Abstract
Coxiella burnetii is an obligate intracellular bacterium that directs biogenesis of a parasitophorous vacuole (PV) for replication. Effectors of PV maturation are likely translocated into the host cytosol by a type IV secretion system (T4SS) with homology to the Dot/Icm apparatus of Legionella pneumophila. Since secreted bacterial virulence factors often functionally mimic the activities of host proteins, prokaryotic proteins with eukaryotic features are considered candidate T4SS substrates. Genes encoding proteins with eukaryotic-type ankyrin repeat domains (Anks) were identified upon genome sequencing of the C. burnetii Nine Mile reference isolate, which is associated with a case of human acute Q fever. Interestingly, recent genome sequencing of the G and K isolates, derived from human chronic endocarditis patients, and of the Dugway rodent isolate revealed remarkable heterogeneity in the Ank gene family, with the Dugway isolate harboring the largest number of full-length Ank genes. Using L. pneumophila as a surrogate host, we identified 10 Dugway Anks and 1 Ank specific to the G and K endocarditis isolates translocated into the host cytosol in a Dot/Icm-dependent fashion. A 10-amino-acid C-terminal region appeared to be necessary for translocation, with some Anks also requiring the chaperone IcmS for secretion. Ectopically expressed Anks localized to a variety of subcellular regions in mammalian cells, including microtubules, mitochondria, and the PV membrane. Collectively, these data suggest that C. burnetii isolates translocate distinct subsets of the Ank protein family into the host cytosol, where they modulate diverse functions, some of which may be unique to C. burnetii pathotypes.
Obligate intracellular Coxiella burnetii is the causative agent of human Q fever. Acute Q fever normally manifests as an influenza-like illness, but approximately 50% of C. burnetii infections are thought to be asymptomatic (52). Chronic Q fever can occur and generally presents as endocarditis. These cases are usually associated with individuals who are immunosuppressed and/or have heart valve defects (52). Host factors are clearly involved in Q fever presentation (36). However, there is also laboratory evidence suggesting that C. burnetii isolates have distinct pathogenic potential. For example, the G endocarditis isolate disseminates less and causes less inflammatory damage in BALB/c mice than the Nine Mile (NM) reference isolate does (50). Additionally, some isolates derived from infected mammals, such as the Priscilla goat isolate and the Dugway rodent isolates, are attenuated relative to NM in a guinea pig infection model of Q fever (26, 51).
In vivo, C. burnetii targets alveolar mononuclear phagocytes and establishes a parasitophorous vacuole (PV) that displays many characteristics of lysosomes (54). Early stages of infection are characterized by phagosome maturation stalling and interaction with the autophagic pathway (18, 37). The maturing PV eventually fuses with lysosomal compartments, as evidenced by acidification to approximately pH 5.0, acquisition of the small GTPase Rab7, and decoration with lysosome-associated membrane proteins 1, 2, and 3 (54). Additionally, the PV contains active lysosomal hydrolases (2). The mature PV can eventually expand to occupy the majority of the host cell cytoplasm (54).
During infection, C. burnetii actively manipulates PV biogenesis and other host cell processes that presumably ensure a stable replication niche for the duration of its lengthy infectious cycle (12). For example, treatment of infected cells with chloramphenicol, a bacterial protein synthesis inhibitor, impedes formation of the large and spacious PV (19), interactions with the autophagy pathway (37), and C. burnetii's antagonism of host cell apoptosis (25, 55). Protein effectors of these processes are likely delivered to the host cytosol by the organism's Dot/Icm type IV secretion system (T4SS). C. burnetii encodes 23 of the 26 Dot/Icm proteins found in Legionella pneumophila (53). C. burnetii Dot/Icm genes are expressed during infection, and four of them complement the corresponding L. pneumophila mutants, including genes encoding the chaperones IcmS and IcmW, indicating functional overlap between the two systems (57, 58). L. pneumophila utilizes its Dot/Icm T4SS to deliver proteins directly to the host cytosol, where their diverse effector activities manipulate multiple host cell processes (47). Over 70 Dot/Icm substrates have been identified for L. pneumophila, with the number continually expanding (30). However, with a few possible exceptions (22), C. burnetii does not encode homologs of these proteins. This observation is consistent with the contrasting endoplasmic reticulum-derived vacuole of L. pneumophila and the lysosome-like compartment of C. burnetii (42).
Bacterial secretion of effector proteins that functionally mimic the activities of eukaryotic proteins is a well-established virulence mechanism used to manipulate a variety of host cell processes, including cell survival signaling and vesicular trafficking (7). Bioinformatic analyses of the C. burnetii NM isolate genome have revealed multiple genes encoding proteins with eukaryotic-type features, such as coiled-coil domains, tetratricopeptide repeats, and ankyrin repeats, that are candidate Dot/Icm substrates (54). Genes encoding proteins with these domains, which are generally rare in prokaryotes, are thought to have arisen by interdomain horizontal gene transfer from a eukaryotic source (15). Eukaryotic ankyrin repeat domain-containing proteins (Anks) mediate protein-protein interactions involved in a multitude of host processes, including cytoskeletal motility, tumor suppression, and transcriptional regulation (27). Anks contain 33-residue repeating motifs consisting of two antiparallel α-helices connected to the next repeat via a loop region (27). A series of repeats are arranged in a curved structure, with protein-protein interactions occurring in the loop regions. In addition to C. burnetii, Ank genes are present in the facultative or obligate intracellular bacteria L. pneumophila (15), Anaplasma phagocytophilum (9), Orientia tsutsugamushi (10), Wolbachia pipientis (21), Rickettsia spp. (32), and Rickettsiella grylli.
Using L. pneumophila as a surrogate host, Dot/Icm-mediated secretion was recently demonstrated for C. burnetii AnkA, -B, -F, and -G from the NM isolate (33). That study also showed transcription of the encoding genes and secretion of native AnkF during C. burnetii infection of Chinese hamster ovary cells and human foreskin fibroblasts, respectively. The initial C. burnetii NM isolate genome study reported the presence of 13 Ank genes (45). However, reannotation of the NM isolate genome, along with compilation of the genomes of the G and K human endocarditis isolates and a Dugway rodent isolate, revealed that many NM Ank genes, including ankB, are disrupted (6). Interestingly, many disrupted NM Ank genes are intact in the G, K, and/or Dugway isolate. Moreover, these isolates encode Anks that are missing in the NM isolate, leading to the intriguing possibility that isolate-specific Dot/Icm secretion of effector proteins results in unique host cell responses.
In this study, we used the calmodulin-activated adenylate cyclase (CyaA) from Bordetella pertussis as an enzymatic reporter (48) to monitor secretion of C. burnetii Anks carried by the Dugway and K isolates. Using L. pneumophila as a surrogate host, we found that most intact C. burnetii Anks are Dot/Icm substrates and that some, but not all, require the chaperone IcmS for translocation into the host cytosol. Moreover, the Ank C-terminal 10 amino acids appear to be critical for secretion. These proteins traffic to a variety of subcellular locations when expressed ectopically in mammalian cells, thereby providing clues about their possible function.
MATERIALS AND METHODS
Mammalian cell culture.
