Abstract
Staphylococcus aureus is a common human cutaneous and nasal commensal and a major life-threatening pathogen. Adaptation to the different environments encountered inside and outside the host is a crucial requirement for survival and colonization. We identified and characterized a eukaryotic-like serine/threonine kinase with three predicted extracellular PASTA domains (SA1063, or Stk1) and its associated phosphatase (SA1062, or Stp1) in S. aureus. Biochemical analyses revealed that Stk1 displays autokinase activity on threonine and serine residues and is localized to the membrane. Stp1 is a cytoplasmic protein with manganese-dependent phosphatase activity toward phosphorylated Stk1. In-frame deletions of the stk1 and stp1 genes were constructed in S. aureus strain 8325-4. Phenotypic analyses of the mutants revealed reduced growth of the stk1 mutant in RPMI 1640 defined medium that was restored when adenine was added to the medium. Furthermore, the stk1 mutant displayed increased resistance to Triton X-100 and to fosfomycin, suggesting modifications in cell wall metabolism. The stk1 mutant was tested for virulence in a mouse pyelonephritis model and found to be strongly reduced for survival in the kidneys (approximately 2-log-unit decrease) compared to the parental strain. Renal histopathological analyses showed severe inflammatory lesions in mice infected with the parental S. aureus SH1000 strain, whereas the Δstk1 mutant led to only minimal renal lesions. These results confirm the important role of Stk1 for full expression of S. aureus pathogenesis and suggest that phosphorylation levels controlled by stk1 are essential in controlling bacterial survival within the host.
Protein phosphorylation has been evolutionarily conserved from Escherichia coli to Homo sapiens and plays a role in a multitude of cellular processes, including proliferation, differentiation, and development. Protein phosphorylation usually results in a functional change of the target protein by modifying enzyme activity, cellular location, or association with other biomolecules (proteins, DNA, or RNA). Phosphorylation/dephosphorylation of serine, threonine, or tyrosine by the respective kinases and phosphatases is used to control many important cellular processes in eukaryotes. In prokaryotes, signal transduction often involves two-component systems, which are composed of a histidine kinase sensing module and its cognate transcriptional regulator. Analysis of the Staphylococcus aureus N315 genome sequence predicted 17 potential two-component systems in S. aureus, only one of which, WalK/WalR, is known to be essential for bacterial viability (12).
An increasing number of reports describe the existence of eukaryotic-type serine/threonine kinases and phosphatases in prokaryotes. Genes encoding these kinases and phosphatases are often linked and have been identified in soil bacteria, such as Myxococcus spp. (31, 32), Streptomyces spp. (39), Corynebacterium glutamicum (16), and Bacillus spp. (30), as well as in bacterial pathogens, such as Mycobacterium tuberculosis (18, 50), Streptococcus agalactiae (41), Streptococcus pneumoniae (14, 35, 45), Streptococcus pyogenes (21), Streptococcus mutans (20), Listeria monocytogenes (1, 2), and Enterococcus faecalis (23).
Global phosphoproteome analyses of Bacillus subtilis revealed several potential targets for Ser/Thr kinases involved in processes such as carbohydrate and energy metabolism, transport, stress, and development (15, 27, 29). These results indicate that phosphorylation on serine and threonine residues is more widespread than previously thought. In prokaryotes, many Ser/Thr kinases have now been characterized and shown to regulate various cellular functions, such as developmental processes (30, 33), secondary metabolism (49), stress response (34), biofilm formation (20), antibiotic resistance (23), virulence (8), cell wall biogenesis, cell division, and central metabolism (50). However, a detailed understanding of their mechanisms of action, as well as their specific substrates and precise roles, remains largely elusive. Only a few substrates of specific Ser/Thr kinases and phosphatases have been identified so far, e.g., B. subtilis elongation factor EF-G (17) and S. agalactiae inorganic pyrophosphatase PpaC (41). Several mycobacterial Stk1 substrates have been identified, including transcription regulators and enzymes of the fatty acid synthase II complex (50). In L. monocytogenes, Stp plays a role in virulence, and two substrates were identified: the elongation factor EF-Tu and Sod, the manganese-dependent superoxide dismutase (1, 2).
S. aureus is a major human pathogen that causes a number of diseases ranging from skin infections to toxic shock syndrome, osteomyelitis, and myocarditis. Recently, a phosphoproteome analysis of S. aureus strain N315 identified nine glycolytic enzymes as substrates of the endogenous Ser/Thr kinase(s) (28).
In the present work, we investigate the roles of the Stk1/Stp1 Ser/Thr kinase and phosphatase of S. aureus. We demonstrate an autophosphorylation activity for Stk1 and show that the Stk1 kinase can be dephosphorylated by the Mn2+-dependent phosphatase Stp1. Interestingly, an stk1 deletion mutant displayed increased resistance to Triton X-100 and to fosfomycin, suggesting modifications in cell wall metabolism, in agreement with the presence of three potential extracellular PASTA (PBP and serine/threonine kinase-associated) domains. We also show that the stk1 mutant is strongly impaired for survival in a murine pyelonephritis model, with renal histopathological analyses showing minimal lesions compared to the severe inflammatory lesions and necrosis seen in mice infected with the parental SH1000 strain, indicating that the Stk1 kinase plays a major role in virulence.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth media.
The S. aureus strains used in this study are listed in Table 1. E. coli K-12 strain DH5α [λ− ö80dlacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rK− mK−) supE44 thi-1 gyrA relA1] (Invitrogen) was used for cloning experiments. E. coli BL21 λ DE3 was used for expression of recombinant proteins (47). The plasmid vector pET15 was purchased from Novagen. The pMAD shuttle vector was used for gene replacement (3).
TABLE 1.
S. aureus strains and plasmids used in this study
| Strain or plasmid | Genotype or description | Source or reference |
|---|---|---|
| Strains | ||
| RN4220 | Restriction-deficient transformation recipient | Laboratory stock |
| 8325-4 | Reference strain rsbU mutant cured of known prophages | 38 |
| SH1000 | 8325-4 rsbU+ | 19 |
| N315 | Methicillin-resistant strain | 24 |
| ST1003 | 8325-4 stp1 | This work |
| ST1004 | 8325-4 stk1 | This work |
| ST1005 | SH1000 stk1 | This work |
| ST1006 | 8325-4 pMK4-Pprot | This work |
| ST1007 | 8325-4 pC2 | This work |
| ST1008 | 8325-4 stk1 pC2 | This work |
| ST1010 | 8325-4 stk1 pMK4-Pprot | This work |
| Plasmids | ||
| pMAD | pE194 derivative with a thermosensitive origin of replication for deletion replacement of genes in gram-positive bacteria | 3 |
| pMK4-Pprot | pMK4 derivative with a constitutive promoter | 1 |
| pET15b | E. coli expression vector | Novagen |
| pC2 | pMK4-Pprot derivative carrying the stk1 gene with an HA tag | This work |
| pET-Stk1 | pET15b derivative for Stk1 overproduction | This work |
| pET-Stp1 | pET15b derivative for Stp1 overproduction | This work |
S. aureus was cultured in Trypticase soy broth (TSB) or agar (TSA) and E. coli in Luria-Bertani medium. Antibiotics were used at the following concentrations: for E. coli, 100 μg/ml ampicillin; for S. aureus, 10 μg/ml chloramphenicol and 1 μg/ml erythromycin.
General DNA techniques.
The oligonucleotides used in this study are shown in Table 2. Standard techniques were used for nucleic acid cloning and restriction analysis (44). Plasmid DNA from E. coli was prepared by rapid alkaline lysis using the Midiprep Plasmid Kit (Qiagen). Genomic DNA from S. aureus was prepared using the MasterPure Gram Positive DNA Purification Kit (Epicentre Biotechnologies). PCR amplifications were carried out with Pwo polymerase as recommended by the manufacturer (Roche). Amplification products were purified using the QIAquick PCR purification kit (Qiagen), and nucleotide sequencing of plasmid constructs was carried out by Genome Express-Cogenics.
TABLE 2.
Oligonucleotides used in this study
| Name | Sequence | Description |
|---|---|---|
| OMD208 | GAAGAATTCGAATGAAGATGCGGGTGG | stk1 deletion |
| OMD224 | CTCCTCGAGTATCATACTTTATCACCTTCA | stk1 deletion |
| OMD227 | CTCCTCGAGGATATTTAAATATAATTGAAG | stk1 deletion |
| OMD228 | GGAGGATCCGTCCAGCTGGCCAAGCTTCTA | stk1 deletion |
| OMD237 | GAAGAATTCATAATGGGTATCGGTGAACC | stp1 deletion |
| OMD238 | CTCCTCGAGTAGCATTTGTCTTTACCTCGT | stp1 deletion/RT-PCR |
| OMD239 | CTCCTCGAGGGTGATAAAGTATGATAGGTA | stp1 deletion |
| OMD240 | GGAGGATCCATCCGTTGCCTCACCTTTTGC | stp1 deletion |
| OMD212 | GCAAATGTCGGTGATTCTAGAGCCTATG | RT-PCR |
| OMD225 | CTCCTCGAGGCCACCGCCGCCAAGCTTATC | RT-PCR |
| OMD263 | GACGGTTCAACTGAAAAAGGTAGTTTCGAC | RT-PCR |
| OMD264 | CGCCATTAACGTCTACTTGATATACCCCAC | RT-PCR |
| OMD289 | CCTGTCAACCATGTTCCAGAAAG | RT-PCR |
| OMD295 | CAACATCCGATAACTGGAGAGTTGGTC | RT-PCR |
| OMD296 | CGCTCGAAATTTTTGTTGACCTTGTTCAAC | RT-PCR |
| OMD298 | CGTGGATTATCACCATGGCCAGTTGC | RT-PCR |
| OMD301 | TGTCATAACACGTTTACGTCCAACAGGTCG | RT-PCR |
| OMD305 | CGGTATCCCTTTTACAGAACGTGCGGACC | RT-PCR |
| OMD314 | AGAAGATCTGTGCGGACCGTATTTTAACAG | RT-PCR |
| OMD331 | CAAAAAGCCCTTGATGCTACAGAAGAGCGC | RT-PCR |
| OMD332 | GTATGTGTAACGACACAAGTAGGTTGTCG | RT-PCR |
| OMD337 | GGAGGATCCATACTCGCGGCTATTGAAGGT | stk1 complementation |
| OMD339 | TGCGGTCGACTTAGGCGTAGTCGGGGACGTTAGGGGTAAATATCATCATAGCTGACTTC | stk1 complementation HA tag |
| OBDk1 | GGAATTCCATATGATAGGTAAAATAATAAATGAAC | Stk1 overproduction |
| OBDk2 | CGCGGATCCTTATACATCATCATAGCTGAC | Stk1 overproduction |
| OBDp1 | GGAATTCCATATGGCAATTACAAAAATCAATGATT | Stp1 overproduction |
| OBDp2 | CGCGGATCCTTATAGTTCAATTTCGTTGTT | Stp1 overproduction |
Expression and purification of recombinant proteins.
