Abstract
Aerobic microorganisms have evolved different strategies to withstand environmental oxidative stresses generated by various reactive oxygen species (ROS). For the facultative anaerobic human oral pathogen Streptococcus mutans, the mechanisms used to protect against ROS are not fully understood, since it does not possess catalase, an enzyme that degrades hydrogen peroxide. In order to elucidate the genes that are essential for superoxide stress response, methyl viologen (MV)-sensitive mutants of S. mutans were generated via ISS1 mutagenesis. Screening of approximately 2,500 mutants revealed six MV-sensitive mutants, each containing an insertion in one of five genes, including a highly conserved hypothetical gene, SMU.1297. Sequence analysis suggests that SMU.1297 encodes a hypothetical protein with a high degree of homology to the Bacillus subtilis YtqI protein, which possesses an oligoribonuclease activity that cleaves nano-RNAs and a phosphatase activity that degrades 3′-phosphoadenosine-5′-phosphate (pAp) and 3′-phosphoadenosine-5′-phosphosulfate (pApS) to produce AMP; the latter activity is similar to the activity of the Escherichia coli CysQ protein, which is required for sulfur assimilation. SMU.1297 was deleted using a markerless Cre-loxP-based strategy; the SMU.1297 deletion mutant was just as sensitive to MV as the ISS1 insertion mutant. Complementation of the deletion mutant with wild-type SMU.1297, in trans, restored the parental phenotype. Biochemical analyses with purified SMU.1297 protein demonstrated that it has pAp phosphatase activity similar to that of YtqI but apparently lacks an oligoribonuclease activity. The ability of SMU.1297 to dephosphorylate pApS in vivo was confirmed by complementation of an E. coli cysQ mutant with SMU.1297 in trans. Thus, our results suggest that SMU.1297 is involved in superoxide stress tolerance in S. mutans. Furthermore, the distribution of homologs of SMU.1297 in streptococci indicates that this protein is essential for superoxide stress tolerance in these organisms.
Streptococcus mutans, a gram-positive bacterium with a low G+C content, is widely considered the primary etiological agent of dental caries, a common human infectious disease (16, 23). S. mutans is also an important agent of infective endocarditis, as a large number of cases of viridans streptococcus-induced endocarditis are caused by S. mutans (18). During colonization of the oral cavity, S. mutans encounters various environmental stresses, including nutritional limitation, temperature fluctuation, osmotic shock, low pH conditions, radiation, toxins, and variations in oxygen tension (21). Despite these harsh conditions, S. mutans has developed multiple mechanisms for successful survival in the human host by forming diverse and densely populated biofilms on the tooth surface (4). The extraordinary ability of S. mutans to adapt and flourish in the diverse and adverse environment of the oral cavity emphasizes the fundamental importance of the need for detailed analyses of the molecular mechanisms of stress tolerance response in this organism.
S. mutans is a facultative anaerobic organism, but it can tolerate aerobic conditions for colonization and survival. Like other streptococci, it does not possess cytochromes and therefore cannot carry out energy-conserving oxidative phosphorylation (2). However, irrespective of the growth conditions, S. mutans derives the energy for growth through fermentation of glucose and other sugars (26). This can lead to unwanted consequences, especially when the organism is exposed to aerobic conditions in the oral cavity. If the molecular oxygen is not fully reduced by the four-electron reduction step to water, it can undergo one- or two-electron reductions to form reactive superoxide radicals, hydroxyl radicals, and hydrogen peroxide, collectively known as reactive oxygen species (ROS) (19). These radicals, when accumulated in large amounts, can trigger oxidation of lipid, protein, and nucleic acid inside the cell, ultimately leading to cellular death (19, 20).
Aerobic bacteria have developed multiple strategies to adapt and protect against ROS insults (19). These strategies include (i) enzymes that scavenge ROS, such as superoxide dismutases (SOD), catalases, and peroxidases; (ii) protein repair systems, such as thioredoxin; (iii) DNA damage repair enzymes such as RecA; and (iv) proteins that regulate intracellular iron level to ameliorate the generation of ROS. Although streptococci contain SOD, NADH oxidase, glutathione reductase, and other proteins to counter ROS threats, they do not contain catalase, a key protective enzyme against oxidative radicals. Therefore, the defense strategy against damage by ROS is significantly different in streptococci than in other bacteria. For example, the growth of S. mutans in planktonic or biofilm mode can influence the respiratory rates as well as the activities of the protective enzymes, such as SOD and NADH oxidase (31).
Apart from studies related to the physiology of oxidative stress in S. mutans, very little information is available on the oxidative-stress response and its regulation in this organism. Many key regulatory genes, including members of the OxyR and SoxR families, which are involved in sensing and responding to ROS attacks, are not encoded in the genome of S. mutans (2). Instead, S. mutans has a PerR homolog, which has been shown to be involved in hydrogen peroxide stress response in this organism (21). The luxS gene of S. mutans, which encodes an enzyme that synthesizes the intercellular signaling molecule AI-2, is also involved in the oxidative-stress response (52). However, the exact mechanism by which LuxS participates in the oxidative-stress response is currently unknown. Furthermore, a recent investigation suggests that a two-component signal transduction system, ScnRK, is necessary for counteracting ROS in S. mutans (11).
The major focus of this study was to identify the genes that are involved in the defense against superoxide stress of S. mutans strain UA159. Toward this end, a library of mutants was generated by insertion mutagenesis, and the mutants were screened for their sensitivity to methyl viologen (MV), a superoxide-generating compound. This study enabled the identification of five loci that are potentially involved in superoxide tolerance. One of the identified loci is SMU.1297, which encodes a protein homologous to YtqI of Bacillus subtilis. The biochemical characterization of SMU.1297 and its role in superoxide stress tolerance response are presented.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Escherichia coli strain Top10 (Invitrogen) and other strains were grown in Luria-Bertani (LB) medium supplemented, when necessary, with ampicillin (Am; 100 μg/ml), erythromycin (Em; 300 μg/ml), and kanamycin (Km; 50 μg/ml). S. mutans UA159 cultures were routinely grown in Todd-Hewitt medium (BBL, Becton Dickinson) supplemented with 0.2% yeast extract (THY). S. mutans was also grown in chemically defined medium (CDM) supplemented with 0.5% glucose, prepared by a modification of previously described methods (22, 24). Briefly, CDM consisted of 58 mM K2HPO4, 15 mM KH2PO4, 10 mM (NH4)2SO4, 35 mM NaCl, 0.1 mM MnCl2, 2 mM MgSO4, and 0.2% (wt/vol) casein hydrolysate. The medium was supplemented with filter-sterilized vitamins and amino acids (1 mM l-arginine HCl, 4 mM l-glutamic acid, and 0.1 mM l-tryptophan). CDM was supplemented with 0.2% l-cysteine HCl for cultures that required cysteine. When necessary, THY or CDM was also supplemented with Em (10 μg/ml) or Km (300 μg/ml). Bacillus subtilis strain 168 and its derivatives were grown in LB medium.
Identification of superoxide-sensitive mutants.
Insertional mutagenesis was performed with the plasmid pGh9:ISS1, according to the method of Maguin et al. (9, 25, 43). Briefly, S. mutans was transformed with pGh9:ISS1, and transformants were selected on THY agar containing Em (THY-Em) and incubated at 30°C. An overnight-grown liquid culture was made from a single transformed colony. Cultures were diluted 100-fold in the same medium, grown for 2 h at 30°C, and then shifted to growth at 37°C for 2.5 h to select for transposition events. Cultures were then plated on THY-Em plates to obtain individual mutants, which were then inoculated into 96-well plates containing THY broth supplemented with Em and grown overnight. The cultures were then spotted on THY agar plates with or without MV (5 mM), using a 48-pin replicator, and incubated at 37°C under microaerophilic conditions. Clones that grew on THY plates but failed to grow on THY+MV plates were identified, cultured overnight in THY-Em at 37°C, and processed for analysis.
