Abstract
Dihydroneopterin aldolase (FolB) catalyzes conversion of dihydroneopterin to 6-hydroxymethyldihydropterin (HMDHP) in the classical folate biosynthesis pathway. However, folB genes are missing from the genomes of certain bacteria from the phyla Chloroflexi, Acidobacteria, Firmicutes, Planctomycetes, and Spirochaetes. Almost all of these folB-deficient genomes contain an unusual paralog of the tetrahydrobiopterin synthesis enzyme 6-pyruvoyltetrahydropterin synthase (PTPS) in which a glutamate residue replaces or accompanies the catalytic cysteine. A similar PTPS paralog from the malaria parasite Plasmodium falciparum is known to form HMDHP from dihydroneopterin triphosphate in vitro and has been proposed to provide a bypass to the FolB step in vivo. Bacterial genes encoding PTPS-like proteins with active-site glutamate, cysteine, or both residues were accordingly tested together with the P. falciparum gene for complementation of the Escherichia coli folB mutation. The P. falciparum sequence and bacterial sequences with glutamate or glutamate plus cysteine were active; those with cysteine alone were not. These results demonstrate that PTPS paralogs with an active-site glutamate (designated PTPS-III proteins) can functionally replace FolB in vivo. Recombinant bacterial PTPS-III proteins, like the P. falciparum enzyme, mediated conversion of dihydroneopterin triphosphate to HMDHP, but other PTPS proteins did not. Neither PTPS-III nor other PTPS proteins exhibited significant dihydroneopterin aldolase activity. Phylogenetic analysis indicated that PTPS-III proteins may have arisen independently in various PTPS lineages. Consistent with this possibility, merely introducing a glutamate residue into the active site of a PTPS protein conferred incipient activity in the growth complementation assay, and replacing glutamate with alanine in a PTPS-III protein abolished complementation.
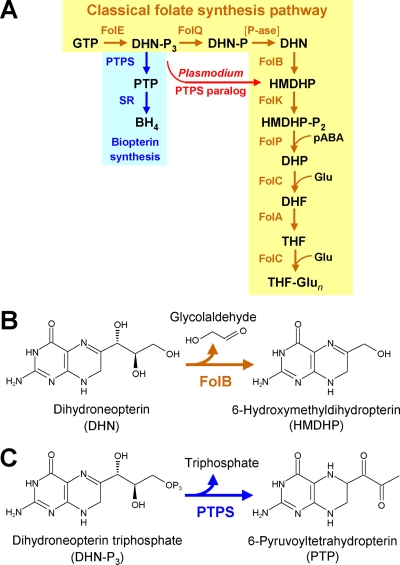
The folate biosynthesis pathway is present in most bacteria, as well as plants, fungi, heterokonts, and certain protozoa (8, 15, 19). The classical pathway (Fig. 1A) involves eight specific enzymes, all of whose genes are known (9, 13, 15). This pathway was until recently thought to be the only one. However, the advent of large-scale genome sequencing has revealed numerous cases of “missing” folate synthesis enzymes (“pathway holes”) in which particular enzymes are lacking in bacterial genomes with otherwise complete pathways (9). This situation implies the existence of either different enzymes with sequences unlike the known ones (“nonorthologous displacement”) or alternative routes (16, 29). One such case is dihydroneopterin aldolase (FolB), which mediates the fourth step in the classical folate pathway, the shortening of the side chain of dihydroneopterin to give 6-hydroxymethyldihydropterin (HMDHP) (Fig. 1B). Comparative genomic analysis showed that the folB gene is missing from diverse bacteria (9), as well as from heterokonts (diatoms and oomycetes) (19).
FIG. 1.
Biosynthesis of tetrahydrofolate and tetrahydrobiopterin. (A) Steps and enzymes of the classical tetrahydrofolate synthesis pathway and the tetrahydrobiopterin synthesis pathway. The bypass reaction catalyzed by the P. falciparum PTPS paralog (PTPS-III type) is in red. Abbreviations: BH4, 5,6,7,8-tetrahydrobiopterin; DHF, 7,8-dihydrofolate; DHN, 7,8-dihydroneopterin; DHP, 7,8-dihydropteroate; Glu, glutamate; GTP, guanosine-5′-triphosphate; HMDHP, 6-hydroxymethyl-7,8-dihydropterin; -P, phosphate; -P2, pyrophosphate; -P3, triphosphate; pABA, p-aminobenzoate; P-ase, nonspecific phosphatase; PTP, 6-pyruvoyl-5,6,7,8-tetrahydropterin; PTPS, 6-pyruvoyl-5,6,7,8-tetrahydropterin synthase; SR, sepiapterin reductase; THF, 5,6,7,8-tetrahydrofolate; THF-Glun, tetrahydrofolate polyglutamates. (B) Reaction mediated by FolB (dihydroneopterin aldolase). (C) Reaction mediated by PTPS (classical PTPS-II type).
Dihydroneopterin aldolase is also missing in the malaria parasite Plasmodium falciparum (14) and was recently proposed from biochemical evidence to be functionally replaced by a paralog of the tetrahydrobiopterin synthesis enzyme 6-pyruvoyltetrahydropterin synthase (PTPS) (10). This paralog forms HMDHP (and a small amount of the normal PTPS product, 6-pyruvoyltetrahydropterin) from the first intermediate of folate synthesis, dihydroneopterin triphosphate (Fig. 1C) and can thus provide a bypass for the missing dihydroneopterin aldolase step (Fig. 1A). Evidence that this bypass operates in vivo was, however, not provided and indeed may be impossible to obtain because disrupting folate synthesis by gene deletion appears to be lethal in P. falciparum even when folate is supplied (38). The P. falciparum PTPS paralog is distinct in having a glutamate residue in place of a cysteine in the active-site region bounded by the three histidine residues that coordinate the essential Zn2+ ion. Mutating the glutamate to cysteine abolished the formation of HMDHP but not 6-pyruvoyltetrahydropterin (10). Interestingly, heterokonts also have a PTPS paralog with an active-site glutamate, which could explain their lack of FolB (19).