DNA fragments corresponding to the entire coding sequences of stk1 (SA1063) and stp1 (SA1062) were produced using genomic DNA of S. aureus strain N315 as a template and the following oligonucleotide pairs: for stk1, OBDk1/OBDk2, and for stp1, OBDp1/OBDp2. The amplified DNA fragments were digested with NdeI and BamHI and cloned into pET15b. The resulting plasmids, pET-Stk1 and pET-Stp1, were introduced into E. coli strain BL21 λ DE3 for protein expression. The six-His-Stk1 and six-His-Stp1 recombinant proteins were purified under native conditions by affinity chromatography on Zn2+ columns according to the manufacturer's recommendations (Novagen). Protein purity was checked by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and protein concentrations were determined using the Coomassie Plus Protein Assay (Pierce).
In vitro phosphorylation assay.
In vitro phosphorylation of about 2 μg of purified six-His-Stk1 protein was performed for 20 min at 37°C in 20 μl of a buffer containing 25 mM Tris-HCl (pH 7.0), 1 mM dithiothreitol, 1 mM EDTA, 5 mM MnCl2, and 200 μCi [γ-32P]ATP/ml. In some assays, MnCl2 was replaced with CaCl2 or ZnCl2. In each case, the reaction was stopped by the addition of an equal volume of 2× sample buffer (25).
Phosphatase assay.
Phosphatase activity was monitored at 37°C by using a continuous method based on the detection of p-nitrophenol formed from p-nitrophenyl phosphate (pNPP). Rates of dephosphorylation were determined by measuring absorbance at 405 nm in a reaction buffer containing Tris-HCl, pH 7.5, 0.1% β-mercaptoethanol, and pNPP at concentrations varying from 0.5 to 40 mM. The amounts of p-nitrophenol released were determined using a range standard established under the same conditions used in the assay.
Stp1 dephosphorylation assay.
In vitro phosphorylation of about 0.5 μg of purified Stk1 protein was performed as described above. After 10 min of incubation, a dephosphorylation assay of Stk1 was carried out with 0.1 μg of purified Stp1 at 37°C for 2 to 30 min in 30 μl of a buffer consisting of 100 mM sodium citrate (pH 6.5) and 1 mM EDTA. The reaction was stopped by the addition of an equal volume of 2× sample buffer, and the mixture was heated at 100°C for 5 min. The Stk1 protein was separated by gel electrophoresis, treated with TCA, and analyzed by autoradiography.
Phosphoamino acid content analysis.
Determination of phosphoresidues was carried out using two-dimensional migration. After phosphorylation, proteins were separated by SDS-PAGE and transferred onto nylon membranes as previously described (13). The radioactive spot was cut out of the membrane and hydrolyzed by 6 M HCl for 1 h at 110°C. The acid-stable phosphoamino acids thus liberated were separated by electrophoresis in the first dimension at pH 1.9 (800 V h−1) in 7.8% acetic acid/2.5% formic acid, followed by ascending chromatography in the second dimension in 2-methyl-1-propanol/formic acid/water (8:3:1). After migration, radioactive molecules were detected by autoradiography. The phosphoamino acid standards, phosphoserine, phosphothreonine, and phosphotyrosine, were separated in parallel and revealed by ninhydrin staining. The pattern of phosphoamino acids revealed by ninhydrin was compared with that revealed by autoradiography, in order to determine the nature of the phosphoamino acid.
stp1 and stk1 mutant construction.
In-frame deletions in stp1 and stk1 were constructed as follows. For each gene inactivation, the upstream and downstream PCR-generated DNA fragments (oligonucleotide pairs OMD237-OMD238 and OMD239-OMD240 for stp1 and OMD208-OMD224 and OMD227-OMD228 for stk1) were digested with XhoI and then ligated using T4 DNA ligase (Biolabs). The ligation product was reamplified using the external primers, purified on an agarose gel using the QIAquick Extraction Kit (Qiagen), digested with the appropriate enzymes (EcoRI and BamHI), and cloned in the thermosensitive shuttle plasmid pMAD (3). Electroporation of S. aureus strains and allelic exchange were performed as previously described (3). In-frame deletion mutants were confirmed by PCR and sequence analysis.
Site-directed mutagenesis.
Site-directed mutagenesis was carried out using the Transformer Site-Directed Mutagenesis Kit from Clontech as recommended by the manufacturer. This procedure was applied directly to the pET-Stk1 construct. The oligonucleotide primers used for mutagenesis were as follows: K39G, 5′-CATTAAAGTTGCAATTGCGGCGATTTTTATAC-3′, and an EcoRI restriction site-eliminating oligonucleotide, 5′-GTTATATCCCGCCGAATTCCACCATCAAAC-3′. The presence of the mutation was verified by DNA sequencing.
Complementation with stk1-HA.
The entire coding sequence of stk1 containing an in-frame hemagglutinin (HA) tag at the 3′ end (stk1-HA) was amplified by PCR from 8325-4 genomic DNA using primers OMD337 and OMD339. After digestion with the appropriate enzymes (SalI and BamHI), the PCR product was cloned into the shuttle vector pMK4-Pprot (1). The construction was verified by sequencing the insert. The resulting plasmid, pC2, was introduced into both the wild-type S. aureus strain 8325-4 and the isogenic Δstk1 strain by electroporation, giving rise to strains ST1007 and ST1008, respectively.
RNA isolation and cDNA synthesis.
Bacteria were grown in TSB (20 ml) to an optical density at 600 nm (OD600) of ∼0.8. The bacteria were pelleted and immediately frozen at −20°C. RNA extractions were then performed as previously described (6), followed by DNase I treatment with the Turbo-DNA-free reagent (Ambion, Austin, TX) in order to eliminate residual contaminating genomic DNA. cDNA synthesis was performed as previously described (12).
Bacterial fractionation and immunoblotting.
S. aureus strain ST1007 was grown to exponential phase in TSB (50 ml), to an OD600of 0.8, and harvested. The supernatant was filtered (0.22 μm) and precipitated with 10% TCA (Sigma). The cell pellet was resuspended in 500 μl of digestion buffer (150 mM NaCl, 20 mM MgCl2, 30% raffinose) containing 0.5 μg ml−1 lysozyme and lysostaphin. The mixture was incubated at 37°C for 30 min until at least 95% of the cells were protoplasted with no detectable lysis seen by phase-contrast microscopy. The protoplasts were pelleted and resuspended in protoplast lysis buffer (100 mM Tris-HCl, pH 7.5, 100 mM NaCl, 10 mM MgCl2) for 15 min at 4°C and disrupted by multiple freeze-thaw cycles. The lysed bacteria were then centrifuged (15 min; 20,000 × g). The pellet (membrane fraction) was resuspended in 500 μl of PBS. The protein concentrations of the supernatant, cell wall, membrane, and cytosolic fractions were determined using the bicinchoninic acid protein assay (Pierce). For analysis of Stk1 production, equivalent amounts of all fractions, representing approximately 1 ml of bacterial culture, were analyzed by immunoblotting. The proteins were boiled in Laemmli sample buffer, separated on an 8% SDS-PAGE gel, and transferred to a nitrocellulose membrane. HA-tagged Stk1 was detected using a monoclonal antibody directed against the HA tag (3F9; Roche), horseradish peroxidase-coupled anti-mouse secondary antibodies (Zymed), and the Western blot Pico chemiluminescence kit (Pierce).
Zymograms.
Fifty-milliliter cultures were grown in TSB at 37°C with aeration to an OD600 of approximately 1, and total protein extracts were prepared by disrupting the cells with a FastPrep cell disintegrator (Bio 101) using the FastPrep FastProtein Blue Protocol (the FastPrep instrument was set at a speed of 6.5 for 30 s). Protein concentrations were determined using the bicinchoninic acid protein assay (Pierce). Equivalent amounts of total protein extracts were subjected to SDS-PAGE with heat-inactivated staphylococcal cells (equivalent to an OD600 of 10) as a substrate in the separation gel (12% acrylamide). Following electrophoresis, the gel was washed in water to remove the SDS and to allow protein refolding by incubation in phosphate buffer (0.1 M NaPO4; pH 7) at 37°C. Protein bands with bacteriolytic activities were detected as clear zones in the opaque gel.
Animal experiments and histopathological analyses.
Eight-week-old female CD1 mice (Janvier) were intravenously inoculated with the S. aureus parental strain and isogenic mutants. Groups of six mice were infected with 108 CFU per mouse in 0.2 ml. The animals were sacrificed 7 days postinoculation, and the kidneys were removed and homogenized for determination of the number of bacterial CFU per kidney following dilution and plating on TSA plates. For blood sampling, 10 μl of blood was collected in a heparin-containing syringe from the tail vein of each mouse at days 3, 5, and 7 postinfection. The blood was then directly plated on TSA plates, which were incubated for 24 h at 37°C before colonies were counted.
