymerase mixture (FINNZYMES, Finland), 1×ROX and 20 pmol of primers. GAPDH was used as an internal control in each procedure. Thermal cycling was done as follows: 10 min at 95°C followed by 40 cycles of 10 sec at 94°C, 30 sec at 53–58°C and 30 sec at 72°C. To verify specific amplification, melting curve analysis was performed (55–95°C, 0.5°C/sec). Relative quantification was performed by the ΔΔCT method.16 All the experiments were repeated twice and the mean value of intensity ratios with the SD was plotted for each case. Primer sequences for TPM3, RPS27, HAX1, PYGO2, CKS1B, ADAM15, CCT3, PRCC, KIF14, ELF3, TGFB2 and AKT3 are available in the Supporting Table 2.
Western blot analysis for KIF14 and TPM3
Tissue lysates (40 μg per lane for TPM3 and 100 μg per lane for KIF14) were electrophoresed in 10% sodium dodecyl sulfate (SDS)-polyacrylamide gel. Separated proteins were transferred to a PVDF membrane, which was blocked with 5% nonfat dried milk in TBST (20 mM Tris-HCl, 150 mM NaCl and 0.1% Tween 20, pH 7.5), and then incubated overnight with anti-KIF14 (Abcam, Cambridge, UK), anti-TPM3 (Abnova, Taipei, Taiwan) and anti β-actin antibody (Sigma, St. Louis, MO). After washing with TBST, the PVDF membrane was incubated with diluted horseradish peroxidase-conjugated anti-rabbit IgG or anti-mouse IgG for 2 hr at room temperature. The blots were detected using an enhanced chemiluminescence (ECL) system (Amersham-Pharmacia Biotech, Braunschweig, Germany).
Functional enrichment analysis
Functional enrichment analysis based on gene ontology was performed for the RARs that are significantly associated with the clinicopathologic characteristics. In brief, gene sets for enrichment analysis were prepared using 17,661 known genes with genomic coordinates downloaded from the UCSC genome browser (2004, May Freeze). Genes were grouped into specific sets using NetAffx Gene Ontology Mining Tools according to functions annotated in public gene databases such as GO (Gene Ontology), KEGG (Kyoto Encyclopedia of Genes and Genomes) and GenMAPP (Gene Map Annotator and Pathway Profiler).17-20 A total of 1,632 gene sets were prepared for enrichment analysis. The functional enrichment analysis was performed using GEAR software (http://www.systemsbiology.co.kr/GEAR/).21 The significance of enrichment was calculated by hypergeometric distribution and p value less than 0.01 was considered significant.
Statistical analysis
To see the association between chromosomal changes such as RARs and total number of altered clones, and clinicopathologic phenotypes, 8 clinical parameters were treated as categorical variables; age (<50 vs. ≥50 years), sex, grade (Grade 1 and 2 as low vs. 3 and 4 as high), stage (Stage I and II as early vs. Stage III and IV as advanced), tumor size (<3.5 vs. ≥3.5 cm) along with the presence or absence of microvascular invasion (MVI), portal vein invasion and encapsulation. Significance of the different distribution of RARs in each category was tested by two-sided Fisher's exact test. The mean number of altered clones in 8 categories and expression levels of genes in different RAR groups were compared by independent t-test. Stata version 7.0 (Stata Corporation, Texas) was used and p value less than 0.05 was considered significant in all statistical analyses.
Results
Profiles of chromosomal alterations
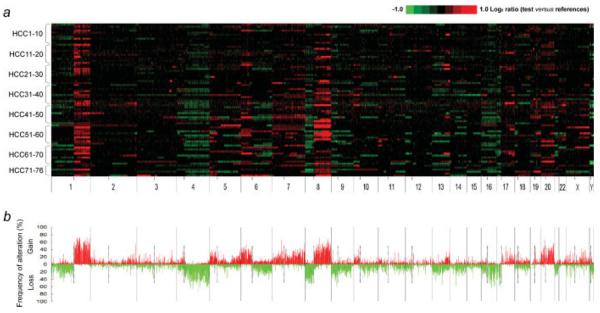
The overall profile of chromosomal alterations identified in the 76 primary HCCs is illustrated in Figure 1a. Chromosomal alterations are not randomly distributed along the genome but clustered in certain chromosomal segments (Fig. 1b). The mean number of altered clones per case is 677.1 (338.7 clones gained and 338.4 clones lost) out of total 2,958 clones. In other words, 22.9% (11.5% gained and 11.4% lost) of the whole genome is altered per each case on an average. The mean numbers of altered clones are significantly higher in high-grade tumors than low-grade ones (769.9 vs. 594.6 clones; p = 0.002), in microinvasion positive cases than negative ones (764.1 vs. 598.9 clones; p = 0.016) and in bigger tumors than smaller ones (756.1 vs. 602.3 clones; p = 0.026). In the case of entire chromosomal arm changes, 5 chromosomal gains in 1q, 6p, 8q, 20p and 20q and 5 losses in 4q, 8p, 9p, 16q and 17p are repetitively observed in over 30% of the samples. The alteration frequency of chromosomal arms is summarized in Supporting Table 3.
Figure 1.
Whole-genome profiles and frequency plot of chromosomal alterations in hepatocellular carcinoma. (a) The genomic alterations in 76 primary human hepatocellular carcinomas are illustrated in individual lanes. A total of 2,958 large insert clones are mapped according to the UCSC genome browser (May, 2004 Freeze) and ordered by chromosomal position from 1pter to Yqter (X axis). Tumor vs. reference intensity ratios (in log2 scale) for individual clones are plotted in different color scales reflecting the extent of copy number gains (red) and losses (green) as indicated in the reference color bar. The boundaries of individual chromosome are indicated by long vertical bars and the locations of centromeres by short vertical bars below the plot. (b) The frequency of chromosomal gains (>3 SD) and losses (<3 SD) for each clone is shown for 76 HCC samples.