The location of the inserted ISS1 element was identified by inverse PCR. Templates generated by self-ligation of HindIII- or EcoRI-digested chromosomal DNAs, isolated from the mutants, were subjected to inverse PCR by using the primers ISS1Rout1 (GATTGTAACGTAGATAATAACCAACAGC) and ISS1Fout1 (GCAAGAACCGAAGAAATGGAACG). The PCR products were sequenced with primer ISS1-Rout2 (AATAGTTCATTGATATATCCTCGCTGTCA) to identify the flanking sequences, which were mapped on the genome of S. mutans UA159 by BLAST search analysis.
Mutants in which the plasmid vector sequence had been excised from the chromosome were obtained by growing the bacteria in plain THY broth at 30°C, which permits plasmid replication (25). Em-sensitive (Ems) colonies, from which the plasmid had been excised, were selected by plating on THY agar and incubation at 37°C. Ems colonies were confirmed for loss of the plasmid sequence by PCR with primers homologous to the flanking regions.
Construction of a ΔSMU.1297 mutant.
The SMU.1297 locus was deleted using a Cre-loxP-based, unmarked gene deletion system as described previously (5). A PCR product, amplified from UA159 chromosomal DNA using the primers Bam-Smu1296 (GCCGGATCCGAATCTGGTTCTATCTTACTT) and Smu1297-R1 (GAAACGGTCGACACGTCCATCTGCTTG), was cloned into the pGEM-T-Easy TA cloning vector (Promega) to generate pIB901. A Km resistance cassette, with flanking loxP sites, was amplified from pUC4ΩKm2 (34) using the primers lox71-Km-F and lox66-Km-R (5) and cloned into HindIII-digested, T4 DNA polymerase-blunted pIB901, yielding pIB904. Plasmid pIB904 was linearized with NotI and transformed into S. mutans UA159 as previously described (7). The transformants were selected on THY plates containing Km; one such transformant was named IBS934 and retained for further analysis.
To excise the loxP-Kmr resistance cassette from the chromosome, IBS934 was transformed with pCrePA (5, 36). This plasmid contains a cre recombinase gene, an Emr gene for selection, and a temperature-sensitive replicon (pWV01). Transformants were grown at 30°C on THY-Em plates, and selected colonies were then grown at 30°C in THY-Em broth and plated on THY-Em plates; colonies from the THY-Em plates were then patched onto THY plates containing Km. Emr and Kms colonies were transferred onto antibiotic-free THY plates by patching and incubated overnight at 37°C to cure pCrePA from the cells, generating Ems cells that were selected for further analysis. Deletion of SMU.1297 was verified by PCR analysis.
Construction of a complementing plasmid for SMU.1297.
A PCR fragment containing the entire SMU.1297 coding region plus a 1.4-kb upstream sequence was amplified from S. mutans UA159 by using the primers Eco-Smu1296-F3 (GGCGAATTCCTCAACTACCTGAACAGCACTC) and Bam-Smu1297-R3 (GGCGGATCCTTGACGACTGACAAGTTTTTATAC), which introduced a unique EcoRI site at the 5′end and a unique BamHI site at the 3′ end. The resulting 2.3-kb fragment was digested with EcoRI plus BamHI and ligated into EcoRI-BamHI-digested pOri23 (38), an Emr shuttle plasmid that replicates in gram-positive bacteria, including S. mutans (8), to create pIB1297.
Physiological characterization of the SMU.1297 mutant strains.
To evaluate the sensitivity of the SMU.1297 mutant strains to superoxide-stress-inducing chemicals, an overnight bacterial culture was collected, washed twice, and resuspended in 0.85% NaCl. The cultures were adjusted to an optical density (A600) of 2.0 or 5.0 and 10-fold serially diluted, and 7.5 μl of each dilution was spotted onto THY agar containing MV (5 mM) or menadione (25 μg/ml). To stimulate oxidative and thermal stresses, the cultures were spotted on THY agar containing 1 mM H2O2 and puromycin (0.75 μg/ml), respectively. To analyze the growth of S. mutans cultures at low pH, the initial pH of the THY agar medium was adjusted, prior to sterilization, to pH 5.5 or 7.0 with HCl (6). Sterile citrate-phosphate buffer (50 mM) of the desired pH was added to the media after sterilization. Different dilutions of S. mutans cultures, prepared as described above, were spotted onto the plates and incubated at 37°C, under microaerophilic conditions.
Cysteine starvation tolerance assay.
For E. coli cysteine starvation tolerance assays, overnight cultures of a cysQ mutant strain, UM285 (27), carrying either pIB1297 or the vector pOri23, were washed and 10-fold serially diluted with 0.85% NaCl. samples (7.5 μl) were spotted onto CDM plates with or without 0.2% cysteine, followed by incubation of the inoculated plates at 37°C under ambient conditions. For S. mutans, the assays were performed in a similar manner, except that the plates were incubated at 37°C under microaerophilic conditions instead of ambient conditions.
Purification of His-tagged SMU.1297.
The entire coding region of SMU.1297 was amplified by PCR using the primers Eco-Smu1297-OF (GCCGAATTCATGACTGCTTTTAAAACTATTCTAGC) and Bam-Smu1297-OR (GCCGGATCCCTATTTTAGCAGATTTTTTAATTC). The amplified fragment was cloned into EcoRI and BamHI sites of pASK-IBA43 (IBA, St. Louis, MO) to generate plasmid pIB900, which expresses an N-terminal His-tagged SMU.1297 (His-SMU.1297) construct. The coding sequence was confirmed by DNA sequence analysis. His-SMU.1297 was purified using a Ni-nitrilotriacetic acid resin column was used following the manufacture's protocol (Qiagen). Purified His-SMU.1297 protein was dialyzed in buffer containing 100 mM Tris·Cl (pH 8.5), with 50 mM NaCl.
pAp phosphatase and oligoribonuclease assay.
To measure the pAp phosphatase activity of His-SMU.1297, 40 μM pAp (Sigma Aldrich, St. Louis, MO) was used as a substrate for 5 μg of protein in P buffer (100 mM Tris·Cl [pH 8.8], 5 mM MnCl2 or MgCl2). The reaction mixtures were incubated at room temperature, and 40-μl aliquots were taken at different time points; the reactions were stopped by the addition of an equal volume of 100 mM EDTA, followed by gentle mixing. The amount of liberated phosphate was measured with a phosphate colorimetric assay kit (BioVision, Mountain View, CA) according to the manufacturer's protocol. Briefly, 10 μl of reaction mixture was added to a 96-well plate, and the volume in each well was adjusted to 200 μl with water. Phosphate reagent (30 μl) was then added to each well, followed by incubation of the plate at room temperature for 30 min. The absorbance was measured at 650 nm using a plate reader. The activity of the test samples was compared against a standard curve obtained using a known amount of phosphate.
Oligoribonuclease activity was assayed using a custom-made 5-mer RNA oligonucleotide (5′Cy5-CCCCC3′), which was labeled at the 5′ end with a fluorescent dye, Cy5. RNA oligonucleotide (1 mM) was incubated with 10 μg of purified His-SMU.1297 protein at 37°C in buffer containing 100 mM Tris·Cl (pH 8.8), 5 mM MnCl2, or MgCl2in a final volume of 100 μl. At 15-min intervals over 60 min, 5-μl aliquots were removed, the reaction was stopped by the addition of 5 μl of sample buffer (4× Tris-borate-EDTA [TBE], 100 mM dithiothreitol, 16% glycerol, 20 mM EDTA), and the mixtures were stored at −20°C until all of the samples were collected. The reaction products were loaded onto a 22% denaturing polyacrylamide gel containing 2× TBE and 7 M urea and electrophoresed in 2× TBE. The gel was visualized under UV light.