For histological observations, groups of three mice were infected with 2 × 107 CFU per mouse in 0.2 ml. The animals were sacrificed 7 days postinoculation, and the kidneys were removed and immediately fixed in 4% neutral buffered formalin for the histopathological analysis. After 48 h of fixation, the kidney was longitudinally sectioned through the hilus in order to obtain the renal cortex, medulla, crest, and pelvis on the same section. These samples were embedded in paraffin, and 5-μm sections were cut and stained with hematoxylin and eosin.
RESULTS
Identification of stk1 and stp1.
Analysis of the S. aureus N315 genome sequence revealed the existence of two adjacent genes, SA1062 and SA1063, encoding proteins sharing similarity with eukaryotic Ser/Thr phosphatases and Ser/Thr kinases, respectively. SA1062 (designated stp1) encodes a 247-amino-acid protein displaying 42% identity with PrpC of B. subtilis (36), 39% identity with Stp of L. monocytogenes (1), 38% identity with Stp1 of S. agalactiae (41), and 22% identity with PP2C of H. sapiens. These proteins belong to the PPM family, whose founding member is the eukaryotic PP2C. The Stp1 sequence shares 14% amino acid sequence identity with the PP2C-like phosphatase from H. sapiens (4). All the motifs required for phosphatase activity, in particular the positions of residues involved in metal binding, such as the aspartate residue, are conserved between Stp1 and PP2C (9). SA1063 (referred to henceforth as stk1) encodes a 664-amino-acid protein with the following characteristic features: an amino-terminal domain homologous to the catalytic domain of Ser/Thr kinases (amino acids [aa] 10 to 268), followed by a transmembrane segment (aa 350 to 372) and three PASTA domains (aa 373 to 440, 441 to 509, and 510 to 575). The important regions in the catalytic domains of STKs are conserved in Stk1, i.e., an ATP-binding site (aa 16 to 39) and a conserved aspartic residue (D126), which is crucial for the catalytic activity of the enzyme. The PASTA domain has also been found in penicillin binding proteins (PBPs), which are involved in cell wall biosynthesis. In PknB, one of several Ser/Thr kinases in M. tuberculosis, the four PASTA domains form the extracellular part of the protein and may represent the signal-binding sensor domain (52).
Stk1 displays autokinase activity.
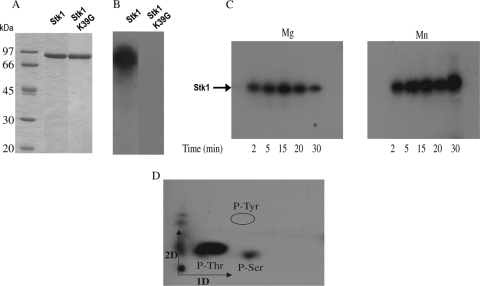
The stk1 gene was amplified and cloned into the expression vector pET15b to allow the synthesis of a recombinant protein with an amino-terminal histidyl tag at the N terminus. The six-His-Stk1 protein was produced following IPTG (isopropyl-β-d-thiogalactopyranoside) induction, and a band of about 75 kDa, consistent with the theoretical molecular mass of the recombinant protein, was purified to homogeneity in a single step using immobilized Zn2+ affinity chromatography (Fig. 1A). We included as a negative control the recombinant Stk1 mutant K39G, which is unable to autophosphorylate (B. Duclos, unpublished data). The protein was then assayed for phosphorylation in vitro. In a protein kinase assay using [γ-32P]ATP as the phosphate donor, recombinant Stk1 was intensively radiolabeled (Fig. 1B), indicating autophosphorylation activity. The intensity of Stk labeling was higher in the presence of manganese than in the presence of magnesium ions (Fig. 1C). To identify the nature of the autophosphorylated residues in Stk1, the γ32P-labeled band was hydrolyzed and the liberated amino acids were analyzed using two-dimensional chromatography and autoradiography. Both phosphothreonine and, to a lesser extent, phosphoserine were detected on the corresponding autoradiogram (Fig. 1D), demonstrating that Stk1 is indeed a Ser/Thr protein kinase.
FIG. 1.
Biochemical properties of recombinant Stk1. (A) SDS-PAGE analysis of purified Stk1 (lane 2) and Stk1 mutant K39G (lane 3) after staining them with Coomassie blue. Molecular mass standards are indicated on the left (lane 1). (B) Autophosphorylation of Stk1 (lane 1) and mutant K39G (lane 2) in the presence of radioactive [γ-32P]ATP. (C) Effects of cations on Stk1 activity in vitro. Autophosphorylation of purified Stk1 incubated in the presence of 5 mM manganese (Mn2+) or magnesium (Mg2+) for the indicated times. (D) Two-dimensional analysis of phosphorylated amino acids in Stk1. The acid-stable phosphoamino acids from γ32P-labeled Stk1 were separated by electrophoresis in the first dimension (1D), followed by ascending chromatography in the second dimension (2D). P-Tyr, phosphotyrosine; P-Ser, phosphoserine; P-Thr, phosphothreonine.
Stp1 is a manganese-dependent phosphatase.
The stp1 gene was amplified and cloned into the pET15b plasmid to allow the synthesis of a recombinant protein with an amino-terminal histidyl tag. The six-His-Stp1 protein was expressed following IPTG induction, and a band of about 33 kDa, consistent with the theoretical molecular mass of the recombinant protein, was purified to homogeneity in a single step using immobilized Zn2+ affinity chromatography (Fig. 2A). The phosphatase activity of six-His-Stp1 was first analyzed for its ability to cleave pNPP. Hydrolysis of pNPP by Stp1 was measured in the presence of various divalent cations: Mn2+, Mg2+, Ca2+, and Zn2+. We found that the catalytic activity of Stp1 using pNPP as a substrate was observed only in the presence of Mn2+ (data not shown). Optimal phosphatase activity was seen at an Mn2+ concentration of 5 mM, pH 8.8, at 37°C. This value is close to that reported for orthologous phosphatases (36). Protein phosphorylation is a reversible process, and we therefore determined whether Stp1 could utilize Stk1 as an endogenous substrate and catalyze its dephosphorylation. To test this hypothesis, γ-32P-radiolabeled Stk1 was incubated in the presence of Stp1. As shown in Fig. 2B, Stp1 could rapidly and extensively dephosphorylate Stk1 in a time-dependent manner. Analysis of the phosphoamino acids liberated during the reaction showed that serine and threonine residues were dephosphorylated (data not shown). These data provide evidence that phosphorylated Stk1 is an endogenous substrate for Stp1.
FIG. 2.
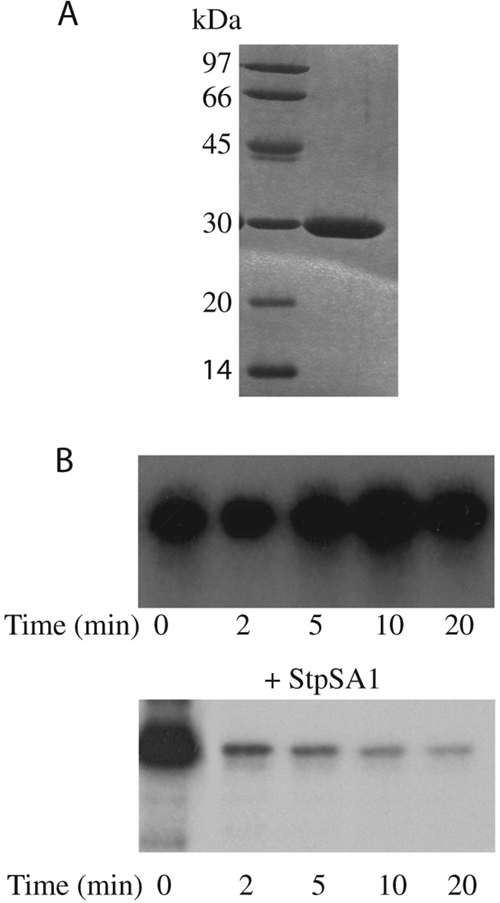
Biochemical properties of recombinant Stp1. (A) SDS-PAGE analysis of purified Stp1 after being stained with Coomassie blue. Molecular mass standards are indicated on the left. (B) Dephosphorylation of Stk1 by StP1. Purified Stk1 phosphorylated in vitro with [γ-32P]ATP was incubated without (top) or with (bottom) Stp1 for the indicated times. Equal amounts of Stk1 were analyzed by electrophoresis, and radioactivity was revealed by autoradiography.
stk1 and stp1 form an operon in S. aureus.