Recurrently altered regions in HCC
Although entire chromosomal arm changes appeared occasionally, vast majority of copy number alterations in HCCs are localized regional changes. To delineate the frequently observed consensus regions, we defined the RARs, which are regional chromosomal alterations observed in at least 15 cases (20% of HCC cases) among the 76 HCCs. In total, 14 RAR gains (RAR-G) and 18 RAR losses (RAR-L) were detected (Table I). Five most frequent RARs are RAR-G1 (64.5%: 1q21.1-1q32.1), RAR-G2 (59.2%: 1q32.1-1q44), RAR-L17 (51.3%: 17p13.3-17p12), RARG9 (48.7%: 8q11.21-8q24.3) and RAR-L5/L8 (both 44.7%: 4q33-4q35.2/8p23.2-8p12). The average size of RAR-Gs (26.6 Mb; ranged 1–94 Mb) is larger than that of RAR-Ls (16.5 Mb; ranged 0.9–43 Mb). Well-known oncogenes such as MYC, FGF, EGFR and CCND3 along with tumor suppressor genes such as TP53, RB1, CDKN2A and CDKN2B are located in identified RARs. The other genes in the RARs are also thought to be potential cancer-related genes contributing to hepatocarcinogenesis directly or indirectly (Table I).
TABLE I.
GENERAL CHARACTERISTICS OF RECURRENTLY ALTERED REGIONS
| RAR | Clone | Chromosome | Map position1 | Size (Mb) | Cytoband | Number of cases | Cancer-related genes |
|---|---|---|---|---|---|---|---|
| G1 | RP11-326G21-RP5-936P19 | 1 | 142391262-183515789 | 41 | 1q21.1-1q32.1 | 49 | PDZK1, MCL1, ARNT, AF1Q, TPM3, ADAR, RPS27, HAX1, PYGO2, CKS1B, ADAM15, MUC1, HDGF, CCT3, PRCC, IFI16, AIM2, USF1, SELP, SELE, LAMC2, TPR, PTGS2, |
| G2 | RP11-572A16-RP11-438H8 | 1 | 198883756-244440465 | 46 | 1q32.1-1q44 | 45 | KIF14, ELF3, MDM4, ATF3, TGFB2, WNT3A, AKT3 |
| G3 | RP11-46C20-CTD-2291F22 | 5 | 27487154-35513594 | 8 | 5p14.1-5p13.2 | 19 | AMACR |
| G4 | CTB-55A14-CTB-73D21 | 5 | 167592449-174643724 | 7 | 5q34-5q35.2 | 16 | FGF18 |
| G5 | RP1-136B1-RP11-524H19 | 6 | 2316508-54856430 | 52 | 6p25.2-6p12.1 | 23 | DEK, ID4, E2F3, PRL, MICA, MICB, HMGA1, NOTCH4, MAPK14, PIM1, TFEB, CCND3, VEGF |
| G6 | RP5-1091E12-RP11-339F13 | 7 | 54851795-55154846 | 1 | 7p11.2 | 16 | EGFR |
| G7 | RP5-1057M1-RP11-212B1 | 7 | 79345845-86861115 | 7 | 7q21.11 | 20 | HGF, DMTF1, ABCB1 |
| G8 | RP5-1059M17-RP11-420H19 | 7 | 100783053-124638919 | 24 | 7q22.1-7q31.33 | 22 | EPO, EPHB4, PIK3CG, CAV1&2, MET, WNT2 |
| G9 | RP11-167E7-RP11-65A5 | 8 | 48736257-141551817 | 94 | 8q11.21-8q24.3 | 37 | PRKDC, MCM4, SNAI2, LYN, MOS, PLAG1, COPS5, TPD52, E2F5, MMP16, NBS1, EIF3S3, MYC, KCNK9, PTK2, EIF2C2, CCNE2 |
| G10 | RP11-472N13-RP11-505N10 | 10 | 31849328-33818450 | 2 | 10p11.22 | 15 | MAP3K8, NRP1, |
| G11 | RP11-95C14-RP11-265C7 | 13 | 91284427-112901415 | 21 | 13q31.3-13q34 | 16 | FGF14, ERCC5, |
| G12 | RP11-515O17-RP11-332H18 | 17 | 50654233-56847074 | 6 | 17q22-17q23.2 | 16 | HLF, MPO, PPM1D, BCAS3, TBX2 |
| G13 | CTD-2043I16-CTC-416D1 | 19 | 33293135-36979856 | 3 | 19q12 | 19 | CCNE1 |
| G14 | RP5-852M4-RP4-563E14 | 20 | 327036-61041280 | 61 | 20p13-20q13.33 | 26 | CDC25B, JAG1, SSTR4, BCL2L1, PLAGL2, DNMT3B, E2F1, MMP24, SRC, TOP1, MYBL2, MMP9, NCOA3, PTPN1, ZNF217, STK6, BMP7 |
| L1 | RP1-37J18-RP11-338N10 | 1 | 4487199-7719107 | 3.3 | 1p36.32-1p36.23 | 18 | CHD5, ICMT, CAMTA1 |
| L2 | RP11-285P3-RP4-560M15 | 1 | 14486429-15354152 | 0.9 | 1p36.21 | 24 | PRDM2, RIZ, CASP9 |
| L3 | RP11-8J9-RP11-412F21 | 1 | 46823997-69303906 | 23 | 1p33-1p31.2 | 17 | RAD54L, FAF1, S(T)IL, CDKN2C, TTC4, JUN, ARHI |
| L4 | RP11-118B23- RP5-1108M17 | 1 | 84667459-103923047 | 19 | 1p22.3-1p21.1 | 16 | BCL10, CLCA2, LMO4, GTF2B, TGFBR3, GFI1, EVI5 |
| L5 | RP11-213L8-RP11-553E4 | 4 | 172094998-190118103 | 18 | 4q33-4q35.2 | 34 | CASP3, FAT |
| L6 | RP1-273N12-RP11-347H8 | 6 | 99385861-102294657 | 3 | 6q16.2-6q16.3 | 17 | CCNC, GRIK2 |