Tetrahydrobiopterin synthesis is known to occur in only a few bacterial groups such as Cyanobacteria and Chlorobia (12, 24) and, predictably, these organisms have PTPS enzymes (designated PTPS-II) that are related to mammalian PTPS and absent from other bacteria (24). However, they also have a second PTPS-like protein (designated PTPS-I) that has no in vivo role in tetrahydrobiopterin synthesis and is widespread in other bacteria (24). PTPS-I mediates an early step in the biosynthesis of the modified tRNA base queuosine (the gene encoding PTPS-I has been named queD) (32), specifically the conversion of dihydroneopterin triphosphate to 6-carboxytetrahydropterin (27). In vitro, Synechocystis and Escherichia coli PTPS-I (QueD) proteins also cleave the side chain of the nonphysiological substrate sepiapterin and also show low PTPS activity (39). Both PTPS-I and PTPS-II have cysteine residues in the active-site region (24).
We report here that bacteria lacking folB have a PTPS-like protein (PTPS-III) with glutamate alone or glutamate plus cysteine in the active site region. Complementation of an E. coli folB deletant, site-directed mutagenesis, and biochemical assays established that these PTPS-III proteins functionally replace FolB via a bypass reaction, as proposed for the P. falciparum protein. We also showed for the first time that the P. falciparum protein can mediate this reaction in vivo.
MATERIALS AND METHODS
Bioinformatics.
Bacterial genomes were analyzed by using the SEED database and its tools (30) at anno-3.nmpdr.org/anno/FIG/subsys.cgi. Sequences were aligned with CLUSTAL W, and phylogenetic analyses were made with MEGA 4 (36).
Pteridines.
Pterin, 6-hydroxymethylpterin, 7,8-dihydroneopterin, and 6-hydroxymethyl-7,8-dihydropterin hydrochloride were obtained from Schircks Laboratories. Near-saturated solutions were freshly prepared in N2-sparged potassium phosphate (10 mM, pH 7.5), excluding light, and titers were determined spectrophotometrically using published extinction coefficients (4, 31). Dihydroneopterin triphosphate was prepared from GTP essentially as detailed previously (23) except that recombinant Synechocystis GTP cyclohydrolase I, purified as described previously (25), replaced the E. coli enzyme. Aliquots of dihydroneopterin triphosphate solution were frozen in liquid N2 and stored at −80°C until use.
Strains and growth conditions.
E. coli strains were grown in Luria-Bertani medium (LB) at 37°C. Growth media were solidified with 15 g of agar/liter for preparation of the plates. Kanamycin (50 μg/ml), carbenicillin (100 μg/ml), thymidine (75 μg/ml), and l-arabinose (0.2% [wt/vol]) were added as required. E. coli K-12 MG1655 was used as the wild-type strain in all experiments. The folB::Kan JW3030-2 strain was from the Keio collection (1). The folB::Kan deletion was transferred to MG1655 by P1 transduction. Clones with the desired deletion were selected on LB plates containing kanamycin and folate-dependent end products, i.e., thymidine (75 μg/ml), methionine (50 μg/ml), glycine (50 μg/ml), adenine (20 μg/ml), and panthothenate (1 μg/ml) (35). Deletion of the folB gene was confirmed by PCR and sequencing. Deletant clones were maintained on LB plates containing thymidine and kanamycin.
Construction of expression vectors.
Genomic DNA of Leptospira interrogans L1-130, Desulfovibrio vulgaris, Acidobacteria bacterium, Dehalococcoides ethenogenes, and Aquifex aeolicus, were obtained from M. Picardeau (Institut Pasteur), D. Stahl (University of Washington), C. Kuske (Los Alamos National Laboratory), S. Zinder (Cornell University), and P. Schimmel (Scripps Research Institute), respectively. Genomic DNA of Legionella pneumophila and Pirellula sp. strain 1 was purchased from the American Type Culture Collection and the German Resource Centre for Biological Material, respectively. The Thermotoga maritima PTPS-III gene was amplified using genomic clone TM0038 (from S. Lesley, Novartis Research Foundation). The E. coli folB gene was amplified from E. coli genomic DNA (strain MG1655). PfPTPS-pET46 (10) was used as a template to amplify the P. falciparum PTPS gene. Clostridium botulinum PTPS-I and PTPS-III, Synthrophobacter fumaroxidans PTPS-I, and Synthrophus aciditrophicus PTPS-III sequences were synthesized by GenScript after optimizing codon usage for expression in E. coli (see Table S1 in the supplemental material). PCR amplifications were carried out using Pfu Turbo DNA polymerase (Stratagene). For functional complementation experiments, all sequences were PCR amplified with a forward primer harboring an NcoI, BspHI, or PciI site and a reverse primer with an XbaI site (see Table S2 in the supplemental material). The amplicons were cloned between the NcoI and XbaI sites of pBAD24 (18) and then introduced into E. coli strain DH5α and sequenced. The expression of PTPS genes in the pBAD24 vector was induced with 0.2% (wt/vol) l-arabinose. For recombinant protein production, PTPS-I and PTPS-III sequences from C. botulinum (Cb) and L. interrogans (Li) were amplified using a forward primer containing an NdeI site and a reverse primer containing a BamHI site (see Table S2 in the supplemental material). The amplicons were cloned into the corresponding sites in pET28b (Novagen) and the resulting constructs (Cb PTPSI-pET28b, Cb PTPSIII-pET28b, Li PTPSI-pET28b, and Li PTPSIII-pET28b) were confirmed by sequencing.
Site-directed mutagenesis.
The Synthrophobacter fumaroxidans PTPS-I and Synthrophus aciditrophicus PTPS-III sequences were mutagenized by using overlap extension PCR (7). In each case, two specific, complementary oligonucleotides were designed (see Table S2 in the supplemental material). The PCR products obtained were cloned into pBAD24 as described above, and the resulting constructs were verified by sequencing.
Functional complementation experiments.
E. coli folB deletant cells were transformed with pBAD24 alone (negative control) or with pBAD24 containing E. coli folB (positive control) or various PTPS genes. Complementation tests were made by streaking transformed cells on LB plates containing appropriate antibiotics and l-arabinose, with or without thymidine. Four independent clones were used for each construct. Plates were incubated for 2 days at 37°C.
Folate analysis.