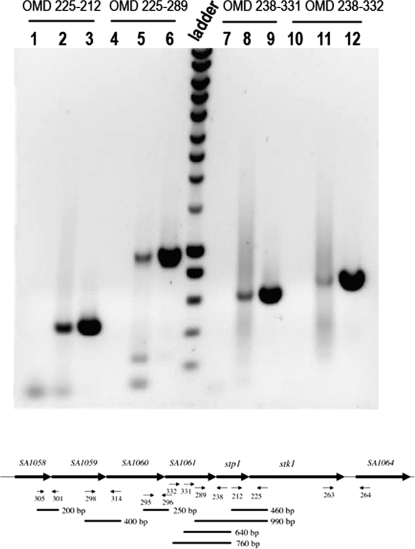
DNA sequence analysis revealed that the last codon of stp1 overlaps with the first codon of stk1 by 1 nucleotide, suggesting that the two genes are cotranscribed and translationally coupled. To demonstrate cotranscription between stp1 and stk1, we performed reverse transcriptase (RT) PCRs using various combinations of primers. The results are summarized in Fig. 3. In all cases, a reaction without RT was also carried out to exclude the presence of contaminating genomic DNA in the RNA preparations. The primer pair OMD212-OMD225, encompassing the 3′ end of stp1 and the 5′ end of stk1, gave an amplified fragment of the expected size of 460 bp. In addition, an upstream primer, OMD289, hybridizing within the 3′ end of the gene upstream from stp1 (SA1061), also gave an amplified fragment of 990 bp with OMD225. This fragment of 990 bp encompasses the 3′ end of SA1061, the entire stp1 gene, and the 5′ end of stk1. A similar experiment was carried out using OMD238 (5′ end of stp1) with either OMD331 or OMD332 (both located in SA1061), giving PCR products of 640 bp and 760 bp, respectively. No PCR product could be detected when an oligonucleotide further upstream (OMD295), located within SA1060, was used. SA1060 and SA1061 (encoding proteins with unknown functions) are separated by only 2 bp. The two genes located immediately upstream from SA1060 and SA1061, SA1058 and SA1059, encode peptidyl deformylase and methionyl tRNA formyl transferase, respectively. Oligonucleotide pairs OMD301-OMD305, OMD298-OMD314, and OMD295-OMD296 were used in similar RT-PCR experiments, giving PCR products of 200 bp, 400 bp, and 250 bp, respectively (Fig. 3 and data not shown). Taken together, these results indicate that stp1 and stk1 are the last two genes of a six-gene operon. No PCR products were detected for the intergenic region between stk1 and the downstream SA1064 gene, indicating that the latter gene is transcribed independently.
FIG. 3.
Physical map of the serine/threonine phosphatase and kinase operon in S. aureus. Shown is RT-PCR analysis of stp1 and stk1 expression in S. aureus. (Top) Cotranscription of stp1 and stk1. DNA PCR products were amplified using the indicated oligonucleotide pairs. Lanes 1, 4, 7, and 10 represent negative controls without RT. Lanes 3, 6, 9, and 12 represent positive controls using chromosomal DNA from strain 8325-4. Lanes 2, 5, 8, and 11 show cotranscriptional analyses of stp1 and stk1. A 1-kb molecular size ladder (Eurogentec) was used. (Bottom) Schematic representation of the stp1 stk1 chromosomal region. The thin arrows located below the genes indicate oligonucleotides used in this analysis. Oligonucleotides OMD225, OMD238, OMD264, OMD296, OMD301, and OMD314 were used to synthesize cDNAs from total RNA using RT. The cDNAs were subsequently amplified by PCR, and the black lines represent the amplified fragments.
Phenotypic analysis of the stp1 and stk1 mutants in S. aureus.
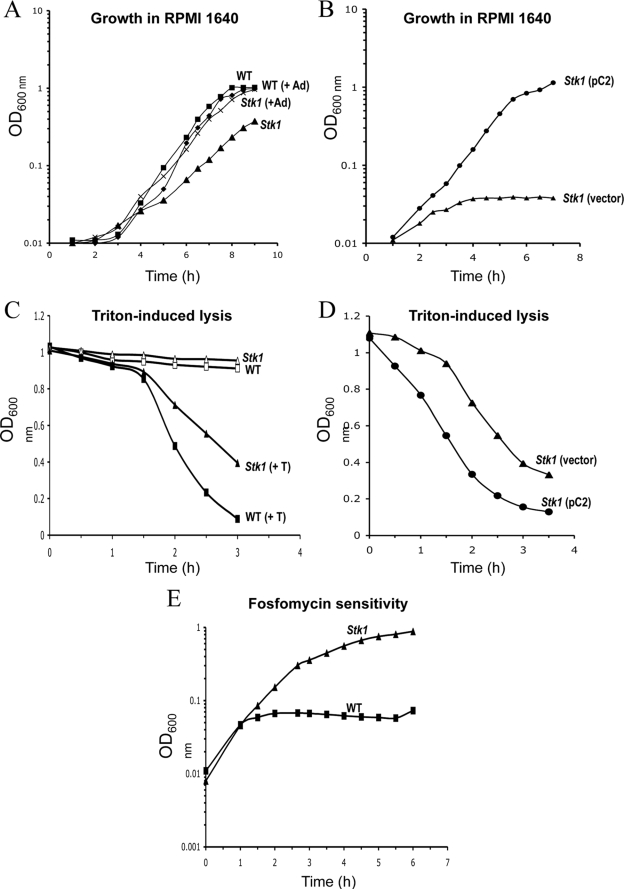
We constructed in-frame deletion mutants in stp1 and stk1 in S. aureus strain 8325-4 (see Materials and Methods). The two isogenic mutants ST1003 (Δstp1) and ST1004 (Δstk1) displayed similar growth characteristics in TSB compared to the parental strain at 37°C (not shown). Analysis of wild-type and mutant cell morphologies by light and scanning electron microscopy did not reveal any major differences. In contrast to the pleiotropic defects of Ser/Thr kinase mutants in other pathogens (14, 20, 21, 45), only a few phenotypes were observed for the Δstk1 mutant. Interestingly, growth curves using the defined RPMI 1640 medium showed reduced growth of the Δstk1 mutant compared to the parental wild-type strain (Fig. 4A). Growth of the Δstp1 mutant was also reduced in RPMI medium, but to a lesser extent than that observed for the Δstk1 mutant (data not shown). Since purine biosynthesis is controlled by a Ser/Thr kinase in S. agalactiae (42), we hypothesized that delayed growth in RPMI 1640 medium could be due to the absence of purines. Indeed, supplementing RPMI 1640 medium with 200 μM adenine relieved the delayed growth of the Δstk1 mutant, consistent with a purine biosynthesis defect in the Δstk1 mutant (Fig. 4A). This is consistent with the results obtained in the accompanying publication, where a transcriptome analysis of the Δstk1 mutant revealed the loss of expression of genes required for purine and pyrimidine biosynthesis, albeit using a different genetic background (10). Given the presence of multiple PASTA domains in Stk1 (see above), we also investigated cell envelope-related phenotypes, such as resistance to antibiotics, autolysis, and sensitivity to lysostaphin- and Triton X-100-induced lysis. Resistance to the following antibiotics was tested on Müller-Hinton plates: β-lactams (penicillin, amoxicillin, cefoxitin, cefotaxim, imipenem, moxalactam, and oxacillin), glycopeptides (vancomycin and teicoplanin), aminoglycosides (kanamycin, tobramycin, netilmycin, and gentamicin), and tetracyclines (tetracycline and minocycline), as well as fusidic acid, rifampicin (rifampin), and fosfomycin. The Δstk1 mutant displayed increased resistance to fosfomycin, an antibiotic inhibiting MurA, which catalyzes the first step in peptidoglycan biosynthesis (Fig. 4B). The fosfomycin MICs for the Δstk1 mutant and the parental wild-type strain were determined in liquid cultures: Δstk1, 64 μg/ml, and 8325-4, 16 μg/ml. The Δstk1 mutant was also more resistant to Triton X-100-induced lysis than the parental strain (Fig. 4C). Complementation of the Δstk1 mutant by introducing a multicopy plasmid carrying the intact stk1 gene fully restored the growth levels of the Δstk1 mutant in RPMI 1640 medium to those of the parental strain, as well as resistance to Triton X-100-induced lysis (Fig. 4B and D) and sensitivity to fosfomycin (data not shown).
FIG. 4.
Phenotypic analyses of the stk1 mutant in S. aureus. (A) Purine auxotrophy of the stk1 deletion mutant strain in S. aureus strain 8325-4. Cell growth was monitored at OD600 in RPMI 1640 synthetic medium at 37°C with or without 200 μM adenine (Ad). ▪, wild-type (WT) strain (8325-4) without adenine; ♦, wild-type strain (8325-4) with 200 μM adenine; ▴, strain ST1004 (Δstk1) without adenine; ×, strain ST1004 (Δstk1) with 200 μM adenine. (B) Purine auxotrophy is corrected in the complemented strain. Cell growth was monitored at OD600 in RPMI 1640 synthetic medium. ▴, strain ST1010 (Δstk1 pMK4-Pprot); •, strain ST1008 (Δstk1 pC2). (C) Triton X-100-induced autolysis of S. aureus strains. Shown are strain 8325-4 without (□) or with (▪) 0.1% Triton X-100 and strain ST1004 (Δstk1) without (▵) or with (▴) 0.1% Triton X-100. (D) Triton X-100-induced autolysis in the complemented strain. ▴, strain ST1010 Δstk1 pMK4-Pprot; •, ST1008 Δstk1 pC2. (E) Sensitivity to fosfomycin. Cultures were grown in TSB medium at 37°C in the presence of fosfomycin (9 μg/ml). ▪, strain 8325-4; ▴, strain ST1004 (Δstk1).
A serine-phosphorylated protein is a target of Stk1.
In order to identify possible protein substrates of the Stk1 kinase, we separated total protein extracts from wild-type strain 8325-4 and its isogenic Δstk1 mutant on SDS-PAGE, transferred the proteins to nitrocellulose membranes, and hybridized them with specific anti-phosphoserine and anti-phosphothreonine antibodies, as described in Materials and Methods. Seven bands were detected in the wild-type strain with the anti-phosphoserine antibody (Fig. 5, left) and no band with the anti-phosphothreonine antibody (Fig. 5, right). In vitro-phosphorylated Stk1 used as a control showed one strong reactive band with the anti-phosphothreonine antibody. No reactive band was detected in the Stk1 K39G mutant, confirming the specificity of the antibody. Only one of the seven serine-phosphorylated proteins, of approximately 120 kDa, was no longer detected as being phosphorylated in the Δstk1 mutant (Fig. 5), suggesting either that these bands correspond to proteins that are phosphorylated on serine residues through the action of other types of kinases, such as PtsK, involved in PTS-dependent sugar transport (43), or RsbW, involved in controlling the activity of sigma B (51), or the possible existence of other, unidentified types of serine kinases in S. aureus strain 8325-4. Genome sequence analysis revealed a second atypical serine threonine kinase gene in the first two sequenced strains of S. aureus, N315 and Mu50 (SA0077 and SAV0081, respectively). We note, however, that although this second gene is also present in the genomes of strains JH1, JH9, Mu3, and MRSA252, it is absent from the genomes of strains NCTC 8325, 8325-4 (used in this study), COL, MW2, Newman, RF122, USA300, and USA300TCH1516.