| L7 | RP1-84N20-RP3-470B24 | 6 | 125389372-168197568 | 43 | 6q22.31-6q27 | 16 | CRSP3, PLAGL1, SASH1, LATS1, IGF2R, UNC93A, MLLT4 |
| L8 | RP11-336N16-RP11-75P13 | 8 | 2898583-34455078 | 31 | 8p23.2-8p12 | 34 | CSMD1, DEFB1, NAT1, NAT2, PSD3, TNFRSF10A, TNFRSF10B, TNFRSF10C, RHOBTB2 |
| L9 | RP11-48M17-RP11-48L13 | 9 | 2136329-29639069 | 27 | 9p24.2-9p21.1 | 26 | SMARCA2, MTAP, CDKN2B, CDKN2A, RECK, PAX5 |
| L10 | RP11-276H19-RP11-65B23 | 9 | 86827119-87654534 | 1 | 9q21.33 | 15 | GAS1, DAPK1 |
| L11 | RP11-92C4-RP11-31J20 | 9 | 98644250-104754734 | 6 | 9q22.33-9q31.1 | 16 | TGFBR1 |
| L12 | RP11-381K7-RP11-338L11 | 10 | 112963138-116971219 | 4 | 10q25.2-10q25.3 | 15 | CASP7 |
| L13 | RP11-153M24-RP11-359P14 | 13 | 27414161-59888914 | 32 | 13q12.2-13q21.2 | 19 | BRCA2, CCNA1, RB1, RFP2, DLEU1, DLEU2, DDX26 |
| L14 | RP11-353N19-RP11-68I8 | 14 | 91389738-99247779 | 8 | 14q32.12-14q32.2 | 15 | BCL11B |
| L15 | RP11-114I12-RP11-142A12 | 16 | 6846342-26727359 | 20 | 16p13.2-16p12.1 | 18 | SOCS1, ERCC4 |
| L16 | RP11-325K4-RP11-514D23 | 16 | 55369591-84922042 | 29 | 16q13-16q24.1 | 28 | CDH1, CDH3, BCAR1, WWOX, CDH13, WFDC1 |
| L17 | RP11-135N5-RP11-219A15 | 17 | 2312021-16718826 | 14 | 17p13.3-17p12 | 39 | TP53 |
| L18 | RP1-270M7-RP5-1031P17 | 21 | 15134621-39788380 | 14 | 21q11.2-21q22.2 | 15 | ADAMTS1 |
The mapping position refers to the UCSC genome browser (http://genome.ucsc.edu/; May 2004 freeze).
RARs and clinical characteristics
Eight types of clinical variables (age of onset, sex, grade, stage, size, portal vein invasion, MVI and encapsulation) were analyzed for their associations with the RARs (Table II). RARs and associated characteristics are as follows: RAR-L11 and -L16 with early onset of age, RAR-G5 and -G8 with male sex, RAR-L5, -L9, -L10, -L11 and -L13 with high tumor grade, especially RAR-L10 showing highly significant association, RAR-L4 with advanced tumor stage, RAR-G9, -G12 and -G13 with MVI, RAR-G13 and -L7 with portal vein invasion in negative direction and RAR-G4 with larger tumor size.
TABLE II.
SIGNIFICANT ASSOCIATIONS BETWEEN RARs AND CLINICAL FEATURES
| Early onset (<50) | Late onset (≥50) | Total | p value | ||
|---|---|---|---|---|---|
| RAR-L11 | − | 23 | 37 | 60 | 0.046 |
| + | 11 | 5 | 16 | ||
| RAR-L16 | − | 23 | 37 | 60 | 0.046 |
| + | 11 | 5 | 16 | ||
|
| |||||
| Female | Male | Total | p value | ||
|
| |||||
| RAR-G5 | − | 11 | 42 | 53 | 0.028 |
| + | 0 | 23 | 23 | ||
| RAR-G8 | − | 11 | 43 | 54 | 0.028 |
| + | 0 | 22 | 22 | ||
|
| |||||
| Low grade (1, 2) | High grade (3, 4) | Total | p value | ||
|
| |||||
| RAR-L5 | − | 30 | 12 | 42 | 0.020 |
| + | 15 | 19 | 34 | ||
| RAR-L9 | − | 36 | 14 | 42 | 0.003 |
| + | 9 | 17 | 26 | ||
| RAR-L10 | − | 42 | 19 | 51 | 0.001 |
| + | 3 | 12 | 15 | ||
| RAR-L11 | − | 40 | 20 | 60 | 0.020 |
| + | 5 | 11 | 16 | ||
| RAR-L13 | − | 38 | 19 | 57 | 0.031 |
| + | 7 | 12 | 19 | ||
|
| |||||
| Stage (I, II) | Stage (III, IV) | Total | p value | ||
|
| |||||
| RAR-L4 | − | 34 | 26 | 60 | 0.047 |
| + | 4 | 12 | 16 | ||
|
| |||||
| Size (<3.5 cm) | Size (≥3.5 cm) | Total | p value | ||
|
| |||||
| RAR-G4 | − | 35 | 25 | 60 | 0.024 |
| + | 4 | 12 | 16 | ||
|
| |||||
| MVI (−) | MVI (+) | Total | p value | ||
|
| |||||
| RAR-G9 | − | 27 | 12 | 39 | 0.005 |
| + | 13 | 24 | 37 | ||
| RAR-G12 | − | 36 | 24 | 60 | 0.022 |
| + | 4 | 12 | 16 | ||
| RAR-G13 | − | 25 | 32 | 57 | 0.009 |
| + | 15 | 4 | 19 | ||
|
| |||||
| PVI (−) | PVI (+) | Total | p value | ||
|
| |||||
| RAR-G13 | − | 43 | 14 | 57 | 0.016 |
| + | 19 | 0 | 19 | ||
| RAR-L7 | − | 46 | 14 | 60 | 0.033 |
| + | 16 | 0 | 16 | ||
MVI, microvascular invasion; PVI, portal vein invasion.