THP-1 human monocyte-like cells (TIB-202; ATCC, Manassas, VA) and HeLa (human epithelioid carcinoma) cells (CCL-2; ATCC) were maintained in RPMI 1640 medium (Invitrogen, Carlsbad, CA) containing 10% fetal calf serum (Invitrogen) at 37°C and 5% CO2. Prior to infection, THP-1 cells were treated with 200 nM phorbol 12-myristate 13-acetate (PMA; EMD Biosciences, San Diego, CA) to promote their differentiation into adherent, macrophage-like cells as previously described (55).
C. burnetii, L. pneumophila, and R. grylli.
The bacteria used in this study are listed in Table 1. The C. burnetii NM phase II, clone 4 (RSA439), G (Q212), K (Q154), and Dugway (5J108-111) isolates were propagated in Vero (African green monkey kidney) cells (CCL-81; ATCC) and purified as previously described (11, 46). L. pneumophila strains were generously provided by H. Shuman (Columbia University) and were cultured on charcoal yeast extract (CYE) agar plates for 2 days prior to genetic transformation and infection of THP-1 cells. For plasmid selection, L. pneumophila transformants were grown in the presence of chloramphenicol (10 μg/ml). Mutant L. pneumophila strains (LELA3118 and GS3001) were also cultured in the presence of 25 μg/ml kanamycin. R. grylli-infected Armadillidum vulgare insects (generously provided by B. Federici, University of California-Riverside) were homogenized, and DNAs were extracted using a High Pure PCR template preparation kit (Roche Applied Science, Indianapolis, IN).
TABLE 1.
Bacteria and plasmids used in this study
| Bacterial isolate or strain or plasmid | Genotype or description | Reference or supplier |
|---|---|---|
| Strains | ||
| C. burnetiiisolates | ||
| NM (RSA439) | Phase II, clone 4, Montana, tick, 1936 | 5 |
| G (Q212) | Phase I, Nova Scotia, heart valve, 1981 | 5 |
| K (Q154) | Phase I, Oregon, heart valve, 1976 | 5 |
| Dugway (5J108-111) | Phase I, Utah, rodent, 1958 | 51 |
| L. pneumophilastrains | ||
| JR32 | Salt-sensitive isolate of AM511 | 41 |
| LELA3118 | JR32 dotA::Tn903dIIlacZ3118 DotA− Kmr | 41 |
| GS3001 | JR32 icmS3001::Km IcmS− Kmr | 44 |
| R. grylli | B. Federici | |
| E. coliTOP10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80dlacZΔM15 ΔlacX74 recA1 araD139 Δ(ara-leu)7697 galU galK rpsL (Strr) endA1 nupG | Invitrogen |
| Plasmids | ||
| pJB2581 | cyaA fusion vector; Cmr | 4 |
| pJB2590 | ralF in pJB2581 | 4 |
| pDHVAnkA | ankA in pJB2581 | This study |
| pDHVAnkB | ankB in pJB2581 | This study |
| pDHVAnkC | ankC in pJB2581 | This study |
| pDHVAnkD | ankD in pJB2581 | This study |
| pDHVAnkF | ankF in pJB2581 | This study |
| pDHVAnkG | ankG in pJB2581 | This study |
| pDHVAnkH | ankH (bp 1927 to 2577) in pJB2581 | This study |
| pDHVAnkI | ankI in pJB2581 | This study |
| pDHVAnkJ | ankJ in pJB2581 | This study |
| pDHVAnkK | ankK in pJB2581 | This study |
| pDHVAnkL | ankL in pJB2581 | This study |
| pDHVAnkM | ankM in pJB2581 | This study |
| pDHVAnkN | ankN in pJB2581 | This study |
| pDHVAnkO | ankO in pJB2581 | This study |
| pDHVAnkP | ankP in pJB2581 | This study |
| pDHVAnkB-K | Isolate K ankB in pJB2581 | This study |
| pDHVAnkI-NM | Isolate NM ankI in pJB2581 | This study |
| pDHVAnkI-82aaFrag | ankI (bp 1932 to 2178) in pJB2581 | This study |
| pDHVAnkI-10 | ankI (bp 1 to 2148) in pJB2581 | This study |
| pDHVAnkI-20 | ankI (bp 1 to 2118) in pJB2581 | This study |
| pDHVAnkI-30 | ankI (bp 1 to 2088) in pJB2581 | This study |
| pDHVAnkI-40 | ankI (bp 1 to 2058) in pJB2581 | This study |
| pDHVAnkI-60 | ankI (bp 1 to 1998) in pJB2581 | This study |
| pDHVAnkI-80 | ankI (bp 1 to 1938) in pJB2581 | This study |
| pDHVLpAnkK | lpg2300 in pJB2581 | This study |
| pDHVRg00379 | rrgr00379 in pJB2581 | This study |
| pDHVRg00660 | rrgr00660 in pJB2581 | This study |
| pDHVRgAnkK | rrgr00938 in pJB2581 | This study |
| pCR2.1-TOPO | TA TOPO vector; Ampr | Invitrogen |
| pENTR-D/TOPO | Gateway entry vector; Kmr | Invitrogen |
| pcDNA6.2/N-mCherry | N-terminal mCherry fusion vector; Ampr | S. Grieshaber |
| pcDNA6.2/C-mCherry | C-terminal mCherry fusion vector; Ampr | S. Grieshaber |
| mChAnkA-N | mCherry::ankA | This study |
| mChAnkA-C | ankA::mCherry | This study |
| mChAnkB-N | mCherry::ankB | This study |
| mChAnkB-C | ankB::mCherry | This study |
| mChAnkF-N | mCherry::ankF | This study |
| mChAnkF-C | ankF::mCherry | This study |
| mChAnkG-N | mCherry::ankG | This study |
| mChAnkG-C | ankG::mCherry | This study |
| mChAnkH-N | mCherry::ankH | This study |
| mChAnkH-C | ankH::mCherry | This study |
| mChAnkI-N | mCherry::ankI | This study |
| mChAnkI-C | ankI::mCherry | This study |
| mChAnkJ-N | mCherry::ankJ | This study |
| mChAnkJ-C | ankJ::mCherry | This study |
| mChAnkM-N | mCherry::ankM | This study |
| mChAnkM-C | ankM::mCherry | This study |
| mChAnkN-N | mCherry::ankN | This study |
| mChAnkN-C | ankN::mCherry | This study |
| mChAnkO-N | mCherry::ankO | This study |
| mChAnkO-C | ankO::mCherry | This study |
| mChAnkP-N | mCherry::ankQ | This study |
| mChAnkP-C | ankQ::mCherry | This study |
Plasmid construction.