RESULTS
Isolation of S. mutans UA159 superoxide stress-sensitive mutants.
ISS1 transposition mutagenesis was used to identify genes that are potentially involved in superoxide stress tolerance, since ISS1 appears to integrate itself randomly into the genome of gram-positive bacteria, including various streptococci (9, 41, 43), and also because it rarely inserts more than once into the genome of the same cell (9, 41). Plasmid pGh9::ISS1, whose replication is temperature sensitive (25), was used to introduce the insertion element into wild-type strain UA159. An Emr transformant containing pGh9::ISS1was isolated and grown overnight at 30°C, and Emr colonies containing the ISS1 element were then isolated after incubation at 37°C. The insertion frequency, which was calculated by dividing the number of Emr colonies by the total number of colonies at 37°C, was found to be below 0.5%, consistent from one independent experiment to the next. This frequency was similar to the frequencies reported for other streptococci (9, 41, 43).
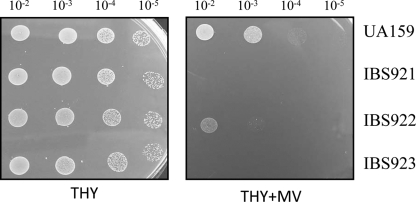
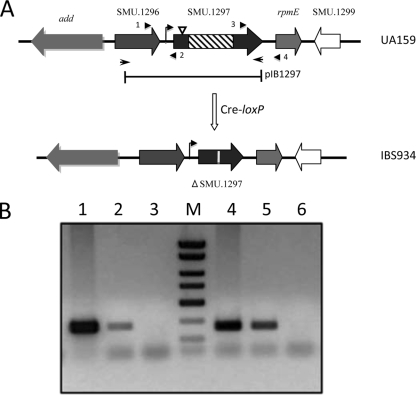
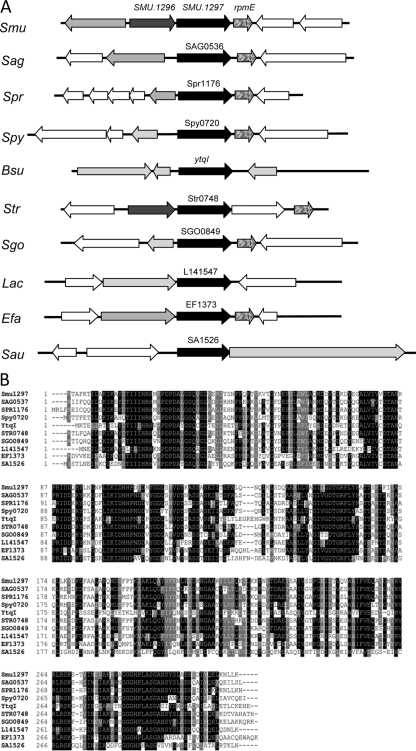
A collection of ∼2,500 mutants were grown in 96-well plates and replica-patched onto THY, with or without MV. A total of six mutants were obtained that displayed an MV-sensitive growth phenotype (Fig. 1; also data not shown). In each case, Southern hybridization showed that ISS1 insertion had occurred at only one location (data not shown). The site of insertion was identified, as described in Materials and Methods, for each of the six MV-sensitive mutants. Two independent insertions occurred in the dnaK gene (SMU.82), which encodes heat shock protein 70. For the remaining four mutants, insertions were mapped to SMU.905 (encoding an ABC transporter), SMU.1128 (encoding the histidine kinase ciaH), SMU.1296 (encoding a putative glutathione S-transferase [GST]), and SMU.1297. With the exception of SMU.1297, all of these genes are known to encode products that are involved in superoxide stress tolerance in other bacteria; therefore, we elected to focus our studies on SMU.1297. SMU.1297 encodes a polypeptide of 311 residues; the ISS1 insertion occurred in this gene at codon position 21. SMU.1296 is found upstream of SMU.1297, while rpmE, which encodes the ribosomal protein L31, is found downstream of SMU.1297. An intergenic region of 70 bp lies between the SMU.1296 and SMU.1297 loci; this region contains a consensus TATA box (TATAAT) at a position 24 bp upstream of the ATG start codon of SMU.1297, indicating that SMU.1297 may be transcribed separately from SMU.1296. To determine if this is the case, linkage PCR analysis was performed using RNA isolated from exponentially grown cultures of UA159. As shown in Fig. 2B, it appears that both SMU.1296 and rpmE are transcriptionally linked to SMU.1297, indicating that SMU.1297 is cotranscribed with these genes.
FIG. 1.
Verification of superoxide-sensitive phenotype. ISS1 transposon mutants that displayed an initial MV-sensitive phenotype were further verified by spotting of 7.5 μl from a 10-fold dilution series, with a starting optical density (A600) of 2.0 made in 0.85% NaCl, onto THY agar plates containing 5 mM MV (THY+MV). As a control, cultures were also spotted on plain THY agar plates with no additions (THY). Experiments were repeated at least three times, and the relevant areas of the representative plates are shown. UA159 is the wild-type strain, while IBS921, IBS922, and IBS923 are independent MV-sensitive mutants.
FIG. 2.
(A) Construction of a SMU.1297 deletion mutant. The direction of transcription of SMU.1297 and adjacent genes in the chromosome of S. mutans is indicated by block arrows. The SMU.1297 deletion strain, IBS934, was constructed using a Cre-loxP-based markerless gene deletion system (see Materials and Methods) that removed the central part of the SMU.1297 gene (hatched box), and inserted a 34-bp lox72 sequence to disrupt the gene (vertical white line). The bar below the chromosome indicates the region of SMU.1297 that was cloned into the complementing plasmid pIB1297. The site of the ISS1 insertion is shown by an inverted triangle. The bent arrow indicates a putative promoter region upstream of SMU.1297 gene. The diagram is not drawn to scale. (B) Transcriptional linkage analysis of the SMU.1297 locus. RNA, isolated from S. mutans UA159, was used as a template to produce cDNA, on which PCR was performed (lanes 2 and 5). RNA (lanes 3 and 6), and chromosomal DNA (lanes 1 and 4) were included as controls. Primers 1 and 2 (5′-GCCGGATCGACTGGACTGAAAGAATCGCTCAG-3′ and 5′-GCCCTCGAGCTGACTGCCTAATGCGTCAGGG-3′, respectively), shown by arrowheads in panel A, were used for the linkage analysis between SMU.1296 and SMU.1297, whereas primers 3 and 4 (5′-GGCCATCCATTAGCAAGTGGTG-3′ and 5′-TTGGTAACCCGTTGAAGTATCC-3′, respectively) were used for the linkage analysis between SMU.1297 and rpmE.
SMU.1297 is involved in superoxide stress tolerance.