FIG. 5.
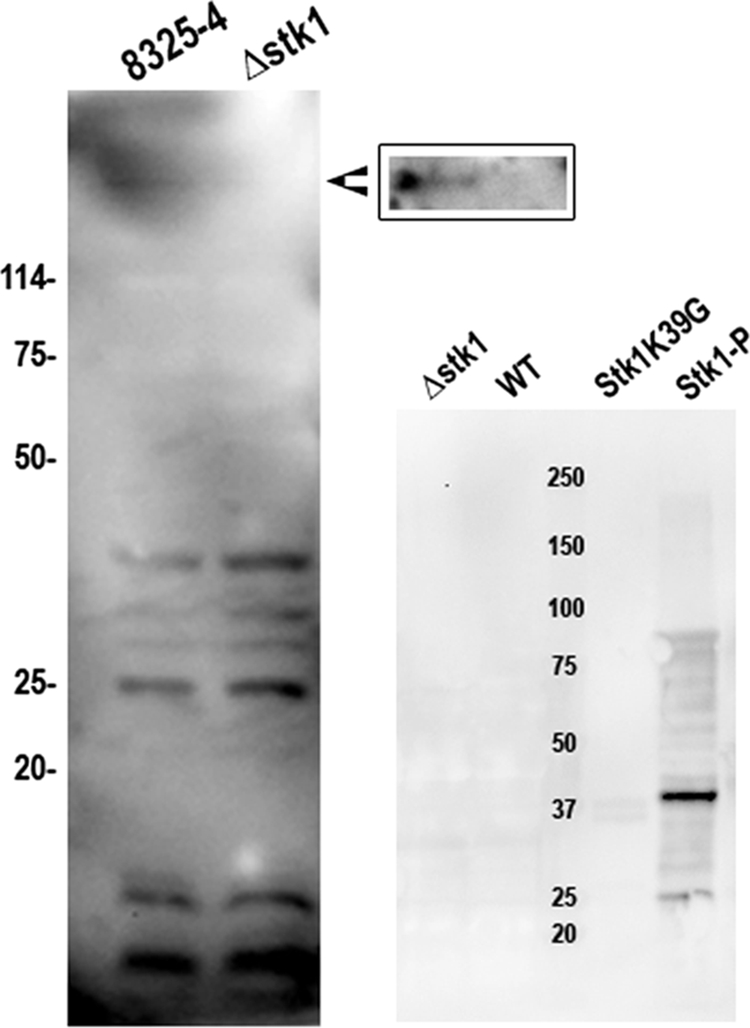
Phosphorylated proteins in S. aureus strain 8325-4 and in the isogenic Δstk1 mutant. (Left) Proteins phosphorylated on serine residues were detected by Western blotting using anti-phosphoserine antibodies following 12% SDS-PAGE of total protein extracts from the wild-type (8325-4) and the Δstk1 strains harvested during exponential phase. Molecular mass standards (kDa) are indicated on the left. The arrowhead indicates the position of a 120-kDa serine-phosphorylated protein, which is absent in the Δstk1 mutant. The inset shows a higher resolution of the missing 120-kDa serine-phosphorylated protein in protein extracts from the Δstk1 mutant separated on a 6% SDS-PAGE gel. (Right) Similar analysis with the anti-phosphothreonine antibody. In vitro-phosphorylated Stk1 used as a control showed one strong reactive band with the anti-phosphothreonine antibody. No reactive band was detected in the Stk1 K39G mutant, confirming the specificity of the antibody. WT, wild type.
Stk1 is essentially membrane bound.
Since Stk1-specific antibodies were not available, we used epitope tagging to localize the Stk1 kinase. For that purpose, we inserted the sequence coding for a 9-aa hemagglutinin at the C-terminal end of stk1 by PCR. The stk1-HA gene was amplified using oligonucleotides OMD337 and OMD339.
The PCR fragment was cloned between the BamHI and SalI sites of the pMK4-pProt vector. The resulting plasmid, pC2, was first introduced into S. aureus strain RN4220 and then used to transform the wild-type strain 8325-4 and the isogenic Δstk1 mutant for complementation. We first analyzed in which bacterial compartment Stk1-HA was located using supernatant, cell wall, membrane, and cytosolic fractions. Stk1 was found to be mostly located in the membrane, but was also present in the cell wall and cytoplasmic fractions (Fig. 6). The cell wall-anchored protein A of S. aureus has affinity for the Fc region of immunoglobulins, especially immunoglobulin G. We repeatedly found a band of about 50 kDa in the cell wall and a band of 45 kDa in the supernatant with all the antibodies tested that could correspond to protein A (Fig. 6). As a control, immunoblotting with antibodies against the conserved chaperone DnaK of E. coli was done and revealed a band of 75 kDa in S. aureus, mostly present in the cytoplasmic fraction, but also in the membrane and cell wall fractions (data not shown). The unexpected presence of DnaK in the cell wall fraction suggests some bacterial lysis may have occurred during preparation of cell wall extracts, and thus, the presence of Stk1 in the cell wall could be an artifact. However, Stk1 is clearly present in the membrane fraction, consistent with the presence of a centrally located transmembrane domain.
FIG. 6.
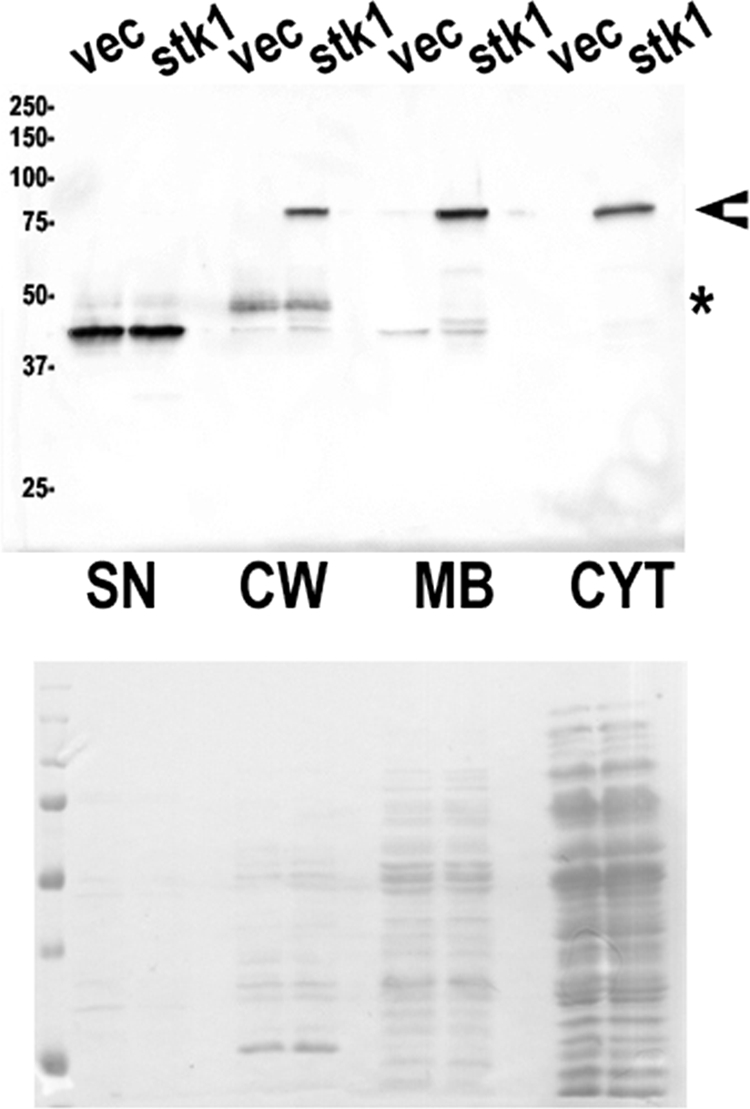
Subcellular localization of Stk1-HA in S. aureus. Exponentially growing cultures of S. aureus strain ST1006 (8325-4 carrying the control vector pMK4-Pprot) (vec) or expressing the stk1-HA gene (strain ST1007) were fractionated into cytosolic (CYT), membrane (MB), cell wall (CW), and supernatant (SN) compartments and immunoblotted with a commercial anti-HA monoclonal antibody. The equivalent of 1 ml of culture was loaded in each well, and molecular mass standards (kDa) are indicated on the left. Equivalent loading was controlled by staining the nitrocellulose membrane with Ponceau red (bottom) before immunoblotting. The asterisk indicates the presence of the likely protein A band, and the arrowhead indicates the position of the Stk1 band.
Stk1 is required for S. aureus virulence in a murine renal-infection model.
To investigate the possible involvement of Stp1 and Stk1 in pathogenesis, we used a murine model of hematogenous pyelonephritis. This model utilizes a localized kidney infection from which bacteria can be recovered and quantified. We infected CD1 mice via the lateral tail vein, removed their kidneys at 7 days postinfection, and carried out a quantification of renal bacterial loads, as well as a histopathological analysis of the infected kidney, to investigate the role of Stk1 in vivo.
