Abstract
We identified a carbohydrate metabolic operon (frz) that is highly associated with extraintestinal pathogenic Escherichia coli (ExPEC) strains. The frz operon codes for three subunits of a phosphoenolpyruvate:carbohydrate phosphotransferase system (PTS) transporter of the fructose subfamily, for a transcriptional activator of PTSs of the MgA family, for two type II ketose-1,6-bisphosphate aldolases, for a sugar-specific kinase (repressor, open reading frame, kinase family [ROK]), and for a protein of the cupin superfamily. We proved that the frz operon promotes bacterial fitness under stressful conditions, such as oxygen restriction, late stationary phase of growth, or growth in serum or in the intestinal tract. Furthermore, we showed that frz is involved in adherence to and internalization in human type II pneumocytes, human enterocytes, and chicken liver cells by favoring the ON orientation of the fim operon promoter and thus acting on the expression of type 1 fimbriae, which are the major ExPEC adhesins. Both the PTS activator and the metabolic enzymes encoded by the frz operon are involved in these phenotypes.
Extraintestinal pathogenic Escherichia coli (ExPEC) strains are particular E. coli strains able to induce diseases that occur in bodily sites outside the gastrointestinal tract. ExPEC strains are phylogenetically and epidemiologically distinct from commensal and intestinal pathogenic strains. Most of the ExPEC strains are found in the B2 and D phylogenetic groups, whereas commensal E. coli strains generally derive from the A and B1 groups and intestinal pathogenic strains are mostly associated with the A, B1, and D groups. ExPEC strains do not produce enteric disease; however, they can asymptomatically colonize the intestinal tract of humans and warm-blooded animals and may be the predominant strains in healthy individuals. ExPEC have acquired various virulence factors that allow the induction, under favorable circumstances, of a wide range of extraintestinal infections in humans and poultry, including bacteremia, urinary tract infections, neonatal meningitidis, respiratory tract infections, and septicemia (19, 28, 41, 47, 48, 54). ExPEC strains of avian origin are phylogenetically linked with strains isolated from normally sterile extraintestinal body sites of humans, with which they share numerous virulence factors. This suggests that ExPEC strains of avian origin could represent a zoonotic risk (43).
Whereas various virulence factors of ExPEC strains of avian origin were described, these factors, present in various combinations among strains, cannot completely explain the entire pathogenicity mechanism, which is complex and multifactorial (10, 11, 13, 14, 19, 24, 26, 36, 37, 49, 53). To improve our knowledge in that domain and identify new virulence factors, a subtractive hybridization between the genomes of the avian ExPEC strain MT512 (02:K1:H7) and of the avirulent avian strain EC79 (O2:K−:H−) was performed in our laboratory. This experiment allowed the identification of the D7 genomic fragment (240 bp, GenBank accession no. AY187873) that is absent in the nonpathogenic strains EC79 and K-12 substrain MG1655. D7 was located near the tRNA locus selC and proved to be present in the genomes of various ExPEC strains of human and animal origin and, notably, in the genome of strain BEN2908, our reference ExPEC strain of avian origin (44, 52). D7 was also found to be highly associated with the E. coli phylogenetic group B2 and to a lesser extent with group D, two phylogenetic groups in which the majority of E. coli strains isolated from normally sterile extraintestinal body sites are clustered (33, 43). The translated D7 DNA sequence of strain MT512 shared 79% similar amino acids with a fructose-like enzyme II component of a phosphoenolpyruvate:carbohydrate phosphotransferase system (PTS) from Listeria sp. strains, suggesting its involvement in metabolism by the strain (52).
In this work, to understand the high level of association of D7 with ExPEC strains, we characterized the D7 genomic environment in the reference strain BEN2908 and analyzed the role of D7 and surrounding genes in the physiology of the strain.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Bacterial strains and plasmids are described in Table 1. The ExPEC strain BEN2908 is a nalidixic acid-resistant derivative of strain MT78 isolated from the trachea of a chicken with a respiratory tract infection (11, 20, 24). BEN2908 belongs to the phylogenetic cluster B2-1 (43). One-hundred eighty-six well-characterized and epidemiologically unrelated strains isolated from chickens in various European countries between 1989 and 2001 were used to analyze the association of the frz operon with the level of virulence of the strains (55). These 186 strains belonged to the four most-frequent serogroups of avian ExPEC strains (14 strains of serogroup O1, 36 strains of serogroup O2, 11 strains of serogroup O18, and 28 strains of serogroup O78) but also to 47 other serogroups. The analyzed collection contains 35 nonpathogenic strains that were isolated from the intestines of healthy chickens presenting no lesions typical of avian colibacillosis. These strains were unable to kill any of the five animals tested in the lethality test with 1-day-old chicks (18). Other strains of this collection were all isolated outside the intestine in chickens presenting lesions typical of colibacillosis. They were able to kill zero (6 strains), one (10 strains), two (6 strains), three (11 strains), four (19 strains), or five (99 strains) of the five chicks tested in the lethality test.
TABLE 1.
Strains and plasmids used in this study
| E. coli strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| Strains | ||
| BEN2908 | ExPEC isolate; frz+fim+iut+ibeA+AGI-3+ O2:K1:H5 Nalr | 11, 20, 24 |
| BEN2908 Δfrz::kan | Isogenic frz deletion mutant of BEN2908; Nalr Kanr | This study |
| BEN2908 ΔORF1frz::kan | Isogenic ORF1frz deletion mutant of BEN2908; Nalr Kanr | This study |
| BEN2908 ΔORF5-8frz::kan | Isogenic ORF5-8frz deletion mutant of BEN2908; Nalr Kanr | This study |
| DHB6501 | Nonpathogenic K-12 strain; F− λ− λs Δlac(MS265) mel supF58 Nalr | 7 |
| BEN3152 | A streptomycin-resistant derivative of DHB6501 Nalr Strr | This study |
| DHB 6500 | DHB6501 λInCh2 Nalr Kanr | 7 |
| BEN3277 | BEN3152 attB::(near ori-bla) Nalr Strr Ampr | This study |
| BEN3278 | BEN3152 attB::(near ori-frz-bla) Nalr Strr Ampr | This study |
| Plasmids | ||
| pKD4 | Plasmid carrying a kanamycin resistance cassette, an oriRγ origin, and the bla (Ampr) gene | 15 |
| pUC18 | High-copy-no. cloning vector containing the bla (Ampr) gene | 45 |
| pGEM-T Easy | High-copy-no. cloning vector containing the bla (Ampr) gene | Promega |
| pUC18-frz | pUC18 containing the frz operon of strain BEN2908 cloned into the EcoRI site | This study |
| pUC18K2 | Derivative of pUC18 carrying the aphA3 kanamycin resistance gene in the SmaI restriction site, creating a nonpolar kanamycin cassette (pUC18K with an additional G residue after the ATG start) | 42; personal communication from A. Allaoui |
Bacterial growth conditions.
Strains were routinely grown in LB broth (10 g/liter tryptone [Becton-Dickinson and Company], 5 g/liter yeast extract [Becton-Dickinson and Company], 10 g/liter NaCl, pH 7.4) at 37°C with agitation and on LB agar plates (1.2% agar). Unless otherwise stated, nalidixic acid (30 μg/ml), kanamycin (50 μg/ml), and ampicillin (100 μg/ml), each at the indicated concentration, were used when necessary.
For cocultures of strain BEN2908 and its isogenic deletion mutants containing a kanamycin resistance cassette, strains BEN3152 and BEN3277, or strains BEN3152 and BEN3278, each strain was first separately incubated overnight in 5 ml of LB broth containing nalidixic acid at 37°C with aeration. Strains from each pair were then inoculated in equivalent numbers in 10 ml of LB containing nalidixic acid. These cocultures were incubated in 50-ml Erlenmeyer flasks at 37°C without agitation (one Erlenmeyer flask was tested per time point). The optical density of the cocultures was measured at 600 nm (OD600). At different times, the contents of the Erlenmeyer flask to be tested were mixed and 10-fold dilutions of the cocultures were plated on LB agar containing nalidixic acid. One hundred colonies were then picked on LB agar plates containing kanamycin (BEN2908 and its isogenic deletion mutants) or 25 μg/ml ampicillin (BEN3152 and BEN3277 or BEN3152 and BEN3278) or on LB agar plates containing nalidixic acid. The proportions of the kanamycin- or ampicillin-resistant strains were then evaluated. For cocultures of BEN2908 and isogenic deletion mutants in chicken serum or in IF0 minimal medium (100 mM NaCl, 5 mM NH4Cl, 2 mM NaH2P04·H20, 0.25 mM NaSO4, 0.05 mM MgCl2, 1 mM KCl, 30 mM triethanolamine·HCl, pH 7.3) containing 5 mM of a sole carbon source, a similar protocol was followed but the overnight cultures were first centrifuged at 4,000 × g for 10 min. Bacteria were then washed three times with PBS (137 mM NaCl, 2.7 mM KCl, 2 mM KH2PO4, 10 mM Na2HPO4, pH 7.4) or IF0 and resuspended in the same volume of PBS or IF0 before being inoculated either in chicken serum (Sigma-Aldrich) previously decomplemented by 30 min of incubation at 56°C and containing nalidixic acid or in IF0. The growth of the cocultures was monitored by plating 10-fold dilutions of the cocultures and counting viable bacteria (chicken serum) or by measuring the OD450 (IF0).
Oxidative-stress susceptibility.
Hydrogen peroxide sensitivity was measured as described by Herren et al. (26). In brief, sterile filter paper discs soaked with 1 or 5% hydrogen peroxide were placed on top of bacteria freshly overlaid on 0.8% agar (5 log10 CFU bacteria taken in the exponential [OD600 = 0.2] and stationary [24 h of culture] growth phase of static and agitated cultures). The diameters of the growth inhibition zones were measured.
Cell lines and cultures.
A549 human type II pneumocytes (ECACC catalog no. 86012804) and Caco-2 human epithelial intestinal cells (ATCC HTB-37) were grown at 37°C with 5% CO2 in Dulbecco's modified Eagle's medium containing 2 mM l-glutamine and 4.5 g/liter glucose (Invitrogen). LMH chicken hepatocytes (CLS, Eppelheim, Germany) were grown at 40°C with 5% CO2 in Dulbecco's modified Eagle's medium-F12 medium (1:1) containing 2 mM l-glutamine and 15 mM HEPES (Invitrogen). Ten or 2.5% decomplemented bovine fetal serum (Invitrogen) was added to all cell media for culture in flasks and plates, respectively. Flasks and plates used for LMH cell cultures were previously incubated for 15 min (4°C) with 0.167% gelatin (wt/vol) in PBS.
Assays of bacterial association to and internalization in eukaryotic cells.