Functional annotation of the clinically significant RARs
We investigated the functional categories of the genes enriched in the RARs, which showed significant associations with the clinical features. Table III lists the enriched functional pathways in tumor grade-associated RARs. Top 5 pathways have interferon-related gene families in common as member genes, because interferon-loci are included in tumor grade-related RARs. As all of these RARs are copy number losses, it can be assumed that the 5 interferon-related pathways are repressed. Cell cycle regulation and angiogenesis pathways are also found to be significantly associated with tumor grade-related RARs. Functional enrichment analysis results for other clinical feature-related RARs are available in the Supporting Table 4.
TABLE III.
- FUNCTIONAL PATHWAYS ENRICHED IN TUMOR GRADE-ASSOCIATED RARs
| Functional annotations | Gene size1 | Observed genes2 | p-value3 | Genes4 |
|---|---|---|---|---|
| Hematopoietin/interferon-class cytokine receptor binding | 20 | 14 | 5.25E-21 | IFNA1, IFNA2, IFNA4, IFNA5, IFNA6, IFNA8, IFNA10, IFNA13, IFNA14, IFNA16, IFNA17, IFNA21, IFNB1, IFNW1 |
| Interferon-alpha/beta receptor binding | 9 | 9 | 1.18E-16 | IFNA1, IFNA2, IFNA4, IFNA10, IFNA13, IFNA16, IFNA17, IFNB1, IFNW1 |
| Response to virus | 66 | 14 | 5.15E-12 | IFN family5 |
| Defense response | 126 | 15 | 4.52E-09 | IFN family, IFNE1 |
| Cytokine activity | 177 | 17 | 1.15E-08 | IFN family, TNFSF11, CER1, IFNE1 |
| Physiological process | 18 | 4 | 0.0002 | INSL4, RLN1, RLN2, INSL6 |
| Regulation of cyclin dependent protein kinase activity | 34 | 4 | 0.0027 | CDKN2A, CDKN2B, CCNA1, RGC32 |
| Condensed chromosome | 6 | 2 | 0.0042 | HMGB1, HMGB2 |
| Angiogenesis | 41 | 4 | 0.0053 | COL15A1, FLT1, VEGFC, HAND2 |
| Pregnancy | 47 | 4 | 0.0086 | FLT1, INSL4, RLN1, RLN2 |
Number of genes in the functionally annotated gene sets.
Number of observed genes in tumor grade-related RARs.
Significance level of enrichment was calculated using hypergeometric distribution and p < 0.01 was considered significant.
Gene symbols for the observed genes.
Includes 14 genes in hematopoietin/interferon-class cytokine receptor binding category. It is used to avoid repeating the gene symbols.
High-level copy number changes
In total, 33 amplifications and 10 homozygous deletions (HD) were identified (see Supporting Table 5). Most high-level copy number changes were observed in a single case, but some of them appeared recurrently. For example, the amplification on 8q11.1-8q24.3 containing MYC and EIF3S3 was observed in 10 cases and the amplification on 11q13.2-11q13.3 containing CCND1, FGF4, FGF3 and ORAOV1 in 5 cases. In addition, the amplifications on 1q31.1-1q43, 1q43-1q44, 13q31.1-13q34 and 17q12-17q25.3 were detected in 4 cases. All HDs were observed in a single case except for 9p21.3 containing CDKN2A and CDKN2B tumor suppressor.
Putative hepatocarcinogenesis related oncogenes in RARs on 1q
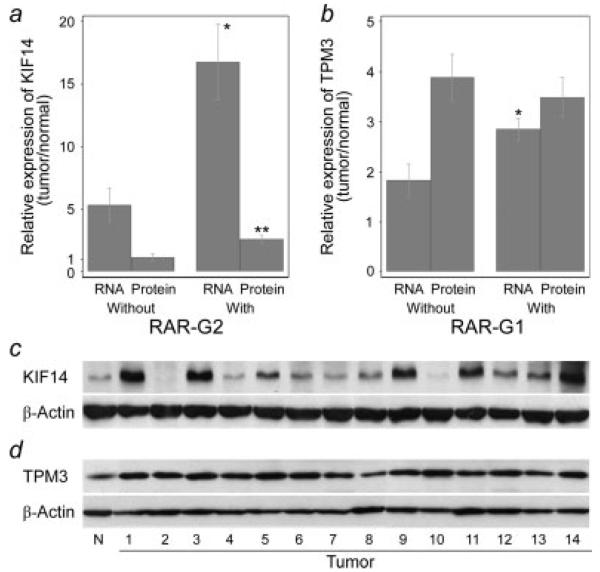
Many of the high-level copy number changes were found within RARs. Sixteen of 33 amplifications and 8 of 10 HDs overlapped with RAR-Gs and -Ls, respectively (see Supporting Table 5). We assumed high-level alterations overlapping with RARs might be more closely related to carcinogenesis. Thus, we examined the RNA profiles of the putative cancer-related genes as follows in RAR-G1 and -G2, which most extensively overlapped with amplifications; TPM3, RPS27, HAX1, PYGO2, CKS1B, ADAM15, CCT3, PRCC, KIF14, ELF3, TGFB2 and AKT3. Total RNA was available for 20 cases out of 76 HCCs, of which 13 cases are RAR-G1 positive and 12 cases are RAR-G2 positive. RNA expression levels of most candidate genes in the HCCs are higher than normal liver control RNA level (see Supporting Fig. 1). Especially, KIF14 (kinesin family member 14) and TPM3 (tropomyosin 3) are highly expressed in HCC and their RNA levels are significantly correlated with copy number status (Figs. 2a and 2b). The mean RNA expression ratio (tumor/normal) of KIF14 in RAR-G2 positive cases was 16.8, whereas 5.3 in RAR-G2 negative cases (p = 0.004). The mean RNA expression ratio of TPM3 was significantly higher in RAR-G1 positive cases than in RAR-G1 negative group (2.85 vs. 1.83, p = 0.026). CKS1B also showed the higher expression in RAR-G1 positive group than negatives (1.42 vs. 2.21), but the difference is not statistically significant. KIF14 and TPM3 protein expression levels (tumor/normal ratio) were also examined by immunoblotting. Both KIF14 and TPM3 were significantly overexpressed compared with normal liver tissue (Figs. 2c and 2d). The mean protein expression ratio (tumor/normal) was 3.63 for TPM3 (p < 0.001) and 1.99 for KIF14 (p = 0.005). When analysis was limited to HCCs, KIF14 protein expression (2.61 in RAR-G2 positive cases vs. 1.15 in RAR-G2 negative cases, p = 0.004) was significantly correlated with copy number status, whereas the correlation was not significant in TPM3 (Figs. 2a and 2b).
