Abstract
Aminoglycoside-2′′-phosphotransferase-IIa [APH(2′′)-IIa] is one of a number of homologous bacterial enzymes responsible for the deactivation of the aminoglycoside family of antibiotics and is thus a major component in bacterial resistance to these compounds. APH(2′′)-IIa produces resistance to several clinically important aminoglycosides (including kanamycin and gentamicin) in both gram-positive and gram-negative bacteria, most notably in Enterococcus species. We have determined the structures of two complexes of APH(2′′)-IIa, the binary gentamicin complex and a ternary complex containing adenosine-5′-(β,γ-methylene)triphosphate (AMPPCP) and streptomycin. This is the first crystal structure of a member of the APH(2′′) family of aminoglycoside phosphotransferases. The structure of the gentamicin-APH(2′′)-IIa complex was solved by multiwavelength anomalous diffraction methods from a single selenomethionine-substituted crystal and was refined to a crystallographic R factor of 0.210 (Rfree, 0.271) at a resolution of 2.5 Å. The structure of the AMPPCP-streptomycin complex was solved by molecular replacement using the gentamicin-APH(2′′)-IIa complex as the starting model. The enzyme has a two-domain structure with the substrate binding site located in a cleft in the C-terminal domain. Gentamicin binding is facilitated by a number of conserved acidic residues lining the binding cleft, with the A and B rings of the substrate forming the majority of the interactions. The inhibitor streptomycin, although binding in the same pocket as gentamicin, is orientated such that no potential phosphorylation sites are adjacent to the catalytic aspartate residue. The binding of gentamicin and streptomycin provides structural insights into the substrate selectivity of the APH(2′′) subfamily of aminoglycoside phosphotransferases, specifically, the selectivity between the 4,6-disubstituted and the 4,5-disubstituted aminoglycosides.
The emergence of bacteria resistant to several important classes of antibiotics has become a major clinical problem over the last few years. Almost every antibacterial compound in clinical use today has associated examples of resistant bacterial isolates (39), including life-threatening strains of Escherichia coli, Mycobacterium tuberculosis, Pseudomonas aeruginosa, and various enterococci. The latter are among the most common antibiotic-resistance bacteria isolated from patients with nosocomial infections in the United States today. The synergistic use of either ampicillin or vancomycin with an aminoglycoside, such as kanamycin or gentamicin, has long been the optimal therapy for serious enterococcal infections; however, many previously susceptible enterococcal strains have since acquired resistance to the aminoglycosides. The mechanisms of resistance are many and varied, although only three are readily understood: (i) mutation of the ribosomal target, (ii) reduced permeability and/or increased efflux of the drug, and (iii) enzymatic deactivation of the drug. Resistance to the aminoglycosides through enzymatic deactivation, although seemingly straightforward, is in reality a complex problem involving three different classes of enzyme. These enzyme classes are the ATP-dependent phosphotransferases (APH) and adenyltransferases (ANT), and the acetyl coenzyme A-dependent N-acetyltransferases (AAC). This area of research has been extensively reviewed in the past few years (2, 4, 13, 29, 39, 47, 52, 53).
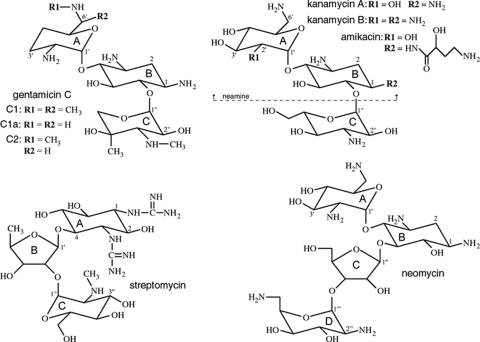
Originally isolated from soil bacteria, including various species of Streptomyces and Micromonospora (20), the aminoglycosides are a family of potent, broad-spectrum antibiotics that includes clinically relevant drugs such as gentamicin, neomycin, amikacin, kanamycin, and streptomycin. The structures of these compounds, with the exception of that of streptomycin, are all similar, consisting of a central aminocyclitol ring (the B ring) with two or three substituted aminoglycan rings (A, C, and in some cases, D) attached at either the 4 and 5 positions (the 4,5-disubstituted aminoglycosides, which include neomycin and lividomycin) or the 4 and 6 positions (the 4,6-disubstituted aminoglycosides, such as gentamicin and kanamycin). Streptomycin, a competitive inhibitor of aminoglycoside-2′′-phosphotransferase-IIa [APH(2′′)-IIa] (45), is an atypical aminoglycoside that does not fall into either the 4,5-disubstituted or 4,6-disubstituted classes. It has a modified ribose (ring B) attached to position 4 on a 1,3-diguanidinium-substituted aminocyclitol ring (ring A) with no substituent at the 5 or 6 position. The structures of gentamicin, kanamycin, neomycin, and streptomycin are shown in Fig. 1. The aminoglycosides are targeted to the 16S rRNA of the bacterial 30S ribosomal subunit, where they selectively bind to the decoding aminoacyl (A) site (31, 51) and stabilize the conformation of the tRNA bound to a cognate mRNA codon. This decreases the dissociation rate of aminoacyl-tRNA and promotes miscoding (28). The structures of a number of the aminoglycosides with either the 30S subunit or oligonucleotides containing minimal A sites are known (51).
FIG. 1.
Structures of gentamicin, kanamycin, streptomycin, and neomycin. Gentamicin and kanamycin are classified as 4,6-disubstituted aminoglycosides, whereas neomycin is an example of a 4,5-disubstituted compound. The three structural variants which comprise gentamicin C are indicated. Amikacin is similar to kanamycin, although the substituent on the N1 amine is a 4-amino-2-hydroxy-1-oxobutyl group. Taken together, the A and B rings of aminoglycosides, such as gentamicin, kanamycin, and neomycin, are commonly known as the neamine moiety.
The enzymes which deactivate the aminoglycosides are named according to the reaction they catalyze and the site on the aminoglycoside at which they act. The APH(2′′) enzymes, which give rise to high-level resistance to gentamicin in enterococci, phosphorylate gentamicin and kanamycin at the 2′′-hydroxyl group of the C ring (Fig. 1). The APH(3′) enzymes, another major subfamily of the phosphotransferases, phosphorylate kanamycin and neomycin at the 3′-hydroxyl on the A ring but cannot deactivate gentamicin, since it has no corresponding 3′-hydroxyl. The individual members of each family can normally bind only a subset of the available drugs, and this difference in drug specificity is known as the resistance profile, designated with a roman numeral and, in some cases, a letter identifying a specific gene. The first APH(2′′) enzyme discovered for enterococci was the bifunctional AAC(6′)-Ie-APH(2′′)-Ia enzyme, which possesses both 6′-acetylating and 2′′-phosphorylating activities (17, 33). Enterococci with the corresponding gene show resistance to almost all clinically relevant aminoglycosides (38). Four additional APH(2′′) enzymes have since been isolated for Enterococcus spp.; they are designated APH(2′′)-Ib (27), APH(2′′)-Ic (11), APH(2′′)-Id (46), and APH(2′′)-Ie (10) and were initially classified as genetic variants of an APH(2′′)-I-type enzyme. Recently, APH(2′′)-Ib, APH(2′′)-Ic, and APH(2′′)-Id have been reclassified as distinct enzymes with different resistance profiles and, more importantly, different nucleotide specificities, such that they are now named APH(2′′)-IIa, APH(2′′)-IIIa, and APH(2′′)-IVa, respectively (44). APH(2′′)-Ie was not included in the latter study, but based upon the very high sequence similarity with APH(2′′)-IVa (93%) (see Table S1 in the supplemental material), it is possible that it is a genetic variant of APH(2′′)-IVa.