For E.coli association and internalization assays, eukaryotic cells cultured in flasks were dissociated with a trypsin solution (0.5 g/liter trypsin, 0.2 g/liter EDTA in Hank's buffered salt solution), suspended in culture medium, and distributed into six-well plates at a density of 2.5 × 105 (LMH), 1.5 × 106 (A549), or 5 × 105 (Caco-2) cells per well. Cells were then incubated overnight in 2 ml of culture medium/well. Culture medium was then removed, and cells were infected with the studied strain at a multiplicity of infection (MOI) of 10. Bacterial inocula were prepared from exponentially growing cultures (LB without antibiotic, at 37°C, with agitation). Upon reaching an OD600 of 0.3, bacteria were diluted to the desired concentration in cell culture medium without bovine fetal serum and immediately used to infect the cells (1.5 ml bacterial dilution/well). The inoculum size was verified by colony plate count for each experiment. After 2 h of infection at 37°C (A549 and Caco-2) or 40°C (LMH), eukaryotic cells were washed with PBS and lysed with cold sterile distilled water for 30 min at 4°C. The growth rates of wild-type and mutant strains in cell culture media were found to be similar during this infection period. Cell-associated bacteria (adherent and intracellular) were enumerated by plating 10-fold serial dilutions of the lysates on LB agar. The percentage of association was calculated [100(number of bacteria recovered after 2 h of infection/number of bacteria inoculated)] and expressed relative to the capacity for association of the wild-type strain BEN2908 (percentage of mutant bacteria to be tested associating with eukaryotic cells versus percentage of BEN2908 bacteria associating with eukaryotic cells). For internalization assays, eukaryotic cells were incubated with bacteria for 2 h as described above. The culture medium was removed, and infected cells were incubated for 90 min at 37°C (A549 and Caco-2) or 40°C (LMH) in culture medium (2 ml/well) containing 2% (Caco-2) or 2.5% (A549 and LMH) bovine fetal serum and 100 μg/ml gentamicin. The eukaryotic cells were then washed with PBS and lysed with cold sterile distilled water. Intracellular bacteria were enumerated by plating 10-fold serial dilutions on LB agar. The percentage of internalization was calculated [100(number of bacteria recovered after the gentamicin treatment/number of bacteria inoculated)] and expressed relative to the capacity for internalization of strain BEN2908 (percentage of mutant bacteria to be tested internalized in eukaryotic cells versus percentage of BEN2908 bacteria internalized in eukaryotic cells). Each assay was performed in triplicate and repeated independently at least two times.
Yeast aggregation assay.
Yeast aggregation titers of E. coli strains were measured as described by Teng et al. (59). Briefly, bacteria prepared as described for eukaryotic cell interaction tests were incubated for 2 h at 37°C with 5% CO2. Bacterial strains were then adjusted to an OD600 of 0.4, subjected to serial twofold dilutions in PBS, and mixed (equal volumes) with commercial baker's yeast cells (Saccharomyces cerevisiae) at 5 mg dry weight per ml. Aggregation was monitored visually in 96-well V-bottom microtitration plates, and the titer was recorded as the highest dilution giving a positive aggregation result.
Chicken intestinal colonization experiments.
Axenic PA12 strain White Leghorn chicks grown according to the method of Le Bars (31) were obtained from the INRA Infectiology Platform. The housing, husbandry, and slaughtering conditions were in conformity with European Union guidelines for the care of laboratory animals. The experimental protocol was approved by the regional ethical committee under number CL2007-43. Animals were reared in isolators, fed ad libitum with a commercial diet sterilized by gamma irradiation (poultry starter HPS; Dietex, France), and supplied with autoclaved tap water. After 18 h of starvation, six 12-day-old axenic chicks were fed with 0.5 ml of a mix of LB static cultures (20 h of culture) of strains BEN2908 and BEN2908 Δfrz::kan (an equivalent amount of each strain, with a total of 109 CFU). Fresh fecal samples were collected for 7 days after bacterial feeding. Viable E. coli cells were analyzed by plating 10-fold fecal dilutions in sterile saline on LB agar plates containing nalidixic acid. One hundred colonies were then picked on LB agar containing kanamycin (for selection of strain BEN2908 Δfrz::kan) and on LB agar containing nalidixic acid plates (for selection of BEN2908 and BEN2908 Δfrz::kan), and the proportion of the frz deletion mutant (kanamycin-resistant strain) was calculated.
Nucleic acid manipulations.
Standard DNA manipulation techniques were carried out as described by Sambrook and Russell (50). Plasmid and E. coli chromosomal DNA were purified with Nucleobond PC100 and Nucleospin tissue kits according to the manufacturer's protocol (Macherey-Nagel). For extraction of total RNA, bacterial cells taken in the mid-exponential phase of growth were first treated with RNA Protect (Qiagen). The stabilized RNAs were then extracted with an RNA Pure Yield kit (Promega). Bacteria were transformed by electroporation following the method of Tung and Chow (62). For Southern blot hybridization, DNA restriction fragments were submitted to electrophoresis and transferred to a Hybond-N+ membrane (Amersham, GE Healthcare Life Sciences). Probes were labeled with peroxidase, and hybridized DNA fragments were revealed by using an enhanced chemiluminescence kit (RPN3000; Amersham Pharmacia Biotech) as described by the manufacturer.
Amplification of DNA sequences by PCR.
Unless otherwise stated, PCR amplification was performed in a mixture with a 50-μl total volume containing 1 μM of the forward and reverse primers, 200 μM of each deoxyribonucleoside triphosphate (Finzyme, Ozyme, France), and 1.25 U of Taq DNA polymerase (New England Biolabs, Inc.) in a PCR buffer containing 10 mM KCl, 10 mM (NH4)2SO4, 2 mM MgSO4, 0.1% Triton X-100, 20 mM Tris-HCl, pH 8.8 (New England Biolabs, Inc.). Amplifications were performed in a Perkin-Elmer thermocycler (GeneAmp 9700; Applied Biosystems) with the following temperature program: 1 cycle of 45 s at 95°C; 30 cycles of 45 s at 95°C, 60 s at 5°C (melting temperature), and 1 min/kb at 72°C; and finally, 1 cycle of 10 min at 72°C. The primers used in this study are listed in Table 2.
TABLE 2.
Oligonucleotide primers used in this study
| Primera | Sequenceb | Location (reference) |
|---|---|---|
| C4488-4as | AAGCGCGGAGAATATTGCTGATTT | ORF1frz |
| C4488-6s (fw) | TCCTGAATGGACGATTGTTGATGTTA | ORF1frz, 3′ end |
| RI 87-88 | GTTATGTCCATTCGTCATACCTCA | ORF2frz, 5′ end, and ORF1-2frz intergenic region |
| C4487-as2 (rv) | AGTCGAGCCTTCCGCTTCG | ORF2frz, 5′ end |
| C4487-s3 (fw) | AACGCTGGATGGCAGTAGCGT | ORF2frz, 3′ end |
| C4486-as4 (rv) | GCACAATACGTTTATTGGTGAAGCG | ORF3frz, 5′ end |
| C4486-as3 (rv) | AGTGATTTTTCGACCTTTTCAA | ORF3frz |
| C4486-s2 (fw) | GGTATTGCTCATACCTATATTGCTC | ORF3frz |
| C4486-as (rv) | CGGCAACACAGACAATATTCA | ORF4frz, 5′ end |
| C4485-s3 (fw) | GTGGAGAGTCCGGTTCCGCA | ORF4frz, 3′ end |
| RI 85-86 | ACCGGCATCTCGTGGATGATA | ORF4frz |
| C4484-3as (rv) | AGATTCCGCGAGCGTTTTAACCA | ORF5frz, 5′ end |
| C4484-4s (fw) | GCGTGCCTCTGGCATTACATG | ORF5frz, 3′ end |
| C4483-3as (rv) | CGCTGGGATGGATGGCAATAA | ORF6frz, 5′ end |
| C4483-4s (fw) | AATACGCCTATTTCCAGAAAGTGCG | ORF6frz, 3′ end |
| C4483-2as (rv) | GGATCTGTGTAGGTCACTTGTGA | ORF6frz |
| C4483-3s (fw) | GAAAGAATTACTGCAACCTACCC | ORF6frz |
| C4482-3as (rv) | GTCCCGCCTAATCTGCTCGATAAG | ORF7frz, 5′ end |
| C4482-4s (fw) | CCCTGGCGTGATTGTGATCG | ORF7frz, 3′ end |
| C4481-as (rv) | GACCAAAATCGGTGATATCCCAAC | ORF8frz, 5′ end |
| YicH | TGGCCTGAATTACCGTTGAC | yicH |
| YicI | GCCCGTTTATGTGCGTGATA | yicI |
| YicH-as-EcoRI | CATGAGAATTCGCGTAGTTTCCAGTGGTGAAGTTAC | yicH |
| YicI-s-EcoRI | GACTGGAATTCACTCCTCAAGGGAATGCGCTG | yicI |
| X1"-P1 | GCGTTCCCCGTTATTAGCGCAAAATTCCATACTCACCGCACAATAACCATGTGTAGGCTGGAGCTGCTTCG | ORF8frz, 5′ end, and ORF8frz-yicH intergenic region |
| X4-a | TGGTTTTGTGTCGTATCCGGTAAATAATCTCACATCTGGTAATAAATATTAGAGCGCTTTTGAAGCTGGG | ORF1frz-yicI intergenic region |
| X1"-a | GCGTTCCCCGTTATTAGCGCAAAATTCCATACTCACCGCACAATAACCATAGAGCGCTTTTGAAGCTGGG | ORF8frz, 5′ end, and ORF8frz-yicH intergenic region |
| E-P1 | AGCGATCAAAAGCTGAAAGATGAAGAGATCACTTTCACCCTCGAATAACCGTGTAGGCTGGAGCTGCTTCG | ORF4frz, 5′ end, and ORF4-5frz intergenic region |
| Reguls-aph3s | GTGAAACACCGAATGAAACGTCGA TCAGTAAATTGTCAGAACGCTACTTTACCATGATTACGAATTCGAGC | ORF1frz, nucleotides 317 to 366 |
| Regulas-aph3as | TTCCGATTCCGGTTGAACAGACCAG TAATACACGTTTACGGGCAATTTGCAGGTCGACTCTAGAGGATCCC | ORF1frz, nucleotides 1297 to 1248 |
| Aph3-2s | CGAGCTATTTTTTGACTTACTGGG | aph3 cassette of pUC18K2 |
| Aph3-2as | CCCACCAGCTTATATACCTTAGCAGGAG | aph3 cassette of pUC18K2 |
| PUC18K2-as | ATTATTCCCTCCAGGTACTAAAAC | aph3 cassette of pUC18K2 |
| PUC18K2-s | CTAAAATGAGAATATCACCGGA | aph3 cassette of pUC18K2 |
| Askana1 | CAGATAGCCCAGTAGCTGACATT | Kanr cassette of pKD4 |
| Askana2 | CCGAAGCCCAACCTTTCATA | Kanr cassette of pKD4 |
| Cat51 | GTGTAGGCTGGAGCTGCTTC | Kanr cassette of pKD4 |
| Skana | CAACCTGCCATCACGAGATT | Kanr cassette of pKD4 |
| Rpos-s | ATTCGTTACAAGGGGAAATC | nlpD |
| RpoS-as | TCAGTCAGATAAAGTTTTAACGCT | rpoS 5′-side intergenic region |
| gal_f | CTTGCTGAGTACGTGAGTTC | gal (7) |
| -att_r | AAGCAGGCTTCAACGGATTC | att (7) |
| -int_R | GGACACCATGGCATCACAGT | int (7) |
| -IG_r | ACGTTGGAGTCCACGTTCTT | M13 I.G. (7) |
| Ch1-F | AGTAATGCTGCTCGTTTTGC | Downstream of fim IE (59) |
| Ch1-R | GACAGAGCCGACAGAACAAC | Upstream of fim IE (59) |
rv, fw; reverse and forward primers, respectively, used for RT-PCR.