Bacteremia was assessed at various time points during the infection process for the Δstp1 and Δstk1 mutants and the parental strain, 8325-4. The results showed a 2-log-unit decrease for the Δstk1 mutant and 1-log-unit decrease for the Δstp1 mutant compared to the wild-type strain, with high variability between mice (data not shown). Since the strain of choice for genetic studies, 8325-4, carries a small deletion in rsbU, which encodes a positive regulator of sigma B activity, we checked the effect of the stk1 deletion in strain SH1000, which is an rsbU+ derivative of 8325-4 (19). As shown in Fig. 7, the stk1 deletion in SH1000 clearly reduced bacterial loads in the kidneys. Interestingly, the parental strain, SH1000, gave higher bacterial loads in kidneys than strain 8325-4 (108 versus 105 CFU, respectively) and less variability between mice. Bacterial loads in the blood were similar between wild-type strains and their isogenic Δstk1 mutant during the course of infection, indicating that the reduced kidney colonization of stk1 deletion mutants was not due to a growth defect or increased sensitivity to immune defenses, such as phagocytosis, antimicrobial peptides, or complement (data not shown). In vitro growth curves in murine blood confirmed the absence of growth defects of the Δstk1 mutant (data not shown). This result indicates that the virulence defect is not due to a general metabolic defect of the mutant in vivo. It also implies that the Δstk1 mutant is able to resist phagocytic killing (reactive oxygen species) as well as the wild-type parental strain. To confirm this point, we tested the survival of the S. aureus wild-type and Δstk1 strains in RAW 264.7 macrophages and did not observe any difference (data not shown). We therefore wondered if Stk1 of S. aureus could modulate adhesion and/or invasion of epithelial kidney cells in vitro using Madin-Darby canine kidney (MDCK) cells (cell line CCL-34). We did not observe reduced adhesion/invasion for the Δstk1 mutant strains compared to the corresponding parental strains (data not shown). Thus, bacterial adhesion to/invasion of host cells, the primary step during kidney infection, is probably not linked to the activity of the serine/threonine kinase in S. aureus. However, it remains possible that MDCK cells do not mimic mouse kidney cells.
FIG. 7.
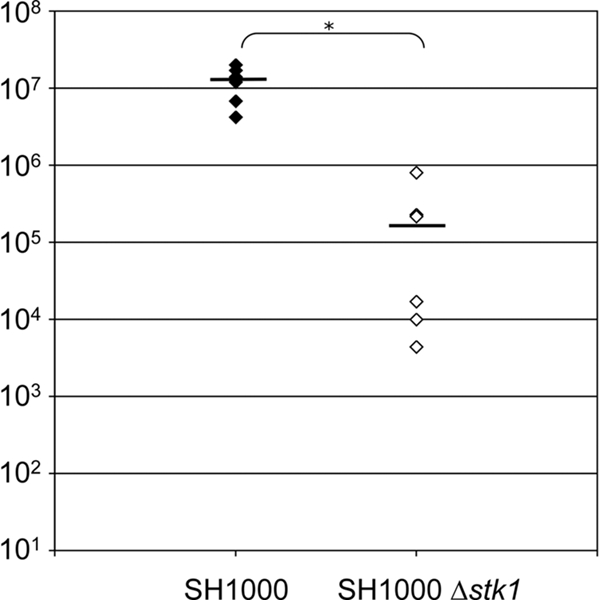
Role of S. aureus Stk1 during infection. Shown is the effect of stk1 deletion in strain SH1000 on S. aureus survival in mice. Bacterial counts of the parental SH1000 and ST1005 (Δstk1 mutant) strains were determined in the kidneys of CD1 mice (n = 6) 7 days after intravenous injection of 108 CFU. Each point is the result for one mouse. The bars represent mean CFU in two kidneys. *, P < 0.001 (Student's t test).
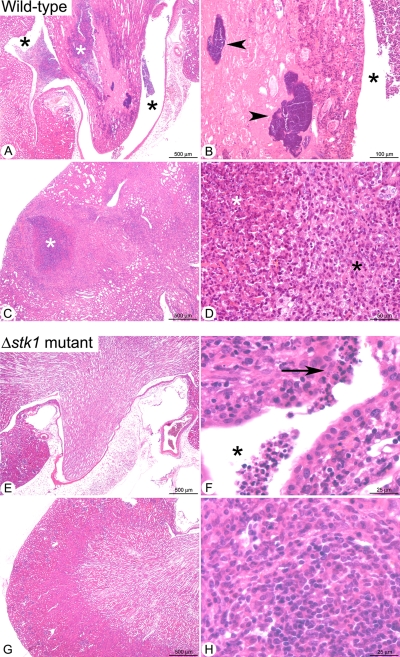
The renal histopathological analysis revealed significant differences between mice infected with the wild-type SH1000 strain and mice infected with the isogenic Δstk1 mutant (Fig. 8). At 7 days postinfection in the kidneys of mice inoculated with the wild-type strain (Fig. 8A to D), we observed multifocal to coalescing inflammatory lesions extending from the renal pelvis to the cortex. These lesions were characterized, in the renal crest, by a severe destruction of the normal tubular organization (Fig. 8A), with collecting tubes either being replaced by an acidophilic and amorphous material containing cell debris (necrosis) and fragmented neutrophils (suppuration) or displaying loss of epithelial cell nuclei associated with a clumping of their cytoplasm but with preservation of the basic outlines of a tubular architecture (ischemic necrosis) (Fig. 8A and B). In the ischemic area, large bacterial colonies were noted (Fig. 8B). Bacteria, associated with neutrophils and cell debris, were also accumulated in the pelvis. These lesions were characteristic of suppurative pyelonephritis. In the cortex, we observed multifocal abscesses measuring up to 1 mm in diameter and displaying a concentric organization (Fig. 8C), characterized from the center to the periphery by (i) an accumulation of fragmented (i.e., karyorrhectic) neutrophils and cell debris (suppuration), circled by (ii) a peripheral rim of numerous macrophages and lymphocytes and plasma cells (Fig. 8D). In the residual renal parenchyma, the interstitial connective tissue was diffusely infiltrated by neutrophils and less numerous mononucleated cells, and a high proportion of tubes in the cortex and the medulla displayed marked dilation of their lumens, filled with cell debris, extracellular proteins, or extravasated erythrocytes (data not shown).
FIG. 8.
Histopathological analyses of kidneys in mice infected with either S. aureus wild-type strain SH1000 (A to D) or its isogenic Δstk1 mutant (E to H). (A) Low magnification of the renal crest (white star) and pelvis (black star), with numerous infiltrates of neutrophils destroying the normal tubular architecture of the tissue. (B) Higher magnification of the renal crest showing bacterial colonies (arrowheads) in a necrotic area (ischemic necrosis). A neutrophilic infiltrate is also observed in the pelvis (black star). (C) Focal abscess in the renal cortex (white star). (D) Higher magnification of the abscess organization, with centrally located fragmented (i.e., karyorrhectic) neutrophils and cell debris (suppuration) (white star) and a peripheral rim of numerous macrophages associated with lymphocytes and plasma cells (black star). (E) Low magnification of the renal crest and pelvis showing no significant histological lesions. (F) Higher magnification of the renal papilli showing a small neutrophilic infiltrate in the pelvis (black star) extending to the overlying epithelium (arrow). (G) Low magnification of the renal cortex with no significant histological lesions. (H) In one mouse, small (less than 150 μm in diameter) infiltrates of macrophages associated with less numerous lymphocytes and plasma cells were observed in the cortex.
In contrast to these severe lesions, mice infected by the Δstk1 mutant displayed only minimal to mild inflammatory lesions (Fig. 8E to H). At low magnification, the medulla appeared morphologically normal (Fig. 8E), but some mice (two of three) displayed small (less than 100-μm) infiltrates of neutrophils in the pelvis or renal crest (Fig. 8F). The cortexes of most mice infected by the Δstk1 mutant (two of three) did not display any inflammatory lesions, but in the remaining mouse, small infiltrates (less than 150 μm in diameter) of macrophages were multifocally observed (Fig. 8H). In contrast to infection with the wild-type strain, bacteria and neutrophils were very rare in the lesions.
DISCUSSION
The availability of an increasing number of bacterial genomes has revealed several eukaryotic-like Ser/Thr protein kinase and phosphatase genes. In this work, we characterized the first genetic locus encoding a eukaryotic-like serine/threonine phosphatase (Stp1) and serine/threonine kinase (Stk1) in S. aureus. We showed that histidyl-tagged recombinant Stk1 is a functional protein kinase that autophosphorylates on threonine and serine residues. We also showed that its cognate protein phosphatase, Stp1, specifically dephosphorylates phosphorylated Stk1 in the presence of manganese ions. As previously described in B. subtilis (30) and S. agalactiae (41), the genes encoding Stp1 and Stk1 were found to be adjacent on the chromosome of S. aureus, and we have shown that they are cotranscribed. Together, these results suggest that Stp1 and Stk1 may act as a functional protein pair with antagonistic roles in vivo.
Stk1 belongs to a family of serine/threonine protein kinases, unique to gram-positive bacteria, with a highly conserved intracellular kinase domain and three extracellular PASTA domains. The PASTA domain (PF03793) is found at the C termini of several penicillin-binding proteins and bacterial Ser/Thr kinases (52). This domain was first identified as associated with a high-molecular-weight type II PBP, PBP2X of S. pneumoniae, whose structure has been solved (37). The PASTA domain is a small globular fold consisting of three beta-sheets and an alpha-helix that binds the beta-lactam stem, involved in sensing d-alanyl-d-alanine, the PBP transpeptidase substrate. Jones and Dyson (22) compared PASTA sequences found in many bacterial genomes and highlighted their wide divergence, which may reflect the diversity and complexity of peptidoglycan structures. They proposed that these serine/threonine protein kinases might play a key role in peptidoglycan remodeling. Recently, an extracellular domain of the B. subtilis PrkC serine/threonine kinase was shown to bind peptidoglycan fragments, thus signaling bacteria to exit dormancy, allowing spore germination (46).
Although we observed that the Δstk1 mutant displayed increased resistance to Triton X-100-induced lysis compared to the parental strain, no difference was observed in the autolytic enzyme profile as analyzed by zymograms (data not shown), and scanning electron microscopy did not reveal any morphological differences from the parental strain.