Structural details are currently known for only two members of the APH(3′) family, APH(3′)-IIIa (5, 18, 23) and APH(3′)-IIa (37). These enzymes share a two-domain structure similar to the catalytic domains of the eukaryotic Ser/Thr and Tyr protein kinases. Moreover, the phosphotransferases and kinases share several important sequence motifs related to nucleotide binding and phosphoryl transfer, most notably the catalytic loop (HXDXXXXN) and the activation segment (GXIDXG), where X is any amino acid. Not surprisingly, the catalytic mechanisms of the phosphotransferases and the kinases are identical, involving the nucleophilic attack by the target hydroxyl on the γ phosphate of ATP, facilitated by a conserved aspartate residue from the catalytic loop (29, 54). A comparison of the known APH(2′′) and APH(3′) sequences shows that the two families of phosphotransferases share these kinase-like motifs, and there appears to be some partial conservation of acidic residues in the substrate binding region. It has been suggested that their structures may be similar (37). Here, we report the first structure of an APH(2′′) enzyme, APH(2′′)-IIa as the binary complex with the preferred substrate gentamicin and the ternary complex with the nonhydrolyzable ATP analog adenosine-5′-(β,γ-methylene)triphosphate (AMPPCP) and the competitive inhibitor streptomycin.
MATERIALS AND METHODS
Protein expression, purification, and crystallization.
Native Enterococcus faecium APH(2′′)-IIa was cloned, expressed, and purified, and the gentamicin complex was prepared and crystallized, as previously described (50). Selenomethionine-labeled APH(2′′)-IIa [SeMet-APH(2′′)-IIa] was produced using a modified protocol based on the inhibition of methionine biosynthesis (15), and the enzyme was purified as described for native APH(2′′)-IIa (50). The gentamicin complex was prepared by the addition of 1 mM gentamicin to a solution of SeMet-substituted enzyme in 10 mM bis-Tris propane (pH 6.5), 150 mM NaCl, 5 mM β-mercaptoenthanol. Crystals were grown by hanging-drop vapor diffusion by mixing equal volumes (1 μl + 1 μl) of the protein solution with unbuffered 12% polyethylene glycol 3350 (PEG 3350) over a reservoir solution comprising unbuffered 16% PEG 3350. The ternary AMPPCP-streptomycin complex was prepared by preincubation of 5 mM AMPPCP, 5 mM MgCl2, and 5 mM streptomycin with apo enzyme (10 mg/ml) in 10 mM bis-Tris propane (pH 6.5), 150 mM NaCl, and 5 mM β-mercaptoenthanol for 24 h at 4°C. Crystals of the resultant complex were grown by hanging-drop vapor diffusion by mixing an equal volume (1 μl + 1 μl) of protein solution with crystallization buffer (30% methoxypolyethylene glycol 5000 (mPEG5000), 0.2 M morpholinosulfonic acid (MOPS)/KOH [pH 7.3]) from the reservoir solution. Additional complexes were also prepared and submitted to crystallization trials, including ternary complexes with the nonhydrolyzable ATP analog adenosine-5′-(β,γ-imido)triphosphate (AMPPNP), substrates gentamicin and kanamycin, and the inhibitor streptomycin and binary complexes with ATP and ADP. Only one of these additional complexes [ADP-APH(2′′)-IIa] gave rise to diffraction-quality crystals.
Diffraction data collection and processing.
The SeMet-APH(2′′)-IIa data were collected at beam line BL9-1 at the Stanford Synchrotron Radiation Laboratory (SSRL) from a crystal frozen in a cryoprotectant composed of 16% PEG 3350 and 20% glycerol. The crystal belonged to space group P21 with the following cell dimensions: a = 78.3 Å, b = 57.5 Å, c = 81.7 Å, and β = 98.2°. The Matthews coefficient (34), assuming two molecules in the asymmetric unit, is 2.7 Å3/Da. The data were collected at three wavelengths equivalent to the peak (0.9790 Å) and the inflection (0.9792 Å) of the selenium absorption edge and a remote energy (0.8377 Å). A total of 180 images were collected for each wavelength, with an oscillation range of 1°. The data were processed with X-ray detector software (XDS) and scaled with XSCALE software (26). Data collection statistics are given in Table 1.
TABLE 1.
SeMet-APH(2′′)-IIa data collection statisticsa
| Parameter | Value at indicated wavelength
|
||
|---|---|---|---|
| Peak | Inflection | Remote | |
| Wavelength (Å) | 0.9794 | 0.9796 | 0.8609 |
| High-resolution limit (Å) | 3.1 | 3.1 | 3.1 |
| No. of observed reflections | 48,986 | 49,026 | 48,742 |
| No. of unique reflections to dminb | 25,161 | 25,172 | 25,148 |
| Rmergec (%) | 8.3 (40.2) | 8.3 (38.1) | 5.8 (29.8) |
| I/σ | 11.5 (2.1) | 11.4 (2.2) | 16.0 (2.9) |
| Avg redundancy | 3.6 | 3.7 | 3.6 |
| Completeness (%) | 97.6 (96.7) | 97.5 (96.4) | 97.0 (86.1) |
Numbers in parentheses relate to the highest-resolution shell, 3.2 to 3.1 Å.
dmin, maximum observed resolution.
Rmerge = Σ|Ii − 〈I〉|/ΣIi, where Ii is the observed intensity and 〈I〉 is the mean intensity.
The native gentamicin-APH(2′′)-IIa data were collected at SSRL beam line BL9-2 from a crystal frozen in a cryoprotectant composed of 16% PEG 3350 and 20% glycerol. A total of 180 images were measured, each with an oscillation angle of 1°. The crystal was isomorphous with the SeMet-APH(2′′)-IIa crystals, with the following cell dimensions: a = 79.7 Å, b = 58.8 Å, c = 81.4 Å, β = 98.4°. The data were processed with XDS and scaled with XSCALE (26). The AMPPCP-streptomycin-APH(2′′)-IIa data were also collected at SSRL beam line BL9-2. A single crystal was transferred in four steps from the crystallization mother liquor to drops containing crystallization buffer (30% mPEG5000, 0.2 M MOPS/KOH [pH 7.3]) mixed with increasing concentrations of cryoprotectant (30% mPEG5000, 0.2 M MOPS/KOH [pH 7.3], and 20% glycerol). The crystal with cell dimensions of a = b = 128.0, c = 57.8 belonged to the space group P31, with a Matthews coefficient (34) of 2.6 Å3/Da, assuming three molecules in the asymmetric unit. A total of 120 images were collected with an oscillation angle of 1°, and these images were indexed, integrated, and scaled with XDS and XSCALE (26). Data collection statistics for the ternary and binary complexes are given in Table 2.
TABLE 2.
Native APH(2′′)-IIa data collection and refinement statisticsa
| Parameter | Value for indicated drug complex
|
|
|---|---|---|
| Gentamicin | AMPPCP-streptomycin | |
| Space group | P21 | P31 |
| Resolution (Å) | 80.6-2.5 | 42.9-2.4 |
| No. of observed reflections/no. of unique reflections | 93,234/25,799 | 154,024/41,774 |
| Rmergeb (%) | 4.2 (44.3) | 4.3 (44.8) |
| Avg redundancy | 3.4 | 3.8 |
| I/σ | 21.4 (2.6) | 20.1 (3.5) |
| Completeness (%) | 98.9 (93.4) | 99.1 (98.8) |
| Resolution limits for refinement (Å) | 20.0-2.5 | 20.0-2.45 |
| Data used in refinementc | 24,499 (1,299) | 39,374 (1,827) |
| R factor/Rfree (%)d | 21.0/27.1 | 21.8/28.4 |
| No. of protein atoms/no. of solvent atoms | 4,978/240 | 7,164/205 |
| Avg B | ||
| Protein (Å2)e | 38.6/38.0 | 42.1/41.9/42.0 |
| Solvent (Å2) | 45.6 | 43.3 |
| Estimated coordinate error, DPI (Å)f | 0.34 | 0.33 |
| RMSD, bonds (Å) | 0.013 | 0.012 |
| RMSD, angles (°) | 1.30 | 1.21 |
Numbers in parentheses relate to the highest-resolution shell, 2.6 to 2.5 Å for the gentamicin complex and 2.5 to 2.45 Å for the AMPPCP-streptomycin complex.