Sequences complementary to the Kanr cassettes are in bold. Tails containing an EcoRI restriction site are underlined.
fim promoter orientation assay.
The orientation of the invertible DNA element (IE) containing the fim promoter was tested as described by Teng et al. (59). In brief, strain BEN2908 and its isogenic deletion mutants were grown overnight in LB medium (37°C) with agitation. The cultures were then diluted 1/100, and bacteria were grown without agitation to the OD600 at which they were tested (10 ml of LB medium containing no antibiotic in a 50-ml Erlenmeyer flask). The cultures were then mixed and centrifuged. After lysis of the pelleted bacteria, the IEs from both phase-ON and phase-OFF (i.e., fimbriated and nonfimbriated) bacteria were amplified by PCR using primers Ch1-F and Ch1-R. The 602-bp fragments amplified were hydrolyzed with SnaB1, and the digested products were separated in a 2% agarose gel. SnaB1 digestion of the IE in the ON orientation yields fragments of 404 and 198 bp, whereas SnaB1 digestion of the IE in the OFF orientation results in fragments of 442 and 160 bp.
RT-PCR.
Reverse transcriptase PCRs (RT-PCRs) were performed as described by Gilot et al. (25). In brief, after treatment with DNase I, total RNA was reverse transcribed with Moloney murine leukemia virus reverse transcriptase (Invitrogen) and one of the reverse primers of interest (Table 2, rv). After inactivation of the reverse transcriptase, the reaction mixtures were used for PCR amplification of the cDNA with the reverse primers and appropriate forward primers (Table 2, fw) as described above for the amplification of DNA sequences by PCR. Control RT-PCRs, omitting reverse transcriptase, were performed to check for DNA contamination of the RNA preparations.
Construction of BEN2908 Δfrz::kan, BEN2908 ΔORF5-8frz::kan, and BEN2908 ΔORF1frz::kan mutants.
The complete frz operon, frz open reading frame 1 (ORF1frz), or frz ORFs 5 to 8 (ORF5-8frz) was deleted from the chromosome of strain BEN2908 by the method of Datsenko and Wanner, with slight modifications (15). Briefly, by using a Red recombination procedure, the above chromosomal sequences were replaced with a kanamycin resistance cassette that was generated by PCR using primers with extensions that are homologous to regions adjacent to the sequences to be inactivated. The complete frz operon or ORF5-8frz was replaced by a kanamycin resistance cassette obtained by PCR amplification from plasmid pKD4, using primers X1"-P1 and X4-a or X1"-a and E-P1, respectively (Table 2). The 5′ extensions of the X1"-P1 and X1"-a primers are homologous to 50 nucleotides covering the last 37 nucleotides of ORF8frz and 13 nucleotides of the yicH-ORF8frz intergenic region; the 5′ extension of the X4-a primer is homologous to 50 nucleotides of the ORF1frz-yicI intergenic region; and the 5′ extension of the E-P1 primer is homologous to 50 nucleotides covering the last 48 nucleotides of ORF4frz and 2 nucleotides of the intergenic region between ORF4frz and ORF5frz. The deletion procedures conserved the putative transcription terminators of yicH and yicI (conservation of 43 and 75 nucleotides just downstream of the stop codons of yicH and yicI, respectively, for the frz deletion mutant and conservation of 43 nucleotides just downstream of the yicH stop codon and the complete yicI-ORF1frz intergenic region for the ORF5 -8frz deletion mutant). The replacement of the complete frz operon or ORF5-8frz was confirmed by PCR using primer pairs YicH/Askana1 and Skana/YicI or YicH/Skana and Askana1/RI85-86 that allow the detection of left and right junctions between the bacterial chromosome and the kanamycin resistance cassette, respectively, for each replacement (Table 2). The junction between the kanamycin cassette and ORF4frz of the BEN2908 ΔORF5-8frz::kan mutant was verified by sequencing (using primer Askana1). Southern blots of EcoRV- or SspI-digested DNA of the mutants with a probe that was generated by PCR amplification of the kanamycin resistance gene (primers Cat51 and Askana2) revealed a 2,334-bp and a 2,750-bp EcoRV DNA fragment of the frz and ORF5-8frz deletion mutant, respectively, or a 1,661- and 3,085-bp SspI DNA fragment of the frz and ORF5-8frz deletion mutant, respectively (Table 2). This indicated that the kanamycin cassette was not also illegitimately inserted into another part of the genome.
The central part (881 bp) of ORF1frz was replaced with a nonpolar aph3 kanamycin resistance cassette obtained by PCR amplification from plasmid pUC18K2 using primers Reguls-aph3s and Regulas-aph3as (Table 2). The 5′ extensions of primers Reguls-aph3s and Regulas-aph3as are homologous to nucleotides 317 to 366 and 1297 to 1248 of ORF1, respectively. This construct generated an early stop in ORF1frz translation and allowed in-frame translation of the remaining 3′ end of the mutated gene, avoiding possible polar effects on the expression of downstream cistrons. Translation of the remaining 3′ end of ORF1frz was made possible by the presence of a consensus ribosome binding site and a start codon at the end of aph3. The replacement of the central part of ORF1frz was confirmed by PCR using primer pairs Aph3-2s/RI87-88 and Aph3-2as/C4488-4as that allowed the detection of left and right junctions between the bacterial chromosome and the aph3 cassette (Table 2). The placement of the ATG downstream from the aph3 gene in frame with the TAA stop codon of ORF1frz was verified by sequencing the PCR-amplified (primers Aph3-2s and RI87-88) junction between the aph3 cassette and the remaining 3′ end of ORF1frz. Transcription of downstream genes was verified by RT-PCR detection of ORF3frz (reverse primer C4486-as3 and forward primer C4486-s2) and ORF6frz (reverse primer C4483-2as and forward primer C4483-3s) transcripts. Southern blotting of FspI- or BseYI-digested DNA of the ORF1frz mutant with a probe that was generated by PCR amplification of the aph3 resistance cassette (primers PUC18K2-as and PUC18K2-s) revealed a 5,818-bp and a 2,452-bp DNA fragment, respectively, indicating that the kanamycin cassette was not also illegitimately inserted into another part of the genome.
Cloning of the frz operon in plasmids pUC18 and pGEM-T Easy.
The whole BEN2908 genomic region from the end of yicH to the end of yicI was amplified by PCR using PfuUltra II fusion HS DNA polymerase (Stratagene) and primers YicH-as-EcoRI and YicI-s-EcoRI containing an EcoRI restriction site tail (Table 2). After digestion with EcoRI, the PCR product was cloned in the EcoRI site of pUC18, generating plasmid pUC18-frz. Identity of the cloned region with the original BEN2908 region was verified by sequencing. The frz operon was then subcloned into the EcoRI site of the pGEM-T Easy plasmid that had previously had one of its two EcoRI sites deleted. In pUC18-frz and pGEM-T Easy-frz, frz genes are oriented in the same direction as the β-galactosidase α-peptide ORF.
Insertion of the frz operon into the chromosome of an E. coli K-12 strain.
The whole BEN2908 genomic region from the end of yicH to the end of yicI was inserted into the chromosome of the K-12 strain BEN3152 by using the λInCh tool as described by Boyd et al. (7). In brief, homology between pUC18-frz (near-ori region and part of the bla gene) and sequences on the λInCh bacteriophage permitted a double recombination which conferred ampicillin resistance on the phage and resulted in pick-up of the frz region. In a second step, the ampicillin resistance gene in the phage and the linked frz region were put on the chromosome of strain BEN3152 by site-specific recombination of the phage into the λ att site. In a third step, most of the λ DNA was deleted by a second homologous recombination event that was made possible because the phage carries a fragment of bacterial DNA from one site of the att site (near-att DNA) in a position such that most of the lambda DNA can be looped out and deleted by a single recombination event. The resulting strain (BEN3278) thus has the near-ori region of pUC18, the frz operon, and the bla gene of pUC18 integrated at the att site. A control strain (BEN3277) was constructed similarly after recombination of pUC18 with λInCh. This strain has the near-ori region and the bla gene of pUC18 integrated at the att site. The structures at the insertion site have been confirmed by PCR with primers gal_f, -att_r, -int_r, and -IG_R as described by Boyd et al. (7) (Table 2).
DNA sequencing and sequence analysis.
PCR-amplified fragments were sequenced on both strands by using Genome Express (Meylan, France). Sequences were assembled and analyzed using the DNA Strider program. Homology searches were performed by using the BLAST server from the National Center for Biotechnology Information website.
Statistical analysis.
Data were analyzed by Student's t test (57). The significance of the differences observed between the mean proportion of the mutant in the bacterial population isolated from chicken feces and the proportion of the mutant in the inoculum was tested by the equation 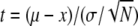 , where μ is the proportion of the mutant in the inoculum; x is the mean proportion of the mutant in chicken feces; σ is the standard deviation of the mean proportion of the mutant in chicken feces; and N is the number of animal tested. The significance of the differences between the mean proportion of the mutant at the onset (time zero) of culture and the mean proportion of the mutant at different times during culture was tested by the equation
, where μ is the proportion of the mutant in the inoculum; x is the mean proportion of the mutant in chicken feces; σ is the standard deviation of the mean proportion of the mutant in chicken feces; and N is the number of animal tested. The significance of the differences between the mean proportion of the mutant at the onset (time zero) of culture and the mean proportion of the mutant at different times during culture was tested by the equation 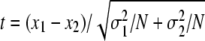 , where x1 is the mean proportion of the mutant at the onset of culture; x2 is the mean proportion of the mutant during culture; σ1 is the standard deviation of the mean proportion of the mutant at the onset of culture; σ2 is the standard deviation of the mean proportion of the mutant during culture; and N is the number of cultures.
, where x1 is the mean proportion of the mutant at the onset of culture; x2 is the mean proportion of the mutant during culture; σ1 is the standard deviation of the mean proportion of the mutant at the onset of culture; σ2 is the standard deviation of the mean proportion of the mutant during culture; and N is the number of cultures.
Nucleotide sequence accession number.
The sequences of the BEN2908 rpoS gene and frz region have been deposited in the EMBL database under accession numbers AM421516 and FM253092, respectively.
RESULTS
Sequencing of the D7 genomic environment of the ExPEC strain BEN2908 and identification of the frz operon.
By sequencing the genomic environment of the D7 genomic fragment of strain BEN2908, we identified a 7,281-bp DNA region inserted between the yicH and yicI genes of the common genomic backbone of E. coli and separated by only two genes (yicI and yicJ) from the tRNA selC locus (Fig. 1) (EMBL accession no. FM253092). This genomic region contains eight ORFs (1,554 bp, 471 bp, 315 bp, 1,092 bp, 852 bp, 861 bp, 966 bp, and 720 bp for ORF1 to -8, respectively) transcribed in the same direction and separated by short intergenic sequences (26 bp, 17 bp, 24 bp, 11 bp, 80 bp, and 14 bp between ORF1 and 2, ORF2 and -3, ORF3 and -4, ORF4 and -5, ORF5 and -6, and ORF6 and -7, respectively) or overlapped on 53 bp (ORF7 and -8). The D7 DNA fragment represents the central part of ORF4.