The genetic organization of the stp1 stk1 locus in S. aureus is strikingly similar to that of the prpC prkC locus of B. subtilis. In B. subtilis, yloQ, the gene immediately downstream from the Ser/Thr kinase gene prkC, was thought to be essential, and the three-dimensional structure of YloQ was recently solved (26). Structural analyses suggest that YloQ is an elongation factor (7). YloQ (also called CpgA) is a GTPase, and analyses of a yloQ conditional mutant suggest a role in cell wall biogenesis and morphogenesis. Interestingly, depletion of YloQ in a conditional yloQ mutant of B. subtilis led to significant sensitivity to fosfomycin (MIC, 8 μg/ml versus 64 μg/ml) (5). We observed the opposite effect for the stk1 deletion mutant strain of S. aureus strain 8325-4, which displayed increased resistance to fosfomycin, which exerts its antibacterial effect through inhibition of MurA, catalyzing the first committed step in peptidoglycan synthesis (11). Thus, it is tempting to speculate that Stp1/Stk1 and YloQ are involved in related but still unclear cellular functions, such as peptidoglycan dynamics, cell division, or ribosome function.
The roles of Stk1 and Stp1 in bacterial pathogenesis were studied by deleting the stp1 and stk1 genes in S. aureus strain 8325-4. Both the Δstp1 and Δstk1 mutants exhibited normal colony morphology and growth in TSB medium. However, using defined RPMI 1640 medium, the stk1 deletion mutant exhibited an extended lag phase during growth compared to the parental wild-type strain. RPMI 1640 medium is formulated for eukaryotic cell culture and contains amino acids, vitamins, inorganic salts, reduced glutathione, and glucose as the sole carbon source. It was shown that the stk1-deficient strain of S. agalactiae is unable to sustain purine biosynthesis and is consequently growth arrested (41). We therefore tested the addition of adenine to RPMI 1640 medium and showed that it restored the growth of the 8325-4 Δstk1 mutant to the level of the parental strain (Fig. 4A). Surprisingly, this delayed growth in RPMI 1640 medium was not observed for the Δstk1 mutant in the rsbU+ genetic background (strain SH1000) (data not shown), suggesting that sigma B may play a role in purine biosynthesis and may modulate the effect of Stk1. It should be noted that RsbU is a PP2C-type serine phosphatase, which could alter the balance of Stk1-phosphorylated proteins, despite the fact that it acts specifically on the phosphorylated form of RsbV (40). In the accompanying paper, Donat et al. (10) performed a transcriptome analysis of the stk1 deletion mutant and showed that expression levels of purine and pyrimidine biosynthesis genes were significantly lower in the mutant, consistent with the observed growth defect in RPMI 1640.
Over the last few years, extensive work has been performed to demonstrate the important role of bacterial serine/threonine kinases and phosphatases in virulence. Examples include major gram-positive pathogens, such as S. agalactiae (41), E. faecalis (23), L. monocytogenes (1), S. pyogenes (21), S. pneumoniae (14), and M. tuberculosis (50). In S. agalactiae, the 50% lethal dose is increased 25-fold for the stk1 deletion mutant and 100-fold for the double stk1 stp1 mutant in a rat neonate model. It was hypothesized that this defect could be due to abnormal cell segregation and altered metabolism (41, 42). In E. faecalis, PrkC modulates inherent resistance to cell envelope active antimicrobials and intestinal persistence (23). Recently, MgrA, a global regulator in S. aureus, was shown to be phosphorylated in a strain overproducing Stk1, leading to increased expression of norA, encoding an efflux pump, and concomitant increased resistance to norfloxacin and ciprofloxacin (48). In S. pneumoniae, cell wall defects were observed under specific conditions for the stkP mutant in vitro (45). The StkP mutant was attenuated for virulence in lungs and blood by approximately 3 log units (14). In all these examples, cell wall defects seemed to be the main cause of attenuation in vivo.
Here, we have shown that the Δstk1 mutant of S. aureus exhibited strongly reduced survival in the kidneys of intravenously infected mice. This result was confirmed in two different backgrounds, 8325-4 and SH1000, indicating that Stk1 is required for full virulence of S. aureus. Interestingly, bacterial loads in the blood were similar for wild-type and isogenic mutant strains. Histopathological analyses of kidneys showed that mice infected by the wild-type S. aureus SH1000 strain displayed marked to severe inflammatory lesions, with numerous intralesional bacteria, in contrast to mice inoculated with the Δstk1 mutant, which had only minimal renal lesions. These results demonstrate that Stk1 is critical for full expression of S. aureus pathogenesis.
Acknowledgments
Work in the group of T. Msadek was supported by research funds from the European Commission (grants from BACELL Health [LSHG-CT-2004-503468], StaphDynamics [LHSM-CT-2006-019064], and BaSysBio [LSHG-CT-2006-037469]), the Centre National de la Recherche Scientifique (CNRS URA 2172), and the Institut Pasteur (GPH no. 9), and work in the group of A. Cozzone was supported by the association Vaincre La Mucoviscidose. G. Jouvion was supported by a Roux Fellowship (Institut Pasteur).
We thank Claire Poyart for help in performing antibiograms; Stéphanie Guadagnini for electron micrographs; Pascale Cossart, in whose laboratory part of this work was carried out; Patrick Trieu-Cuot for helpful discussion; and Sarah Dubrac for critical reading of the manuscript.
Footnotes
Published ahead of print on 24 April 2009.
REFERENCES
- 1.Archambaud, C., E. Gouin, J. Pizarro-Cerda, P. Cossart, and O. Dussurget. 2005. Translation elongation factor EF-Tu is a target for Stp, a serine-threonine phosphatase involved in virulence of Listeria monocytogenes. Mol. Microbiol. 56383-396. [DOI] [PubMed] [Google Scholar]
- 2.Archambaud, C., M. A. Nahori, J. Pizarro-Cerda, P. Cossart, and O. Dussurget. 2006. Control of Listeria superoxide dismutase by phosphorylation. J. Biol. Chem. 28131812-31822. [DOI] [PubMed] [Google Scholar]
- 3.Arnaud, M., A. Chastanet, and M. Debarbouille. 2004. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl. Environ. Microbiol. 706887-6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bork, P., N. P. Brown, H. Hegyi, and J. Schultz. 1996. The protein phosphatase 2C (PP2C) superfamily: detection of bacterial homologues. Protein Sci. 51421-1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campbell, T. L., D. M. Daigle, and E. D. Brown. 2005. Characterization of the Bacillus subtilis GTPase YloQ and its role in ribosome function. Biochem. J. 389843-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chastanet, A., M. Prudhomme, J. P. Claverys, and T. Msadek. 2001. Regulation of Streptococcus pneumoniae clp genes and their role in competence development and stress survival. J. Bacteriol. 1837295-7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cladiere, L., K. Hamze, E. Madec, V. M. Levdikov, A. J. Wilkinson, I. B. Holland, and S. J. Seror. 2006. The GTPase, CpgA(YloQ), a putative translation factor, is implicated in morphogenesis in Bacillus subtilis. Mol. Genet. Genomics 275409-420. [DOI] [PubMed] [Google Scholar]
- 8.Cozzone, A. J. 2005. Role of protein phosphorylation on serine/threonine and tyrosine in the virulence of bacterial pathogens. J. Mol. Microbiol. Biotechnol. 9198-213. [DOI] [PubMed] [Google Scholar]
- 9.Das, A. K., N. R. Helps, P. T. Cohen, and D. Barford. 1996. Crystal structure of the protein serine/threonine phosphatase 2C at 2.0 Å resolution. EMBO J. 156798-6809. [PMC free article] [PubMed] [Google Scholar]
- 10.Donat, S., K. Streker, T. Schirmeister, and K. Ohlsen. 2009. Transcriptome and functional analysis of the eukaryotic-type Ser/Thr kinase PknB in Staphylococcus aureus. J. Bacteriol. doi: 10.1128/JB0117-09. [DOI] [PMC free article] [PubMed]
- 11.Du, W., J. R. Brown, D. R. Sylvester, J. Huang, A. F. Chalker, C. Y. So, D. J. Holmes, D. J. Payne, and N. G. Wallis. 2000. Two active forms of UDP-N-acetylglucosamine enolpyruvyl transferase in gram-positive bacteria. J. Bacteriol. 1824146-4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dubrac, S., I. G. Boneca, O. Poupel, and T. Msadek. 2007. New insights into the WalK/WalR (YycG/YycF) essential signal transduction pathway reveal a major role in controlling cell wall metabolism and biofilm formation in Staphylococcus aureus. J. Bacteriol. 1898257-8269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duclos, B., S. Marcandier, and A. J. Cozzone. 1991. Chemical properties and separation of phosphoamino acids by thin-layer chromatography and/or electrophoresis. Methods Enzymol. 20110-21. [DOI] [PubMed] [Google Scholar]
- 14.Echenique, J., A. Kadioglu, S. Romao, P. W. Andrew, and M. C. Trombe. 2004. Protein serine/threonine kinase StkP positively controls virulence and competence in Streptococcus pneumoniae. Infect. Immun. 722434-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eymann, C., D. Becher, J. Bernhardt, K. Gronau, A. Klutzny, and M. Hecker. 2007. Dynamics of protein phosphorylation on Ser/Thr/Tyr in Bacillus subtilis. Proteomics 73509-3526. [DOI] [PubMed] [Google Scholar]