Figure 2.
Expression levels of KIF14 and TPM3 by copy number status. Mean RNA and protein expression levels (tumor/normal ratio) of KIF14 (a and c) and TPM3 (b and d) were calculated by real-time qRT-PCR and Western blotting in the presence of RAR. Human GAPDH gene and β-actin was used as an internal control for qRT-PCR and immunoblotting, respectively. KIF14 RNA level was significantly higher in samples with RAR-G2 (n = 12) than without (n = 8) (*p = 0.004), and KIF14 protein level was also significantly higher in samples with RAR-G2 (n = 8) than without (n = 6) (** p = 0.004). TPM3 RNA level was significantly higher in samples with RAR-G1 (n = 13) than without (n = 7) (*p = 0.026), but protein expression was not significantly different between RAR-G1 positives and negatives (n = 9 and 5, respectively). Tumor 1, 3, 5, 7, 8, 9, 10, 12 and 14: RAR-G1+; Tumor 2, 4, 6, 11 and 13: RAR-G1−. Tumor 1, 3, 7, 8, 9, 11, 12 and 14: RAR-G2+; Tumor 2, 4, 5, 6, 10 and 13: RAR-G2−. X axis represents RAR status and Y axis represents mean expression ratio. Error bars represent mean ± standard error of mean. N, normal.
Discussion
In our study, we analyzed the genome-wide copy number alteration profiles in HCC using BAC arrays with 1Mb resolution. Frequently observed entire arm chromosomal changes (>30% of samples) are largely consistent with previous observations in HCCs of Asians and Caucasians.5,6,10,11 Some of the discrepancies might be caused by using the populations or criteria different from our study. The relatively frequent gains of 20p and 20q might have been observed due to the increased resolution of array platform we used.10
We delineated the RARs on the assumption that alterations shared by significant number of cases may contain genes essential to hepatocarcinogenesis. Among the 32 RARs identified in our study, 18 RARs (RAR-G1,-2,-5,-6,-9,-11,-12 and -14 along with RAR-L1,-2,-5,-7,-8,-9,-11,-13,-17 and -18) including the 5 most frequent RARs are consistent with those previously reported in HCC.22-24 The other 14 RARs have not been identified in other studies. But, some of them such as RAR-G7 (7q21.11), RAR-G8 (7q22.1–7q31.33) and RAR-L16 (16q13–16q24.1) appeared in more than 20 cases in our study, which suggests that they are unlikely to be false positives. These potentially novel RARs may give a clue to understand hepatocarcinogenesis. For example, copy number gain of 7q21.11(RAR-G7), frequently observed in our study, could explain overexpression of HGF (hepatocyte growth factor) in HCCs. Higher expression of HGF, which is known to promote angiogenesis, has been frequently reported in HCC25 and highlighted as a potential biomarker for the progression of HCC.26,27 Another example is EPO located in 7q 22.1 (RAR-G8). EPO is also known to be involved in angiogenesis in HCC,28 but copy number gain of EPO gene has been rarely reported in HCC.
In high-level copy number changes identified in our study, many well-known cancer related genes such as MYC, MDM4, EIF3S3, CCND1, FGF4, FGF3, CDKN2A and CDKN2B were found11,29,30 along with other genes unexplored in HCCs. Twenty-four regions of amplifications or HDs were found to overlap with RARs across diverse chromosomes. It can be assumed that the genes located in this overlapped region may contribute to hepatocarcinogenesis more directly. Because this overlapping phenomenon was most apparent on chromosome 1q, we examined the RNA profile of the putative cancer-related genes on the 8 overlapped regions on 1q. Of the 12 candidate genes listed in the result, RNA levels of KIF14 and TPM3 were elevated in all tested cases except for 1 and showed the significant correlations with the copy number status. Similarly, both KIF14 and TPM3 proteins were significantly overexpressed in HCCs compared with normal liver tissue. Increased expression of TPM3 protein was observed in all the HCCs tested regardless of copy number status, which suggests the possibility of TPM3 as a HCC-biomarker. KIF14 expression was elevated in DNA dosage-dependent manner in both RNA and protein levels. Those results suggest that copy number gains on 1q (RAR-G1 and 2) induce overexpression of TPM3 and KIF14, subsequently influencing hepatocarcinogenesis. Although copy number gain on 1q31-q32 where KIF14 is located has been reported in diverse cancers,31 this has not been reported in HCC yet.