FIG. 1.
Schematic representation of the frz region of Escherichia coli BEN2908. The frz region of E. coli BEN2908 was sequenced on both strands (EMBL accession no. FM253092). Open reading frames labeled consecutively from 1 to 8 (open arrows), a potential Fnr/Cap binding site (open ellipse), and ribosome (open rectangles) binding sites were identified with the DNA Strider program. Putative σ70 transcriptional promoters (bent arrows) and transcriptional terminators (Ω) were predicted with the BPROM (SoftBerry, Inc.) and the mfold (www.bioinfo.rpi.edu/∼zukerm/rna/) programs, respectively. Direct repeats (DR) were identified with the RePuter program (http://bibiserv.techfak.uni-bielefeld.de/reputer/). Genomic regions belonging to the common conserved genomic backbone of E. coli are represented in gray, using symbols similar to those described above. The selC tRNA coding region is represented by a gray triangle. Predicted functions of the proteins encoded are indicated, and the avian pathogenic E. coli genomic island 3 (AGI-3) is represented by a black bold line.
Homology searches indicated that these eight genes are potentially implicated in carbohydrate metabolism. Three of them (ORF2-4) encode proteins homologous to subunits IIA, IIB, and IIC of PTS transporters of the fructose subfamily. In E. coli, the first transporter of this subfamily to be discovered was Fru, the fructose transporter. Homologs further identified were then named Frv to Fry (58). The transporter encoded by ORF2-4 was named Frz to follow this nomenclature. ORF1 codes for a protein with PTS regulatory domains (PRDs) that is homologous to the transcriptional activator of the PTS of the multiple gene regulator of group A streptococcus (MgA) family (27, 56). ORF5 and -6 code for two proteins homologous to type II ketose-1,6-bisphosphate aldolases (8, 23); ORF7 codes for a homolog of the sugar-specific kinase of the repressor, ORF, kinase (ROK) family (60); and ORF8 codes for a hypothetical protein that is a member of the DUF1498 family and the cupin superfamily (21).
The yicI-ORF1 and yicH-ORF8 intergenic regions are 294 bp and 43 bp long, respectively. Correctly conserved and positioned ribosome binding sites were identified upstream of all ORFs with the exception of ORF8 (35). Correctly spaced −35 and −10 sequences (3 and 4 conserved nucleotides with the respective consensus sequences) of a putative σ70 transcriptional promoter were identified before ORF1 (35). Hairpin structures potentially acting as transcriptional terminators for yicI [a 14-bp stem with a 6-bp loop followed by a poly(U) stretch] or yicH and ORF8 (an 11-bp stem with an 8-bp loop) were predicted on the RNA encoded by the yicI-ORF1 and yicH-ORF8 intergenic regions (35). Potential Fnr/Cap binding sites were also identified in a palindromic region located 70 bp upstream of the −35 promoter sequence of ORF1 (22, 35, 38) (Fig. 1) (EMBL accession no. FM253092).
The genetic organization of the 7,281-bp DNA region inserted between the yicI and yicH genes suggests that mRNA of ORF1 to ORF8 is transcribed in one piece and forms an operon under the possible regulation of the closely related Fnr/Cap protein(s) involved in adaptation to oxygen-restricted conditions and catabolite repression, respectively. By RT-PCR performed with reverse primers localized at the 5′ end of each ORF and forward primers annealing at the 3′ end of an upstream ORF, we confirmed the presence of cotranscripts between these eight ORFs, and thus, we named this DNA region the frz operon (Fig. 2).
FIG. 2.
Cotranscription of frz open reading frames. Cotranscription of frz ORFs was revealed by RT-PCR performed with reverse primers localized at the 5′ end of each ORF (from ORF2 to ORF8) and forward primers annealing at the 3′ end of an upstream ORF. Amplified cotranscripts between ORF1 and -2 (lanes 2 and 3; primers C4487-as2 and C4488-6s), ORF2 and -3 (lanes 4 and 5; primers C4487-s3 and C4486-as4), ORF4 and -5 (lanes 6 and 7; primers C4484-3as and C4485-s3), ORF5 and -6 (lanes 8 and 9; primers C4483-3as and C4484-4s), ORF6 and -7 (lanes 10 and 11; primers C4482-3as and C4483-4s), ORF7 and -8 (lanes 12 and 13; primers C4481-as and C4482-4s), ORF1 and -3 (lanes 16 and 17; C4488-6s and C4486-as4), and ORF2 and -4 (lanes 18 and 19; primers C4487-s3 and C4486-as) were electrophoresed in a 1% agarose gel containing ethidium bromide and visualized under UV light (260 nm). RT-PCRs were performed in the absence of reverse transcriptase to check for DNA contamination (lanes 3, 5, 7, 9, 11, 13, 17, and 19). Molecular weight markers (Smart DNA ladder; Eurogentec) of the indicated sizes are in lanes 1, 14, and 15.
Finally, 9-bp direct repeats (with one mismatch) were also found in the yicI-ORF1 and yicH-ORF8 intergenic regions (Fig. 1) (EMBL accession no. FM253092).
BlastN searches indicated that an frz operon is present in the sequenced genomes of B2 phylogenetic group strains of human (uropathogenic strains CFT073, F11, UT198, and 536; neonatal meningitis-causing strains RS218 and S88; enteropathogenic strain E2348/69; and fecal strain ED1A) and avian (avian ExPEC strain APEC 01) origin. The frz operon is also present in the sequenced genomes of two phylogenetic group D strains: IAI39, a uropathogenic strain; and SMS-3-5, a strain isolated from an industrially polluted aquatic environment that is closely associated with avian extraintestinal E. coli. However, frz is absent from the sequenced genomes of O157:H7 enterohemorrhagic strains Sakai, EDL933, and EC4115; enterotoxigenic strain E24377A; enteroaggregative strain 55989; clonal group A uropathogenic strain UMN026; and nonpathogenic strains HS, SE11, IAI1, K-12 substrain MG1655, K-12 substrain W3310, K-12 substrain DH10B, and C substrain ATCC 8739. BlastN searches also revealed the presence of a homolog of the frz operon that is truncated of ORF7 and -8 in the genome of the enterobacterial phytopathogen Erwinia caratovora subsp. atroseptica SCRI1043 (“Pectobacterium atrosepticum”), whereas BlastP searches revealed the presence of a homolog of frz truncated of ORF7 and -8 or of ORF8 in the genomes of several Listeria sp. strains and in the genome of Clostridium sp. strain SS2/1 (a member of the human gut microbiome) respectively.
The similarities between the frz operon of strain BEN2908 and those identified in sequenced E. coli strains of human and avian origin are very high (between 99.4 and 100% identical nucleotides), suggesting a similar role for the frz operon in those strains, whatever the origin of the isolates.
Relationship between the presence of the frz operon and the pathogenicity of the strain.
A relationship between the presence of frz and the pathogenicity of the strain is suggested by the high association of this operon with ExPEC strains and its absence in the genomes of the above-cited nonpathogenic strains. To better characterize this association, we analyzed a highly characterized and epidemiologically unrelated collection of 151 ExPEC and 35 nonpathogenic strains of avian origin (55). These strains were divided into seven classes depending on their level of pathogenicity for 1-day-old chicks, and the presence of the frz operon in their genomes was searched by hybridization with the ORF4frz sequence, as described by Schouler et al. (52). As shown in Fig. 3, the results indicate that the frz operon is very rarely found among nonpathogenic strains of avian origin (5% of the strains) and that its association with ExPEC strains increases with their virulence for 1-day-old chicks, reaching an association of 75% with the strains of the most virulent group. Taken together, these results demonstrate an association of the frz operon with both clinical and increased virulence of avian E. coli.
FIG. 3.
Association of the frz operon with the level of virulence of the strains. One hundred eighty-six strains isolated from chickens were divided into seven classes depending on their level of pathogenicity (n, total number of strains in each class). The level 0 class contains nonpathogenic strains that were isolated from the intestines of healthy chickens presenting no lesions typical of avian colibacillosis. These strains were unable to kill any of the five animals tested in the 1-day-old-chick lethality test. Classes of levels 1 to 6 contain pathogenic strains. They were all isolated outside the intestine in chickens presenting lesions typical of colibacillosis. They were able to kill zero, one, two, three, four, or five of the five chicks tested in the lethality test, respectively. The proportions of the strains containing the frz operon in each class were assessed by hybridization of their DNA with the D7 fragment (central part of ORF4frz).
The frz operon is involved in the survival of BEN2908 in Luria-Bertani medium during the late stationary growth phase under oxygen-restricted conditions.
To test whether the frz metabolic operon has an impact on the growth capacity of E. coli, we replaced the complete frz operon of BEN2908 with a kanamycin resistance cassette. Static growth of the wild-type strain BEN2908 and of its isogenic mutant BEN2908 Δfrz::kan in LB medium was then monitored. Under oxygen-restricted conditions that are close to those found in the intestinal tract (the primary habitat of ExPEC), the two growth curves were nearly indistinguishable (results not shown). Cocultures of strains BEN2908 and BEN2908 Δfrz::kan were then performed in LB medium under oxygen-restricted conditions (static growth), and the proportion of each strain was examined during 336 h. Under these conditions, the wild-type strain BEN2908 strongly outcompeted its isogenic deletion mutant BEN2908 Δfrz::kan during the late stationary but not during the exponential growth phase, indicating an involvement of the frz operon in the survival mechanism of the strain during the late stationary growth phase in Luria-Bertani medium (Fig. 4A). We also performed similar experiments in minimal medium containing different carbohydrates as a sole carbon source. In none of these media did the wild-type strain strongly outcompete its isogenic deletion mutant during the late stationary growth phase, although a slight effect was observable with three (d-psicose, d-tagatose, and d-sorbose) of the eight potential substrates of ketose-1,6-bisphosphate aldolases (Fig. 4B). This suggests that a molecule present in LB medium and absent in the minimal media tested is necessary for promoting fitness of an frz strain during the late stationary growth phase.
FIG. 4.
Involvement of the frz operon in the survival of strain BEN2908 during the late stationary phase of static growth. (A and C) Strains BEN2908 and BEN2908 Δfrz::kan were inoculated in equivalent numbers into LB medium containing nalidixic acid. These cultures were incubated at 37°C without (A) or with (C) agitation. The cultures were measured at OD600, and growth curves were traced. The proportion of strain BEN2908 Δfrz::kan in the cultures was analyzed during 336 h. (B) Cocultures were incubated for 168 h without agitation in a minimal medium containing different carbohydrates as the sole carbon source (A, glucose; B, mannitol; C, mannose; D, 3-O-β-d-galactopyranosyl-d-arabinose; E, d-fructose; F, l-fructose; G, d-psicose; H, l-psicose; I, d-sorbose; J, l-sorbose; K, d-tagatose; and L, l-tagatose). Results are presented as means ± standard deviations for three independent cocultures. Growth in minimal medium was estimated by measuring the OD after 168 h (−, OD450 = 0.07; ±, OD450 = 0.35; +, OD450 = 0.6). The proportion of the mutant in the inoculum (starting ratio) is represented by the dotted line. The standard deviation of the inoculum ratio is less than 5%. The significance of the difference between the proportion of the mutant in the inoculum and during culture was tested with the Student t test (*, 0.05 > P > 0.02; **, 0.02 > P > 0.01; ***, P < 0.001).