The complete open reading frames of Ank genes were amplified from C. burnetii, L. pneumophila, and R. grylli genomic DNAs by PCR, using Accuprime Taq polymerase (Invitrogen) and gene-specific primers (Integrated DNA Technologies, Coralville, IA), where the 5′ primer incorporated a BamHI or BglII site and the 3′ primer incorporated a SalI or PstI site (Table 2). Difficulty in cloning the entire C. burnetii ankH open reading frame necessitated PCR-based cloning of a fragment encoding the C-terminal 216 residues of the protein. The same cloning strategy was used to generate C. burnetii AnkI deletion clones. Amplified gene products were cloned into pCR2.1-TOPO (Invitrogen) and digested with either BamHI-SalI or BamHI-PstI (New England Biolabs, Ipswich, MA), and the resulting fragments were ligated with similarly digested pJB2581, using the Ligate-IT system (U.S. Biologicals, Cleveland, OH), to generate gene fusions encoding Anks N-terminally fused to the C terminus of CyaA. To generate plasmids encoding Ank-mCherry fusion proteins, C. burnetii genes were amplified by PCR, using gene-specific primers, and the amplicons were directionally cloned into pENTR-D/TOPO (Invitrogen). Forward primers contained CACC at the 5′ end for directional cloning and a 5′ Kozak sequence (ATGGGC) for mammalian expression. Ank genes were subsequently subcloned via homologous recombination into pcDNA6.2/N-mCherry or pcDNA6.2/C-mCherry (generously provided by S. Grieshaber, University of Florida), using LR Clonase II (Invitrogen). All plasmids encoding CyaA and mCherry fusion proteins were sequenced to confirm in-frame cloning and are listed in Table 1.
TABLE 2.
Primers used in this study
| Primer name and use | Sequencea |
|---|---|
| Primers for pJB2581 cloning | |
| AnkA-F | CATGCGGATCCATGTTGCTTAGCTTAATGGCC |
| AnkA-R | CGCATGCGTCGACTTAAAACAGTCCGGGGCCTG |
| AnkB-F | CATGCAGATCTATGTTTAACCAATTGGAAATAGTG |
| AnkB-R | CGCATGCGTCGACTTACATGTGCTTACCCGGGG |
| AnkC-F | CATGCGGATCCATGTTAATCCCGAGCCTCCAATC |
| AnkC-R | CGCATGCGTCGACTTATTGCTGGGTTTCGTGATTGAC |
| AnkD-F | CATGCGGATCCATGCGTAAAAGTGAGTTACGAAAG |
| AnkD-R | CGCATGCGTCGACTTACATAACCAGACAGTCCTCTG |
| AnkF-F | CATGCGGATCCATGAGACAGCGTGAAATTAATG |
| AnkF-R | CGCATGCCTGCAGTTACTACCGCTGGAAGCCGC |
| AnkG-F | CATGCGGATCCATGAGTAGACGTGAGACTCCC |
| AnkG-R | CGCATGCGTCGACTTATCACCGAGGACTAGACAG |
| AnkH-25-F | CATGCGGATCCTTGGCTTGCGGTGATGGAACGACG |
| AnkH-R | CGCATGCGTCGACTTAGATTAACGTGCGCGTATAATG |
| AnkI-F | CATGCGGATCCATGAGAGAATCATCAGAAAATC |
| AnkI-R | CGCATGCGTCGACTTACTAAATTCCAAAAGAACCC |
| AnkJ-F | CATGCGGATCCATGGCGAAATTTACTATACGTTTAG |
| AnkJ-R | CGCATGCCTGCAGTTACGCAGCGCGCATGGTTTGTCG |
| AnkK-F | CATGCGGATCCATGAATTTATCAGACACGATTATC |
| AnkK-R | CGCATGCGTCGACTTAAAATACCTCGTCCAGCTC |
| AnkL-F | CATGCGGATCCATGCGGCCCGACCCCGC |
| AnkL-R | CGCATGCGTCGACTTACTAATTTTCTTTAACATCCTC |
| AnkM-F | CATGCGGATCCATGGGTAATTTATTGAGTAAACG |
| AnkM-R | CGCATGCCTGCAGTTACTAACCACACCGAAGCAG |
| AnkN-F | CATGCGGATCCATGAGCCGGACCTACAACGC |
| AnkN-R | CGCATGCGTCGACTTAAATGCAAACCACCCGATTC |
| AnkO-F | CATGCGGATCCATGGAAATAATTTCCTTAATGAAG |
| AnkO-R | CGCATGCGTCGACTTAATAACGATTTTTTGTTTCATAAAC |
| AnkP-F | CATGCGGATCCATGGTGGGACAAAATACAACG |
| AnkP-R | CGCATGCGTCGACTTAAGCTAAGCAAGGGGTATTAC |
| AnkB-K-F | CATGCGGATCCATGAGACATCCCATAATTCACAAG |
| AnkB-K-R | CGCATGCGTCGACTTATCAAAAAATTAGGACGCCATC |
| AnkI-NM-F | CATGCGGATCCATGAGAGAATCATCAGAAAATCAAAAAAAG |
| AnkI-NM-R | CGCATGCGTCGACTTATTGCTTCAATGGAGGTACAC |
| AnkI-82aa-F | GGATCCATGAGAGAATCATCAGAAAATCAAAAAAG |
| AnkI-10-R | CTGCAGCTATTGGGATGAGGCGCTCGGA |
| AnkI-20-R | CTGCAGCTATAACTTTTCTAAGCTTTTCCAAG |
| AnkI-30-R | CTGCAGCTATTGTTTTTTTCCCCAAAATGCG |
| AnkI-40-R | CTGCAGCTATGAGAGTTTCCTACATTTACTAATCG |
| AnkI-60-R | CTGCAGCTATATTGCTTCAATGGAGGTACACTC |
| AnkI-80-R | CTGCAGCTAGAAGGCGGGGCTATGAAC |
| LpAnkK-F | CATGCGGATCCATGAGTATTGCAAACGATATTATC |
| LpAnkK-R | CGCATGCGTCGACTTATAGGCCTGTCGCAACAGG |
| Rg00379-F | CATGCGGATCCATGACCGCAATTAATATGTTAC |
| Rg00379-R | CGCATGCGTCGACTTATGAAAGAAACGTTGATTCAATTGG |
| Rg00660-F | CATGCGGATCCATGGATGTGCATGCCACTGATGCG |
| Rg00660-R | CGCATGCGTCGACTTAAAATTTATAGGAAAGTTTTTTAC |
| RgAnkK-F | CATGCGGATCCATGAATCATACGTTTGCACATG |
| RgAnkK-R | CGCATGCGTCGACTTATAGCGTTACAACAACCCC |
| Primers for pENTR cloning | |
| mChAnkA-F | CACCATGGGCTTGCTTAGCTTAATGGCCAGC |
| mChAnkA-R | AAACAGTCCGGGGCCTGAATAATTG |
| mChAnkB-F | CACCATGGGCTTTAACCAATTGGAAATAGTG |
| mChAnkB-R | CATGTGCTTACCCGGGG |
| mChAnkF-F | CACCATGGGCAGACAGCGTGAAATTAATGATGAAG |
| mChAnkF-R | CCGCTGGAAGCCGCGATTATTTC |
| mChAnkG-F | CACCATGGGCAGTAGACGTGAGACTCCCAC |
| mChAnkG-R | CCGAGGACTAGACAGACAAGAGAG |
| mChAnkH-F | CACCATGGGCTCAGAATTAGGAGGGGATGTTATTATC |
| mChAnkH-R | GATTAACGTGCGCGTATAATGAGACTC |
| mChAnkI-F | CACCATGGGCAGAGAATCATCAGAAAATCAAAAAAG |
| mChAnkI-R | AATTCCAAAAGAACCCGGCTC |
| mChAnkJ-F | CACCATGGGCGCGAAATTTACTATACGTTTAG |
| mChAnkJ-R | CGCAGCGCGCATGGTTTGTCG |
| mChAnkM-F | CACCATGGGCGGTAATTTATTGAGTAAACGTCG |
| mChAnkM-R | ACCACACCGAAGCAGAGAAGCCC |
| mChAnkN-F | CACCATGGGCAGCCGGACCTACAACGCAGC |
| mChAnkN-R | AATGCAAACCACCCGATTCTTCC |
| mChAnkO-F | CACCATGGGCGAAATAATTTCCTTAATGAAGGC |
| mChAnkO-R | ATAACGATTTTTTGTTTCATAAACTTTC |
| mChAnkP-F | CACCATGGGCGTGGGACAAAATACAACGAGG |
| mChAnkP-R | AGCTAAGCAAGGGGTATTACCTGC |
Underlining indicates restriction sites in primers.