Rmerge = S(Ii - 〈I〉)/SIi where Ii is the observed intensity of a given reflection and 〈I〉 is the mean intensity for all observations of that reflection.
Numbers in parentheses are the numbers of reflections used for the Rfree calculation. They comprise 5% of the unique reflections chosen at random.
R = S‖Fo| - |Fc‖/|Fo|, where Fo is the observed structure factor amplitude and Fc is the calculated structure factor.
The numbers refer to the average values for the independent molecules in the asymmetric unit of each complex.
Cruikshank diffraction precision index estimated from Rfree.
Structure solution and refinement.
The gentamicin SeMet-APH(2′′)-IIa structure was solved by multiwavelength anomalous diffraction methods using data to a 3.1-Å resolution. The program SOLVE (42) gave the positions of all 12 selenium atoms (6 in each molecule, including the N-terminal residue), resulting in a mean figure of merit of 0.38. Maximum likelihood density modification and refinement of the phases and noncrystallographic symmetry were carried out by RESOLVE (41), giving a mean figure of merit of 0.67 and leading to the automated building of 380 residues out of 598 into the electron density map (40). Interactive model building with COOT (16) was used to add the majority of the remaining residues and produce a partial model that was used for refinement. The initial crystallographic R factor prior to the first round of refinement was 0.45, with an Rfree of 0.48. Subsequent model building with COOT completed the structure and added 240 solvent molecules (modeled as water) and two gentamicin molecules. Residual electron density observed in the experimentally phased electron density maps in the vicinity of the proposed catalytic aspartate, Asp192, was modeled as gentamicin in both molecules. Maximum likelihood refinement with REFMAC (36), incorporating individual temperature factor and torsion-libration-screw (TLS) refinement at a 2.5-Å resolution, gave a final model with a crystallographic R factor of 0.210 and an Rfree of 0.271. The two molecules in the asymmetric unit are related in a tail-to-tail fashion by a noncrystallographic twofold rotation axis, with approximately 550 Å2 (3.5%) of surface area buried per monomer. This dimer is most probably not physiologically relevant given the small area of contact and the nonspecificity of the surface, although there is no biochemical either way. The final model comprises 598 amino acid residues, 2 gentamicin molecules, and 240 water molecules. The final refinement statistics are given in Table 2.
The structure of the AMPPCP-streptomycin complex of APH(2′′)-IIa was determined by molecular replacement with PHASER (35) using a search model derived from the P21 gentamicin-APH(2′′)-IIa structure with the gentamicin and water molecules removed. The starting R factor and Rfree for refinement were 0.512 and 0.508, respectively. Strong residual electron density was seen in all three molecules in the region identified as the nucleotide binding site and, to a lesser extent, in the substrate binding site. An AMPPCP molecule was added to all three molecules early in the refinement, and the substrate density was modeled as streptomycin in all molecules during later stages of refinement. After iterative cycles of refinement with REFMAC, incorporating TLS for the final rounds, and manual model building with COOT, the final R factor was 0.218 (Rfree = 0.284 for 5% of the unique reflections) at a 2.45-Å resolution. The final model comprises 879 amino acid residues (molecule A, 1 to 274 and 278 to 299; molecule B, 1 to 299; and molecule C, 1 to 159, 165 to 274, and 285 to 299), three AMPPCP molecules, three Mg2+ ions, three streptomycin molecules, and a total of 205 water molecules. Refinement statistics are given in Table 2.
Data programs.
Superpositions were performed using the secondary-structure matching procedure (32) implemented in COOT and using LSQKAB in the CCP4 suite (12). Figures were generated using PyMOL (14).
Protein Data Bank accession numbers.
The atomic coordinates for gentamicin-APH(2′′)-IIa and AMPPCP-streptomycin-APH(2′′)-IIa were deposited in the Protein Data Bank (3) with accession codes 3ham and 3hav, respectively.
RESULTS
Overall monomer structure.
The APH(2′′)-IIa molecule is divided into two major domains (Fig. 2a). The N-terminal domain (residues 1 to 90) is composed of a five-stranded β sheet (strands β1 to β5) flanked by a long α helix (α2), with a short α helix (α1) at the N terminus. A short linker (residues 85 to 89) connects this domain with the C-terminal domain, the latter comprising two subdomains, the core subdomain (residues 91 to 134 and 185 to 250) based upon four α helices (α3, α4, α7, and α8) and four β strands (β6 to β9) along with two active site loops (β6 to β7 and β8 to β9) and a helical subdomain (residues 135 to 184 and 251 to 299) formed from four helices (α5, α6, α9, α10). A negatively charged cleft between the core and the helical subdomains forms the aminoglycoside binding site, with the nucleotide binding site sandwiched between the N-terminal domain and the core subdomain (Fig. 2a) and linked to the “top” of the aminoglycoside binding cleft.
FIG. 2.
Crystal structure of APH(2′′)-IIa. (a) Stereo view of the APH(2′′)-IIa monomer showing the location of the substrate binding site, indicated by a gentamicin molecule in yellow, and the nucleotide binding site, indicated by a partially transparent AMP molecule (upper panel). The N-terminal domain is at the top of the figure (pale green), with the core subdomain in the center (bright green) and the helical subdomain at the bottom (dark green). The numbering of the secondary structure elements is indicated. (b) Stereo view of the superposition of the gentamicin-APH(2′′)-IIa complex (green ribbons) and the kanamycin-APH(3′)-IIa complex (red ribbons) based upon residues from the core subdomain. The similarity of the N domain and the core subdomain can be seen, along with the differences in the helical subdomain. The location of the substrate binding site is indicated by a yellow gentamicin molecule and a kanamycin molecule in thinner white sticks. (c) Stereo view of the close-up view of the area inside the black box in panel b, showing the substrate binding site with the “top” and the “base” of the binding cleft indicated. The three rings of the gentamicin (yellow sticks) are labeled in boldface type (C-B-A). The kanamycin molecule as it is bound to APH(3′)-IIa is shown as thin white sticks with the three rings labeled in italics (A-B-C).
The APH(2′′)-IIa structure is essentially identical in the two complexes, although nucleotide and substrate binding results in small reorientations of both the N-terminal domain and the helical subdomain. In the ternary streptomycin-AMPPCP complex, although the N-terminal domain and the core subdomain of all three molecules in the asymmetric unit are generally well ordered, there is a high degree of disorder in the helical subdomains, resulting in an inability to accurately model some of this region in two of the molecules and resulting in the observed difference of over 7% between the crystallographic R factor and the Rfree. Superposition of the two structures based on the core subdomain gives a root-mean-square difference (RMSD) of 0.57 Å for 113 matching Cα atoms. Subsequent superposition of the rotated AMPPCP-streptomycin-APH(2′′)-IIa model onto the gentamicin-APH(2′′)-IIa complex based on the N-terminal domain (RMSD, 0.7 Å; 81 Cα) and the helical subdomain (RMSD, 0.6 Å; 91 Cα) gives an estimate of the amount of movement of these domains. The helical subdomain has moved, on average, by about 6°, and while this could be due to the replacement of the gentamicin by the slightly more bulky streptomycin molecule, crystal packing interactions in the two different space groups are markedly different and could also account for this difference. When AMPPCP is present, the N-terminal domain moves away from the core domain by approximately 5°, on average. In the nucleotide-free complex, the loop between strands β1 and β2 closes over the empty binding site. In the nucleotide-bound structure, this loop swings away from the nucleotide binding pocket such that the outer edge of the loop is approximately 5 Å from the equivalent location in the nucleotide-free complex. The mobility of this loop in the protein kinases is critical for catalysis, and conformational changes of this region have been observed for these enzymes (1) and also for the two APH(3′) enzymes (23, 37).