Wild-type and folB deletant strains harboring pBAD24 alone or containing PTPS-I or PTPS-III genes from L. interrogans were grown at 37°C in 50 ml of LB medium containing thymidine, l-arabinose, and appropriate antibiotics. Cells were pelleted when A600 reached 1.0 ± 0.1 and stored at −80°C until use. Pellets were resuspended in 10 ml of 50 mM HEPES-CHES [2-(N-cyclohexylamino)ethanesulfonic acid] (pH 7.8) containing 2% (wt/vol) ascorbic acid and 10 mM β-mercaptoethanol and then sonicated, boiled for 10 min, and centrifuged (13,000 × g, 10 min). The pellets were reextracted the same way, and the combined extracts were treated for 2 h at 37°C with 2 ml of dialyzed rat plasma to deglutamylate folates. Samples were then boiled for 10 min, centrifuged as described above, and filtered. Folates were isolated by using 2-ml folate affinity columns (17). Samples of the eluate were analyzed by high-pressure liquid chromatography (HPLC) with electrochemical detection (2). Detector response was calibrated with standards from Merck Eprova. The folate content of wild-type cells was in the normal range for E. coli (34); note that methionine, present at a high level in LB medium, represses 5,10-methylenetetrahydrofolate reductase (metF) so that little, if any, 5-methyltetrahydrofolate is formed (21).
Expression and purification of recombinant proteins.
E. coli BL21-CodonPlus (DE3)-RIPL competent cells (Stratagene) were transformed with hsPTPS-pET28a, PfPTPS-pET46 (10), CbPTPSI-pET28b, CbPTPSIII-pET28b, LiPTPSI-pET28b, or LiPTPSIII-pET28b constructs, all of which introduced an N-terminal His tag. Cells were grown at 37°C in LB medium containing appropriate antibiotics. When A600 reached 0.7, IPTG (isopropyl-β-d-thiogalactopyranoside) was added to a final concentration of 1 mM, and growth was continued for 3 h. Subsequent steps were carried out at 4°C. The cells were harvested by centrifugation, resuspended in 50 mM Tris-HCl (pH 8.0), and disrupted by sonication. The cleared supernatant was loaded onto a Ni2+-nitrilotriacetic acid agarose resin column (Qiagen) equilibrated with the sonication buffer. After the column was washed with 50 mM NaH2PO4, 300 mM NaCl, and 20 mM imidazole (pH 8.0), the recombinant protein was eluted with the same buffer containing 250 mM imidazole and dialyzed overnight against 100 mM Tris-HCl (pH 8.0) containing 2 mM dithiothreitol. Protein was estimated by the Bradford method (5) using bovine serum albumin as a standard. Purified proteins were frozen in liquid N2 and stored at −80°C after the addition of 10% (vol/vol) glycerol. Freezing was shown not to affect activity.
Enzyme assays.
Reactions (50 μl) were performed in 100 mM Tris-HCl (pH 8.0), containing 2 mM dithiothreitol, 9 μM dihydroneopterin triphosphate, 10 mM MgCl2, and 75 μg of PTPS enzyme as described previously (10). After incubation for 1 h at 37°C, the reaction products were oxidized for 1 h in the dark at 4°C by the addition of 10 μl of 1% I2-2% KI (wt/vol) in 0.1 M HCl, after which the excess iodine was oxidized by adding 10 μl of 10% (wt/vol) sodium ascorbate. Pterins were separated on a 4-μm, 250-by-4.6-mm Synergi Fusion-RP 80 column (Phenomenex) that was eluted isocratically with 10 mM sodium phosphate (pH 6.0) at 1.5 ml/min. Peaks were detected by fluorescence (350-nm excitation, 450-nm emission) and identified relative to standards.
RESULTS
Bacteria lacking FolB have unusual PTPS-like proteins.
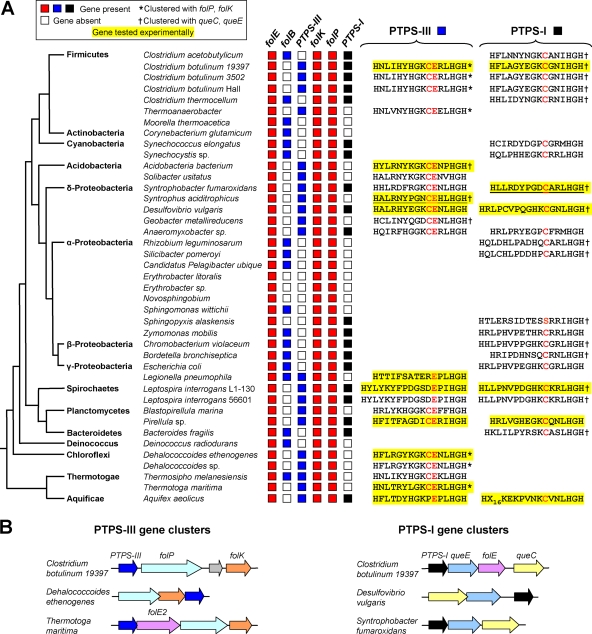
We first extended a comparative genomics analysis of the folate biosynthesis pathway (9) to hundreds of newly available bacterial genomes. The results are summarized in Fig. 2A and are available in full at the SEED server (theseed.uchicago.edu) in the subsystem titled “Experimental-PTPS.” We found 40 highly diverse genomes that lack FolB but have the other enzymes of de novo folate synthesis (represented for simplicity in Fig. 2A by those on either side of the FolB step, namely, FolE, FolK, and FolP). Thirty-eight of these genomes (95%) specified, in addition to a typical PTPS-I, a PTPS-like protein with an active-site glutamate but no cysteine or, alternatively, a PTPS-like protein with both glutamate and cysteine (Fig. 2A). Both types of glutamate-containing proteins were designated PTPS-III. In sharp contrast, of 355 genomes having FolB as well as a PTPS-like protein, only 10 (3%) were of the PTPS-III type, i.e., containing an active-site glutamate. The occurrence of PTPS-III is thus strongly anticorrelated with that of FolB, constituting a strong a priori case that PTPS-III functionally replaces FolB in the folate pathway. Moreover, PTPS-III genes are often situated in folate synthesis gene clusters, whereas PTPS-I genes are frequently in queuosine synthesis gene clusters (Fig. 2B).
FIG. 2.