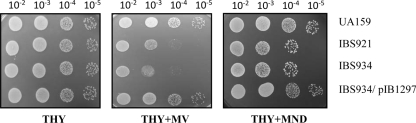
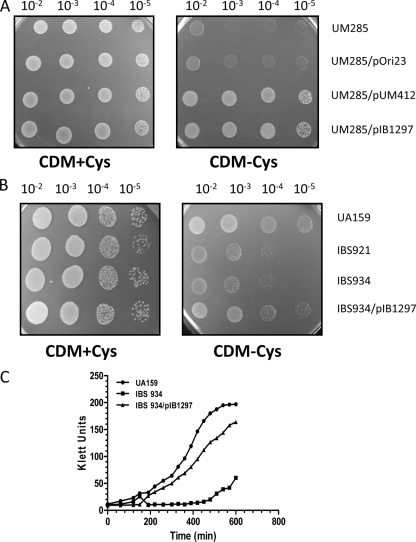
To confirm that the observed phenotype of the SMU.1297 ISS1 insertion mutation did not result from additional spontaneous mutations elsewhere in the genome, a marker-free deletion mutation of SMU.1297 was constructed in strain UA159 using a Cre-loxP-based system, which was shown to work efficiently in S. mutans (5). No obvious growth defects were observed in the SMU.1297 deletion mutant strain, IBS934, relative to the wild-type UA159 (data not shown). We also tested its ability to form a biofilm on polystyrene and glass surfaces using crystal violet staining (10). We found that the biomass of the biofilm formed by IBS934 on various abiotic surfaces was similar to that of the wild-type parent (data not shown). IBS934 was tested for its ability to withstand exposure to MV. As shown in Fig. 3, IBS934 displayed the same degree of sensitivity to MV exposure as the original ISS1 insertion mutant, IBS921. Complementation analysis was then performed to confirm that this effect was due to deletion of SMU.1297. A DNA fragment including SMU.1297 along with its potential promoter region was cloned into plasmid pOri23 (38), which replicates in S. mutans, to generate pIB1297, which was then introduced into IBS934 (Fig. 2). As shown in Fig. 3A, transformation of IBS934 with pIB1297 eliminated the growth defect of the mutant strain during exposure to MV. To confirm that SMU.1297 is indeed involved in superoxide stress tolerance, the wild-type and SMU.1297 mutant strains were exposed to menadione, a quinone compound that generates superoxide anions in bacteria. As shown in Fig. 3B, both IBS921 and IBS934 were more susceptible to menadione exposure than the isogenic wild-type parent strain. The menadione-induced growth defect of IBS934 was alleviated after complementation with pIB1297. Taken together, the results suggest that SMU.1297 is involved in superoxide stress tolerance.
FIG. 3.
Phenotypic characterization of the SMU.1297 deletion mutants. Overnight cultures of the wild type (UA159), the ISS1 insertion mutant (IBS921), the clean Cre-loxP deletion mutant (IBS934), and the complemented strain (IBS934/pIB1297) were grown in THY medium with appropriate antibiotics. Different dilutions of the overnight culture, with a starting optical density (A600) of 5.0 made in 0.85% NaCl, were spotted on THY agar (THY) or THY agar supplemented with 5 mM MV (THY+MV) or 25 μg/ml menadione (THY+MND) and incubated overnight under microaerophilic conditions at 37°C. Experiments were repeated at least three times, and relevant areas of the representative plates are shown.
SMU.1297 does not take part in other stress responses.
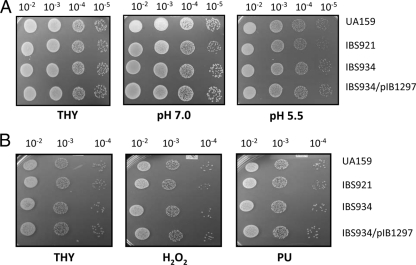
In various bacteria, a gene responsible for a specific stress response can also confer cross-protection against other stresses (13, 39). To identify potential cross-resistance to other stresses, UA159 and IBS934 were exposed to oxidative, thermal, and acid stress conditions. H2O2 was used to induce oxidative stress, while puromycin, a compound that induces premature chain termination during protein synthesis, was used to mimic thermal stress conditions. For acid stress, bacterial growth of the two strains in THY media buffered at pH 5.5 and pH 7.0 was compared. As shown in Fig. 4, there were no significant differences between UA159 and IBS934, indicating that SMU.1297 is not involved in the response to oxidative, thermal, or acid stress conditions.
FIG. 4.
SMU.1297 is not required for general stress tolerance in S. mutans. (A) Acid stress. Dilutions of fresh overnight cultures were spotted on THY agar plates in the absence (THY) or presence of sodium citrate buffer to generate a pH of 5.5 to mimic acid stress; the pH 7.0 culture served as a control. (B) Oxidative and thermal stress. Cultures were prepared and spotted as described above, on THY plates containing 1 mM hydrogen peroxide (H2O2), for oxidative stress, or on plates containing 0.75 μg/ml puromycin (PU), for thermal stress. Plates were incubated overnight under microaerophilic conditions at 37°C. Experiments were repeated at least three times, and the relevant areas of the representative plates are shown.
SMU.1297 is required for cysteine biosynthesis.
Analysis of the SMU.1297 sequence using a BLAST search against the microbial genome database at the National Center for Biotechnology Information website indicates that the gene is present in many bacteria, including Bacillus and Streptococcus (Fig. 5A). In Bacillus subtilis, the homolog, known as YtqI, has been characterized by Mechold et al. (27). Those investigators demonstrated that YtqI possesses dual activities: oligoribonuclease, which can cleave small RNAs (less than 5-mer), and 3′-phosphoadenosine-5-phosphate (pAp) phosphatase. The latter activity of YtqI is similar to that of the E. coli CysQ protein, which is required for sulfur assimilation (27). Mechold et al. (27) have also shown that the expression of ytqI can restore the growth defect of an E. coli cysQ mutant, which cannot grow in the absence of cysteine. To test whether SMU.1297 has any physiological similarity to CysQ, E. coli strain UM285, which lacks a functional cysQ, was transformed with pIB1297, which expresses SMU.1297 from the P23 promoter (UM285/pIB1297). As a negative control, UM285 was transformed with pOri23 (UM285/pOri23), while UM285 transformed with plasmid pUM412, which expresses YtqI (UM285/pUM412), was used as a positive control. These strains were grown overnight in LB medium, washed thoroughly, and serially diluted in CDM lacking cysteine. Diluted cultures were then spotted onto CDM agar plates with or without cysteine and incubated overnight at 37°C. As shown in Fig. 6A, all of the strains grew well when the CDM was supplemented with cysteine, while neither UM285 nor UM285/pOri23 could grow on CDM lacking cysteine. On the other hand, both UM285/pIB1297 and the positive control, UM285/pUM412, grew very well on CDM without cysteine. This suggests that under in vivo conditions, SMU.1297 functions as a CysQ homolog, similar to YtqI.
FIG. 5.
Genomic context and sequence alignment of SMU.1297. (A) Organization of open reading frames in the SMU.1297 region in various members of the Firmicutes. The SMU.1297 locus (black arrow) appears to be variable in different Firmicutes. However, the gene immediately downstream of SMU.1297, rpmE, appears to be conserved in streptococci. A putative GST gene (dark gray arrows) is located immediately upstream of SMU.1297 in S. mutans and S. thermophilus only. Smu. S. mutans; Sag, Streptococcus agalactiae; Spr, Streptococcus pneumoniae; Spy, S. pyogenes; Bsu, B. subtilis; Str, S. thermophilus; Sgo, Streptococcus gordonii; Lac, Lactococcus lactis; Efa, Enterococcus faecalis; and Sau, S. aureus. (B) Comparison of the amino acid sequence of SMU.1297 with homologous proteins identified by BLASTP search. Clustal-W was used to align the sequences from different Firmicutes.
FIG. 6.
SMU.1297 is required for growth under cysteine starvation conditions. (A) Complementation of an E. coli cysQ mutant by the expression of SMU.1297. E. coli UM285, a cysQ-deficient strain, was transformed with vector plasmid pOri23 or pOri23 carrying the full-length wild-type SMU.1297 gene (pIB1297). As a positive control, UM285 containing pUM412, which contains the B. subtilis ytqI gene, was also included. Cultures were prepared as described in Materials and Methods, 10-fold serially diluted, and spotted on a semisynthetic minimum agar medium (CDM) with or without 0.2% cysteine. Plates were incubated overnight at 37°C under aerobic conditions. Experiments were repeated three times, and relevant areas of the representative plates are shown. (B) Growth of S. mutans under cysteine-depleted conditions. Fresh overnight cultures were washed with 0.85% NaCl, 10-fold serially diluted, and spotted on CDM with or without 0.2% cysteine. Plates were incubated 48 h at 37°C under microaerophilic conditions. Experiments were repeated four times, and relevant areas of the representative plates are shown. (C) Growth curve of S. mutans strains in CDM broth containing 0.075% cysteine.