Since a putative binding site for the Fnr protein involved in the adaptation of E. coli to oxygen-restricted conditions was predicted upstream of the transcriptional promoter of frz, we tested whether oxygenation has an impact on the effect of frz on the survival of the strain during the late stationary phase of growth. To that end, cocultures of strains BEN2908 and BEN2908 Δfrz::kan were performed in highly oxygenated LB medium (growth with agitation), and the proportion of each strain was examined during 336 h. Under these conditions, the proportions of strains BEN2908 and BEN2908 Δfrz::kan did not change during the entire experiment, indicating that oxygenation has a strong influence on the effect of frz on the survival of the strain during the late stationary growth phase (Fig. 4C).
We also tested the impact of other environmental factors on the survival capacity of the BEN2908 and BEN2908 Δfrz::kan strains. To that end, LB medium was buffered at pH 5.0 (100 mM morpholineethanesulfonic acid [MES]), pH 7.0 (100 mM HEPES), or pH 9.0 (100 mM TAPS; Sigma) and the NaCl concentration was modified to 50, 100, or 300 mM. Under all these conditions, the wild-type strain BEN2908 strongly outcompeted its isogenic deletion mutant BEN2908 Δfrz::kan during the late stationary static growth phase but not during the exponential growth phase (results not shown). The oxidative stress sensitivities of strain BEN2908 and of its isogenic frz deletion mutant were also compared by measuring the diameters of growth inhibition zones around paper discs soaked with hydrogen peroxide placed on top of bacteria overlaid on agar plates. No significant differences were observed between the two strains (17.3 ± 0.5 mm [mean ± standard deviation] and 17.7 ± 1.0 mm around discs soaked in 1% H202 for BEN2908 and BEN2908 Δfrz::kan, respectively, and 24.8 ± 0.4 mm and 24.3 ± 0.5 mm around discs soaked in 5% H202 for BEN2908 and BEN2908 Δfrz::kan, respectively). Saline, pH, or oxidative stress thus has little or no influence on the expression of the frz phenotype. Finally, by sequencing the rpoS genes of the wild-type strain (EMBL accession no. AM421516) and of the frz deletion mutant, we verified that mutations in the alternative σS factor involved in the regulation of several genes implicated in stress response during the stationary phase of growth are not responsible for the different capacities of the two strains to survive in Luria-Bertani medium during the late stationary growth phase under oxygen-restricted conditions.
The frz operon is sufficient to give an advantage to an Escherichia coli strain during the late stationary growth phase under oxygen-restricted conditions.
By cloning the whole DNA region present between the end of the yicH and yicI genes of BEN2908 into the EcoRI site of plasmid pUC18 and introducing this recombinant plasmid into the mutant strain BEN2908 Δfrz::kan, we were able to restore the wild-type phenotype of the strain (see Fig. S1 in the supplemental material).
Whereas the results of the experiment described above proved that the reduced survival capacity of strain BEN2908 Δfrz::kan is due to the deletion of the frz operon and not to a mutation arising in another part of the genome, they do not prove that other specific BEN2908 genes in conjunction with the frz operon are not implicated in the generation of this phenotype. We thus inserted the frz operon at the lambda phage attachment site (att site) of the nonpathogenic K-12 strain BEN3152, generating strain BEN3278. As the genetic mechanism used to insert the frz operon at the att site also generated the insertion of the near-ori and the bla gene of pUC18, we constructed the control strain BEN3277 by inserting only the near-ori and the bla gene in the att site of BEN3152. In cocultures performed in LB medium under oxygen-restricted conditions (static growth), strain BEN3278 (frz+) strongly outcompeted strain BEN3152 (frz negative) during the late stationary growth phase. However, the proportions of strains BEN3277 (frz negative) and BEN3152 (frz negative) remained constant during the entire incubation time of the cocultures (Fig. 5). These data indicate that the frz operon is sufficient to favor an E. coli strain during the late stationary phase of growth under oxygen-restricted conditions.
FIG. 5.
The frz operon is sufficient to favor an Escherichia coli strain during the late stationary phase of growth in oxygen-restricted conditions. Strains BEN3152 and BEN3277 (□) or BEN3152 and BEN3278 (░⃞) were inoculated in equivalent numbers into LB medium containing nalidixic acid. These cocultures were incubated at 37°C without agitation. After 24 h, 72 h, and 168 h of incubation, the proportion of the bla strains (Ampr) in the cocultures was evaluated. Results are presented as means ± standard deviations for three independent cocultures. The proportion of the mutant in the inoculum (starting ratio) is represented by a dotted line. The standard deviation of the inoculum ratio is less than 5%. The significance of the results was tested with the Student t test (*, P < 0.001).
The PTS activator and the enzymes encoded by the frz operon are involved in the survival of BEN2908 in Luria-Bertani medium during the late stationary growth phase under oxygen-restricted conditions.
To learn if the metabolic enzymes and the PTS activator encoded by the frz operon are involved in the survival phenotype described above, we replaced ORF1frz and ORF5-8frz with the nonpolar aph3 and the pKD4 kanamycin resistance cassette, respectively. Like the BEN2908 Δfrz::kan mutant, mutants BEN2908 ΔORF1frz::kan and BEN2908 ΔORF5-8frz::kan were always outcompeted by the wild-type strain during the late stationary growth phase of static cocultures (Fig. 6). The PTS activator and the metabolic enzymes are thus both implicated in the survival of strain BEN2908 during the late stationary phase of static growth.
FIG. 6.
Involvement of ORF1frz and ORF5-8frz in the survival of strain BEN2908 during the late stationary phase of static growth under oxygen-restricted conditions. (A and B) Strains BEN2908 and BEN2908 ΔORF1frz::kan (A) or BEN2908 and BEN2908 ΔORF5-8frz::kan (B) were inoculated in equivalent numbers into LB medium containing nalidixic acid. These cocultures were incubated at 37°C without agitation. The cocultures were measured at OD600, and growth curves were traced. The proportion of strain BEN2908 ΔORF1frz::kan (A) or BEN2908 ΔORF5-8frz::kan (B) in the cultures was analyzed during 336 h. Results are presented as means ± standard deviations for three independent cocultures. The proportion of the mutant in the inoculum (starting ratio) is represented by a dotted line. The standard deviation of the inoculum ratio is less than 5%. The significance of the difference between the proportion of a mutant in the inoculum and during culture was tested with the Student t test (*, P < 0.001).
The frz operon promotes fitness in host and eukaryotic cell invasion.
In its primary habitat, the intestine, strain BEN2908 encounters stressful conditions, notably, an oxygen-restricted situation. We thus tested whether the frz operon also promotes bacterial fitness in the intestine. To this end, we fed 12-day-old axenic White Leghorn chicks with equivalent numbers of strains BEN2908 and BEN2908 Δfrz::kan. We then examined the proportions of these strains in the fecal content of the animals during seven days. The data presented in Fig. 7 indicate that the frz region gives an advantage to strain BEN2908 for the colonization of the chick intestinal tract.
FIG. 7.
Colonization of the intestinal tract of chicks with a coculture of strains BEN2908 and BEN2908 Δfrz::kan. Six 12-day-old axenic White Leghorn chicks were fed a coculture of strains BEN2908 and BEN2908 Δfrz::kan. The proportion of the mutant in feces of the chicks was analyzed during 7 days. The proportion of the mutant in the inoculum was 51% (dotted line). Results are presented as means ± standard deviations for the six animals analyzed. The significance of the difference between the proportion of the mutant in the inoculum and in chick feces was tested with the Student t test (*, 0.01 > P > 0.001; **, P < 0.001).
Septicemia being an important step in ExPEC pathogenesis, we also tested whether the frz operon promotes bacterial fitness in serum. To that end, static cocultures of strains BEN2908 and BEN2908 Δfrz::kan were performed in chicken serum. The proportion of each strain was then examined during 336 h. Under these conditions, the two strains remained in similar proportions until 8 h of incubation. After that time, strain BEN2908 strongly outcompeted its isogenic deletion mutant BEN2908 Δfrz::kan (Fig. 8A). Similar results were also obtained during cocultures of BEN2908 and mutants strains BEN2908 ΔORF1frz::kan or BEN2908 ΔORF5-8frz::kan, indicating an involvement of this metabolic region in bacterial fitness in serum (Fig. 8B and C).
FIG. 8.
Involvement of the frz operon, ORF1frz, and ORF5-8frz in the survival of strain BEN2908 in chicken serum. (A to C) Strains BEN2908 and BEN2908 Δfrz::kan (A), BEN2908 and BEN2908 ΔORF1frz::kan (B), or BEN2908 and BEN2908 ΔORF5-8frz::kan (C) were inoculated in equivalent numbers into chicken serum containing nalidixic acid. These cultures were incubated at 37°C without agitation. Due to the turbidity of serum, the growth of the cocultures was followed by plating 10-fold dilutions of the cocultures and counting viable bacteria (CFU/ml). The proportion of strain BEN2908 Δfrz::kan (A), BEN2908 ΔORF1frz::kan (B), or BEN2908 ΔORF5-8frz::kan (C) in each coculture was analyzed during 336 h. Results are presented as means ± standard deviations for three independent cocultures. The proportion of the mutant in the inoculum (starting ratio) is represented by a dotted line. The standard deviation of the inoculum ratio is less than 5%. The significance of the difference between the proportion of a mutant in the inoculum and during culture was tested with the Student t test (*, P < 0.001).
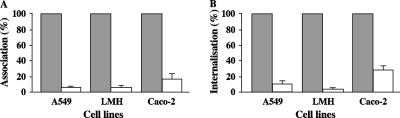
Finally, as strain BEN2908 is able to enter diverse eukaryotic cells and survive for several days (N. K. Chanteloup, unpublished results), we tested whether the frz operon is involved in the ability of the strain to survive in chicken liver cells (LMH), human type II pneumocytes (A549), and human intestinal cells (Caco-2). Whereas BEN2908 Δfrz::kan survived as well as BEN2908 in these cell types (results not shown), we observed that the mutant strain is severely impaired in its ability to adhere to and enter eukaryotic cells (Fig. 9). Similarly, we found that the mutant strains BEN2908 ΔORF1frz::kan and BEN2908 ΔORF5-8frz::kan were also severely impaired in their ability to adhere to and enter eukaryotic cells (Fig. 10).
FIG. 9.
Involvement of the frz operon in the interaction with eukaryotic cells. (A and B) Strains BEN2908 (░⃞) and BEN2908 Δfrz::kan (□) were inoculated independently into cell culture wells containing A549, LMH, or Caco-2 cells (MOI = 10). After 2 h of incubation at 37°C (A549 and Caco-2) or at 40°C (LMH), cells were washed and lysed. Lysates were plated on LB agar, and cell-associated bacteria were enumerated (A). Cells from other wells were then incubated for 1 h 30 min in the presence of gentamicin, washed, and lysed. These lysates were also plated on LB agar, and internalized bacteria were enumerated (B). Each test was made in three different wells in two independent experiments. Percentages of association and internalization were calculated with respect to the number of bacteria inoculated into the culture wells. Values for the mutant strain are represented relative to the value for the wild-type strain. All differences between the wild-type and the mutant strain are significant at a P value of <0.001 (the Student t test).