Comparative genomic evidence implying that PTPS-III can replace FolB in bacteria. (A) Phylogenetic distribution of genes encoding FolB, PTPS-III, and PTPS-I among representative bacteria. The tree shows the standard branching order for each group and is adapted from Barrick and Breaker (3), with the addition of Planctomycetes and Aquificae (6). Apart from folB, which is missing in some cases, all species shown have complete sets of folate synthesis genes as exemplified by folE, folK, and folP. The active-site regions of PTPS-III and PTPS-I proteins, bounded by three His residues coordinating the essential Zn2+ ion, are shown on the right. Asterisks indicate that the corresponding gene is clustered on the chromosome with folK and folP; daggers indicate clustering with the queuosine synthesis genes queC and queE. Yellow highlighting indicates that the corresponding gene was tested experimentally. Underlining denotes the two sequences selected for mutagenesis. (B) Clustering of genes specifying PTPS-III with folate synthesis genes, and clustering of genes specifying PTPS-I with queuosine synthesis genes. Matching colors correspond to orthologous genes; gray arrows are unrelated genes. Arrows show transcriptional direction; overlap denotes translational coupling.
Genes encoding PTPS-III proteins complement the deletion of E. coli folB.
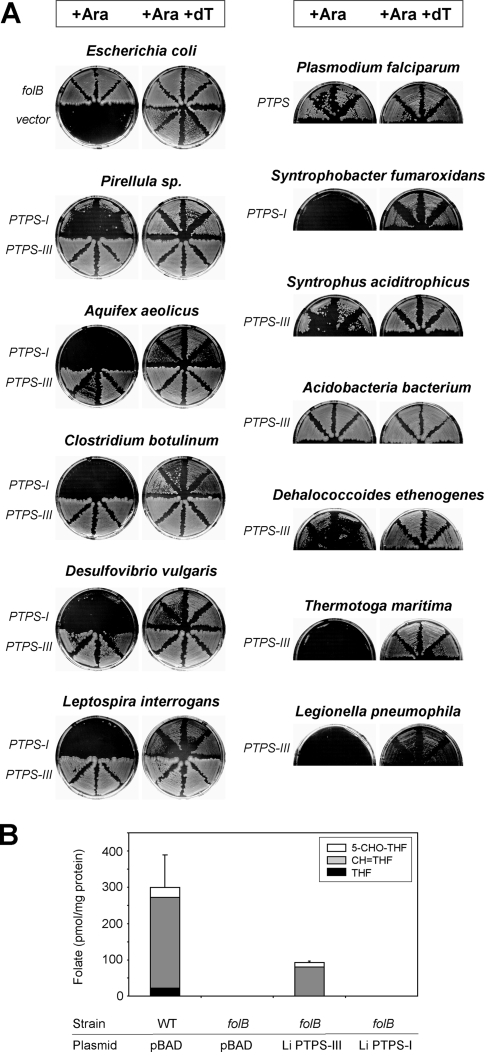
A folB deletion was transferred by P1 transduction from Keio collection strain JW3030-2 to the K-12 strain MG1655. Like other folate synthesis mutants (20, 22), the resulting deletant strain showed little or no growth on rich medium unless it was supplied with thymidine. The deletant was transformed with the arabinose-inducible plasmid vector pBAD24 containing PTPS-I or PTPS-III genes from diverse bacteria or the PTPS gene of P. falciparum. The native E. coli folB gene served as a positive control. The transformants were plated on LB medium supplemented with arabinose or arabinose plus thymidine. As expected from the biochemical evidence (10), the P. falciparum gene conferred thymidine prototrophy, thereby validating the complementation assay (Fig. 3A).
FIG. 3.
Functional complementation of the E. coli folB deletant by genes specifying PTPS-III but not PTPS-I. (A) The E. coli folB deletant strain harboring pBAD24 alone or containing PTPS genes from various sources was plated on LB medium containing 0.2% l-arabinose (Ara) with or without 75 μg of thymidine (dT)/ml, 100 μg of carbenicillin/ml, and 50 μg of kanamycin/ml. Plates were incubated at 37°C for 2 days. (B) Folate analysis from 50-ml liquid cultures of wild-type (WT) and folB deletant strains harboring pBAD24 alone or containing PTPS-I or PTPS-III genes from L. interrogans (Li). The values are expressed as the means and standard error from three replicates. The medium was LB plus 75 μg of thymidine/ml, 0.2% l-arabinose, and 100 μg of carbenicillin/ml. Cells were harvested when the A600 reached 1.0 ± 0.1. No folates were detected in folB deletant cells or in deletant cells expressing L. interrogans PTPS-I; detection limits for individual folates were 0.2 to 0.5 pmol/mg of protein. THF, tetrahydrofolate; CH=THF, 5,10-methenyltetrahydrofolate; 5-CHO-THF, 5-formyltetrahydrofolate.
In the absence of thymidine, five of the six PTPS-I genes showed no growth, and the sixth (from Pirellula sp.) showed very little. In contrast, 8 of the 10 bacterial PTPS-III genes allowed vigorous growth, as did the folB control (Fig. 3A). Of the two inactive PTPS-III genes, one came from a hyperthermophile (Thermotoga maritima) whose proteins may not be active in E. coli at 37°C (28). The other was from Legionella pneumophila, which—unlike the sources of the other PTPS-III genes—has a folB gene and so presumably needs no functional alternative. All strains grew in the presence of thymidine, confirming that none of the PTPS genes is toxic.
To substantiate the results from complementation of the growth phenotype, we analyzed folates in the wild-type strain and the folB deletant harboring either vector alone or vector plus representative PTPS-I or PTPS-III sequences (from L. interrogans) (Fig. 3B). As expected, no folates were detectable in the folB deletant. Consistent with the growth complementation data, expression of PTPS-III substantially restored folate levels, whereas the expression of PTPS-I did not.
PTPS-III proteins catalyze the conversion of dihydroneopterin triphosphate to HMDHP.