The ability of SMU.1297 to complement a cysQ mutation in E. coli led to the investigation of the cysteine auxotroph phenotype of SMU.1297-deficient strains of S. mutans. To do so, two approaches were used: an agar plate-based method, similar to the technique used for the E. coli complementation assay, and a liquid-based assay. As shown in Fig. 6B, the insertion mutant, IBS921, and the targeted-deletion mutant, IBS934, failed to grow well in the absence of cysteine. Growth of strain IBS934 in cysteine-free medium was restored after transformation with pIB1297.
To confirm that the SMU.1297-deficient strains are unable to grow in the absence of an external source of cysteine, bacterial growth was observed in liquid CDM. The wild-type strain is unable to grow in CDM completely devoid of cysteine; the minimum concentration of cysteine that can support growth of UA159 in CDM was found to be 0.075%. Therefore, CDM supplemented with 0.075% cysteine was used to monitor the growth of UA159, IBS934, and IBS934/pIB1297. As expected, IBS934 showed a drastic growth defect compared to the wild-type and complemented mutant strains. Taken together, these results support the results of the BLAST analysis that indicate that SMU.1297 encodes a protein with a function similar to that of CysQ and YtqI in vivo.
Since SMU.1297 was able to complement cysQ deficiency in E. coli, we asked whether the E. coli cysQ mutant is able to withstand MV exposure. We also included a B. subtilis ytqI mutant in the study, since YtqI and CysQ seem to be homologous proteins. We observed that both the E. coli cysQ and the B. subtilis ytqI mutant strains displayed the same degree of tolerance to MV exposure as their isogenic wild-type parental strains (data not shown). We also observed that, unlike S. mutans, the wild-type E. coli and B. subtilis strains failed to grow on plates containing 5 mM MV but grew if the MV concentration was less than 0.5 mM (data not shown). Thus, our results indicate that the stress-sensitive phenotype observed is unique to S. mutans.
SMU.1297 displays pAp phosphatase activity in vitro.
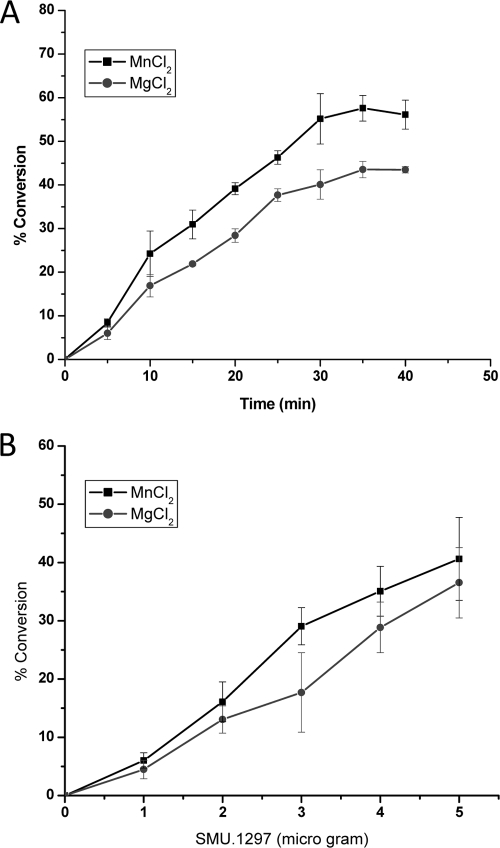
Under in vitro conditions, both YtqI and CysQ display a pAp phosphatase activity that converts pAp to AMP and inorganic phosphate (27). To determine whether SMU.1297 can degrade pAp in vitro, an N-terminally His-tagged SMU.1297 (His-SMU.1297) construct was generated and purified to near homogeneity using a Ni-nitrilotriacetic acid affinity column. After purification, the potential dephosphorylation activity of His-SMU.1297 was tested using pAp as a substrate. Since YtqI, CysQ, and other pAp phosphatases have maximum activity between pH 8.5 and 9.5, pH 8.8 was used for our assay. As shown in Fig. 7, His-SMU.1297 catalyzes a time- and concentration-dependent phosphatase activity when pAp was used as a substrate. This phosphatase activity was dependent on the presence of divalent metal ions, since no activity was observed in their absence (data not shown). As anticipated, the protein was most active with Mn2+ and Mg2+, which are the preferred cofactors for members of the phosphomonoesterase family (53). Under the conditions tested, the specific activity of SMU.1297 was estimated to be about 0.08 nmol/μg/min. This activity is approximately 75- and 400-fold lower than those of the YtqI and CysQ proteins, respectively (27). Similar to that of other CysQ homologs, the phosphatase activity of SMU.1297 was very specific for pAp, since no enzymatic activity was observed when ADP, ATP, GDP, or GTP was used as a substrate. Taken together, the results strongly suggest that SMU.1297 encodes a pAp phosphatase that is involved in sulfur metabolism in this organism.
FIG. 7.
pAp phosphatase activity of S. mutans SMU.1297 protein. (A) pAp phosphatase activity as a function of time. Reaction mixtures containing 5 μg of protein and 40 μM pAp were incubated for various lengths of time at room temperature. The reaction buffer contained either MnCl2 or MgCl2, as described in the text. Reactions were stopped by the addition of an equal volume of 100 mM EDTA, the liberated phosphates were quantified using a phosphate colorimetric assay kit (malachite green method) as described in Materials and Methods, and the absorbance was measured using a BioTek Synergy HT reader, at 600 nm. (B) pAp phosphatase activity as a function of protein concentration. The assay for phosphatase activity was carried out for 20 min, at room temperature, in the presence of increasing amounts of His-SMU.1297 protein and 40 μM pAp. The liberated phosphate was quantified as described for panel A. For both the graphs, the y axis shows percent conversion of pAp to AMP and inorganic phosphate.
Oligoribonuclease activity is absent in SMU.1297.
While both E. coli CysQ and B. subtilis YtqI exhibit pAp phosphatase activity, YtqI also demonstrates an oligoribonuclease activity in vitro. Since SMU.1297 shows more similarity to YtqI than to CysQ, it was speculated that SMU.1297 may also possess an oligoribonuclease activity. To this end, a custom-made 5-mer RNA oligonucleotide (5′-Cy5-CCCCC-3′) was used as a substrate, similar to what was used for the measurement of the oligoribonuclease activity of YtqI (27); the activity was assayed in a buffer containing divalent metal ions of either Mn2+ or Mg2+. No oligoribonuclease activity was observed, even in the presence of 10 μg of His-SMU.1297 and incubation up to 60 min (data not shown). Since YtqI is also able to degrade 24-mer RNA (27), a 20-mer RNA was tested as a substrate for His-SMU.1297; as with the 5-mer, no oligoribonuclease activity was observed. Thus, it appears that SMU.1297 does not have any obvious oligoribonuclease activity.