Transformation of L. pneumophila.
L. pneumophila JR32 (wild type), LELA3118 (DotA−), and GS3001 (IcmS−) were cultured on CYE agar. Bacteria were washed from agar plates into 1 ml of DNase- and RNase-free water, washed two more times in water, and then aliquoted for use in transformations. For each transformation, 0.5 μg of CyaA-Ank-encoding plasmid was added to 50 μl of an L. pneumophila suspension, and the mixture was electroporated in 0.1-cm cuvettes at 1.8 kV/cm, 100 Ω, and 25 μF. Following electroporation, cultures were incubated in ACES buffered yeast extract (AYE) broth in the absence of antibiotics for 2 h at 37°C with shaking and then subcultured on CYE agar containing the appropriate antibiotic for 4 to 5 days. Plasmids were isolated from colonies, and clone-specific restriction enzyme digests were performed to confirm the presence of the expected restriction fragment.
Immunoblot analysis.
Expression of CyaA-Ank fusion proteins by wild type, DotA−, and IcmS− strains of L. pneumophila was verified by immunoblotting (data not shown). Protein synthesis was induced by adding 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG; ICN Biomedicals, Costa Mesa, CA) to overnight cultures of L. pneumophila transformants and incubating them for 2 h at 37°C with shaking. One milliliter of culture was centrifuged at 13,000 × g for 5 min, the cell pellet was resuspended in sodium dodecyl sulfate-polyacrylamide gel electrophoresis sample buffer, and then the proteins were separated via 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Proteins were transferred to an Immobilon-P membrane (Millipore, Bedford, MA), and membranes were blocked for 1 h at room temperature in Tris-buffered saline (TBS; 150 mM NaCl, 100 mM Tris-HCl, pH 7.6) containing 0.1% Tween 20 and 5% nonfat milk. Membranes were then incubated for 1 h at room temperature in TBS plus Tween 20 containing a mouse monoclonal antibody directed against CyaA (clone 3D1; Santa Cruz Biotechnology, Santa Cruz, CA). Membranes were washed and incubated for 1 h at room temperature in TBS plus Tween 20 containing anti-mouse immunoglobulin G secondary antibody conjugated to horseradish peroxidase (Pierce, Rockford, IL). Reacting proteins were detected via chemiluminescence using ECL Pico reagent (Pierce).
CyaA translocation assay.
L. pneumophila transformants were cultured overnight in AYE broth containing the appropriate antibiotic. PMA-differentiated THP-1 cells were plated in 24-well plates (1 × 106 cells/well) and infected with L. pneumophila expressing CyaA-Ank fusion proteins at a multiplicity of infection of approximately 20. Bacteria were added to wells, and the plate was centrifuged for 5 min at 180 × g to initiate contact between organisms and THP-1 cells. After a 30-min incubation at 37°C, cells were washed three times in phosphate-buffered saline (PBS; 1 mM KH2PO4, 155 mM NaCl, 3 mM Na2HPO4, pH 7.4) and lysed in 500 μl of a solution containing 50 mM HCl and 0.1% Triton X-100. Samples were boiled for 5 min, and 30 μl of 0.5 M NaOH was added to neutralize the acid. One milliliter of 95% ethanol was added to samples, which were incubated on ice for 5 min, dried under a vacuum, and resuspended in assay buffer (0.05 M sodium acetate [pH 5.8], 0.02% bovine serum albumin). The level of cyclic AMP (cAMP) in dried samples was determined using a cAMP enzyme immunoassay (GE Healthcare, Piscataway, NJ) according to the manufacturer's protocol. CyaA-Ank fusions were considered positive for translocation when the increase in cAMP over the level with CyaA alone (negative control) was at least 2.5-fold and this increase was abolished following expression of fusion proteins in the L. pneumophila DotA− mutant (LELA3118). A CyaA fusion to RalF, a well-characterized L. pneumophila Dot/Icm effector (29), was used as a positive control. cAMP levels resulting from translocation of CyaA-RalF were similar to those reported by Bardill et al. (4).
Transfection and fluorescence microscopy.
Confluent HeLa cells on 12-mm glass coverslips were infected with C. burnetii NM phase II organisms for 24 h. At 24 h postinfection, cells were transfected with plasmids encoding Ank-mCherry constructs, using Effectene reagent (Qiagen, Valencia, CA) according to the manufacturer's protocol. At 18 h posttransfection, cells were washed three times in cold PBS and fixed in 100% methanol for 3 min at room temperature. Following fixation, cells were washed three times in cold PBS and incubated with DRAQ5 (Alexa Corporation, Lausanne, Switzerland) for 5 min to stain host and bacterial DNA. Where indicated, cells were incubated with mouse monoclonal antibodies directed against β-tubulin (Sigma-Aldrich, St. Louis, MO) or CD63 (BD Biosciences, San Jose, CA) or with a rabbit polyclonal antibody directed against COXIV (Cell Signaling, Danvers, MA). Binding of primary antibodies was detected using anti-mouse or anti-rabbit secondary antibodies conjugated to Alexa Fluor 488 (Molecular Probes, Carlsbad, CA). Confocal fluorescence microscopy was performed using a modified Perkin-Elmer UltraView spinning disk confocal system connected to a Nikon Eclipse TE2000-S microscope. Images were acquired with a ×60 oil immersion objective (1.4 numerical aperture; Nikon, Melville, NY) and a Photometrics Cascade II:512 digital camera (Princeton Instruments, Trenton, NJ) using Metamorph software (Molecular Devices, Downingtown, PA).