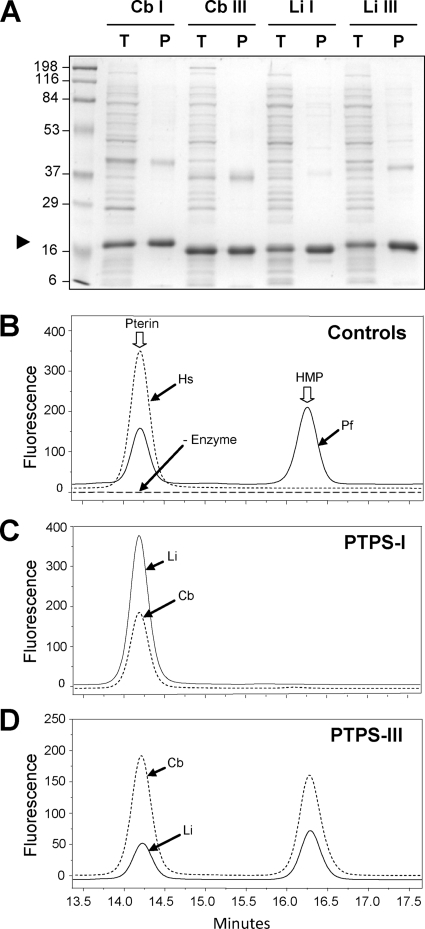
To reinforce the complementation results, we tested the ability of bacterial PTPS-III and PTPS-I proteins to mediate formation of HMDHP from dihydroneopterin triphosphate in vitro. PTPS-III and PTPS-I from the representative species C. botulinum and L. interrogans were overexpressed as N-terminally His-tagged proteins in E. coli and affinity purified; the products were judged to be ≥90% homogeneous by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (Fig. 4A). The purified proteins were incubated with dihydroneopterin triphosphate generated from GTP using recombinant GTP cyclohydrolase I. Purified P. falciparum PTPS and human PTPS were included as controls (10). The reaction mixtures were oxidized by treatment with acidic iodine before HPLC separation. The oxidation step converts HMDHP to 6-hydroxymethylpterin and 6-pyruvoyltetrahydropterin to pterin, allowing sensitive fluorescence detection of the reaction products. As reported by Dittrich et al. (10), 6-hydroxymethylpterin and pterin were, respectively, the major and minor products of the P. falciparum enzyme, and pterin was the sole product of the human enzyme (Fig. 4B). Both bacterial PTPS-I proteins gave pterin, as reported for E. coli PTPS-I (10, 39) (Fig. 4C), whereas the bacterial PTPS-III proteins gave 6-hydroxymethylpterin, as well as pterin (Fig. 4D). Collectively, these data are consistent with the view that PTPS-III proteins catalyze HMDHP formation from dihydroneopterin triphosphate and that PTPS-I proteins do not. It should be noted that the bacterial proteins gave rise to other fluorescent peaks besides pterin and 6-hydroxymethylpterin; these presumably corresponded, for PTPS-I proteins at least, to 6-carboxytetrahydropterin or its breakdown products (27). The bacterial, P. falciparum, and human proteins all appeared to lack significant dihydroneopterin aldolase activity since they formed only traces of HMDHP (detected after oxidation as 6-hydroxymethylpterin) when dihydroneopterin was used as the substrate (data not shown). This trace HMDHP formation was no greater in PTPS-III reactions than in PTPS-I reactions, supporting its physiological insignificance.
FIG. 4.
Purification of PTPS proteins and HPLC analysis of the products they form from dihydroneopterin triphosphate. (A) SDS-polyacrylamide gel electrophoresis of 15 μg of total proteins of IPTG-induced E. coli cells (T lanes), and 5 μg of Ni2+-affinity-purified PTPS-I or PTPS-III proteins (P lanes). Staining was performed with Coomassie blue. The positions of molecular markers (in kilodaltons) are indicated. The PTPS monomer bands are indicated with an arrowhead. Li, L. interrogans; Cb, C. botulinum. (B to D) HPLC analyses of reaction products. No products were formed in blank reactions without dihydroneopterin triphosphate substrate. Reaction products were subjected before chromatography to iodine oxidation, which converts 6-pyruvoyltetrahydropterin to pterin and HMDHP to 6-hydroxymethylpterin, and products were detected fluorometrically. The normalized retention times of pterin and 6-hydroxymethylpterin (HMP) standards added to each sample are marked with arrows. Frame B shows data for controls using human (Hs) PTPS-II (which forms only 6-pyruvoyltetrahydropterin) and P. falciparum PTPS (Pf) (which forms mainly HMDHP with some 6-pyruvoyltetrahydropterin). Frames C and D show data for PTPS-I or PTPS-III proteins from L. interrogans and C. botulinum.
Phylogenetic and mutational analyses of the origin of PTPS-III proteins.
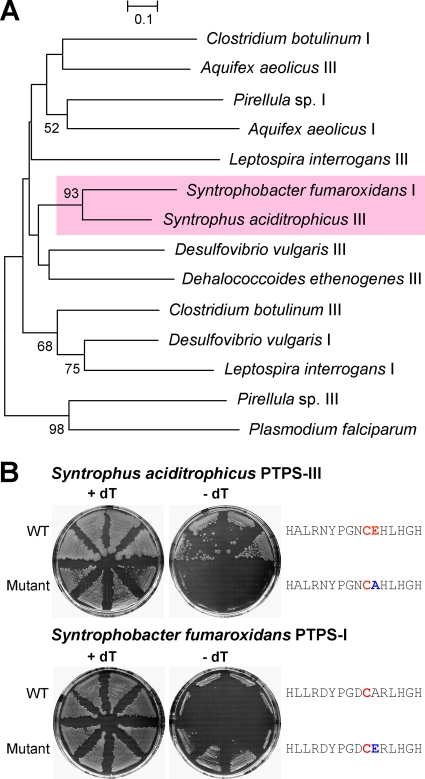
The evolutionary origin of PTPS-III proteins was investigated first by phylogenetic analysis of the representative PTPS-III and PTPS-I proteins that had been tested experimentally. The resulting tree (Fig. 5A) showed an intermingling of PTPS-III and PTPS-I sequences. The absence of distinct PTPS-III and PTPS-I clades implies that these proteins diverged at various times in various lineages. This inference is corroborated by the strong bootstrap support for the nodes linking C. botulinum PTPS-III with two PTPS-I proteins, and Syntrophus aciditrophicus PTPS-III with Syntrophobacter fumaroxidans PTPS-I.
FIG. 5.