FIG. 10.
Involvement of ORF1frz and ORF5-8frz in the interaction with human type II pneumocytes. (A and B) Strains BEN2908, BEN2908 ΔORF1frz::kan, and BEN2908 ΔORF5-8frz::kan were inoculated independently in cell culture wells containing A549 cells (MOI = 10). After 2 h of incubation at 37°C, cells were washed and lysed. Lysates were plated on LB agar, and cell-associated bacteria were enumerated (A). Cells from other wells were then incubated for 1 h 30 min in the presence of gentamicin, washed, and lysed. These lysates were also plated on LB agar, and internalized bacteria were enumerated (B). Each test was made in three different wells in two independent experiments. Percentages of association and internalization were calculated with respect to the number of bacteria inoculated into the culture wells. Values for mutant strains are represented relative to the value for the wild-type strain. Differences between the wild-type and the mutant strains were tested with the Student t test and are significant at 0.05 < P < 0.02 (*), 0.02 < P < 0.01 (**) or P < 0.001 (***).
As type 1 fimbriae are the major adhesins of strain BEN2908, and since type 1 fimbriated bacteria are capable of aggregating yeast cells through their ability to bind mannosylated glycoproteins present on the surface of yeasts, we compared BEN2908 and its isogenic mutants for their ability to aggregate yeast cells (13, 39, 59). To that end, bacteria prepared as for eukaryotic cell interaction tests were recovered from cell culture medium and incubated with yeast cells. Whereas BEN2908 was able to aggregate yeasts at up to a 1/8 dilution, none of the three mutants could aggregate yeast cells, either diluted or undiluted. This finding suggests that the decreased interaction of the deletion mutants with eukaryotic cells is due to a decrease in the expression of type 1 fimbriae. Transcription of the fim operon, coding for proteins involved in the biosynthesis and assembly of the diverse subunits of type 1 fimbriae, is mainly regulated by the switch of an IE containing the fim promoter. Transcription is possible when this IE is in the correct orientation (ON) (6). Since the proportion of BEN2908 bacteria with IE in the ON orientation gradually increases during time in static culture (results not shown), we compared the orientation of the IE of strains BEN2908 and of its isogenic Δfrz, ΔORF1frz, and ΔORF5-8frz mutants harvested at an OD600 of 1 or after 24 h of growth (OD600 = 2.9). For this aim, the IEs were amplified by PCR and their orientations were revealed by an asymetric digestion with SnaB1. The digestion patterns shown in Fig. 11 indicate that the proportion of IE in the OFF orientation is higher in the mutant populations than in the population of the wild-type BEN2908. By cloning the frz operon into plasmid pGEM-T Easy and introducing the recombinant plasmid into the mutant strain BEN2908 Δfrz::kan, we were able to restore the wild-type IE status of the bacterial population (see Fig. S2 in the supplemental material). These results indicate that the frz operon promotes the expression of type 1 fimbriae by favoring the ON orientation of the fim operon promoter.
FIG. 11.
Orientation of the type 1 fimbria IE in strains BEN2908, BEN2908 ΔORF1frz::kan, and BEN2908 ΔORF5-8frz::kan. (A and B) Strain BEN2908 (lanes 2) and its isogenic deletion mutants BEN2908 Δfrz (lanes 3), BEN2908 ΔORF1frz::kan (lanes 4), and BEN2908 ΔORF5-8frz::kan (lanes 5) were grown without agitation to an OD600 of 1 (A) or for 24 h (B) in LB medium containing no antibiotic. The IEs from both phase-ON and phase-OFF bacteria were then amplified by PCR. The amplified fragments were hydrolyzed with SnaB1, and the digested products were separated on a 2% agarose gel. Lanes 1 and 6 contain molecular weight standards of 200, 400, and 600 bp.
DISCUSSION
To survive and compete in a changing environment, bacteria have developed complex molecular mechanisms to sense and respond to environmental stimuli (3, 29, 30, 34, 61, 63, 64). We identified an ExPEC metabolic operon (the frz operon) that is involved in bacterial fitness under stressful conditions, such as oxygen restriction and late stationary phase of growth in LB medium and serum (but not in diverse minimal media) or growth in the intestinal tract (Fig. 4 to 8). This operon was also shown, by modifying the expression of type 1 fimbriae, the major adhesins of ExPEC strains (Fig. 9 to 11), to be involved in adherence to and entry into eukaryotic cells. An attractive hypothesis for explaining all these phenotypes would be that the PTS transporter (Frz) encoded by the frz operon is a sensor of the environment that is involved not only in the regulation of bacterial genes implicated in the expression of type 1 fimbriae but also in the regulation of genes involved in the protection of the bacteria from the particular environmental stresses encountered during both nutritional deprivation (late stationary phase of growth) and oxygen restriction. However, we cannot completely exclude the possibility that fitness of the frz-positive strain BEN2908 under stressful conditions also reflects the energetic gain obtained by the entry and the significant metabolization of the substrate of the PTS transporter at the beginning of the stationary phase of growth under oxygen-restricted conditions. Hybridization of RNAs isolated from strain BEN2908 and its isogenic frz deletion mutants to microarrays covering the genomes of several ExPEC strains was undertaken to answer this question. These experiments allowed the identification of several genes whose expression is significantly modified in the frz mutants (unpublished results). We are now confirming these data by using quantitative RT-PCR.
We showed that the frz operon is involved in the expression of type 1 fimbriae by favoring the ON orientation of the fim operon promoter (fim switch). The synthesis of type 1 fimbriae is dependent on an operon of seven genes (fimA, -I, -C, -D, -F, -G, and -H) coding for proteins involved in the biosynthesis and assembly of its diverse subunits. The FimH subunit, protruding outside the bacteria, is responsible for the adherence to the mannosyl residues present on the surface of yeast cells, red blood cells, and eukaryotic cells (9). In humans, type 1 fimbriae are involved in the colonization of the oropharynx of newborns (a first step for developing meningitis and septicemia in newborns) and of the urinary tract (5, 40). In chickens, they were described as being involved in the adherence of the bacteria to the trachea, the air sacs, and the lungs (39). Two genes located upstream of the fim operon code for recombinases (FimB and FimE) involved in the inversion of a DNA fragment (IE) containing the transcriptional promoter of the fim operon. Transcription of the fim operon is possible when this fragment is in the correct orientation (ON) (6). The expression of type 1 fimbriae is highly regulated. FimB and FimE catalyze the inversion of the IE in the ON-OFF orientation, but the inversion in the OFF-ON orientation is principally due to FimB. Other proteins modifying DNA bending, such as Lrp (leucine responsive protein), H-NS (histone-like nucleoid structuring protein), and IHF (integration host factor), and other recombinases, such as IpuA, IpuB, and IpbA, whose roles are less known, participate in this inversion mechanism. Several environmental factors were described as being involved in the inversion of the fim operon promoter and the expression of type 1 fimbriae: temperature, osmolarity, pH, growth phase, leucine and alanine concentration, and the presence of certain carbohydrates. Notably, it was shown that the nanC (located upstream of fimB) and fimB genes are coordinately regulated via the Dam methylase and via the NanR and NagC regulators, whose expression depends on the N-acetylglucosamine (GlcNAc) and N-acetyl neuraminic acid (Neu5Ac) concentration, respectively (6). It was also reported that the ppGpp and pppGpp alarmones are involved in IE inversion by favoring the orientation of the fim promoter in the ON orientation (1). We also reported recently that the IbeA and IbeT proteins, predicted to be involved in carbon metabolism, are involved in the expression of type 1 fimbriae by favoring the position of the fim promoter of BEN2908 in the ON orientation (13). By a yet-unknown mechanism, the Frz machinery thus participates in this complex and highly regulated process by which the bacterial population is continuously balanced between a fimbriated stage that allows eukaryotic cell adherence and invasion and an afimbriated stage that avoids the risk of recognition of type 1 fimbriae by the host immune system but allows a smaller possibility of interaction with eukaryotic cells.
PTS transporters and their specific regulators, with PRDs, are absent from eukaryotic organisms and are only present in some bacterial species, including E. coli. Their main function is the translocation of carbohydrates inside bacteria, but more and more data from the literature indicate that these systems are also, as two-component systems, environmental sensors affecting diverse physiological aspects of the bacteria, notably, the expression of their virulence (2, 17). Based on homology of sequences used for the classification of PTS transporters, the Frz transporter belongs to the fructose-mannitol family (Fru; Transport Classification Database transport classification no. 4.A.2) and to the fructose subfamily. Four other PTS transporters of the fructose subfamily were previously described for E. coli K-12: FruAB, a functional fructose transporter; FrvAB, a probable inactive transporter; Frw, a transporter putatively involved in carbon metabolism during anaerobiosis whose expression is under the regulation of the AI-2 autoinducer involved in quorum sensing; Frx, a porter putatively involved in mannoside transport; and Fry (also named HrsA or MngA), a functional 2-O-α-mannosyl d-glycerate porter. The nitrogen-related enzyme IIA (IIANtr) encoded by the ptsN gene of the rpoN operon is also homologous to the IIA domains of the Frz transporter and of the other transporters of the fructose subfamily. It was suggested that IIANtr is only involved in regulation mechanisms (16, 51, 58).
Phosphoryl relay, which energizes PTS transporters, proceeds sequentially from phosphoenolpyruvate to enzyme I; heat-stable histidine protein (Hpr); subunits IIA, IIB, and IIC of the transporters; and finally, the incoming sugar which is transported across the internal membrane via the integral membrane IIC porter. Sugars transported by PTS transporters of the fructose subfamily are internalized inside E. coli either as a 1- or 6-phospho sugar, depending on the transporter involved. The level of phosphorylation of PTS components thus depends on the presence of their substrates in the environment of the bacteria and has a direct impact on the modulation of phosphoryl group transfers to PRDs of PRD regulators. Depending on the phosphorylation level of these regulators, operons encoding PTS transporters and PRD protein-regulated genes, which affect diverse aspects of the bacteria, are activated or not (2, 17).
Some bacterial metabolic enzymes (other than those of the PTS) were also described as being involved in gene regulation by the acquisition of a DNA or RNA binding domain (12). Other metabolic enzymes were found to control the activity of transcription factors. This is notably the case for the Lac.D1 class I tagatose-1,6-biphosphate aldolase of Streptococcus pyogenes that functions as an environmental sensor. By binding to its substrates, Lac.D1 is able to sequester a transcriptional inhibitor of the virulence gene speB (12). We showed that the metabolic enzymes encoded by the frz operon are involved in the fitness of BEN2908 under stressful conditions and in the expression of type 1 fimbriae (Fig. 6, 8, 10, and 11). As no DNA or RNA binding domains could be identified in the sequences of these enzymes, it is probable that one or several frz-encoded enzymes use a mechanism similar to the S. pyogenes Lac.D1 aldolase to regulate genes involved in the phenotypes described above. The regulatory role of the Frz system is nevertheless not exclusively due to the metabolic enzymes, as we also showed that a nonpolar mutant of the PRD regulator gene possesses the same phenotypic defect as the Δfrz or the ΔORF5-8frz mutant (Fig. 6, 8, 10, and 11). The PRD activator of the frz operon is thus also involved in the mechanisms that are related to bacterial fitness under stressful conditions and the expression of type 1 fimbriae. It is one of the first targets that should be tested for possible sequestration by frz-encoded metabolic enzymes. Such experiments are now being undertaken in our laboratory.