RESULTS
C. burnetii Ank genes display extensive heterogeneity among isolates derived from disparate sources.
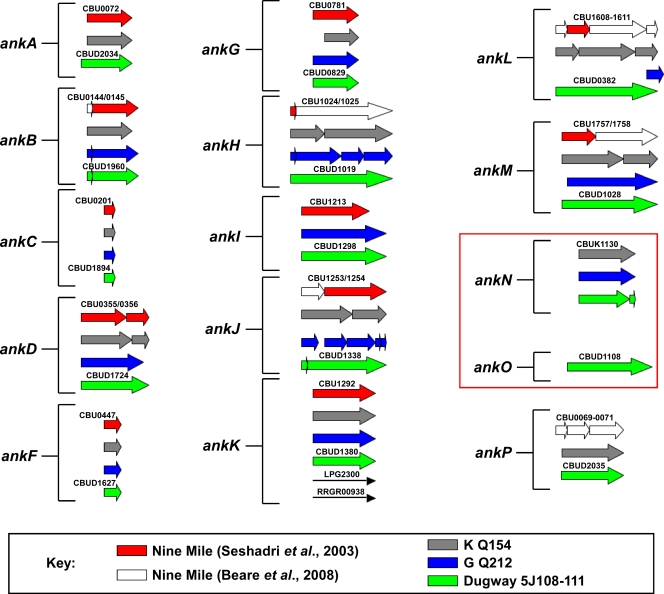
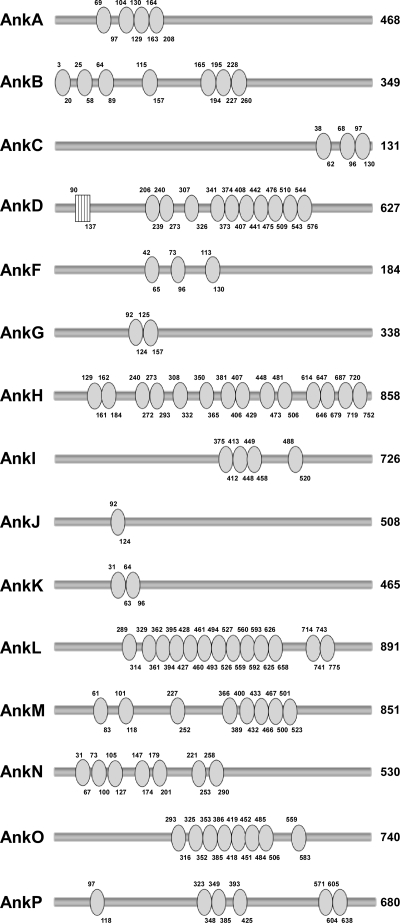
The original GenBank entry detailing the C. burnetii NM reference isolate genome reported the presence of 13 Ank genes (45). However, reannotation of this genome, along with new genome sequencing and annotation of the C. burnetii G, K, and Dugway isolates, resulted in multiple changes in the NM Ank gene family (6) (Fig. 1). Immediately downstream of NM ankA is the newly annotated gene ankP, which has a frameshift mutation. In addition to ankP, NM ankH, -L, and -M are now recognized as fragmented genes. ankD (CBU0355) and the previously annotated downstream gene ankE (CBU0356) are now known to constitute a single frameshift mutant gene that is denoted ankD. Lineups of the C. burnetii Ank gene family revealed that the Dugway isolate encodes 11 intact Anks (encoded by ankA, -C, -D, -F, -G, -H, -I, -K, -M, -O, and -P), while the NM isolate encodes only 5 (encoded by ankA, -C, -F, -G, and -K). Ank genes were considered disrupted if they contained a frameshift(s) or a 3′ deletion relative to a full-length ortholog carried by another C. burnetii isolate and were named according to the nomenclature of Pan et al. (33). ankD, -H, and -O are intact only in the Dugway isolate, with ankO being unique to Dugway. ankA, -I, -M, and -P are intact in Dugway and either the G or K isolate. ankN is intact only in the G and K endocarditis isolates. Although at first glance ankL of the Dugway isolate appears to be a full-length gene, a small ankL gene fragment in the G isolate extends beyond the 3′ end of the Dugway ortholog, suggesting that Dugway ankL is truncated at the 3′ end. Along with ankL, ankB and ankJ appear to be disrupted in all isolates, with both genes containing frameshifts that result in small gene fragments at the 5′ end. However, these fragments do not encode proteins with ankyrin repeat domains, leading to the possibility that the larger fragments of ankB and ankJ, which do encode ankyrin repeats, may be transcribed into functional proteins. Interestingly, only ankC, -F, and -K appear to be conserved as full-length genes in all four isolates. Although ankG from the K isolate contains a 5′ truncation, this truncation is not the result of a frameshift, suggesting that this gene is still functional. The number of repeats in each C. burnetii Ank was highly variable and ranged from 1 in AnkJ to 14 in AnkH (Fig. 2). At least two neighboring repeats are required for a functional protein-protein interaction domain (27); therefore, AnkJ is predicted to contain a nonfunctional repeat. Only AnkD contained an additional eukaryotic motif, with an F-box domain located in its N terminus (Fig. 2).
FIG. 1.
C. burnetii Anks display considerable heterogeneity among isolates derived from disparate sources. Ank genes from the NM, G, K, and Dugway isolates were compared and placed into 15 groups. The Ank gene orthologs present in each isolate are designated by colors, as defined in the key. Arrows represent either whole genes or a gene fragment. White arrows denote Ank gene fragments that were discovered through reannotation of the NM isolate genome sequence. The NM isolate contains the fewest intact Ank genes (5), while the Dugway isolate carries the most intact Ank genes (11). Disrupted Ank genes from the NM isolate typically contained frameshift mutations resulting in truncations. ankN and ankO (boxed area) are absent as full-length or fragmented genes in the NM isolate.
FIG. 2.
C. burnetii Anks contain various numbers of ankyrin repeats. Anks were examined using Pfam and Interpro to identify repeat regions. Each circle represents one ankyrin repeat, and the numbers above and below the circle denote amino acid residues. The box indicates an F-box domain present in AnkD. The number to the right of each Ank indicates the total number of amino acid residues in each protein. All Anks examined were from the Dugway isolate, except for AnkN, which was from the K isolate. C. burnetii Anks contained as few as 1 (AnkJ) and as many as 14 (AnkH) repeats.
Multiple C. burnetii Anks are translocated into the host cytosol in a Dot/Icm-dependent manner.
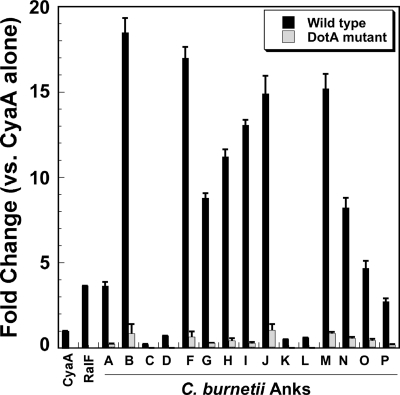
Methods for gene-specific genetic manipulation of C. burnetii are currently not available. Therefore, we used L. pneumophila as a surrogate host to examine Dot/Icm-directed translocation of C. burnetii Anks. Unless otherwise indicated, all Anks examined in the current study were from the Dugway isolate, except for AnkN, which was from the K isolate. Based on reports indicating that the translocation signal for most L. pneumophila Dot/Icm substrates is located at the C terminus (8, 28), plasmids were constructed to encode C. burnetii Anks N-terminally fused to the C terminus of CyaA from Bordetella pertussis. Due to difficulty in expressing full-length AnkH, the C-terminal 216 residues (∼25% of the intact protein) were expressed as a CyaA fusion protein. PMA-differentiated THP-1 macrophage-like cells were infected with L. pneumophila strains producing each fusion protein, and intracellular cAMP levels were measured. As shown in Fig. 3, cAMP levels were substantially increased relative to that in L. pneumophila expressing CyaA alone (negative control) when cells were infected with bacteria producing AnkA, -B, -F, -G, -H, -I, -J, -M, -N, -O, and -P, indicating that these Anks are delivered to the host cytosol. Translocation of Dugway AnkA, -B, -F, and -G was consistent with the results of Pan et al. (33), who reported translocation of the orthologous proteins from the NM isolate. However, unlike the case in that report, we also observed translocation of AnkJ.