Phylogenetic and mutational evidence on the origin of PTPS-III proteins. (A) Phylogenetic tree of PTPS proteins from representative bacteria and P. falciparum. Sequences were taken from the SEED database. The tree was constructed by the neighbor-joining method; bootstrap values are indicated only for nodes with >50% support. The tree is drawn to scale; evolutionary distances are in units of number of amino acid substitutions per site. The two sequences selected for mutagenesis are highlighted in rose. (B) Effects of mutations in the active-site region of Syntrophus aciditrophicus PTPS-III or Syntrophobacter fumaroxidans PTPS-I on functional complementation of the E. coli folB deletant. The deletant strain harboring pBAD24 containing wild-type (WT) or mutated PTPS genes was plated on LB medium containing 0.2% l-arabinose with or without 75 μg of thymidine (dT)/ml, and appropriate antibiotics. Plates were incubated at 37°C for 2 days. Active-site regions are shown on the right; the predicted catalytic cysteine and glutamate residues in the wild type are colored red, and the mutated residues are colored blue.
The high overall sequence similarity (75%) of the Syntrophus aciditrophicus PTPS-III and Syntrophobacter fumaroxidans PTPS-I sequences, especially that of the active site region (Fig. 2A), prompted us to use site-directed mutagenesis to investigate the role of the active-site glutamate residue in determining enzyme activity (Fig. 5B). This residue in Syntrophus aciditrophicus PTPS-III (E27) was accordingly changed to alanine, and the equivalent residue in Syntrophobacter fumaroxidans PTPS-I (A27) was changed to glutamate. Replacing the glutamate residue abolished the ability to complement the E. coli folB mutation, confirming that this amino acid is necessary for PTPS-III activity, as is the case for the P. falciparum enzyme (10). Conversely, adding a glutamate residue conferred incipient complementation activity in the plate assay (Fig. 5B). To corroborate this result, we measured the growth in liquid medium (LB plus 0.2% l-arabinose, 100 μg of carbenicillin/ml, and 50 μg of kanamycin/ml) of 10 independent clones expressing mutant (A27E) or wild-type Syntrophobacter fumaroxidans PTPS-I. The former grew significantly (P < 0.05) more than the latter, yielding a difference in mean A600 values of 1.2 after 20.5 h.
DISCUSSION
Our findings solve the enigma (9) of how numerous bacteria dispense with the essential folate synthesis enzyme FolB and show that they do this by an expedient adopted by P. falciparum and perhaps heterokonts: the use of a variant form of PTPS. FolB (dihydroneopterin aldolase) thus joins a growing list of steps in the classical folate synthesis pathway for which there exist alternative, completely different enzymes. Other examples include FolE (11), FolA (26), and FolC (9). In fact, the FolB case is more radical than the others since it involves a bypass that replaces three steps, the other two being those mediated by FolQ (dihydroneopterin triphosphate pyrophosphatase) and a nonspecific phosphatase (Fig. 1A). We can therefore predict that FolQ is also missing from genomes in which PTPS-III replaces FolB, but this cannot be verified from genome sequences alone because FolQ proteins are heterogeneous and belong to the large and almost omnipresent Nudix family (13, 23).
Our functional complementation and folate analysis data provide the first genetic evidence for replacement of FolB by PTPS-III type proteins, since prior evidence on this point—for the P. falciparum protein—was solely biochemical (10). Our biochemical data indicate that bacterial PTPS-III proteins, like their P. falciparum counterpart (10), mediate the formation of HMDHP from dihydroneopterin triphosphate. Note, however, that since HMDHP was not detected directly, the data do not exclude the possibility that the native PTPS-III product is hydroxymethyltetrahydropterin or another pterin that yields 6-hydroxymethylpterin after acidic iodine treatment (33). Our biochemical data also leave open the question of the other product(s) of the reaction and consequently of the mechanism involved. A two-carbon fragment must be released from the pterin side chain, as in the FolB reaction (Fig. 1B), and it will interesting to determine whether this is accompanied by the unusual elimination of triphosphate that characterizes the normal PTPS reaction leading to 6-pyruvoyltetrahydropterin (Fig. 1C) (37).
Our results bear also on the evolutionary origin of bacterial PTPS-III proteins. Their relative rarity and sporadic taxonomic distribution compared to the far more common PTPS-I proteins suggest a likely origin from PTPS-I proteins. Phylogenetic analysis further suggests that PTPS-III proteins may have arisen multiple times in various PTPS-I lineages, several PTPS-III proteins being more similar to various PTPS-I proteins than to each other. The idea of multiple origins of PTPS-III proteins is supported by the finding that, for closely related PTPS proteins, a single glutamate residue immediately downstream of the catalytic cysteine can determine the presence or absence of PTPS-III activity. It is thus reasonable to suppose that evolution of PTPS-III proteins began with a mutation that introduced a glutamate residue into a PTPS-I protein. Such mutations could in some instances have been a single base change, as in the case of the alanine-to-glutamate conversion (GCG→GAG) in Syntrophobacter fumaroxidans PTPS-I (Fig. 5B). Subsequent mutations could have increased the efficiency of HMDHP production, eventually making FolB redundant. It is also reasonable to suppose that adding a glutamate residue does not abolish PTPS-I activity. Syntrophus aciditrophicus and several other bacteria that lack FolB have a single glutamate- and cysteine-containing PTPS (which in the case of Syntrophus aciditrophicus has PTPS-III activity) whose gene is clustered with genes of queuosine biosynthesis (Fig. 2A), the clustering implying that the protein remains a functional PTPS-I (QueD) that participates in queuosine biosynthesis (32). In contrast to these cases of putatively bifunctional PTPS-III/PTPS-I proteins, there are several genomes (e.g., Aquifex aeolicus) that specify a PTPS-III protein with no active-site cysteine, plus a PTPS-I protein with cysteine only. In such cases the PTPS-III protein may have become specialized and lost the ancestral PTPS-I function. A final point about the acquisition of PTPS-III proteins is that the phylogenetic tree (Fig. 5A) suggests the possibility of horizontal gene transfer in the clade containing the planctomycete bacterium Pirellula and eukaryotic P. falciparum proteins.
Although the details of PTPS-III evolution necessarily remain conjectural, there is no mistaking the overall pattern. It is seemingly facile for a PTPS-I protein to acquire PTPS-III activity, and this probably occurred independently in various lineages so that PTPS-III proteins provide striking instances of convergent evolution. Their occurrence in pathogenic bacteria (Clostridia, Spirochaetes), and probably also in the oomycete pathogen Phytophthora (19), could make PTPS-III proteins targets for novel antifolate compounds.