DISCUSSION
The lifestyle of S. mutans is quite unique compared with that of other pathogenic streptococci, such as S. pyogenes or S. pneumoniae. The human oral cavity, the natural niche of this pathogen, is a very dynamic environment that undergoes rapid changes in temperature, pH, and osmotic and oxygen tension. To counteract various environmental fluctuations and growth-limiting conditions, like many other streptococci, S. mutans has also developed multiple strategies that allow it to successfully colonize and maintain a dominant presence in the oral cavity (21). While most of the previous studies were focused on understanding the mechanisms of acid tolerance and oxidative-stress responses, our knowledge of the mechanisms of superoxide stress tolerance and response remains limited. To gain further insight into this process, a collection of random insertion mutants of S. mutans UA159 was screened to select clones with high sensitivity to MV. This approach allowed us to identify genes that may be responsible for superoxide stress defense, without prior knowledge of the genes' function(s). In this study, we screened only ∼2,500 such mutants; therefore, the screening process was by no means exhaustive. There were other drawbacks associated with the approach used. An insertion in an essential gene would not have been identified in this study, and mutants with a weak phenotype could potentially be overlooked by our screening process. Among the five loci that were identified in our analysis, two of the genes, dnaK and ciaH, were previously reported as important players in the oxidative-stress response in streptococcus and other bacteria (1, 14, 32, 37, 56), signifying that the screening method used here is a viable approach. However, we did not identify the sodA gene, which encodes the SOD activity, nor did we identify mreD, rodA, pbp2b, or other genes that were isolated by Thibessard et al., who attempted to identify superoxide stress-responsive loci in Streptococcus thermophilus (43). Our inability to identify these genes is likely due to the nonexhaustive nature of the screening process. On the other hand, three unique loci were identified by our search: an ABC transporter-encoding gene (SMU.905), a GST gene (SMU.1296), and SMU.1297. In this study, we further characterized the SMU.1297 locus.
SMU.1297 shows strong homology to the B. subtilis YtqI protein, which is a functional homolog of the E. coli CysQ protein (27). In E. coli, the CysQ protein contains a pAp phosphatase activity and is required for sulfur metabolism (30, 42). In the sulfur assimilation pathway, adenosine 5′-phosphosulfate (APS), an active intermediate, is phosphorylated by APS kinase to form the universal sulfuryl group donor 3′phosphoadenosine-5′-phosphosulfate (pApS) or reduced by APS reductase to form sulfite (12, 30, 35). The sulfuryl group is then transferred from pApS by sulfuryl transferases to various metabolites to form sulfated metabolites and pAp. This transfer sometimes involves thioredoxin- or glutaredoxin-bound intermediates (45, 46). pApS is also converted by pApS reductase to pAp and sulfite, which is later incorporated into sulfur-containing metabolites. CysQ is the key protein that regulates the intracellular concentration of pApS, which is highly cytotoxic at high concentration (30, 33). CysQ, in addition to pAp degradation, can also convert pApS to APS (29, 42), thereby controlling the intracellular level of pApS. We believe that, similar to B. subtilis YtqI, SMU.1297 is also a functional homolog of CysQ, since it can complement a cysQ-deficient strain of E. coli in vivo, and it displays a pAp phosphatase activity under in vitro conditions. However, the pAp phosphatase activity displayed by SMU.1297 was one or two orders of magnitude lower than that of YtqI and CysQ. This difference in activity could be due to the incorporation of the histidine tag in the SMU.1297 protein, which could interfere with the phosphatase activity, or to the different assay conditions used in this study. Nevertheless, the phosphatase activity was sufficient to complement a cysQ-deficient strain of E. coli. Therefore, SMU.1297 probably participates in sulfur assimilation and controls the intracellular level of pAp and pApS in S. mutans.
YtqI of B. subtilis is one of the two gram-positive pAp phosphatases that have been biochemically characterized (27), the other being Rv2131c of Mycobacterium tuberculosis (17). However, unlike Rv2131c, YtqI has an additional oligoribonuclease activity, similar to the E. coli oligoribonuclease activity encoded by orn (15, 54). A study by Mechold et al. suggested that the dual activity contained by various YtqI homologs could be present in other members of the Firmicutes, including streptococci (27). Although the substrate and conditions used in our study were similar to the conditions used to assay YtqI activity, we were unable to detect any oligoribonuclease activity in the purified SMU.1297 protein, and there are several possible reasons for this observation. The substrate, a 5-mer RNA fragment, may not be the ideal substrate for SMU.1297. On the other hand, the in vitro assay conditions may not have been optimum for its activity. Alternatively, the biochemical properties and activity of SMU.1297 may be different from YtqI, since the lifestyles of B. subtilis and S. mutans are very different: the former is a strictly aerobic organism, while the latter is a facultative anaerobic organism. A BLAST search using YtqI as the query against the S. mutans UA159 genome also identified a putative protein, the product of SMU.2140, that shares a high degree of similarity with YtqI (an E value of 5e-09). The gene SMU.2140 encodes a hypothetical protein of 657 residues. Whether SMU.1297 and SMU.2140 possess any oligoribonuclease activity remains to be explored.
Sequence analysis of the SMU.1297 protein indicates that residues 10 to 153 constitute a highly conserved domain with strong homology (E value = 2.3e-42) to the DHH family (Pfam ID, PF01368) of proteins (3). The DHH family includes diverse proteins such as the Drosophila prune gene product, yeast exopolyphosphatase, YtqI, recombinational exonuclease RecJ, inorganic pyrophosphatase, and many other hypothetical proteins (3, 27, 40, 44, 47). The active residues of this domain are distributed among the four motifs (I to IV); motif III is the most important, since it contains the signature DHH sequence that constitutes the catalytic site (3). SMU.1297 also contains another highly conserved domain (residues 248 to 307), known as DHHA1 (Pfam ID, PF02272). This DHHA1 domain, which is about 60 residues long with a conserved GGG motif, is generally found adjacent to the DHH domain and is characteristic of members belonging to the DHH subfamily 1 (3). Because this domain is also found in alanyl-tRNA synthetase, it may have an RNA binding function. Of note, SMU.2140 also contains both the DHH and the DHHA1 domains; therefore, it is possible that SMU.2140 may have biochemical properties similar to those exhibited by SMU.1297.
The molecular mechanisms by which SMU.1297 confers superoxide stress tolerance are currently unclear. Since SMU.1297 appears to be involved in the sulfur assimilation pathway, one could imagine that pApS would accumulate in higher concentrations in SMU.1297-deficient cells. Since high intracellular levels of pApS can inhibit the function of many enzymes (33), it is possible that increased amounts of pApS interferes with the function of proteins involved in superoxide stress response in S. mutans. Alternatively, SMU.1297 could also modulate intracellular levels of pAp and regulate biosynthesis of reduced sulfur-containing metabolites, since pAp is a competitive inhibitor of pApS reductase (29, 33). Intracellular accumulation of pAp also negatively interferes with many enzymes, including oligoribonuclease (28) and phosphopantetheinyl-transferase, an enzyme involved in fatty acid biosynthesis and production of secondary metabolites (51). Therefore, high levels of pApS or pAp could inhibit the activity of a key protein involved in superoxide stress tolerance. SOD is one of the key enzymes involved in the superoxide stress tolerance response, by catalyzing the dismutation of the superoxide radical into O2 and H2O2, an important function required for the defense against oxygen toxicity (19). To verify whether the activity of SOD is altered in the SMU.1297-deficient strains, we measured the SOD activity by preparing crude cellular extracts from IBS934 and UA159; we found no difference in the SOD activity in these two strains (data not shown). Thus, it is unlikely that the intracellular concentration of pApS or pAp modulates the SOD activity.
The observed phenotype is probably unique to S. mutans, since the E. coli cysQ and B. subtilis ytqI mutants did not display a superoxide stress-sensitive phenotype. Since SMU.1296 is transcriptionally linked to SMU.1297, it is tempting to speculate that the SMU.1296 protein could be another target whose activity could be altered in SMU.1297-deficient cells. SMU.1296 shows strong homology to the bacterial GST proteins, and the gene is also annotated as a GST-encoding gene in the National Center for Biotechnology Information database. Of note, we also isolated an independent insertion in the SMU.1296 locus that showed increased sensitivity to the presence of superoxide radicals. The physiological role of bacterial GST is not very well defined, and not all bacteria contain GST (49, 50). In general, GST can use 1-chloro-2,4-dinitrobenzene (CDNB) as a substrate and can bind to glutathione affinity matrices (50). Moreover, some bacterial GSTs, such as FosB of Staphylococcus epidermidis, are required for phosphomycin resistance (55). When we tested crude cell lysates isolated from exponentially grown cultures of S. mutans for GST activity, no detectable activity against CDNB was observed. Furthermore, the wild type and the SMU.1297 mutant exhibited the same degree of resistance against phosphomycin (data not shown). Thus, these activities do not appear to be required for superoxide stress tolerance. Another possibility is that SMU.1297 is directly involved in counteracting superoxide radical toxicity, by sequestering the superoxide anion or converting it to a less toxic or nontoxic compound. Further characterization is required to understand the mechanisms by which SMU.1297 confers superoxide stress resistance in this organism.