FIG. 3.
Multiple C. burnetii Anks are translocated into the host cytosol in a Dot/Icm-dependent manner. Intracellular levels of cAMP were determined following infection of THP-1 cells with L. pneumophila strains expressing CyaA-Ank fusion proteins. The results are expressed as changes in the cAMP level compared to that resulting from infection with L. pneumophila expressing CyaA alone. L. pneumophila expressing the characterized Dot/Icm substrate RalF fused to CyaA was used as a positive control. All Anks examined were from the Dugway isolate, except for AnkN, which was from the K isolate. Elevated levels of cAMP were observed after infection by L. pneumophila cells expressing 11 different C. burnetii Anks (black bars), indicating translocation into the host cytosol. Basal levels of cAMP were observed when these Anks were expressed in DotA-deficient L. pneumophila (gray bars), indicating that secretion requires a functional Dot/Icm T4SS. Experiments were performed in triplicate, and error bars represent standard deviations from the means.
Wild-type L. pneumophila JR32 encodes non-Dot/Icm secretion systems (43). Therefore, to verify that C. burnetii Anks are translocated in a Dot/Icm-dependent fashion, constructs that resulted in positive CyaA translocation were introduced into a DotA− mutant of L. pneumophila, which is deficient in Dot/Icm type IV secretion (13, 39-41), and the CyaA assay was repeated. As shown in Fig. 3, cAMP levels approximated that of the negative control in the absence of DotA, indicating that translocation is directed by Dot/Icm.
L. pneumophila and R. grylli homologs of C. burnetii AnkK are not secreted by L. pneumophila.
AnkK, which is full length in all four C. burnetii isolates, was not translocated by the L. pneumophila Dot/Icm T4SS. Proteins that are 41% and 39% identical to AnkK at the amino acid level are present in L. pneumophila and R. grylli, respectively. R. grylli is an obligate intracellular pathogen of insects, arachnids, and isopods that is closely related to C. burnetii (38) and carries 19 of the 26 L. pneumophila Dot/Icm components (23). To ascertain whether either C. burnetii AnkK homolog is secreted, each protein was tested in the CyaA assay. Neither protein was translocated by L. pneumophila (data not shown), suggesting that their function is not associated with secretion. Moreover, two additional R. grylli Anks, Rg00379 and Rg00660, were not secreted by L. pneumophila (data not shown). A lack of L. pneumophila AnkK secretion is consistent with results reported by de Felipe et al. (15).
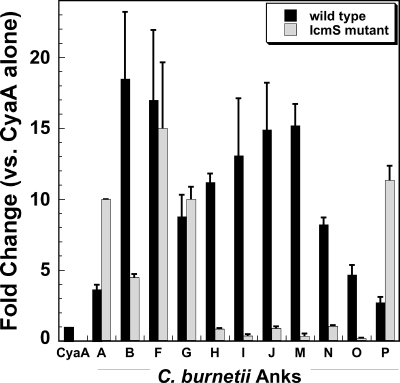
A subset of C. burnetii Anks requires the Dot/Icm chaperone IcmS for translocation.
Chaperone proteins are required for efficient delivery of some effectors to the Dot/Icm apparatus prior to translocation into the host cytosol (30). IcmS is a well-characterized chaperone of L. pneumophila Dot/Icm effector proteins (4, 8), and C. burnetii icmS complements L. pneumophila IcmS− mutants (57, 58). To examine whether IcmS is required for translocation of C. burnetii Anks, CyaA-positive Ank constructs were introduced into an IcmS-deficient L. pneumophila mutant and tested for secretion in the CyaA assay. As shown in Fig. 4, translocation of AnkB, -H, -I, -J, -M, -N, and -O, but not that of AnkA, -F, -G, or -P, was dramatically inhibited in the absence of functional IcmS. These data indicate that IcmS is required for translocation of some, but not all, C. burnetii Anks.
FIG. 4.
A subset of C. burnetii Anks require the chaperone IcmS for translocation. Each C. burnetii translocation-positive Ank was expressed in wild-type L. pneumophila or an IcmS-deficient strain, and cAMP levels were determined following infection of THP-1 cells. AnkB, -H, -I, -J, -M, -N, and -O required functional IcmS for translocation. In contrast, AnkA, -F, -G, and -P were delivered to the cytosol in the absence of IcmS. Experiments were performed in triplicate, and error bars represent the standard deviations from the means.
The C. burnetii Ank Dot/Icm translocation signal is located in the C terminus.
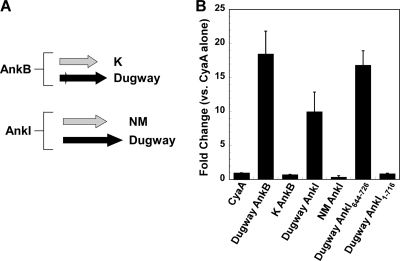
Multiple studies indicate that the C terminus of L. pneumophila Dot/Icm substrates contains a signal required for secretion (8, 28). To determine if this requirement extends to C. burnetii effector proteins, AnkB and AnkI from the K and NM isolates, respectively, were tested for secretion. These Anks each have a 69-amino-acid C-terminal truncation relative to the Dugway orthologs (Fig. 5A). As shown in Fig. 5B, neither truncated Ank was translocated into the host cytosol by L. pneumophila. To further define the Dugway isolate AnkI translocation signal, CyaA fusions to the AnkI protein missing 10, 20, 30, 40, 60, or 80 amino acids from its C terminus were tested for translocation. None of these fusion proteins were translocated (Fig. 5B and data not shown). Conversely, a CyaA fusion to the C-terminal 82 amino acids of AnkI was translocated (Fig. 5B). Collectively, these data indicate that the C-terminal 82 amino acids of AnkI are sufficient for secretion, with the C-terminal 10 amino acids being required.
FIG. 5.
The Ank translocation signal is located in the C terminus. (A) AnkB from the K isolate and AnkI from the NM isolate lack 69 amino acids at their C termini compared to the Dugway orthologs. (B) Dugway isolate AnkB and AnkI, but not K and NM isolate AnkB and AnkI, respectively, were translocated as CyaA fusion proteins by wild-type L. pneumophila. CyaA-AnkI644-762, a fusion protein containing the C-terminal 82 amino acids of Dugway AnkI, was also translocated, whereas CyaA-AnkI1-716, a fusion protein lacking the C-terminal 10 amino acids of Dugway AnkI, was not translocated. With the exception of those with the Dugway AnkI truncations, experiments were performed in triplicate and error bars represent the standard deviations from the means. The results of Dugway AnkI truncation experiments are representative of those obtained from two experiments, and error bars indicate the standard errors from the means.