Supplementary Material
Acknowledgments
This project was supported by U.S. Department of Energy grant DE-FG02-07ER64498 (to V.D.C.-L.); by U.S. Department of Agriculture Cooperative State Research, Education, and Extension Service National Research Initiative grant 2008-35318-04589 (to A.D.H.); by National Institutes of Health grant R01 GM70641-01 (to V.D.C.-L.); by Wellcome Trust grant 073896 (to J.E.H.); and by an endowment from the C. V. Griffin, Sr., Foundation.
We thank J. F. Gregory III for help with the folate analyses, C. Kuske for Acidobacteria bacterium DNA, M. Picardeau for L. interrogans L1-130 DNA, D. Stahl for Desulfovibrio vulgaris DNA, S. Zinder for Dehalococcoides ethenogenes DNA, P. Schimmel for Aquifex aeolicus DNA, S. Lesley for Thermotoga maritima genomic clone TM0038, and Y. Park for hsPTPS-pET28a and sGTPc-pET15b plasmids.
Footnotes
Published ahead of print on 24 April 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Baba, T., T. Ara, M. Hasegawa, Y. Takai, Y. Okumura, M. Baba, K. A. Datsenko, M. Tomita, B. L. Wanner, and H. Mori. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 22006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bagley, P. J., and J. Selhub. 2000. Analysis of folate form distribution by affinity followed by reversed-phase chromatography with electrochemical detection. Clin. Chem. 46404-411. [PubMed] [Google Scholar]
- 3.Barrick, J. E., and R. R. Breaker. 2007. The distributions, mechanisms, and structures of metabolite-binding riboswitches. Genome Biol. 8R239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blakley, R. L. 1969. Chemical and physical properties of pterins and folate derivatives, p. 58-105. In R. L. Blakley (ed.), The biochemistry of folic acid and related pteridines. Wiley, New York, NY.
- 5.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72248-254. [DOI] [PubMed] [Google Scholar]
- 6.Ciccarelli, F. D., T. Doerks, C. von Mering, C. J. Creevey, B. Snel, and P. Bork. 2006. Toward automatic reconstruction of a highly resolved tree of life. Science 3111283-1287. [DOI] [PubMed] [Google Scholar]
- 7.Cormack, B. 1997. Directed mutagenesis using the polymerase chain reaction, p. 8.5.1-8.5.10. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. Wiley, New York, NY. [DOI] [PubMed]
- 8.Cossins, E. A., and L. Chen. 1997. Folates and one-carbon metabolism in plants and fungi. Phytochemistry 45437-452. [DOI] [PubMed] [Google Scholar]
- 9.de Crécy-Lagard, V., B. El Yacoubi, R. D. de la Garza, A. Noiriel, and A. D. Hanson. 2007. Comparative genomics of bacterial and plant folate synthesis and salvage: predictions and validations. BMC Genomics 8245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dittrich, S., S. L. Mitchell, A. M. Blagborough, Q. Wang, P. Wang, P. F. Sims, and J. E. Hyde. 2008. An atypical orthologue of 6-pyruvoyltetrahydropterin synthase can provide the missing link in the folate biosynthesis pathway of malaria parasites. Mol. Microbiol. 67609-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.El Yacoubi, B., S. Bonnett, J. N. Anderson, M. A. Swairjo, D. Iwata-Reuyl, and V. de Crécy-Lagard. 2006. Discovery of a new prokaryotic type I GTP cyclohydrolase family. J. Biol. Chem. 28137586-37593. [DOI] [PubMed] [Google Scholar]
- 12.Forrest, H. S., and C. Van Baalen. 1970. Microbiology of unconjugated pteridines. Annu. Rev. Microbiol. 2491-108. [DOI] [PubMed] [Google Scholar]
- 13.Gabelli, S. B., M. A. Bianchet, W. Xu, C. A. Dunn, Z. D. Niu, L. M. Amzel, and M. J. Bessman. 2007. Structure and function of the Escherichia coli dihydroneopterin triphosphate pyrophosphatase: a Nudix enzyme involved in folate biosynthesis. Structure 151014-1022. [DOI] [PubMed] [Google Scholar]
- 14.Gardner, M. J., N. Hall, E. Fung, O. White, M. Berriman, R. W. Hyman, J. M. Carlton, A. Pain, K. E. Nelson, S. Bowman, I. T. Paulsen, K. James, J. A. Eisen, K. Rutherford, S. L. Salzberg, A. Craig, S. Kyes, M. S. Chan, V. Nene, S. J. Shallom, B. Suh, J. Peterson, S. Angiuoli, M. Pertea, J. Allen, J. Selengut, D. Haft, M. W. Mather, A. B. Vaidya, D. M. Martin, A. H. Fairlamb, M. J. Fraunholz, D. S. Roos, S. A. Ralph, G. I. McFadden, L. M. Cummings, G. M. Subramanian, C. Mungall, J. C. Venter, D. J. Carucci, S. L. Hoffman, C. Newbold, R. W. Davis, C. M. Fraser, and B. Barrell. 2002. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 419498-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Green, J. M., and R. G. Mathews. March 2007, posting date. Chapter 3.6.3.6, Folate biosynthesis, reduction, and polyglutamylation and the interconversion of folate derivatives. In A. Böck, R. Curtiss III, J. B. Kaper, F. C. Neidhardt, T. Nyström, K. E. Rudd, and C. L. Squires (ed.), EcoSal—Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC. http://www.ecosal.org/ecosal/index.jsp.