The overall fold of APH(2′′)-IIa is similar to that of the two APH(3′) enzymes despite an extremely low sequence identity [6% and 9% for APH(3′)-IIa and APH(3′)-IIIa, respectively] between these enzymes (see Table S1 in the supplemental material). Superposition of APH(2′′)-IIa onto these two structures gives RMSDs of 3.3 Å and 3.2 Å for 260 matching Cα positions for APH(3′)-IIIa and APH(3′)-IIa, respectively. An analysis of the superimposed structures (Fig. 2b) shows that the N-terminal domains and the core domains are spatially and topologically similar in these two subfamilies, and although the helical subdomain has the same topology, this subdomain as a whole is in a different orientation relative to the core subdomain and is approximately 30 residues longer. The additional residues comprise an extra helix (α10), which runs antiparallel to helix α9, and a final short piece of 310 helix (α10′) at the C terminus (Fig. 2b). Superposition of the individual domains gives significantly lower RMSDs, 2.0 Å, 2.1 Å and 2.9 Å for the N-terminal domain, the core subdomain, and the helical subdomain, respectively.
Substrate binding site.
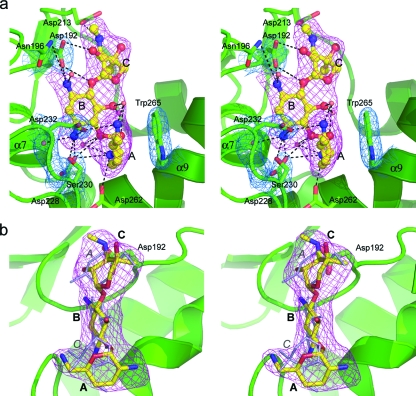
The gentamicin molecule is oriented in the binding cleft such that the site of phosphorylation (the 2′′-OH on the C ring) is approximately 3.0 Å from the Oδ1 atom of Asp192, the putative catalytic base. This residue is conserved in all aminoglycoside-modifying enzymes (see Fig. S1 in the supplemental material) and is part of a conserved motif equivalent to the protein kinase subdomain VIb (21, 22), also known as the catalytic loop (25). It has been shown to be essential for activity in the APH(3′) enzymes (23, 30) and is responsible for orienting the hydroxyl group for an optimum nucleophilic attack on the γ phosphate (29) and as a proton acceptor or a “proton trap” in the latter stages of the reaction process, once cleavage of the phosphorus-oxygen bond has begun (48). The B ring of the gentamicin is coplanar with the C ring (Fig. 3a) and nestled against the core subdomain, and a network of potential hydrogen bonding interactions anchors this part of the substrate in the binding cleft (for potential hydrogen bonding interactions, see Table S2 in the supplemental material). The A ring projects across the substrate binding cleft and makes two potential interactions with the helical subdomain, a hydrogen bond between the N2′ atom and the Oδ1 atom of Asp262 from helix α9 and a nonpolar stacking interaction between the A ring and the indole ring of Trp265 one turn further along helix α9. The C2′ and C5′ substituents on the A ring are in equatorial positions which facilitate this face-to-face stacking. This type of nonpolar interaction has also been observed between the A site of 16S RNA and the A rings of both paromomycin (7) and gentamicin (19). The α9 helix has a pronounced kink at residues 265 and 266, which causes the C-terminal half of the helix to bend upwards by over 45° (Fig. 3a). The α7-α8 loop, along with helix α9, forms the “base” of the substrate binding site.
FIG. 3.
Gentamicin binding site. (a) Stereo view of gentamicin in the binary gentamicin-APH(2′′)-IIa complex showing composite omit 2Fo-Fc electron density (pink; 1.0 σ) for the gentamicin substrate (yellow ball-and-stick representation). The rings are labeled in boldface type (C-B-A). The hydrogen bonding interactions with the substrate are indicated as dashed lines. The kinked helix α9 is at the lower right. The final 2Fo-Fc density (blue; 1.0 σ) is shown for some of the protein residues. (b) View in panel a rotated vertically by 90° and showing the gentamicin bound in the refined position (yellow ball-and-stick representation) and in an alternative “upside-down” orientation (thinner gray sticks). The rings are labeled in boldface type for the correct orientation (C-B-A), and for the alternative orientation, only the A and C rings are labeled in gray italics for clarity.
If the binding of kanamycin to the APH(3′) enzymes is taken as the paradigm for aminoglycoside binding to the phosphotransferases, then gentamicin binding to APH(2′′)-IIa can be considered as being “upside-down” in comparison. Superposition of kanamycin-APH(3′)-IIa and gentamicin-APH(2′′)-IIa shows that the two molecules are bound in different orientations (Fig. 2c). In the APH(3′) enzymes, the kanamycin molecule is bound in an A-B-C configuration, with the A ring containing the 3′-OH at the “top” of the binding cleft where it joins the nucleotide binding site, the central B ring projecting across the cleft and interacting with residues from the acidic loop and the DExF motif, and the C ring at the “base” of the cleft interacting with the core subdomain. In APH(2′′)-IIa, the gentamicin molecule adopts a C-B-A configuration, with the C ring containing the 2′′-OH at the “top” of the cleft, the B ring anchored to the core subdomain, and the A ring at the “base” of the cleft angled toward the helical subdomain. Binding of gentamicin in an A-B-C orientation was also tested using a composite omit map at the end of refinement, and it was observed that although the central B ring could fit adequately into the density, there were major issues with both the other rings, with unaccounted-for density and substituents projecting out of the density (Fig. 3b). The C ring in particular could not be modeled well into the available electron density, with the final best fit resulting in a steric clash between the O5′′ atom and the O5 atom of the B ring.
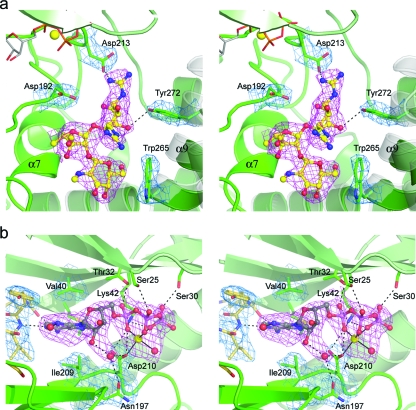
The streptomycin is bound in an A-B-C configuration, and although there are fewer interactions with the enzyme, the majority occur with the B and C rings. The C ring lies at the “base” of the binding cleft and projects across toward the helical subdomain and is involved in a hydrophobic packing interaction with Trp265 that is similar but not identical to the interaction of the gentamicin A ring in the gentamicin-APH(2′′)-IIa complex. Two of the APH(2′′)-IIa molecules in the asymmetric unit have the A ring projecting toward the helical subdomain, with the subsequent formation of a new hydrogen bonding interaction between the Oη of Tyr272 and the O5 atom. The A ring is further anchored by a potential electrostatic interaction between the guanidinium group at position 1 and the side chain of Asp213 (Fig. 4a). In this orientation of the A ring, although the O6 atom comes within 3.4 Å of the Oδ1 atom of the catalytic aspartate (Asp192), it points down toward the B ring and is thus oriented incorrectly for attack on the γ phosphate. In the third APH(2′′)-IIa molecule, the inhibitor has moved approximately 1.5 Å toward the helical subdomain, and the A ring has rotated up toward the nucleotide binding pocket. The guanidinium group at position 1 interacts with a water molecule associated with the β phosphate of the AMPPCP. It has been recently shown that streptomycin is a competitive inhibitor of APH(2′′)-IIa, albeit a rather poor inhibitor with a Ki of around 170 μM (45), whereas gentamicin binds very tightly, with a Km of around 0.55 μM (Marta Toth, personal communication). In all three molecules, the streptomycin binds in the same location as the substrate, yet there is no hydroxyl group appropriately oriented for phosphorylation, so the molecule would not be turned over. Furthermore, the bulky guanidinium groups on the A ring project toward the nucleotide, interfering with an extended triphosphate moiety.