C. burnetii Anks localize to distinct subcellular regions when expressed ectopically in HeLa cells.
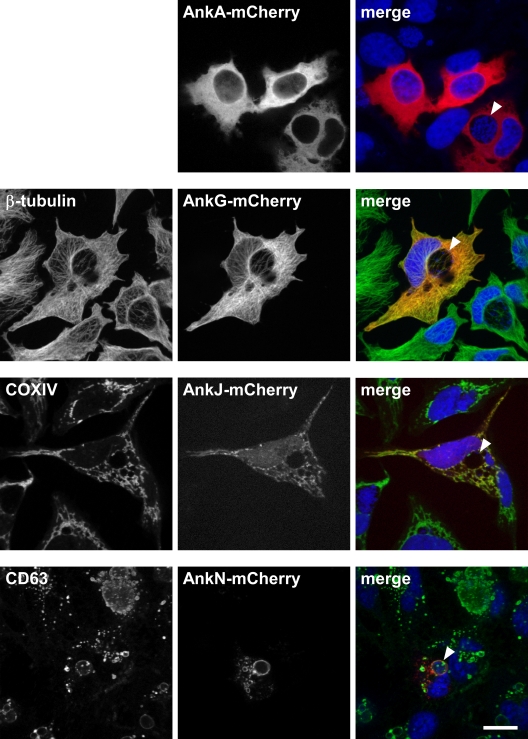
L. pneumophila Dot/Icm substrates commonly localize to distinct intracellular sites/organelles, including the pathogen-containing vacuole, where they can modulate host cell functions associated with the respective subcellular compartment (14, 56). Therefore, to provide insight into C. burnetii Ank function, each translocation-positive Ank was expressed ectopically in C. burnetii-infected HeLa cells as an N- or C-terminal fusion to red fluorescent mCherry, and localization was visualized by confocal fluorescence microscopy. The orientation of the mCherry fusion tag did not demonstrably alter intracellular Ank trafficking. AnkA, -B, -F, -H, -I, -M, and -P showed diffuse localization in the host cytoplasm (Fig. 6 and data not shown). Conversely, AnkG localized to cytoskeleton-like filamentous structures (Fig. 6). Immunostaining with a monoclonal antibody directed against β-tubulin identified these structures as microtubules. AnkJ trafficked to mitochondria, as evidenced by colocalization with COXIV, a host protein that resides in the inner leaflet of the mitochondrial membrane (Fig. 6). AnkN and AnkO localized to the C. burnetii PV membrane and to vesicular bodies that colabeled with the lysosomal marker CD63 (Fig. 6 and data not shown).
FIG. 6.
AnkG, -J, and -N localize to distinct subcellular regions in HeLa cells. Each translocation-positive Ank was expressed ectopically in HeLa cells as an N- or C-terminal fusion to mCherry. Trafficking of ectopically expressed C-terminal fusion proteins is shown. HeLa cells infected with C. burnetii for 30 h were transfected with each construct for 18 h. Cells were subsequently fixed and processed for confocal fluorescence microscopy. In each merged panel, mCherry fluorescence is shown in red, DNA stained with DRAQ5 is shown in blue, and antibody-stained structures are shown in green. AnkA-mCherry localized diffusely throughout the host cytoplasm. AnkG-mCherry colocalized with microtubules, as evidenced by fluorescence overlap with antibody-stained β-tubulin. AnkJ-mCherry colocalized with mitochondria, as shown by fluorescence overlap with antibody-stained COXIV. AnkN-mCherry localized to lysosomal compartments, including the C. burnetii PV membrane, as evidenced by fluorescence overlap with antibody-stained CD63. Arrowheads denote C. burnetii PV. Bar, 10 μm.
DISCUSSION
The Ank gene family displays striking heterogeneity between C. burnetii isolates, with the naturally attenuated Dugway isolate containing the most intact Ank genes. Here we demonstrated that most Dugway Anks and AnkN of the K endocarditis isolate are translocated into the host cytosol by L. pneumophila in a Dot/Icm-dependent fashion. We also show that Anks traffic to distinct host cell regions when expressed ectopically, suggesting that they modulate diverse cellular functions through interactions with host factors at these subcellular sites.
Anks were originally described for eukaryotes as mediators of diverse protein-protein interactions. However, genome sequencing recently revealed Ank genes in at least nine bacterial species that replicate inside host cells (10, 17, 21, 24, 32), with O. tsutsugamushi encoding 50 Anks (10). The common occurrence of Anks in intracellular pathogens suggests a critical role for this protein family in successful host cell parasitism. Indeed, two recent reports demonstrated a role for three L. pneumophila Anks during pathogen growth in human monocyte-derived macrophages (3, 17). Moreover, five L. pneumophila Anks are translocated into the host cytosol, among which AnkX disrupts microtubule-mediated trafficking events (3, 33). Three distinct cellular activities have been described for AnkA of A. phagocytophilum during growth of the pathogen in neutrophils (20, 24, 34). One activity involves binding of eukaryotic chromatin to potentially influence host gene expression (34). The second activity involves binding to Abl interactor 1, an adaptor protein that engages the eukaryotic tyrosine kinase Abl-1 (24). This interaction results in phosphorylation of AnkA and is important for the infectious process of A. phagocytophilum. Finally, the third activity involves direct binding to the eukaryotic phosphatase SHP-1 that may alter cellular signaling early during A. phagocytophilum infection (20).
Only C. burnetii AnkK has apparent bacterial homologs, with L. pneumophila and R. grylli proteins exhibiting approximately 40% identity. Habyarimana et al. recently showed that AnkK-deficient L. pneumophila is impaired for growth in human monocyte-derived macrophages (17). Here we demonstrated that AnkK homologs are not secreted by L. pneumophila, suggesting that these proteins function in a conserved bacterial process not requiring delivery to the host cytosol. Our findings agree with those of de Felipe et al., who showed that L. pneumophila AnkK is not secreted during infection of Dictyostelium cells (15), but differ from those of Pan et al., who demonstrated low levels of protein translocation during L. pneumophila infection of Chinese hamster ovary cells (33). In addition to AnkK, R. grylli encodes eight unique Anks. Two of these proteins, Rg00379 and Rg00660, are also not translocated by L. pneumophila. R. grylli Anks may be bona fide T4SS substrates but may lack the translocation signal and/or chaperone interactions required for secretion by L. pneumophila. Moreover, given the incomplete nature of the R. grylli Dot/Icm gene complement relative to those of L. pneumophila and C. burnetii, it is unclear whether this system constitutes a functional T4SS. The absence of the other C. burnetii Anks in other bacterial species suggests that they modulate host cell events specific to C. burnetii infection.
The L. pneumophila chaperones IcmS and IcmW play a critical role in efficient delivery of some effectors to the Dot/Icm apparatus (4, 8). Indeed, IcmW has been used as a bait in yeast two-hybrid assays to identify novel L. pneumophila Dot/Icm substrates, indicating the direct binding of this protein to effectors (31). C. burnetii AnkA, -F, -G, and -P, but not AnkB, -H, -I, -J, -M, -N, and -O, are translocated into the cytosol by an L. pneumophila IcmS− mutant. These data suggest that other chaperone complexes, such as IcmQ/IcmR, may be used for translocation of these effectors. In support of this hypothesis, recent evidence shows that some L. pneumophila effectors are translocated in an IcmS-independent fashion (8). Although C. burnetii lacks IcmR, an IcmR functional homolog, CoxigA (CBU1634a), directly interacts with IcmQ and may function in an IcmR-like manner (16).