Analysis of the genomic context suggests that in streptococci, there is a topological association between SMU.1297 and rpmE, which encodes the 50S ribosomal protein L31; our transcriptional analysis indicates that these two genes are transcriptionally linked as well. Therefore, it is possible that SMU.1297 may be directly or indirectly involved in protein translation, likely during conditions of stress. This topological association was not observed in B. subtilis or Staphylococcus aureus, both of which are aerobic organisms. Therefore, it is tempting to speculate that SMU.1297 and its homologs in other streptococcal species have evolved to perform a genus-specific function. A recent DNA microarray analysis in Streptococcus pyogenes indicates that Spy0720, the SMU.1297 homolog, is strongly induced during infection (48); therefore, in addition to superoxide stress tolerance, these proteins also appear to be involved in pathogenesis. Further analysis will be required to determine if this is the case in S. mutans or other pathogens.
Acknowledgments
We thank Undine Mechold (France) for kindly providing us with the B. subtilis ytqI and the E. coli cysQ mutant strains as well as the E. coli complementing plasmid. We thank members of the Biswas laboratory for technical assistance and for critically reading the manuscript.
This work was supported by an NIDCR grant (DE016686) and an NCRR COBRE project grant (P20RR01644) to I.B.
Footnotes
Published ahead of print on 1 May 2009.
REFERENCES
- 1.Ahn, S. J., Z. T. Wen, and R. A. Burne. 2006. Multilevel control of competence development and stress tolerance in Streptococcus mutans UA159. Infect. Immun. 741631-1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ajdic, D., W. M. McShan, R. E. McLaughlin, G. Savic, J. Chang, M. B. Carson, C. Primeaux, R. Tian, S. Kenton, H. Jia, S. Lin, Y. Qian, S. Li, H. Zhu, F. Najar, H. Lai, J. White, B. A. Roe, and J. J. Ferretti. 2002. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc. Natl. Acad. Sci. USA 9914434-14439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aravind, L., and E. V. Koonin. 1998. A novel family of predicted phosphoesterases includes Drosophila prune protein and bacterial RecJ exonuclease. Trends Biochem. Sci. 2317-19. [DOI] [PubMed] [Google Scholar]
- 4.Banas, J. A. 2004. Virulence properties of Streptococcus mutans. Front. Biosci. 91267-1277. [DOI] [PubMed] [Google Scholar]
- 5.Banerjee, A., and I. Biswas. 2008. Markerless multiple-gene-deletion system for Streptococcus mutans. Appl. Environ. Microbiol. 742037-2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biswas, I., L. Drake, D. Erkina, and S. Biswas. 2008. Involvement of sensor kinases in the stress tolerance response of Streptococcus mutans. J. Bacteriol. 19068-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Biswas, I., L. Drake, S. Johnson, and D. Thielen. 2007. Unmarked gene modification in Streptococcus mutans by a cotransformation strategy with a thermosensitive plasmid. BioTechniques 42487-490. [DOI] [PubMed] [Google Scholar]
- 8.Biswas, I., J. K. Jha, and N. Fromm. 2008. Shuttle expression plasmids for genetic studies in Streptococcus mutans. Microbiology 1542275-2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Biswas, I., and J. R. Scott. 2003. Identification of rocA, a positive regulator of covR expression in the group A streptococcus. J. Bacteriol. 1853081-3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Biswas, S., and I. Biswas. 2006. Regulation of the glucosyltransferase (gtfBC) operon by CovR in Streptococcus mutans. J. Bacteriol. 188988-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, P. M., H. C. Chen, C. T. Ho, C. J. Jung, H. T. Lien, J. Y. Chen, and J. S. Chia. 2008. The two-component system ScnRK of Streptococcus mutans affects hydrogen peroxide resistance and murine macrophage killing. Microbes Infect. 10293-301. [DOI] [PubMed] [Google Scholar]
- 12.Crane, B. R., and E. D. Getzoff. 1996. The relationship between structure and function for the sulfite reductases. Curr. Opin. Struct. Biol. 6744-756. [DOI] [PubMed] [Google Scholar]
- 13.Duwat, P., S. D. Ehrlich, and A. Gruss. 1999. Effects of metabolic flux on stress response pathways in Lactococcus lactis. Mol. Microbiol. 31845-858. [DOI] [PubMed] [Google Scholar]
- 14.Echave, P., M. A. Esparza-Ceron, E. Cabiscol, J. Tamarit, J. Ros, J. Membrillo-Hernandez, and E. C. Lin. 2002. DnaK dependence of mutant ethanol oxidoreductases evolved for aerobic function and protective role of the chaperone against protein oxidative damage in Escherichia coli. Proc. Natl. Acad. Sci. USA 994626-4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghosh, S., and M. P. Deutscher. 1999. Oligoribonuclease is an essential component of the mRNA decay pathway. Proc. Natl. Acad. Sci. USA 964372-4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hamada, S., and H. D. Slade. 1980. Biology, immunology, and cariogenicity of Streptococcus mutans. Microbiol. Rev. 44331-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hatzios, S. K., A. T. Iavarone, and C. R. Bertozzi. 2008. Rv2131c from Mycobacterium tuberculosis is a CysQ 3′-phosphoadenosine-5′-phosphatase. Biochemistry 475823-5831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horaud, T., and F. Delbos. 1984. Viridans streptococci in infective endocarditis: species distribution and susceptibility to antibiotics. Eur. Heart J. 5(Suppl. C)39-44. [DOI] [PubMed] [Google Scholar]
- 19.Imlay, J. A. 2008. Cellular defenses against superoxide and hydrogen peroxide. Annu. Rev. Biochem. 77755-776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Imlay, J. A. 2002. How oxygen damages microbes: oxygen tolerance and obligate anaerobiosis. Adv. Microb. Physiol. 46111-153. [DOI] [PubMed] [Google Scholar]
- 21.Lemos, J. A., J. Abranches, and R. A. Burne. 2005. Responses of cariogenic streptococci to environmental stresses. Curr. Issues Mol. Biol. 795-107. [PubMed] [Google Scholar]
- 22.Li, Y. H., P. C. Lau, N. Tang, G. Svensater, R. P. Ellen, and D. G. Cvitkovitch. 2002. Novel two-component regulatory system involved in biofilm formation and acid resistance in Streptococcus mutans. J. Bacteriol. 1846333-6342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Loesche, W. J. 1986. Role of Streptococcus mutans in human dental decay. Microbiol. Rev. 50353-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loo, C. Y., D. A. Corliss, and N. Ganeshkumar. 2000. Streptococcus gordonii biofilm formation: identification of genes that code for biofilm phenotypes. J. Bacteriol. 1821374-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maguin, E., H. Prevost, S. D. Ehrlich, and A. Gruss. 1996. Efficient insertional mutagenesis in lactococci and other gram-positive bacteria. J. Bacteriol. 178931-935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marquis, R. E. 1995. Oxygen metabolism, oxidative stress and acid-base physiology of dental plaque biofilms. J. Ind. Microbiol. 15198-207. [DOI] [PubMed] [Google Scholar]