FIG. 4.
Binding sites in the ternary complex. (a) Stereo view of streptomycin in the ternary AMPPCP-streptomycin-APH(2′′)-IIa complex showing composite omit 2Fo-Fc density (pink; 1.0 σ) for the streptomycin inhibitor from molecule A (cyan ball-and-stick representation). The equivalent streptomycin conformation from molecule B is overlaid as thin bonds (gray). The location of helices α9 and α10 from the binary gentamicin-APH(2′′)-IIa complex are shown as semitransparent gray coils. (b) Stereo view of the AMPPCP (gray ball-and-stick representation) in the ternary AMPPCP-streptomycin-APH(2′′)-IIa complex showing composite omit 2Fo-Fc density (pink; 1.0σ). The magnesium ion is shown as a yellow sphere surrounded by the chelating AMPPCP triphosphate moiety. The hydrogen-bonding interactions with the AMPPCP are indicated by dashed black lines. Final 2Fo-Fc densities (blue; 1.0 σ) for surrounding protein residues are shown, along with the 2Fo-Fc density for the interdomain linker (gold bonds, left).
The electron density for the streptomycin (Fig. 4a) is somewhat weaker than that observed for the gentamicin complex, which might be suggestive of lower site occupancy. The weaker electron density for the streptomycin could be consistent with the lower affinity for streptomycin, although since the inhibitor is being observed in the context of a crystal structure, crystal packing interactions could play a role and either limit accessibility to the site or cause structural changes in the enzyme which restricts binding. The observation that the three streptomycin inhibitors in the three independent APH(2′′)-IIa molecules are bound in a similar yet not identical fashion in the binding pocket suggests that the binding pocket has evolved to specifically bind aminoglycosides with a gentamicin-like structure; although streptomycin is able to bind, it does so in an essentially nonspecific manner, in all likelihood constantly moving between the two major configurations represented in the crystal structure.
Nucleotide binding site.
The nucleotide interacts with residues from both the N-terminal domain and the core subdomain (Fig. 4b). The pocket for the adenine ring is formed by strands β3, β7, and β8 and the interdomain linker. The ring is sandwiched between the four hydrophobic side chains, Leu24, Val40, Ile199, and Ile209, although the only specific interactions it makes with the protein are two potential hydrogen bonds to the carbonyl oxygen of Lys86 and the amide nitrogen of Ile88 from the linker peptide. The ribose moiety makes no interactions with the protein. The triphosphate group is anchored to the N-terminal domain through interactions with the side chains of Ser25, Ser30, Lys42 (highly conserved in all APH enzymes), and Thr32. The triphosphate adopts a compact tridentate chelate conformation and wraps around a magnesium ion such that an oxygen atom from each phosphate group coordinates with the metal (Fig. 4b). The magnesium ion is further coordinated to the side chain of Asp210. The chelate conformation of the triphosphate group is unusual and in marked contrast to the typical extended conformation seen with the majority of ATP-enzyme complexes.
A superposition of the AMPPNP complex of APH(3′)-IIIa (5) and the ternary APH(2′′)-IIa complex, based upon residues from the N domain and the core subdomain, shows that the adenine moieties of the nucleotides are in almost identical locations in the two enzymes. In the APH(3′) enzymes, the adenine is sandwiched between an isoleucine [equivalent to Ile209 in APH(2′′)-IIa] and a highly conserved aromatic residue from strand β3 [Phe48 in APH(3′)-IIa and Tyr42 in APH(3′)-IIIa]. Rotations about the N9-C1* and the C5*-O5* bonds in the APH(2′′)-bound nucleotide put the ribose and the α-phosphate groups approximately 2.4 Å and 3.0 Å away from their respective positions in APH(3′)-IIIa (5), displaced from the catalytic loop. In APH(3′)-IIIa, there is a single direct interaction between the triphosphate and the core subdomain through Asp208 [APH(3′)-IIIa residue numbering]. This interaction is retained in APH(2′′)-IIa, despite the difference in conformation of the triphosphate, facilitated by an almost 90° rotation of the Asp210 side chain. This aspartate residue is part of the conserved sequence motif GvIDFG in the β8-β9 loop, equivalent in location to but significantly shorter than the protein kinase conserved subdomain VII (21, 22) or the activation segment (25).
DISCUSSION
The crystal structures of the binary gentamicin and the ternary AMPPCP-streptomycin complexes of APH(2′′)-IIa represent the first structures determined for a member of this family, the second-largest family of aminoglycoside phosphotransferases. The APH(2′′)-IIa enzyme is structurally and topologically similar to the APH(3′′) enzymes and also to the catalytic domains of the eukaryotic Ser/Thr and Tyr protein kinases. The major differences between these enzymes arise in the substrate binding site and, to a lesser extent, the nucleotide binding site. In the adenine binding pocket, APH(2′)-IIa lacks the conserved aromatic residue on strand β3, having a valine in this position; this residue is either a valine or an isoleucine in the other members of the APH(2′′) family (see Figure S1 in supplemental material). In this regard, both the APH(2′′) and the APH(3′) families differ markedly from the protein kinases, which invariably have an alanine at this position (21). It has been suggested that this sequence variation may give rise to differences in the way the adenine moiety is bound in the two enzyme groups and that this difference could be exploited in the design of an APH(3′)-specific inhibitor (4). Although the lack of the aromatic residue in APH(2′′)-IIa suggests that an inhibitor designed for the APH(3′) enzymes might not be effective against the APH(2′′) enzymes, the similarities in the locations of the adenine group in the ternary complex shows that this end of the nucleotide is anchored in much the same way in the two enzymes despite the sequence differences. A leucine residue (Leu24) from the neighboring strand β1, and a tyrosine residue (Tyr87) from the interdomain linker, both project inward toward the adenine pocket and overlap slightly the position occupied by the aromatic side chain in the APH(3′) enzymes, and this combination of residues could provide a similar platform for adenine binding.