- 16.Green, M. L., and P. D. Karp. 2004. A Bayesian method for identifying missing enzymes in predicted metabolic pathway databases. BMC Bioinform. 576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gregory, J. F., and J. P. Toth. 1988. Chemical synthesis of deuterated folate monoglutamate and in vivo assessment of urinary excretion of deuterated folates in man. Anal. Biochem. 17094-104. [DOI] [PubMed] [Google Scholar]
- 18.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose pBAD promoter. J. Bacteriol. 1774121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hyde, J. E., S. Dittrich, P. Wang, P. F. Sims, V. de Crécy-Lagard, and A. D. Hanson. 2008. Plasmodium falciparum: a paradigm for alternative folate biosynthesis in diverse microorganisms? Trends Parasitol. 24502-508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jönsson, M., and G. Swedberg. 2005. Hydroxymethyldihydropterin pyrophosphokinase from Plasmodium falciparum complements a folK-knockout mutant in Escherichia coli when expressed as a separate polypeptide detached from dihydropteroate synthase. Mol. Biochem. Parasitol. 140123-125. [DOI] [PubMed] [Google Scholar]
- 21.Katzen, H. M., and J. M. Buchanan. 1965. Enzymatic synthesis of the methyl group of methionine. VIII. Repression-derepression, purification, and properties of 5,10-methylenetetrahydrofolate reductase from Escherichia coli. J. Biol. Chem. 240825-835. [PubMed] [Google Scholar]
- 22.Klaus, S. M., E. R. Kunji, G. G. Bozzo, A. Noiriel, R. D. de la Garza, G. J. Basset, S. Ravanel, F. Rébeillé, J. F. Gregory III, and A. D. Hanson. 2005. Higher plant plastids and cyanobacteria have folate carriers related to those of trypanosomatids. J. Biol. Chem. 28038457-38463. [DOI] [PubMed] [Google Scholar]
- 23.Klaus, S. M., A. Wegkamp, W. Sybesma, J. Hugenholtz, J. F. Gregory III, and A. D. Hanson. 2005. A nudix enzyme removes pyrophosphate from dihydroneopterin triphosphate in the folate synthesis pathway of bacteria and plants. J. Biol. Chem. 2805274-5280. [DOI] [PubMed] [Google Scholar]
- 24.Kong, J. S., J. Y. Kang, H. L. Kim, O. S. Kwon, K. H. Lee, and Y. S. Park. 2006. 6-Pyruvoyltetrahydropterin synthase orthologs of either a single or dual domain structure are responsible for tetrahydrobiopterin synthesis in bacteria. FEBS Lett. 5804900-4904. [DOI] [PubMed] [Google Scholar]
- 25.Lee, S. W., H. W. Lee, H. J. Chung, Y. A. Kim, Y. J. Kim, Y. Hahn, P. H. Chung, and Y. S. Park. 1999. Identification of the genes encoding enzymes involved in the early biosynthetic pathway of pteridines in Synechocystis sp. PCC 6803. FEMS Microbiol. Lett. 176169-176. [DOI] [PubMed] [Google Scholar]
- 26.Levin, I., M. Giladi, N. Altman-Price, R. Ortenberg, and M. Mevarech. 2004. An alternative pathway for reduced folate biosynthesis in bacteria and halophilic archaea. Mol. Microbiol. 541307-1318. [DOI] [PubMed] [Google Scholar]
- 27.McCarty, R. M., A. Somogyi, and V. Bandarian. 2009. Escherichia coli QueD is a 6-carboxy-5,6,7,8-tetrahydropterin synthase. Biochemistry 482301-2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miyazaki, K. 2005. Hyperthermophilic α-l-arabinofuranosidase from Thermotoga maritima MSB8: molecular cloning, gene expression, and characterization of the recombinant protein. Extremophiles 9399-406. [DOI] [PubMed] [Google Scholar]
- 29.Osterman, A., and R. Overbeek. 2003. Missing genes in metabolic pathways: a comparative genomics approach. Curr. Opin. Chem. Biol. 7238-251. [DOI] [PubMed] [Google Scholar]
- 30.Overbeek, R., T. Begley, R. M. Butler, J. V. Choudhuri, H. Y. Chuang, M. Cohoon, V. de Crécy-Lagard, N. Diaz, T. Disz, R. Edwards, M. Fonstein, E. D. Frank, S. Gerdes, E. M. Glass, A. Goesmann, A. Hanson, D. Iwata-Reuyl, R. Jensen, N. Jamshidi, L. Krause, M. Kubal, N. Larsen, B. Linke, A. C. McHardy, F. Meyer, H. Neuweger, G. Olsen, R. Olson, A. Osterman, V. Portnoy, G. D. Pusch, D. A. Rodionov, C. Ruckert, J. Steiner, R. Stevens, I. Thiele, O. Vassieva, Y. Ye, O. Zagnitko, and V. Vonstein. 2005. The subsystems approach to genome annotation and its use in the project to annotate 1,000 genomes. Nucleic Acids Res. 335691-5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pfleiderer, W. 1985. Chemistry of naturally occurring pteridines, p. 43-114. In R. L. Blakley and S. J. Benkovic (ed.), Folates and pterins, vol. 2. Wiley, New York, NY. [Google Scholar]
- 32.Reader, J. S., D. Metzgar, P. Schimmel, and V. de Crécy-Lagard. 2004. Identification of four genes necessary for biosynthesis of the modified nucleoside queuosine. J. Biol. Chem. 2796280-6285. [DOI] [PubMed] [Google Scholar]
- 33.Rembold, H., and R. Gyure. 1972. Biochemistry of the pteridines. Angew. Chem. Int. Ed. Engl. 111061-1072. [DOI] [PubMed] [Google Scholar]
- 34.Rohlman, C. E., and R. G. Matthews. 1990. Role of purine biosynthetic intermediates in response to folate stress in Escherichia coli. J. Bacteriol. 1727200-7210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Singer, S., R. Ferone, L. Walton, and L. Elwell. 1985. Isolation of a dihydrofolate reductase-deficient mutant of Escherichia coli. J. Bacteriol. 164470-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 241596-1599. [DOI] [PubMed] [Google Scholar]
- 37.Thöny, B., G. Auerbach, and N. Blau. 2000. Tetrahydrobiopterin biosynthesis, regeneration and functions. Biochem. J. 3471-16. [PMC free article] [PubMed] [Google Scholar]
- 38.Wang, P., Q. Wang, T. V. Aspinall, P. F. Sims, and J. E. Hyde. 2004. Transfection studies to explore essential folate metabolism and antifolate drug synergy in the human malaria parasite Plasmodium falciparum. Mol. Microbiol. 511425-1438. [DOI] [PubMed] [Google Scholar]
- 39.Woo, H. J., Y. K. Hwang, Y. J. Kim, J. Y. Kang, Y. K. Choi, C. G. Kim, and Y. S. Park. 2002. Escherichia coli 6-pyruvoyltetrahydropterin synthase ortholog encoded by ygcM has a new catalytic activity for conversion of sepiapterin to 7,8-dihydropterin. FEBS Lett. 523234-238. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.