- 27.Mechold, U., G. Fang, S. Ngo, V. Ogryzko, and A. Danchin. 2007. YtqI from Bacillus subtilis has both oligoribonuclease and pAp phosphatase activity. Nucleic Acids Res. 354552-4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mechold, U., V. Ogryzko, S. Ngo, and A. Danchin. 2006. Oligoribonuclease is a common downstream target of lithium-induced pAp accumulation in Escherichia coli and human cells. Nucleic Acids Res. 342364-2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murguia, J. R., J. M. Belles, and R. Serrano. 1995. A salt-sensitive 3′(2′),5′-bisphosphate nucleotidase involved in sulfate activation. Science 267232-234. [DOI] [PubMed] [Google Scholar]
- 30.Neuwald, A. F., B. R. Krishnan, I. Brikun, S. Kulakauskas, K. Suziedelis, T. Tomcsanyi, T. S. Leyh, and D. E. Berg. 1992. cysQ, a gene needed for cysteine synthesis in Escherichia coli K-12 only during aerobic growth. J. Bacteriol. 174415-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nguyen, P. T., J. Abranches, T. N. Phan, and R. E. Marquis. 2002. Repressed respiration of oral streptococci grown in biofilms. Curr. Microbiol. 44262-266. [DOI] [PubMed] [Google Scholar]
- 32.Okano, S., Y. Shibata, T. Shiroza, and Y. Abiko. 2006. Proteomics-based analysis of a counter-oxidative stress system in Porphyromonas gingivalis. Proteomics 6251-258. [DOI] [PubMed] [Google Scholar]
- 33.Peng, Z., and D. P. Verma. 1995. A rice HAL2-like gene encodes a Ca2+-sensitive 3′(2′),5′-diphosphonucleoside 3′(2′)-phosphohydrolase and complements yeast met22 and Escherichia coli cysQ mutations. J. Biol. Chem. 27029105-29110. [DOI] [PubMed] [Google Scholar]
- 34.Perez-Casal, J., M. G. Caparon, and J. R. Scott. 1991. Mry, a trans-acting positive regulator of the M protein gene of Streptococcus pyogenes with similarity to the receptor proteins of two-component regulatory systems. J. Bacteriol. 1732617-2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pinto, R., J. S. Harrison, T. Hsu, W. R. Jacobs, Jr., and T. S. Leyh. 2007. Sulfite reduction in mycobacteria. J. Bacteriol. 1896714-6722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pomerantsev, A. P., R. Sitaraman, C. R. Galloway, V. Kivovich, and S. H. Leppla. 2006. Genome engineering in Bacillus anthracis using Cre recombinase. Infect. Immun. 74682-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qi, F., J. Merritt, R. Lux, and W. Shi. 2004. Inactivation of the ciaH gene in Streptococcus mutans diminishes mutacin production and competence development, alters sucrose-dependent biofilm formation, and reduces stress tolerance. Infect. Immun. 724895-4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Que, Y. A., J. A. Haefliger, P. Francioli, and P. Moreillon. 2000. Expression of Staphylococcus aureus clumping factor A in Lactococcus lactis subsp. cremoris using a new shuttle vector. Infect. Immun. 683516-3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rallu, F., A. Gruss, S. D. Ehrlich, and E. Maguin. 2000. Acid- and multistress-resistant mutants of Lactococcus lactis: identification of intracellular stress signals. Mol. Microbiol. 35517-528. [DOI] [PubMed] [Google Scholar]
- 40.Rantanen, M. K., L. Lehtio, L. Rajagopal, C. E. Rubens, and A. Goldman. 2007. Structure of the Streptococcus agalactiae family II inorganic pyrophosphatase at 2.80 A resolution. Acta Crystallogr. D 63738-743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spellerberg, B., B. Pohl, G. Haase, S. Martin, J. Weber-Heynemann, and R. Lutticken. 1999. Identification of genetic determinants for the hemolytic activity of Streptococcus agalactiae by ISS1 transposition. J. Bacteriol. 1813212-3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spiegelberg, B. D., J. P. Xiong, J. J. Smith, R. F. Gu, and J. D. York. 1999. Cloning and characterization of a mammalian lithium-sensitive bisphosphate 3′-nucleotidase inhibited by inositol 1,4-bisphosphate. J. Biol. Chem. 27413619-13628. [DOI] [PubMed] [Google Scholar]
- 43.Thibessard, A., F. Borges, A. Fernandez, B. Gintz, B. Decaris, and N. Leblond-Bourget. 2004. Identification of Streptococcus thermophilus CNRZ368 genes involved in defense against superoxide stress. Appl. Environ. Microbiol. 702220-2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Timmons, L., and A. Shearn. 1996. Germline transformation using a prune cDNA rescues prune/killer of prune lethality and the prune eye color phenotype in Drosophila. Genetics 1441589-1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsang, M. L. 1981. Assimilatory sulfate reduction in Escherichia coli: identification of the alternate cofactor for adenosine 3′-phosphate 5′-phosphosulfate reductase as glutaredoxin. J. Bacteriol. 1461059-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsang, M. L., and J. A. Schiff. 1976. Sulfate-reducing pathway in Escherichia coli involving bound intermediates. J. Bacteriol. 125923-933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ugochukwu, E., A. L. Lovering, O. C. Mather, T. W. Young, and S. A. White. 2007. The crystal structure of the cytosolic exopolyphosphatase from Saccharomyces cerevisiae reveals the basis for substrate specificity. J. Mol. Biol. 3711007-1021. [DOI] [PubMed] [Google Scholar]
- 48.Virtaneva, K., S. F. Porcella, M. R. Graham, R. M. Ireland, C. A. Johnson, S. M. Ricklefs, I. Babar, L. D. Parkins, R. A. Romero, G. J. Corn, D. J. Gardner, J. R. Bailey, M. J. Parnell, and J. M. Musser. 2005. Longitudinal analysis of the group A Streptococcus transcriptome in experimental pharyngitis in cynomolgus macaques. Proc. Natl. Acad. Sci. USA 1029014-9019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vuilleumier, S. 1997. Bacterial glutathione S-transferases: what are they good for? J. Bacteriol. 1791431-1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vuilleumier, S., and M. Pagni. 2002. The elusive roles of bacterial glutathione S-transferases: new lessons from genomes. Appl. Microbiol. Biotechnol. 58138-146. [DOI] [PubMed] [Google Scholar]
- 51.Walsh, C. T., A. M. Gehring, P. H. Weinreb, L. E. Quadri, and R. S. Flugel. 1997. Post-translational modification of polyketide and nonribosomal peptide synthases. Curr. Opin. Chem. Biol. 1309-315. [DOI] [PubMed] [Google Scholar]
- 52.Wen, Z. T., and R. A. Burne. 2004. LuxS-mediated signaling in Streptococcus mutans is involved in regulation of acid and oxidative stress tolerance and biofilm formation. J. Bacteriol. 1862682-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.York, J. D., J. W. Ponder, and P. W. Majerus. 1995. Definition of a metal-dependent/Li+-inhibited phosphomonoesterase protein family based upon a conserved three-dimensional core structure. Proc. Natl. Acad. Sci. USA 925149-5153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang, X., L. Zhu, and M. P. Deutscher. 1998. Oligoribonuclease is encoded by a highly conserved gene in the 3′-5′ exonuclease superfamily. J. Bacteriol. 1802779-2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zilhao, R., and P. Courvalin. 1990. Nucleotide sequence of the fosB gene conferring fosfomycin resistance in Staphylococcus epidermidis. FEMS Microbiol. Lett. 56267-272. [DOI] [PubMed] [Google Scholar]
- 56.Zotta, T., K. Asterinou, R. Rossano, A. Ricciardi, M. Varcamonti, and E. Parente. 2008. Effect of inactivation of stress response regulators on the growth and survival of Streptococcus thermophilus Sfi39. Int. J. Food Microbiol. 129211-220. [DOI] [PubMed] [Google Scholar]