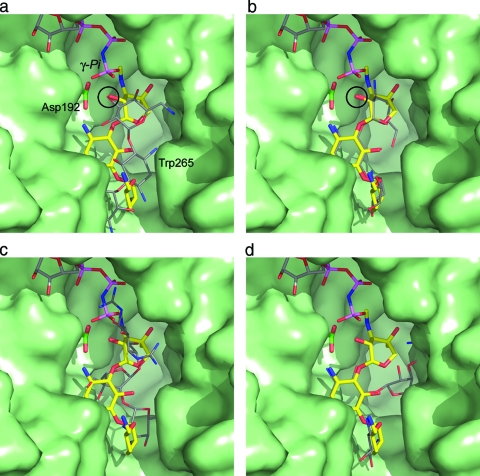
When the nucleotide from the AMPPCP-streptomycin-APH(2′′)-IIa structure is modeled into the gentamicin-APH(2′′)-IIa complex, the γ-phosphate group is approximately 7.5 Å from the 2′′ hydroxyl group of the bound gentamicin. This distance is far too long to facilitate the transfer of the phosphate group to the deprotonated hydroxyl, and such an arrangement of the triphosphate would certainly give rise to an unproductive complex were it to adopt this conformation in the presence of a substrate such as gentamicin. Conversely, an extended triphosphate conformation [based on the modeling of the AMPPNP from the superimposed AMPPNP-APH(3′)-IIIa complex (5) into the nucleotide binding site in the gentamicin-APH(2′′)-IIa complex] would put the γ phosphate approximately 2.6 Å from the 2′′ hydroxyl of the gentamicin, within range for effective phosphoryl transfer (Fig. 5a). However, the question as to why the triphosphate has adopted the observed uncommon and unproductive chelate conformation still remains. An analysis of the structures in the Protein Data Bank which contain an AMPPCP shows that while very few have been reported, most are complexes with kinase domains, either from FGF receptor 2 (FGFR2) (8, 9) or human RSK-1 (24). Inspection of the bound AMPPCP in five FGFR2 structures shows that in all cases, the triphosphate adopts an extended conformation, interacting with two magnesium ions. In the case of human RSK-1, the triphosphate adopts a loose chelated structure around a single magnesium. In the preparation of the FGFR2 complexes, a fivefold excess of Mg2+ was used, whereas in the AMPPCP-RSK-1 and the ternary AMPPCP-streptomycin-APH(2′′)-IIa complex, AMPPCP and MgCl2 were in a 1:1 ratio. In both of these cases, the presence of a chelated triphosphate is most likely an artifact of the limiting amount of magnesium rather than a disruption of any potential hydrogen bonding pattern caused by the methylene bridge (6).
FIG. 5.
Surface representation of the substrate binding site in APH(2′′)-IIa (green) showing the bound gentamicin (yellow sticks) and an AMPPNP molecule (colored sticks; top) from APH(3′)-IIIa modeled into the structure based upon the superposition of the catalytic aspartate loop [residues 189 to 197 in APH(2′′)-IIa; 187 to 195 in APH(3′)-IIIa]. (a) The kanamycin from APH(3′)-IIa is shown as thin gray bonds. APH(3′)-IIa was superimposed on APH(2′′)-IIa based upon the catalytic aspartate loop. The location of this residue (Asp192) is indicated. When the structures are superimposed in this way, the hydroxyl group which is phosphorylated on both substrates is in essentially the same position (indicated by the black circle). (b) The kanamycin molecule modeled in the same conformation as gentamicin. In this orientation, the 2′′-OH of the kanamycin overlaps the equivalent atom on gentamicin (black circle). (c) The superimposed streptomycin from molecule B of the ternary AMPPCP-streptomycin-APH(2′′)-IIa complex, shown as thin gray sticks. The streptomycin occupies approximately the same location as the gentamicin. One of the guanidinium groups on streptomycin A ring projects upwards toward the nucleotide binding site and would clearly interfere with the β- and γ-phosphate groups of an extended ATP triphosphate. (d) A neomycin molecule modeled in the gentamicin binding site such that the A and B rings occupy approximately the same position as the corresponding rings in gentamicin. The C projects toward the helical subdomain, and in this orientation, the D ring of neomycin is hidden behind the helical subdomain.
Despite being in almost identical locations with respect to the catalytically relevant residues (the HXDXXXXN and GXIDXG kinase fingerprint motifs), the substrate binding sites differ markedly between the two phosphotransferase subfamilies, primarily due to differences in the helical subdomain. Eleven out of the twelve potential hydrogen bonding interactions with gentamicin are with residues from the core subdomain, and the helical subdomain plays only a minor role in substrate binding. This is in distinct contrast to the case for the kanamycin binding site in the APH(3′′) enzymes, where more than half of the interactions with the substrate involve residues from the helical subdomain (18, 37). The long acidic loop which in the APH(3′) enzyme connects the α5 and α6 helices and serves as part of the kanamycin binding site (18, 37) is missing in APH(2′′)-IIa. The corresponding loop is very short and is approximately 10 Å from the gentamicin binding site, playing no role in substrate binding. An important difference between the two families of enzymes occurs at the C terminus. The APH(3′) enzymes end with an α helix [helix α8 in APH(3′)-IIa] and a highly conserved motif with the consensus sequence DExF (see Figure S1 in the supplemental material), with the terminal phenylalanine residue and the conserved aspartate playing key roles in substrate binding (37, 43). The corresponding helix in APH(2′′)-IIa (helix α9) is significantly longer and forms two sides of the substrate binding cleft. This creates a much more restricted binding pocket in APH(2′′)-IIa and limits this enzyme to binding only the 4,6-disubstitued aminoglycosides with three rings, whereas APH(3′)-IIIa is able to accommodate both classes, including the bulky four-ringed neomycin (18).
As noted above, in APH(2′′)-IIa, the gentamicin molecule is “upside-down” relative to the kanamycin. However, modeling kanamycin into the APH(2′′)-IIa substrate binding site in the same conformation as gentamicin shows that kanamycin is readily able to adopt a configuration similar to that seen for gentamicin (Fig. 5b), and this may account for the similarity in activity toward these two substrates. APH(2′′)-IIa shows activity toward a number of 4,6-disubstituted aminoglycosides, including kanamycin, tobramycin, netilmicin, dibekacin, amikacin, arbekacin, and isepamicin, but not the 4,5-disubstituted aminoglycosides molecules. The rates of reaction for the first four substrates (around 2 × 107 M−1 s−1) (45) are only about an order of magnitude lower than the diffusion limit, with the latter three substrates having rates ranging from 3 × 106 to 1 × 105 M−1 s−1. These molecules all have similar structures, particularly with respect to the A and B rings (the neamine group) (49; Fig. 1), whereas their C rings are either of the gentamicin type, with both a methyl group and a hydroxyl at the 4′′ carbon and a 3′′ secondary amine, or the kanamycin type, with hydroxyl groups at the 4′′ and 5′′ positions and a primary amine on the 3′′ carbon (Fig. 1). The lack of specific interactions with the C ring substituents makes APH(2′′)-IIa easily able to accommodate the different ring structures, and the differences in the substituents on the C ring do not affect the catalytic viability. This lack of specificity for the C ring explains why APH(2′′)-IIa is so promiscuous with respect to 2′′ hydroxyl phosphorylation.
A comparison of the complexes of APH(2′′)-IIa with gentamicin and streptomycin gives some insight as to why the 4,6-disubstituted aminoglycosides are substrates whereas the 4,5-disubstituted aminoglycosides are not (45). Although streptomycin is not a typical aminoglycoside, if the ribose ring is taken as the central ring, then streptomycin resembles a 4,5-disubstituted aminoglycoside, such as neomycin. In the ternary complex, streptomycin binds in a similar location to gentamicin, albeit “upside-down.” The B and C rings of streptomycin and the neamine moiety of gentamicin form a rigid unit which binds in a specific orientation at the “base” of the aminoglycoside cleft. The angular distance between the A and C rings then becomes the critical factor in determining whether the C ring (or the A ring in streptomycin) can be brought close enough to the catalytic aspartate. In gentamicin (and the 4,6-disubstituted aminoglycosides in general), the molecule is easily able to adopt an extended conformation with a large angular spacing between the A and C rings, whereas in streptomycin, where the A and C rings are attached to adjacent positions on the central modified ribose, there is a much smaller range of motion of the A ring and consequently it must sit further from the core subdomain (Fig. 5c). The same would be true of the 4,5-disubstituted aminoglycosides, such as neomycin, assuming the molecules were to bind in a fashion similar to that of gentamicin, with the neamine moiety attached to the “base” of the cleft. The C ring would project toward the helical domain (Fig. 5d) and although it would be possible for the 5′′ hydroxyl to come within hydrogen bonding distance of the catalytic aspartate, this orientation would place the C and B rings too close together, such that it would be a sterically unfavored conformation.