- 16.Fiuza, M., M. J. Canova, I. Zanella-Cleon, M. Becchi, A. J. Cozzone, L. M. Mateos, L. Kremer, J. A. Gil, and V. Molle. 2008. From the characterization of the four serine/threonine protein kinases (PknA/B/G/L) of Corynebacterium glutamicum toward the role of PknA and PknB in cell division. J. Biol. Chem. 28318099-18112. [DOI] [PubMed] [Google Scholar]
- 17.Gaidenko, T. A., T. J. Kim, and C. W. Price. 2002. The PrpC serine-threonine phosphatase and PrkC kinase have opposing physiological roles in stationary-phase Bacillus subtilis cells. J. Bacteriol. 1846109-6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greenstein, A. E., C. Grundner, N. Echols, L. M. Gay, T. N. Lombana, C. A. Miecskowski, K. E. Pullen, P. Y. Sung, and T. Alber. 2005. Structure/function studies of Ser/Thr and Tyr protein phosphorylation in Mycobacterium tuberculosis. J. Mol. Microbiol. Biotechnol. 9167-181. [DOI] [PubMed] [Google Scholar]
- 19.Horsburgh, M. J., J. L. Aish, I. J. White, L. Shaw, J. K. Lithgow, and S. J. Foster. 2002. σB modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4. J. Bacteriol. 1845457-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hussain, H., P. Branny, and E. Allan. 2006. A eukaryotic-type serine/threonine protein kinase is required for biofilm formation, genetic competence, and acid resistance in Streptococcus mutans. J. Bacteriol. 1881628-1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jin, H., and V. Pancholi. 2006. Identification and biochemical characterization of a eukaryotic-type serine/threonine kinase and its cognate phosphatase in Streptococcus pyogenes: their biological functions and substrate identification. J. Mol. Biol. 3571351-1372. [DOI] [PubMed] [Google Scholar]
- 22.Jones, G., and P. Dyson. 2006. Evolution of transmembrane protein kinases implicated in coordinating remodeling of gram-positive peptidoglycan: inside versus outside. J. Bacteriol. 1887470-7476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kristich, C. J., C. L. Wells, and G. M. Dunny. 2007. A eukaryotic-type Ser/Thr kinase in Enterococcus faecalis mediates antimicrobial resistance and intestinal persistence. Proc. Natl. Acad. Sci. USA 1043508-3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K. Aoki, Y. Nagai, J. Lian, T. Ito, M. Kanamori, H. Matsumaru, A. Maruyama, H. Murakami, A. Hosoyama, Y. Mizutani-Ui, N. K. Takahashi, T. Sawano, R. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu. 2001. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet 3571225-1240. [DOI] [PubMed] [Google Scholar]
- 25.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227680-685. [DOI] [PubMed] [Google Scholar]
- 26.Levdikov, V. M., E. V. Blagova, J. A. Brannigan, L. Cladiere, A. A. Antson, M. N. Isupov, S. J. Seror, and A. J. Wilkinson. 2004. The crystal structure of YloQ, a circularly permuted GTPase essential for Bacillus subtilis viability. J. Mol. Biol. 340767-782. [DOI] [PubMed] [Google Scholar]
- 27.Levine, A., F. Vannier, C. Absalon, L. Kuhn, P. Jackson, E. Scrivener, V. Labas, J. Vinh, P. Courtney, J. Garin, and S. J. Seror. 2006. Analysis of the dynamic Bacillus subtilis Ser/Thr/Tyr phosphoproteome implicated in a wide variety of cellular processes. Proteomics 62157-2173. [DOI] [PubMed] [Google Scholar]
- 28.Lomas-Lopez, R., P. Paracuellos, M. Riberty, A. J. Cozzone, and B. Duclos. 2007. Several enzymes of the central metabolism are phosphorylated in Staphylococcus aureus. FEMS Microbiol. Lett. 27235-42. [DOI] [PubMed] [Google Scholar]
- 29.Macek, B., I. Mijakovic, J. V. Olsen, F. Gnad, C. Kumar, P. R. Jensen, and M. Mann. 2007. The serine/threonine/tyrosine phosphoproteome of the model bacterium Bacillus subtilis. Mol. Cell. Proteomics 6697-707. [DOI] [PubMed] [Google Scholar]
- 30.Madec, E., A. Laszkiewicz, A. Iwanicki, M. Obuchowski, and S. Seror. 2002. Characterization of a membrane-linked Ser/Thr protein kinase in Bacillus subtilis, implicated in developmental processes. Mol. Microbiol. 46571-586. [DOI] [PubMed] [Google Scholar]
- 31.Munoz-Dorado, J., S. Inouye, and M. Inouye. 1991. A gene encoding a protein serine/threonine kinase is required for normal development of M. xanthus, a gram-negative bacterium. Cell 67995-1006. [DOI] [PubMed] [Google Scholar]
- 32.Munoz-Dorado, J., S. Inouye, and M. Inouye. 1990. Nucleoside diphosphate kinase from Myxococcus xanthus. II. Biochemical characterization. J. Biol. Chem. 2652707-2712. [PubMed] [Google Scholar]
- 33.Nariya, H., and S. Inouye. 2005. Identification of a protein Ser/Thr kinase cascade that regulates essential transcriptional activators in Myxococcus xanthus development. Mol. Microbiol. 58367-379. [DOI] [PubMed] [Google Scholar]
- 34.Neu, J. M., S. V. MacMillan, J. R. Nodwell, and G. D. Wright. 2002. StoPK-1, a serine/threonine protein kinase from the glycopeptide antibiotic producer Streptomyces toyocaensis NRRL 15009, affects oxidative stress response. Mol. Microbiol. 44417-430. [DOI] [PubMed] [Google Scholar]
- 35.Novakova, L., L. Saskova, P. Pallova, J. Janecek, J. Novotna, A. Ulrych, J. Echenique, M. C. Trombe, and P. Branny. 2005. Characterization of a eukaryotic type serine/threonine protein kinase and protein phosphatase of Streptococcus pneumoniae and identification of kinase substrates. FEBS J. 2721243-1254. [DOI] [PubMed] [Google Scholar]
- 36.Obuchowski, M., E. Madec, D. Delattre, G. Boel, A. Iwanicki, D. Foulger, and S. J. Seror. 2000. Characterization of PrpC from Bacillus subtilis, a member of the PPM phosphatase family. J. Bacteriol. 1825634-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pares, S., N. Mouz, Y. Petillot, R. Hakenbeck, and O. Dideberg. 1996. X-ray structure of Streptococcus pneumoniae PBP2x, a primary penicillin target enzyme. Nat. Struct. Biol. 3284-289. [DOI] [PubMed] [Google Scholar]
- 38.Peng, H. L., R. P. Novick, B. Kreiswirth, J. Kornblum, and P. Schlievert. 1988. Cloning, characterization, and sequencing of an accessory gene regulator (agr) in Staphylococcus aureus. J. Bacteriol. 1704365-4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petrickova, K., and M. Petricek. 2003. Eukaryotic-type protein kinases in Streptomyces coelicolor: variations on a common theme. Microbiology 1491609-1621. [DOI] [PubMed] [Google Scholar]
- 40.Price, C. W. 2002. General stress response, p. 369-384. In A. L. Sonenshein, J. A. Hoch, and R. M. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, DC.
- 41.Rajagopal, L., A. Clancy, and C. E. Rubens. 2003. A eukaryotic type serine/threonine kinase and phosphatase in Streptococcus agalactiae reversibly phosphorylate an inorganic pyrophosphatase and affect growth, cell segregation, and virulence. J. Biol. Chem. 27814429-14441. [DOI] [PubMed] [Google Scholar]
- 42.Rajagopal, L., A. Vo, A. Silvestroni, and C. E. Rubens. 2005. Regulation of purine biosynthesis by a eukaryotic-type kinase in Streptococcus agalactiae. Mol. Microbiol. 561329-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reizer, J., C. Hoischen, F. Titgemeyer, C. Rivolta, R. Rabus, J. Stulke, D. Karamata, M. H. Saier, Jr., and W. Hillen. 1998. A novel protein kinase that controls carbon catabolite repression in bacteria. Mol. Microbiol. 271157-1169. [DOI] [PubMed] [Google Scholar]
- 44.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 45.Saskova, L., L. Novakova, M. Basler, and P. Branny. 2007. Eukaryotic-type serine/threonine protein kinase StkP is a global regulator of gene expression in Streptococcus pneumoniae. J. Bacteriol. 1894168-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shah, I. M., M. H. Laaberki, D. L. Popham, and J. Dworkin. 2008. A eukaryotic-like Ser/Thr kinase signals bacteria to exit dormancy in response to peptidoglycan fragments. Cell 135486-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Studier, F. W., and B. A. Moffatt. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189113-130. [DOI] [PubMed] [Google Scholar]
- 48.Truong-Bolduc, Q. C., Y. Ding, and D. C. Hooper. 2008. Posttranslational modification influences the effects of MgrA on norA expression in Staphylococcus aureus. J. Bacteriol. 1907375-7381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Umeyama, T., P. C. Lee, and S. Horinouchi. 2002. Protein serine/threonine kinases in signal transduction for secondary metabolism and morphogenesis in Streptomyces. Appl. Microbiol. Biotechnol. 59419-425. [DOI] [PubMed] [Google Scholar]
- 50.Wehenkel, A., M. Bellinzoni, M. Grana, R. Duran, A. Villarino, P. Fernandez, G. Andre-Leroux, P. England, H. Takiff, C. Cervenansky, S. T. Cole, and P. M. Alzari. 2008. Mycobacterial Ser/Thr protein kinases and phosphatases: physiological roles and therapeutic potential. Biochim. Biophys. Acta 1784193-202. [DOI] [PubMed] [Google Scholar]
- 51.Yang, X., C. M. Kang, M. S. Brody, and C. W. Price. 1996. Opposing pairs of serine protein kinases and phosphatases transmit signals of environmental stress to activate a bacterial transcription factor. Genes Dev. 102265-2275. [DOI] [PubMed] [Google Scholar]
- 52.Yeats, C., R. D. Finn, and A. Bateman. 2002. The PASTA domain: a beta-lactam-binding domain. Trends Biochem. Sci. 27438. [DOI] [PubMed] [Google Scholar]