An in silico crossover between the two direct repeats identified in the intergenic regions of yicH-ORF8frz and yicI-ORF1frz allowed the deletion of the frz operon from the genome of strain BEN2908 and the conservation of 53 base pairs from the intergenic regions between the yicH and yicI genes. These 53 base pairs were aligned (58% identical nucleotides) with the yicH-yicI intergenic region of the nonpathogenic E. coli K-12 substrain MG1655, a strain whose genome was sequenced and for which it was previously shown that the D7 DNA fragment was absent (Fig. 12). The data described above, the fact that the G+C content of the frz operon (48.7%) is close to the G+C content (50.8%) (4) of the whole E. coli genome, and the absence of insertion element remnants (integrase or transposase genes or plasmidic replication origin) indicate that the frz operon was probably not acquired by horizontal transfer by commensal E. coli strains but, rather, was initially present in the ancestor of commensal and ExPEC strains and was then deleted during evolution in most of the commensal strains. We showed that the frz operon is highly associated with E. coli clonal groups B2 and D (43; our BlastN searches described herein). Interestingly, group B2 is considered by some to be the first E. coli group to emerge, and then came group D, the sister group of the A+B1 clade, in which A is the sister group of B1 (32). It is thus plausible that the frz operon and the yicI-yicJ genes that are transcribed in the same direction as the frz operon and that also code for proteins involved in carbohydrate metabolism (glycosyl hydrolase and a transporter of the galactose-pentose-hexuronate family) originally formed a unique functional metabolic unit. This hypothesis is supported by the fact that some transcripts of the yicI-yicJ genes of the ExPEC strain BEN2908 pass through the transcriptional terminator identified in the yicI-ORF1frz intergenic region and form cotranscripts with frz genes (results not shown).
FIG. 12.
In silico deletion of the frz operon of strain BEN2908 by recombination between two direct repeats. An in silico crossover between two direct repeats identified in the yicH-ORF8frz and yicI-ORF1frz intergenic regions was performed. This allowed the deletion of the frz operon from the genome of strain BEN2908 and the conservation of 53 base pairs from the intergenic region between the yicH and yicI genes. These 53 base pairs were aligned with the yicH-yicI intergenic region of the nonpathogenic strain K-12 substrain MG1655. The sequence of the conserved direct repeat is boxed.
The substrate of the Frz transporter is actually unknown. It could neither be revealed by competition growth tests in several minimal media nor by comparing the wild-type strain and the frz deletion mutant for their ability to metabolize 102 different carbohydrates present on Biolog plates and API 50H strips. While the latter test identified two carbohydrates (3-O-β-d-galactopyranosyl-d-arabinose and glucose) as being metabolized in smaller amounts by the mutant strain than by the wild-type strain in the three experiments made, none of these sugars allowed strain BEN2908 to outcompete the frz deletion mutant during growth competition tests in minimal medium (results not shown). It is worth noting that the strong outcompetition of the frz deletion mutant by the wild-type strain during the late stationary phase of growth in low-oxygen-content cocultures was only observed in LB medium and in serum and was not discernible in minimal medium containing different carbohydrates tested as sole carbon sources, although a slight effect was observable with d-psicose, d-tagatose, and d-sorbose (Fig. 4B). This suggests that the molecule recognized by the Frz transporter is present in some media and physiological environments (LB medium, serum, cell culture medium, and the intestine) but is not any of 12 carbohydrates tested during growth competition tests in minimal medium. The data described above also indicate that the molecule recognized by the Frz transporter is not released by the lysis of part of the bacterial population during long-term culture. In bacteria, genes involved in the same pathway are generally found in a proximate region of the genome. Based on this assumption, it can be speculated that the substrate of the Frz transporter is a disaccharide. This disaccharide would be translocated inside the bacteria as a 6-phosphate disaccharide, through the IIC subunit of the Frz transporter, and would then be hydrolyzed into a 6-phosphate hexose and an unphosphorylated hexose by a glycosyl hydrolase encoded by a gene of the E. coli genetic core. This hydrolase could be the product of the yicI gene that codes for a family 31 glycosyl hydrolase with a low α-glycosidase activity (46). The resulting unphosphorylated hexose would then be phosphorylated on carbon 6 by the sugar-specific kinase of the ROK family (ORF7frz). The two hexose phosphates thus generated would then be phosphorylated on carbon 1 by a 6-phospho-hexose kinase encoded by a gene of the E. coli genetic core. Each of these 1,6-hexose biphosphates would then be transformed in 3-phospho-glycerol and dihydroxy-acetone-phosphate by the aldolases coded by the frz operon (ORF5-6frz). These aldolases belong to the class II aldolases that are known to include enzymes with high substrate specificity (8, 23). Finally, dihydroxy-acetone-phosphate and 3-phospho-glycerol would be able to enter into the process of glycolysis and produce phosphoenolpyruvate that will again energize the Frz transporter. The eight potential substrates of class II aldolases were tested in growth competition tests in minimal medium. Although a slight effect of outcompetition was observed when three of these substrates were tested (d-psicose, d-sorbose, and d-tagatose), these three d-ketoses do not allow (or allow very slightly) the growth of strain BEN2908 (Fig. 4B). The presence of d-psicose, d-sorbose, or d-tagatose in some environments should thus activate metabolic pathways allowing BEN2908 to survive without multiplication.
In conclusion, our analysis of the role of the frz metabolic operon in the physiology of the ExPEC strain BEN2908 revealed interesting connections between bacterial adhesin expression, fitness in diverse environments, and metabolic capacities of the bacteria. As several family of proteins encoded by the frz operon are not present in higher eukaryotes (PTS transporter, PRD activator, and class II aldolases), the understanding of the Frz system's mechanism should open interesting perspectives both on the comprehension of bacterial host adaptation and pathogenicity mechanisms and on the development of new antibacterial prevention approaches. Such work is now being undertaken in our laboratory.
Supplementary Material
Acknowledgments
This work was supported by the Era-NET PathoGenoMics European program (grant ANR-06-PATHO-002-01). G. Rouquet is a predoctoral fellow of the INRA(MICA)/Région Centre (France).
A. Allaoui (Free University of Brussels) is acknowledged for the gift of the pUC18K2 plasmid and D. Boyd (Harvard Medical School, Boston, MA) for the gift of the lambda Inch plasmid-chromosome shuttle system. We thank A. Brée for technical help provided during chicken intestinal colonization experiments. Maryvonne Moulin-Schouleur and P. Germon are acknowledged for fruitful discussions.
Footnotes
Published ahead of print on 17 April 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Aberg, A., V. Shingler, and C. Balsalobre. 2006. (p)ppGpp regulates type 1 fimbriation of Escherichia coli by modulating the expression of the site-specific recombinase FimB. Mol. Microbiol. 601520-1533. [DOI] [PubMed] [Google Scholar]
- 2.Barabote, R. D., and M. H. Saier, Jr. 2005. Comparative genomic analyses of the bacterial phosphotransferase system. Microbiol. Mol. Biol. Rev. 69608-634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bejerano-Sagie, M., and K. B. Xavier. 2007. The role of small RNAs in quorum sensing. Curr. Opin. Microbiol. 10189-198. [DOI] [PubMed] [Google Scholar]
- 4.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 2771453-1474. [DOI] [PubMed] [Google Scholar]
- 5.Bloch, C. A., and P. E. Orndorff. 1990. Impaired colonization by and full invasiveness of Escherichia coli K1 bearing a site-directed mutation in the type 1 pilin gene. Infect. Immun. 58275-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blomfield, I. C., and M. Van der Woude. 13September2007. Regulation of fimbrial expression. In R. Curtiss III et al. (ed.), EcoSal—Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC. http://www.ecosal.org. [DOI] [PubMed]
- 7.Boyd, D., D. S. Weiss, J. C. Chen, and J. Beckwith. 2000. Towards single-copy gene expression systems making gene cloning physiologically relevant: lambda InCh, a simple Escherichia coli plasmid-chromosome shuttle system. J. Bacteriol. 182842-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brinkkotter, A., A. Shakeri-Garakani, and J. W. Lengeler. 2002. Two class II D-tagatose-bisphosphate aldolases from enteric bacteria. Arch. Microbiol. 177410-419. [DOI] [PubMed] [Google Scholar]
- 9.Capitani, G., O. Eidam, R. Glockshuber, and M. G. Grutter. 2006. Structural and functional insights into the assembly of type 1 pili from Escherichia coli. Microbes Infect. 82284-2290. [DOI] [PubMed] [Google Scholar]
- 10.Caza, M., F. Lepine, S. Milot, and C. M. Dozois. 2008. Specific roles of the iroBCDEN genes in virulence of an avian pathogenic Escherichia coli O78 strain and in production of salmochelins. Infect. Immun. 763539-3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chouikha, I., P. Germon, A. Bree, P. Gilot, M. Moulin-Schouleur, and C. Schouler. 2006. A selC-associated genomic island of the extraintestinal avian pathogenic Escherichia coli strain BEN2908 is involved in carbohydrate uptake and virulence. J. Bacteriol. 188977-987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Commichau, F. M., and J. Stulke. 2008. Trigger enzymes: bifunctional proteins active in metabolism and in controlling gene expression. Mol. Microbiol. 67692-702. [DOI] [PubMed] [Google Scholar]
- 13.Cortes, M. A., J. Gibon, N. K. Chanteloup, M. Moulin-Schouleur, P. Gilot, and P. Germon. 2008. Inactivation of ibeA and ibeT results in decreased expression of type 1 fimbriae in extraintestinal pathogenic Escherichia coli strain BEN2908. Infect. Immun. 764129-4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crepin, S., M. G. Lamarche, P. Garneau, J. Seguin, J. Proulx, C. M. Dozois, and J. Harel. 2008. Genome-wide transcriptional response of an avian pathogenic Escherichia coli (APEC) pst mutant. BMC Genomics 9568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 976640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeLisa, M. P., C. F. Wu, L. Wang, J. J. Valdes, and W. E. Bentley. 2001. DNA microarray-based identification of genes controlled by autoinducer 2-stimulated quorum sensing in Escherichia coli. J. Bacteriol. 1835239-5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deutscher, J., C. Francke, and P. W. Postma. 2006. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol. Mol. Biol. Rev. 70939-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dho, M., and J. P. Lafont. 1984. Adhesive properties and iron uptake ability in Escherichia coli lethal and nonlethal for chicks. Avian Dis. 281016-1025. [PubMed] [Google Scholar]