KIF14 is essential for cytokinesis and chromosome segregation.32 Its upregulation was previously observed in malignancies and suggested as a predictor for poor prognosis.33 As TPM3 plays similar roles in cytokinesis,34 it can be assumed that both molecules may be involved in carcinogenesis including the development of HCC. In our unpublished data, siRNA knockdown of KIF14 and TPM3 repressed the growth of a HCC cell line. However, in our study, we could not observe any significant association between their expression levels and invasiveness (MVI and PVI), tumor grade, size and survival (data not shown). It might be due to limited number of samples (20 for qRT-PCR and 14 for immunoblotting). Further large-scale studies will be required to validate this possibility in HCCs.
TPM3, located in 1q21.3, is known to be involved in hematopoietic tumorigenesis by forming TPM3-ALK fusion through (1;2) translocation.35 TPM3-ALK fusion gene is also known to be involved in transformation, proliferation, invasion and metastasis.36 When we examined the existence of TPM3-ALK fusion by real-time qRT-PCR for the HCCs showing TPM3 overexpression as previously described,35 none of them showed fusion-specific signal (data not shown). But, the absence of the fusion protein in our samples cannot exclude the potential role of TPM3 in hepatocarcinogenesis. To our knowledge, this is the first report to suggest oncogenic potential of copy number gains related to overexpression of KIF14 and TPM3 in hepatocarcinogenesis.
Some RARs are significantly associated with clinical features. Especially, RAR-L9 and -L10 (9p21) and RAR-G9 (8q24) showed the most significant associations with the higher tumor grade and MVI, respectively. This result is consistent with previous reports showing that copy number losses on 9p21 and 8q24 amplification have been frequently associated with poor prognosis of HCC.37,38 Our result indicates that these copy number alterations can affect prognosis of HCC.
To see functional implication of copy number alterations, we applied functional enrichment analysis on RARs associated with clinical features. Gene sets significantly enrichment in with phenotype-correlated RARs might represent functionally related genes, which are likely to work collaboratively in hepatocarcinogenesis. For example, cytokine receptor binding and defense response to virus pathways are enriched in recurrent copy number deletion regions (RAR-L5,-9,-10,-11 and -13), which are associated with the high tumor grade. This is comparable with previous studies reporting that the progression of liver disease and hepatocarcinogenesis is not only due to direct cytopathic effects of viruses but also due to repressed host immune response to viral proteins.39,40 Given that more than 90% of HCC patients examined in our study are HBV or HCV positive, it seems reasonable to observe the significant correlation between the recurrent deletion of defense response to virus pathway and the high tumor grade. Also, cell cycle regulation and angiogenesis pathways were found to be enriched in the RAR-Ls associated with high grade. Disruption of cell cycle regulation in aggressive HCC has been consistently reported in HCC.41
Our study has some limitations. The resolution of arrays used is not high enough to detect microcopy number changes and to determine exact boundaries for altered regions. Further studies using higher resolution arrays and target region-focusing subarrays will be required to discover novel microalterations. Because of limited availability of cases for RNA and protein assays, potential roles of the RAR-associated genes in hepatocarcinogenesis could not be validated reliably. Further large scale study will be necessary to explore these genes. Lastly, because most of the study subjects are HBV or HCV positive, it is difficult to generalize our results to nonviral HCCs.
In conclusion, we successfully identified copy number changes including novel ones in 76 primary HCCs using whole-genome array-CGH and proposed KIF14 and TPM3 as potential oncogenes in HCC. We also demonstrated the relationship between RARs and tumor characteristics. Through functional enrichment analysis, we suggested several functional pathways likely to be involved in hepatocarcinogenesis. Taken together, our results and the strategy of analysis will help to elucidate pathogenesis of HCC and to develop biomarkers for predicting the behaviors of HCC.
Supplementary Material
Acknowledgements
The authors thank The Wellcome Trust Sanger Institute Microarray Facility for printing BAC array slides.
Footnotes
Additional Supporting Information may be found in the online version of this article.
Grant sponsor: Ministry of Science and Technology in Korea (21C Frontier Functional Human Genome Project); Grant number: FG06-12-01.
The first two authors contributed equally to this work.