The neamine moiety of the gentamicin (and related aminoglycosides) is responsible for recognition of the target RNA A site (49). Gentamicin binds in the major groove of the A site, where the A ring stacks against G15 (equivalent to G1491 in the 30S ribosomal A site) and is anchored by a pseudo-base pair to A30 (A1408) (19). The bases A16 and A17 (A1492 and A1493) are forced out of the helix. The B ring forms two hydrogen bonds, with G18 and U19 (G1494 and U1495). These interactions are identical to those made by paromomycin in complex with the 30S ribosomal subunit (7). The majority of the aminoglycoside-modifying enzymes target functionalities on the neamine ring, and this two-ring group is thus the basis for the decreased binding affinity of the modified drugs. The APH(2′′) enzymes, however, modify on the C ring, but since this ring interacts with the phosphate backbone of the RNA, a phosphate group on the 2′′ hydroxyl would destabilize the interaction, giving rise to the decreased binding affinity for drugs modified at this position. The similarity of the binding orientations of the aminoglycosides to the target A site and the deactivating enzymes, such as APH(2′′)-IIa could create an obstacle in the development of novel aminoglycosides unaffected by phosphorylation. Any changes made in the neamine ring, for example, would clearly affect binding to the enzyme but would also conceivably have a detrimental effect on binding to the ribosome. One possibility to circumvent the activity of at least the APH(2′′) family might be to add a bulky group, such as a guanidinium functionality (as in streptomycin), to the C ring. A neamine-containing aminoglycoside modified in this way would most likely still bind to the recognition site at the “base” of the APH(2′′)-IIa binding cleft, and the added guanidinium moiety could restrict efficient interaction between potential sites of phosphorylation on the substrate and the γ phosphate of ATP.
Supplementary Material
Acknowledgments
This work was supported by a program grant from the Health Research Council of New Zealand (E.N.B., P.M., and C.A.S.) and by National Institutes of Health grant AI057393 (S.B.V.). The SSRL Structural Molecular Biology Program is supported by the Department of Energy (Office of Basic Energy Sciences, Biological and Environmental Research) and by the National Institutes of Health (National Center for Research Resources, Biotechnology Training Program, National Institute of General Medical Sciences). SSRL is a national user facility operated by Stanford University on behalf of the U.S. Department of Energy, Office of Basic Energy Sciences. Project support was provided by grant 5 P41 RR001209 from the National Center for Research Resources. L.J.B. was supported by a Summer Undergraduate Laboratory Internship.
Footnotes
Published ahead of print on 1 May 2009.
Supplemental data for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Aimes, R. T., W. Hemmer, and S. S. Taylor. 2000. Serine-53 at the tip of the glycine-rich loop of cAMP-dependent protein kinase: role in catalysis, P-site specificity, and interaction with inhibitors. Biochemistry 398325-8332. [DOI] [PubMed] [Google Scholar]
- 2.Azucena, E., and S. Mobashery. 2001. Aminoglycoside-modifying enzymes: mechanisms of catalytic processes and inhibition. Drug Resist. Updates 4106-117. [DOI] [PubMed] [Google Scholar]
- 3.Berman, H. M., J. Westbrook, Z. Feng, G. Gilliland, T. N. Bhat, H. Weissig, I. N. Shindyalov, and P. E. Bourne. 2000. The Protein Data Bank. Nucleic Acids Res. 28235-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burk, D. L., and A. M. Berghuis. 2002. Protein kinase inhibitors and antibiotic resistance. Pharmacol. Ther. 93283-292. [DOI] [PubMed] [Google Scholar]
- 5.Burk, D. L., W. C. Hon, A. K. Leung, and A. M. Berghuis. 2001. Structural analysis of nucleotide binding to an aminoglycoside phosphotransferase. Biochemistry 408756-8764. [DOI] [PubMed] [Google Scholar]
- 6.Bystrom, C. E., D. W. Pettigrew, S. J. Remington, and B. P. Branchard. 1997. ATP analogs with non-transferable groups in the γ-position as inhibitors of glycerol kinase. Bioorg. Med. Chem. Lett. 72613-2616. [Google Scholar]
- 7.Carter, A. P., W. M. Clemons, D. E. Broderson, R. J. Morgan-Warren, B. T. Wimberly, and V. Ramakrishnan. 2000. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature 407340-348. [DOI] [PubMed] [Google Scholar]
- 8.Chen, H., J. Ma, W. Li, A. V. Eliseenkova, C. Xu, T. A. Neubert, W. T. Miller, and M. Mohammadi. 2007. A molecular brake in the kinase hinge region regulates the activity of receptor tyrosine kinases. Mol. Cell 27717-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, H., C. F. Xu, J. Ma, A. V. Eliseenkova, W. Li, P. M. Pollock, N. Pitteloud, W. T. Miller, T. A. Neubert, and M. Mohammadi. 2008. A crystallographic snapshot of tyrosine trans-phosphorylation in action. Proc. Natl. Acad. Sci. USA 10519660-19665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, Y.-G., T.-T. Qu, Y.-S. Yu, J.-Y. Zhou, and L.-J. Li. 2006. Insertion sequence ISEcp1-like element connected with a novel aph(2′′) allele [aph(2′′)-Ie] conferring high-level gentamicin resistance and a novel streptomycin adenylyltransferase gene in Enterococcus. J. Med. Microbiol. 551521-1525. [DOI] [PubMed] [Google Scholar]
- 11.Chow, J. W., M. J. Zervos, S. A. Lerner, L. A. Thal, S. M. Donabedian, D. D. Jaworski, S. Tsai, K. J. Shaw, and D. B. Clewell. 1997. A novel gentamicin resistance gene in Enterococcus. Antimicrob. Agents Chemother. 41511-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collaborative Computing Project, Number 4. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. 50(Pt. 5)760-763. [DOI] [PubMed] [Google Scholar]
- 13.Davies, J., and G. D. Wright. 1997. Bacterial resistance to aminoglycoside antibiotics. Trends Microbiol. 5234-240. [DOI] [PubMed] [Google Scholar]
- 14.DeLano, W. L. 2002. The PyMOL molecular graphics system. DeLano Scientific, San Carlos, CA.
- 15.Doublié, S. 1997. Preparation of selenomethionyl proteins for phase determination. Methods Enzymol. 276523-530. [PubMed] [Google Scholar]
- 16.Emsley, P., and K. Cowtan. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 602126-2132. [DOI] [PubMed] [Google Scholar]
- 17.Ferretti, J. J., K. S. Gilmore, and P. Courvalin. 1986. Nucleotide sequence analysis of the gene specifying the bifunctional 6′-aminoglycoside acetyltransferase 2′′-aminoglycoside phosphotransferase enzyme in Streptococcus faecalis and identification and cloning of gene regions specifying the two activities. J. Bacteriol. 167631-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fong, D., and A. M. Berghuis. 2002. Substrate promiscuity of an aminoglycoside antibiotic resistance enzyme via target mimicry. EMBO J. 212323-2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.François, B., R. J. Russell, J. B. Murray, F. Aboul-ela, B. Masquida, Q. Vicens, and E. Westhof. 2005. Crystal structures of complexes between aminoglycosides and decoding A site oligonucleotides: role of the number of rings and positive charges in the specific binding leading to miscoding. Nucleic Acids Res. 335677-5690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Greenwood, D. 1995. Antimicrobial chemotherapy, 3rd ed., p. 32-48. Oxford University Press, Oxford, United Kingdom.