- 19.Dho-Moulin, M., and J. M. Fairbrother. 1999. Avian pathogenic Escherichia coli (APEC). Vet. Res. 30299-316. [PubMed] [Google Scholar]
- 20.Dozois, C. M., N. Chanteloup, M. Dho-Moulin, A. Bree, C. Desautels, and J. M. Fairbrother. 1994. Bacterial colonization and in vivo expression of F1 (type 1) fimbrial antigens in chickens experimentally infected with pathogenic Escherichia coli. Avian Dis. 38231-239. [PubMed] [Google Scholar]
- 21.Dunwell, J. M., A. Culham, C. E. Carter, C. R. Sosa-Aguirre, and P. W. Goodenough. 2001. Evolution of functional diversity in the cupin superfamily. Trends Biochem. Sci. 26740-746. [DOI] [PubMed] [Google Scholar]
- 22.Eiglmeier, K., N. Honore, S. Iuchi, E. C. Lin, and S. T. Cole. 1989. Molecular genetic analysis of FNR-dependent promoters. Mol. Microbiol. 3869-878. [DOI] [PubMed] [Google Scholar]
- 23.Gavalda, S., R. Braga, C. Dax, A. Vigroux, and C. Blonski. 2005. N-Sulfonyl hydroxamate derivatives as inhibitors of class II fructose-1,6-diphosphate aldolase. Bioorg. Med. Chem. Lett. 155375-5377. [DOI] [PubMed] [Google Scholar]
- 24.Germon, P., Y. H. Chen, L. He, J. E. Blanco, A. Bree, C. Schouler, S. H. Huang, and M. Moulin-Schouleur. 2005. IbeA, a virulence factor of avian pathogenic Escherichia coli. Microbiology 1511179-1186. [DOI] [PubMed] [Google Scholar]
- 25.Gilot, P., Y. Jossin, and J. Content. 2000. Cloning, sequencing and characterisation of a Listeria monocytogenes gene encoding a fibronectin-binding protein. J. Med. Microbiol. 49887-896. [DOI] [PubMed] [Google Scholar]
- 26.Herren, C. D., A. Mitra, S. K. Palaniyandi, A. Coleman, S. Elankumaran, and S. Mukhopadhyay. 2006. The BarA-UvrY two-component system regulates virulence in avian pathogenic Escherichia coli O78:K80:H9. Infect. Immun. 744900-4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hondorp, E. R., and K. S. McIver. 2007. The Mga virulence regulon: infection where the grass is greener. Mol. Microbiol. 661056-1065. [DOI] [PubMed] [Google Scholar]
- 28.Hooton, T. M., and W. E. Stamm. 1997. Diagnosis and treatment of uncomplicated urinary tract infection. Infect. Dis. Clin. N. Am. 11551-581. [DOI] [PubMed] [Google Scholar]
- 29.Jenal, U., and J. Malone. 2006. Mechanisms of cyclic-di-GMP signaling in bacteria. Annu. Rev. Genet. 40385-407. [DOI] [PubMed] [Google Scholar]
- 30.Lamarche, M. G., B. L. Wanner, S. Crepin, and J. Harel. 2008. The phosphate regulon and bacterial virulence: a regulatory network connecting phosphate homeostasis and pathogenesis. FEMS Microbiol. Rev. 32461-473. [DOI] [PubMed] [Google Scholar]
- 31.Le Bars, J. 1976. Mise au point d'un protocole pour l'obtention de poulets axéniques. Ann. Rech. Vétér. 7383-396. [PubMed] [Google Scholar]
- 32.Lecointre, G., L. Rachdi, P. Darlu, and E. Denamur. 1998. Escherichia coli molecular phylogeny using the incongruence length difference test. Mol. Biol. Evol. 151685-1695. [DOI] [PubMed] [Google Scholar]
- 33.Le Gall, T., O. Clermont, S. Gouriou, B. Picard, X. Nassif, E. Denamur, and O. Tenaillon. 2007. Extraintestinal virulence is a coincidental by-product of commensalism in B2 phylogenetic group Escherichia coli strains. Mol. Biol. Evol. 242373-2384. [DOI] [PubMed] [Google Scholar]
- 34.Lengeler, J. W. 1993. Carbohydrate transport in bacteria under environmental conditions, a black box? Antonie van Leeuwenhoek 63275-288. [DOI] [PubMed] [Google Scholar]
- 35.Lewin, B. 2000. Genes VII. Oxford University Press, New York, NY.
- 36.Li, G., C. Ewers, C. Laturnus, I. Diehl, K. Alt, J. Dai, E. M. Antao, K. Schnetz, and L. H. Wieler. 2008. Characterization of a yjjQ mutant of avian pathogenic Escherichia coli (APEC). Microbiology 1541082-1093. [DOI] [PubMed] [Google Scholar]
- 37.Lymberopoulos, M. H., S. Houle, F. Daigle, S. Leveille, A. Bree, M. Moulin-Schouleur, J. R. Johnson, and C. M. Dozois. 2006. Characterization of Stg fimbriae from an avian pathogenic Escherichia coli O78:K80 strain and assessment of their contribution to colonization of the chicken respiratory tract. J. Bacteriol. 1886449-6459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lynch, A. S., and E. C. C. Lin. 1996. Responses to molecular oxygen, p. 1526-1538. In F. C. Neidhardt (ed.), Escherchia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, DC.
- 39.Marc, D., P. Arne, A. Bree, and M. Dho-Moulin. 1998. Colonization ability and pathogenic properties of a fim- mutant of an avian strain of Escherichia coli. Res. Microbiol. 149473-485. [DOI] [PubMed] [Google Scholar]
- 40.Martinez, J. J., M. A. Mulvey, J. D. Schilling, J. S. Pinkner, and S. J. Hultgren. 2000. Type 1 pilus-mediated bacterial invasion of bladder epithelial cells. EMBO J. 192803-2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.May, M., A. J. Daley, S. Donath, and D. Isaacs. 2005. Early onset neonatal meningitis in Australia and New Zealand, 1992-2002. Arch. Dis. Child. Fetal Neonatal Ed. 90F324-F327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Menard, R., P. J. Sansonetti, and C. Parsot. 1993. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J. Bacteriol. 1755899-5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moulin-Schouleur, M., M. Reperant, S. Laurent, A. Bree, S. Mignon-Grasteau, P. Germon, D. Rasschaert, and C. Schouler. 2007. Extraintestinal pathogenic Escherichia coli strains of avian and human origin: link between phylogenetic relationships and common virulence patterns. J. Clin. Microbiol. 453366-3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moulin-Schouleur, M., C. Schouler, P. Tailliez, M. R. Kao, A. Bree, P. Germon, E. Oswald, J. Mainil, M. Blanco, and J. Blanco. 2006. Common virulence factors and genetic relationships between O18:K1:H7 Escherichia coli isolates of human and avian origin. J. Clin. Microbiol. 443484-3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Norrander, J., T. Kempe, and J. Messing. 1983. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene 26101-106. [DOI] [PubMed] [Google Scholar]
- 46.Okuyama, M., H. Mori, S. Chiba, and A. Kimura. 2004. Overexpression and characterization of two unknown proteins, YicI and YihQ, originated from Escherichia coli. Protein Expr. Purif. 37170-179. [DOI] [PubMed] [Google Scholar]
- 47.Russo, T. A., and J. R. Johnson. 2006. Extraintestinal isolates of Escherichia coli: identification and prospects for vaccine development. Expert Rev. Vaccines 545-54. [DOI] [PubMed] [Google Scholar]
- 48.Russo, T. A., and J. R. Johnson. 2003. Medical and economic impact of extraintestinal infections due to Escherichia coli: focus on an increasingly important endemic problem. Microbes Infect. 5449-456. [DOI] [PubMed] [Google Scholar]
- 49.Sabri, M., M. Caza, J. Proulx, M. H. Lymberopoulos, A. Bree, M. Moulin-Schouleur, R. Curtiss III, and C. M. Dozois. 2008. Contribution of the SitABCD, MntH, and FeoB metal transporters to the virulence of avian pathogenic Escherichia coli O78 strain chi7122. Infect. Immun. 76601-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 51.Sampaio, M. M., F. Chevance, R. Dippel, T. Eppler, A. Schlegel, W. Boos, Y. J. Lu, and C. O. Rock. 2004. Phosphotransferase-mediated transport of the osmolyte 2-O-alpha-mannosyl-D-glycerate in Escherichia coli occurs by the product of the mngA (hrsA) gene and is regulated by the mngR (farR) gene product acting as repressor. J. Biol. Chem. 2795537-5548. [DOI] [PubMed] [Google Scholar]
- 52.Schouler, C., F. Koffmann, C. Amory, S. Leroy-Setrin, and M. Moulin-Schouleur. 2004. Genomic subtraction for the identification of putative new virulence factors of an avian pathogenic Escherichia coli strain of O2 serogroup. Microbiology 1502973-2984. [DOI] [PubMed] [Google Scholar]
- 53.Schouler, C., A. Taki, I. Chouikha, M. Moulin-Schouleur, and P. Gilot. 2009. A genomic island of an extraintestinal pathogenic Escherichia coli strain enables the metabolism of fructooligosaccharides, which improves intestinal colonization. J. Bacteriol. 191388-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smith, J. L., P. M. Fratamico, and N. W. Gunther. 2007. Extraintestinal pathogenic Escherichia coli. Foodborne Pathog. Dis. 4134-163. [DOI] [PubMed] [Google Scholar]
- 55.Stordeur, P., D. Marlier, J. Blanco, E. Oswald, F. Biet, M. Dho-Moulin, and J. Mainil. 2002. Examination of Escherichia coli from poultry for selected adhesin genes important in disease caused by mammalian pathogenic E. coli. Vet. Microbiol. 84231-241. [DOI] [PubMed] [Google Scholar]
- 56.Stulke, J., M. Arnaud, G. Rapoport, and I. Martin-Verstraete. 1998. PRD—a protein domain involved in PTS-dependent induction and carbon catabolite repression of catabolic operons in bacteria. Mol. Microbiol. 28865-874. [DOI] [PubMed] [Google Scholar]
- 57.Swinscow, T. 1978. Statistics at square one, 4th ed. The Mendip Press, Bath, England.
- 58.Tchieu, J. H., V. Norris, J. S. Edwards, and M. H. Saier, Jr. 2001. The complete phosphotranferase system in Escherichia coli. J. Mol. Microbiol. Biotechnol. 3329-346. [PubMed] [Google Scholar]
- 59.Teng, C. H., Y. Xie, S. Shin, F. Di Cello, M. Paul-Satyaseela, M. Cai, and K. S. Kim. 2006. Effects of ompA deletion on expression of type 1 fimbriae in Escherichia coli K1 strain RS218 and on the association of E. coli with human brain microvascular endothelial cells. Infect. Immun. 745609-5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Titgemeyer, F., J. Reizer, A. Reizer, and M. H. Saier, Jr. 1994. Evolutionary relationships between sugar kinases and transcriptional repressors in bacteria. Microbiology 1402349-2354. [DOI] [PubMed] [Google Scholar]
- 61.Tobe, T. 2008. The roles of two-component systems in virulence of pathogenic Escherichia coli and Shigella spp. Adv. Exp. Med. Biol. 631189-199. [DOI] [PubMed] [Google Scholar]
- 62.Tung, W. L., and K. C. Chow. 1995. A modified medium for efficient electrotransformation of E. coli. Trends Genet. 11128-129. [DOI] [PubMed] [Google Scholar]
- 63.Typas, A., G. Becker, and R. Hengge. 2007. The molecular basis of selective promoter activation by the sigmaS subunit of RNA polymerase. Mol. Microbiol. 631296-1306. [DOI] [PubMed] [Google Scholar]
- 64.Wick, L. M., and T. Egli. 2004. Molecular components of physiological stress responses in Escherichia coli. Adv. Biochem. Eng. Biotechnol. 891-45. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.