References
- 1.Blum HE. Hepatocellular carcinoma: therapy and prevention. World J Gastroenterol. 2005;11:7391–400. doi: 10.3748/wjg.v11.i47.7391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.El-Serag HB. Hepatocellular carcinoma: recent trends in the United States. Gastroenterology. 2004;127:S27–S34. doi: 10.1053/j.gastro.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 3.Llovet JM, Burroughs A, Bruix J. Hepatocellular carcinoma. Lancet. 2003;362:1907–17. doi: 10.1016/S0140-6736(03)14964-1. [DOI] [PubMed] [Google Scholar]
- 4.Vissers LE, Veltman JA, van Kessel AG, Brunner HG. Identification of disease genes by whole genome CGH arrays. Hum Mol Genet. 2005;14:R215–R223. doi: 10.1093/hmg/ddi268. [DOI] [PubMed] [Google Scholar]
- 5.Hashimoto K, Mori N, Tamesa T, Okada T, Kawauchi S, Oga A, Furuya T, Tangoku A, Oka M, Sasaki K. Analysis of DNA copy number aberrations in hepatitis C virus-associated hepatocellular carcinomas by conventional CGH and array CGH. Mod Pathol. 2004;17:617–22. doi: 10.1038/modpathol.3800107. [DOI] [PubMed] [Google Scholar]
- 6.Moinzadeh P, Breuhahn K, Stützer H, Schirmacher P. Chromosome alterations in human hepatocellular carcinomas correlate with aetiology and histological grade–results of an explorative CGH meta-analysis. Br J Cancer. 2005;92:935–41. doi: 10.1038/sj.bjc.6602448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zimonjic DB, Keck CL, Thorgeirsson SS, Popescu NC. Novel recurrent genetic imbalances in human hepatocellular carcinoma cell lines identified by comparative genomic hybridization. Hepatology. 1999;29:1208–14. doi: 10.1002/hep.510290410. [DOI] [PubMed] [Google Scholar]
- 8.Pinkel D, Segraves R, Sudar D, Clark S, Poole I, Kowbel D, Collins C, Kuo WL, Chen C, Zhai Y, Dairkee SH, Ljung BM, et al. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat Genet. 1998;20:207–11. doi: 10.1038/2524. [DOI] [PubMed] [Google Scholar]
- 9.Solinas-Toldo S, Lampel S, Stilgenbauer S, Nickolenko J, Benner A, Döhner H, Cremer T, Lichter P. Matrix-based comparative genomic hybridization: biochips to screen for genomic imbalances. Genes Chromosomes Cancer. 1997;20:399–407. [PubMed] [Google Scholar]
- 10.Patil MA, Gütgemann I, Zhang J, Ho C, Cheung ST, Ginzinger D, Li R, Dykema KJ, So S, Fan ST, Kakar S, Furge KA, et al. Array-based comparative genomic hybridization reveals recurrent chromosomal aberrations and Jab1 as a potential target for 8q gain in hepatocellular carcinoma. Carcinogenesis. 2005;26:2050–7. doi: 10.1093/carcin/bgi178. [DOI] [PubMed] [Google Scholar]
- 11.Schlaeger C, Longerich T, Schiller C, Bewerunge P, Mehrabi A, Toedt G, Kleeff J, Ehemann V, Eils R, Lichter P, Schirmacher P, Radlwimmer B. Etiology-dependent molecular mechanisms in human hepatocarcinogenesis. Hepatology. 2008;47:511–20. doi: 10.1002/hep.22033. [DOI] [PubMed] [Google Scholar]
- 12.Kim TM, Yim SH, Lee JS, Kwon MS, Ryu JW, Kang HM, Fiegler H, Carter NP, Chung YJ. Genome-wide screening of genomic alterations and their clinicopathologic implications in non-small cell lung cancers. Clin Cancer Res. 2005;11:8235–42. doi: 10.1158/1078-0432.CCR-05-1157. [DOI] [PubMed] [Google Scholar]
- 13.Kim MY, Yim SH, Kwon MS, Kim TM, Shin SH, Kang HM, Lee C, Chung YJ. Recurrent genomic alterations with impact on survival in colorectal cancer identified by genome-wide array comparative genomic hybridization. Gastroenterology. 2006;131:1913–24. doi: 10.1053/j.gastro.2006.10.021. [DOI] [PubMed] [Google Scholar]
- 14.Fiegler H, Carr P, Douglas EJ, Burford DC, Hunt S, Scott CE, Smith J, Vetrie D, Gorman P, Tomlinson IP, Carter NP. DNA microarrays for comparative genomic hybridization based on DOP-PCR amplification of BAC and PAC clones. Genes Chromosomes Cancer. 2003;36:361–74. doi: 10.1002/gcc.10155. [DOI] [PubMed] [Google Scholar]
- 15.Kim SY, Nam SW, Lee SH, Park WS, Yoo NJ, Lee JY, Chung YJ. ArrayCyGHt: a web application for analysis and visualization of array-CGH data. Bioinformatics. 2005;21:2554–5. doi: 10.1093/bioinformatics/bti357. [DOI] [PubMed] [Google Scholar]
- 16.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔ C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 17.Cheng J, Sun S, Tracy A, Hubbell E, Morris J, Valmeekam V, Kimbrough A, Cline MS, Liu G, Shigeta R, Kulp D, Siani-Rose MA. NetAffx gene ontology mining tool: a visual approach for microarray data analysis. Bioinformatics. 2004;20:1462–3. doi: 10.1093/bioinformatics/bth087. [DOI] [PubMed] [Google Scholar]
- 18.Dahlquist KD, Salomonis N, Vranizan K, Lawlor SC, Conklin BR. GenMAPP, a new tool for viewing and analyzing microarray data on biological pathways. Nat Genet. 2002;31:19–20. doi: 10.1038/ng0502-19. [DOI] [PubMed] [Google Scholar]
- 19.Lomax J, Ashburner M, Foulger R, Eilbeck K, Lewis S, Marshall B, Mungall C, Richter J, Rubin GM, Blake JA, Bult C, Dolan M, et al. The gene ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32:D258–D261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanehisa M, Goto S, Kawashima S, Nakaya A. The KEGG databases at genomenet. Nucleic Acids Res. 2002;30:42–6. doi: 10.1093/nar/30.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim TM, Jung YC, Rhyu MG, Jung MH, Chung YJ. GEAR: genomic enrichment analysis of regional DNA copy number changes. Bioinformatics. 2008;24:420–1. doi: 10.1093/bioinformatics/btm582. [DOI] [PubMed] [Google Scholar]