- 21.Hanks, S. K., and T. Hunter. 1995. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J. 9576-596. [PubMed] [Google Scholar]
- 22.Hanks, S. K., A. M. Quinn, and T. Hunter. 1988. The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science 24142-52. [DOI] [PubMed] [Google Scholar]
- 23.Hon, W. C., G. A. McKay, P. R. Thompson, R. M. Sweet, D. S. C. Yang, G. D. Wright, and A. M. Berghuis. 1997. Structure of an enzyme required for aminoglycoside antibiotic resistance reveals homology to eukaryotic protein kinases. Cell 89887-895. [DOI] [PubMed] [Google Scholar]
- 24.Ikuta, M., M. Kornienko, N. Byrne, J. C. Reid, S. Mizuarai, H. Kotani, and S. K. Munshi. 2007. Crystal structures of the N-terminal kinase domain of human RSK1 bound to three different ligands: Implications for the design of RSK1 specific inhibitors. Protein Sci. 162626-2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson, L. N., M. E. M. Noble, and D. J. Owen. 1996. Active and inactive protein kinases: structural basis for regulation. Cell 85149-158. [DOI] [PubMed] [Google Scholar]
- 26.Kabsch, W. 1993. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 26795-800. [Google Scholar]
- 27.Kao, S. J., I. You, D. B. Clewell, S. M. Donabedian, M. J. Zervos, J. Petrin, K. J. Shaw, and J. W. Chow. 2000. Detection of the high-level aminoglycoside resistance gene aph(2′′)-Ib in Enterococcus faecium. Antimicrob. Agents Chemother. 442876-2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karimi, R., and M. Ehrenberg. 1994. Dissociation rate of cognate peptidyl-tRNA from the A-site of hyper-accurate and error-prone ribosomes. Eur. J. Biochem. 226355-360. [DOI] [PubMed] [Google Scholar]
- 29.Kim, C., and S. Mobashery. 2005. Phosphoryl transfer by aminoglycoside 3′-phosphotransferases and manifestation of antibiotic resistance. Bioorg. Chem. 33149-158. [DOI] [PubMed] [Google Scholar]
- 30.Kocabiyik, S., and M. H. Perlin. 1992. Site-specific mutations of conserved C-terminal residues in aminoglycoside 3′-phosphotransferase II: phenotypic and structural analysis of mutant enzymes. Biochem. Biophys. Res. Commun. 185925-931. [DOI] [PubMed] [Google Scholar]
- 31.Kotra, L. P., J. Haddad, and S. Mobashery. 2000. Aminoglycosides: perspectives on mechanisms of action and resistance and strategies to counter resistance. Antimicrob. Agents Chemother. 443249-3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krissinel, E., and K. Henrick. 2004. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D 602256-2268. [DOI] [PubMed] [Google Scholar]
- 33.Martel, A., M. Masson, N. Moreau, and F. LeGoffic. 1983. Kinetic studies of aminoglycoside acetyltransferase and phosphotransferase from Staphylococcus aureus RPAL: relationship between the two activities. Eur. J. Biochem. 133515-521. [DOI] [PubMed] [Google Scholar]
- 34.Matthews, B. W. 1968. Solvent contents of protein crystals. J. Mol. Biol. 33491-497. [DOI] [PubMed] [Google Scholar]
- 35.McCoy, A. J., R. W. Grosse-Kunstleve, L. C. Storoni, and R. J. Read. 2005. Likelihood-enhanced fast translation functions. Acta Crystallogr. D 61458-464. [DOI] [PubMed] [Google Scholar]
- 36.Murshudov, G. N., A. A. Vagin, and E. J. Dodson. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53240-255. [DOI] [PubMed] [Google Scholar]
- 37.Nurizzo, D., S. C. Shewry, M. H. Perlin, S. A. Brown, J. N. Dholakia, R. L. Fuchs, T. Deva, E. N. Baker, and C. A. Smith. 2003. The crystal structure of aminoglycoside-3′-phosphotransferase-IIa, an enzyme responsible for antibiotic resistance. J. Mol. Biol. 327491-506. [DOI] [PubMed] [Google Scholar]
- 38.Shaw, K. J., P. N. Rather, R. S. Hare, and G. H. Miller. 1993. Molecular genetics of aminoglycoside resistance genes and familial relationships of the aminoglycoside-modifying enzymes. Microbiol. Rev. 57138-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith, C. A., and E. N. Baker. 2002. Aminoglycoside antibiotic resistance by enzymatic deactivation. Curr. Drug Targets Infect. Disord. 2143-160. [DOI] [PubMed] [Google Scholar]
- 40.Terwilliger, T. C. 2003. Automated main-chain model building by template matching and iterative fragment extension. Acta Crystallogr. D 5938-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Terwilliger, T. C. 2000. Maximum likelihood density modification. Acta Crystallogr. D 56965-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Terwilliger, T. C., and J. Berendzen. 1999. Automated MAD and MIR structure solution. Acta Crystallogr. D 55849-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thompson, P. R., J. Schwartzenhauer, D. W. Hughes, A. M. Berghuis, and G. D. Wright. 1999. The COOH terminus of aminoglycoside phosphotransferase(3′)-IIIa is critical for antibiotic recognition and resistance. J. Biol. Chem. 27430697-30706. [DOI] [PubMed] [Google Scholar]
- 44.Toth, M., J. W. Chow, S. Mobashery, and S. B. Vakulenko. 2009. Source of phosphate in the enzymatic reaction as a point of distinction among aminoglycoside 2′′-phosphotransferases. J. Biol. Chem. 2846690-6696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Toth, M., J. Zajicek, C. Kim, J. W. Chow, C. A. Smith, S. Mobashery, and S. Vakulenko. 2007. Kinetic mechanism of enterococcal aminoglycoside phosphotransferase 2′′-Ib. Biochemistry 465570-5578. [DOI] [PubMed] [Google Scholar]
- 46.Tsai, S., M. J. Zervos, D. B. Clewell, S. M. Donabedian, D. F. Sahm, and J. W. Chow. 1998. A new high-level gentamicin resistance gene, aph(2′′)-Id, in Enterococcus spp. Antimicrob. Agents Chemother. 421229-1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vakulenko, S. B., and S. Mobashery. 2003. Versatility of aminoglycosides and prospects for their future. Clin. Microbiol. Rev. 16430-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Valiev, M., R. Kawai, J. A. Adams, and J. H. Weare. 2003. The role of the putative catalytic base in the phosphoryl transfer reaction in a protein kinase: first-principles calculations. J. Am. Chem. Soc. 1259926-9927. [DOI] [PubMed] [Google Scholar]
- 49.Vicens, Q., and E. Westhof. 2003. Molecular recognition of aminoglycoside antibiotics by ribosomal RNA and resistance enzymes: an analysis of x-ray crystal structures. Biopolymers 7042-57. [DOI] [PubMed] [Google Scholar]
- 50.Walanj, R., P. Young, H. M. Baker, E. N. Baker, P. Metcalf, J. W. Chow, S. Lerner, S. Vakulenko, and C. A. Smith. 2005. Purification, crystallization and preliminary X-ray analysis of Enterococcus faecium aminoglycoside-2′′-phosphotransferase-Ib [APH(2′′)-Ib]. Acta Crystallogr. F 61410-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wirmer, J., and E. Westhof. 2006. Molecular contacts between antibiotics and the 30S ribosomal particle. Methods Enzymol. 415180-202. [DOI] [PubMed] [Google Scholar]
- 52.Wright, G. D. 1999. Aminoglycoside-modifying enzymes. Curr. Opin. Microbiol. 2499-503. [DOI] [PubMed] [Google Scholar]
- 53.Wright, G. D., A. M. Berghuis, and S. Mobashery. 1998. Aminoglycoside antibiotics: structures, functions, and resistance. Adv. Exp. Med. Biol. 45627-69. [PubMed] [Google Scholar]
- 54.Wright, G. D., and P. R. Thompson. 1999. Aminoglycoside phosphotransferases: proteins, structure and mechanism. Front. Biosci. 4D9-D21. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.