- 22.Yasui K, Arii S, Zhao C, Imoto I, Ueda M, Nagai H, Emi M, Inazawa J. TFDP1, CUL4A, and CDC16 identified as targets for amplification at 13q34 in hepatocellular carcinomas. Hepatology. 2002;35:1476–84. doi: 10.1053/jhep.2002.33683. [DOI] [PubMed] [Google Scholar]
- 23.Fang W, Piao Z, Simon D, Sheu JC, Huang S. Mapping of a minimal deleted region in human hepatocellular carcinoma to 1p36.13-p36.23 and mutational analysis of the RIZ (PRDM2) gene localized to the region. Genes Chromosomes Cancer. 2000;28:269–75. [PubMed] [Google Scholar]
- 24.Guan XY, Sham JS, Tai LS, Fang Y, Li H, Liang Q. Evidence for another tumor suppressor gene at 17p13.3 distal to TP53 in hepatocellular carcinoma. Cancer Genet Cytogenet. 2003;140:45–8. doi: 10.1016/s0165-4608(02)00654-4. [DOI] [PubMed] [Google Scholar]
- 25.Spangenberg HC, Thimme R, Blum HE. Serum markers of hepatocellular carcinoma. Semin Liver Dis. 2006;26:385–90. doi: 10.1055/s-2006-951606. [DOI] [PubMed] [Google Scholar]
- 26.Vejchapipat P, Tangkijvanich P, Theamboonlers A, Chongsrisawat V, Chittmittrapap S, Poovorawan Y. Association between serum hepatocyte growth factor and survival in untreated hepatocellular carcinoma. J Gastroenterol. 2004;39:1182–8. doi: 10.1007/s00535-004-1469-8. [DOI] [PubMed] [Google Scholar]
- 27.Horiguchi N, Takayama H, Toyoda M, Otsuka T, Fukusato T, Merlino G, Takagi H, Mori M. Hepatocyte growth factor promotes hepatocarcinogenesis through c-Met autocrine activation and enhanced angiogenesis in transgenic mice treated with diethylnitrosamine. Oncogene. 2002;21:1791–9. doi: 10.1038/sj.onc.1205248. [DOI] [PubMed] [Google Scholar]
- 28.Ribatti D, Marzullo A, Gentile A, Longo V, Nico B, Vacca A, Dammacco F. Erythropoietin/erythropoietin-receptor system is involved in angiogenesis in human hepatocellular carcinoma. Histopathology. 2007;50:591–6. doi: 10.1111/j.1365-2559.2007.02654.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okamoto H, Yasui K, Zhao C, Arii S, Inazawa J. PTK2 and EIF3S3 genes may be amplification targets at 8q23-q24 and are associated with large hepatocellular carcinomas. Hepatology. 2003;38:1242–9. doi: 10.1053/jhep.2003.50457. [DOI] [PubMed] [Google Scholar]
- 30.Reeves ME, DeMatteo RP. Genes and viruses in hepatobiliary neoplasia. Semin Surg Oncol. 2000;19:84–93. doi: 10.1002/1098-2388(200009)19:2<84::aid-ssu2>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 31.Corson TW, Huang A, Tsao MS, Gallie BL. KIF14 is a candidate oncogene in the 1q minimal region of genomic gain in multiple cancers. Oncogene. 2005;24:4741–53. doi: 10.1038/sj.onc.1208641. [DOI] [PubMed] [Google Scholar]
- 32.Gruneberg U, Neef R, Li X, Chan EH, Chalamalasetty RB, Nigg EA, Barr FA. KIF14 and citron kinase act together to promote efficient cytokinesis. J Cell Biol. 2006;172:363–72. doi: 10.1083/jcb.200511061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Corson TW, Gallie BL. KIF14 mRNA expression is a predictor of grade and outcome in breast cancer. Int J Cancer. 2006;119:1088–94. doi: 10.1002/ijc.21954. [DOI] [PubMed] [Google Scholar]
- 34.Gunning PW, Schevzov G, Kee AJ, Hardeman EC. Tropomyosin isoforms: divining rods for actin cytoskeleton function. Trends Cell Biol. 2005;15:333–41. doi: 10.1016/j.tcb.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 35.Lamant L, Dastugue N, Pulford K, Delsol G, Mariame B. A new fusion gene TPM3-ALK in anaplastic large cell lymphoma created by a (1;2)(q25;p23) translocation. Blood. 1999;93:3088–95. [PubMed] [Google Scholar]
- 36.Armstrong F, Duplantier MM, Trempat P, Hieblot C, Lamant L, Espinos E, Racaud-Sultan C, Allouche M, Campo E, Delsol G, Touriol C. Differential effects of X-ALK fusion proteins on proliferation, transformation, and invasion properties of NIH3T3 cells. Oncogene. 2004;23:6071–82. doi: 10.1038/sj.onc.1207813. [DOI] [PubMed] [Google Scholar]
- 37.Jou YS, Lee CS, Chang YH, Hsiao CF, Chen CF, Chao CC, Wu LS, Yeh SH, Chen DS, Chen PJ. Clustering of minimal deleted regions reveals distinct genetic pathways of human hepatocellular carcinoma. Cancer Res. 2004;64:3030–6. doi: 10.1158/0008-5472.can-03-2320. [DOI] [PubMed] [Google Scholar]
- 38.Wang Y, Wu MC, Sham JS, Zhang W, Wu WQ, Guan XY. Prognostic significance of c-myc and AIB1 amplification in hepatocellular carcinoma. A broad survey using high-throughput tissue microarray. Cancer. 2002;95:2346–52. doi: 10.1002/cncr.10963. [DOI] [PubMed] [Google Scholar]
- 39.Herzer K, Sprinzl MF, Galle PR. Hepatitis viruses: live and let die. Liver Int. 2007;27:293–301. doi: 10.1111/j.1478-3231.2006.01422.x. [DOI] [PubMed] [Google Scholar]
- 40.Radaeva S, Jaruga B, Hong F, Kim WH, Fan S, Cai H, Strom S, Liu Y, El-Assal O, Gao B. Interferon-α activates multiple STAT signals and down-regulates c-Met in primary human hepatocytes. Gastroenterology. 2002;122:1020–34. doi: 10.1053/gast.2002.32388. [DOI] [PubMed] [Google Scholar]
- 41.Greenbaum LE. Cell cycle regulation and hepatocarcinogenesis. Cancer Biol Ther. 2004;3:1200–7. doi: 10.4161/cbt.3.12.1392. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.