The translocation signal of L. pneumophila Dot/Icm substrates is thought to reside in the effector protein C terminus (8, 28). This region contains a disordered segment that may be stabilized by effector interactions with IcmS/IcmW (8). A different mechanism is proposed for A. phagocytophilum AnkA, where a C-terminal translocation signal consisting of basic amino acid residues (arginine and lysine) may provide a hydrophilic “tail” for delivery to the secretion complex (24). The translocation signal for C. burnetii Anks also appears to be located C-terminally. AnkB and AnkI of the K and NM isolates, respectively, which lack 69 amino acids at their C termini relative to the full-length orthologs in the Dugway isolate, are not secreted. Moreover, removal of as little as 10 amino acids from the C terminus of Dugway AnkI abolishes translocation. This 10-amino-acid motif (NTSEPGSFGI) is not conserved among C. burnetii Anks; thus, the disordered signal hypothesis (8) may apply for the subset of Anks, such as AnkB and AnkI, that rely on IcmS for translocation.
Secreted bacterial virulence factors often traffic to distinct cellular locations, where they modulate host processes during infection (14, 49). Ectopically expressed AnkG, -J, -N, and -O localize to specific cellular sites, while the other C. burnetii Anks disperse throughout the cytoplasm. AnkG localizes to host microtubules, suggesting that this effector modulates a microtubule-mediated function, such as PV movement to the perinuclear region or delivery of vesicular cargo to the vacuole. A similar function has been demonstrated for the Salmonella enterica type III secretion system effector SseF, which interacts with the microtubule motor protein dynein to promote proper positioning of the S. enterica-containing vacuole during epithelial cell infection (1). Interestingly, disruption of HeLa cell microtubules with nocodazole inhibits formation of the large and spacious PV, indicating a critical role for microtubules in vacuole biogenesis (D. E. Voth and R. A. Heinzen, unpublished data). ankG is conserved as an intact gene in the NM, G, and Dugway isolates, while ankG from the K isolate contains a 5′ truncation. However, the absence of a frameshift mutation suggests that ankG from the K isolate encodes a functional protein. Thus, AnkG activity may absolutely be required for successful C. burnetii infection. Interestingly, AnkC and AnkK are also conserved among all four isolates but are not secreted. However, the possibility exists that these Anks are false-negative proteins that are not translocated by L. pneumophila in the CyaA assay. AnkJ localizes to mitochondria, where it may modulate the apoptotic functions of this organelle. Indeed, members of our laboratory and others recently demonstrated that C. burnetii potently inhibits mitochondrion-mediated apoptosis (25, 55). AnkN and AnkO localize to the PV membrane, where they might mediate vesicular fusion events required for PV maturation. A similar activity is associated with the L. pneumophila Dot/Icm substrates SidC and SdcA, which attach to the membrane of the pathogen-containing vacuole and mediate recruitment of the endoplasmic reticulum membrane (35, 56).
In summary, identification of secreted C. burnetii virulence factors has been difficult due to a lack of systems to genetically manipulate the organism. By taking advantage of the evolutionarily related Dot/Icm T4SSs of L. pneumophila and C. burnetii, we have identified a large and diverse family of Ank proteins that are Dot/Icm substrates. Based on ectopic expression results, native Anks are predicted to traffic to specific subcellular sites, where they likely mimic or disrupt the activities of host factors to promote pathogen growth. The NM isolate, with the fewest intact Ank genes, is significantly more virulent in a guinea pig infection model of acute Q fever than the Dugway isolate, which contains the most intact Ank genes (51; J. E. Samuel, unpublished data). Thus, it is intriguing to speculate that Ank gene content plays a role in C. burnetii pathotype-specific virulence. Determining the definitive role for Anks in C. burnetii pathogenesis awaits gene knockout studies and subsequent animal challenge.
Acknowledgments
We thank Gary Hettrick for graphic illustrations.
This research was supported by the Intramural Research Program of the National Institutes of Health National Institute of Allergy and Infectious Diseases.
Footnotes
Published ahead of print on 1 May 2009.
REFERENCES
- 1.Abrahams, G. L., P. Muller, and M. Hensel. 2006. Functional dissection of SseF, a type III effector protein involved in positioning the Salmonella-containing vacuole. Traffic 7950-965. [DOI] [PubMed] [Google Scholar]
- 2.Akporiaye, E. T., J. D. Rowatt, A. A. Aragon, and O. G. Baca. 1983. Lysosomal response of a murine macrophage-like cell line persistently infected with Coxiella burnetii. Infect. Immun. 401155-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Al-Khodor, S., C. T. Price, F. Habyarimana, A. Kalia, and Y. Abu Kwaik. 2008. A Dot/Icm-translocated ankyrin protein of Legionella pneumophila is required for intracellular proliferation within human macrophages and protozoa. Mol. Microbiol. 70908-923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bardill, J. P., J. L. Miller, and J. P. Vogel. 2005. IcmS-dependent translocation of SdeA into macrophages by the Legionella pneumophila type IV secretion system. Mol. Microbiol. 5690-103. [DOI] [PubMed] [Google Scholar]
- 5.Beare, P. A., J. E. Samuel, D. Howe, K. Virtaneva, S. F. Porcella, and R. A. Heinzen. 2006. Genetic diversity of the Q fever agent, Coxiella burnetii, assessed by microarray-based whole-genome comparisons. J. Bacteriol. 1882309-2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beare, P. A., N. Unsworth, M. Andoh, D. E. Voth, A. Omsland, S. D. Gilk, K. P. Williams, B. W. Sobral, J. J. Kupko, III, S. F. Porcella, J. E. Samuel, and R. A. Heinzen. 2009. Comparative genomics reveal extensive transposon-mediated genomic plasticity and diversity among potential effector proteins within the genus Coxiella. Infect. Immun. 77642-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bruggemann, H., C. Cazalet, and C. Buchrieser. 2006. Adaptation of Legionella pneumophila to the host environment: role of protein secretion, effectors and eukaryotic-like proteins. Curr. Opin. Microbiol. 986-94. [DOI] [PubMed] [Google Scholar]
- 8.Cambronne, E. D., and C. R. Roy. 2007. The Legionella pneumophila IcmSW complex interacts with multiple Dot/Icm effectors to facilitate type IV translocation. PLoS Pathog. 3e188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caturegli, P., K. M. Asanovich, J. J. Walls, J. S. Bakken, J. E. Madigan, V. L. Popov, and J. S. Dumler. 2000. ankA: an Ehrlichia phagocytophila group gene encoding a cytoplasmic protein antigen with ankyrin repeats. Infect. Immun. 685277-5283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cho, N. H., H. R. Kim, J. H. Lee, S. Y. Kim, J. Kim, S. Cha, S. Y. Kim, A. C. Darby, H. H. Fuxelius, J. Yin, J. H. Kim, J. Kim, S. J. Lee, Y. S. Koh, W. J. Jang, K. H. Park, S. G. Andersson, M. S. Choi, and I. S. Kim. 2007. The Orientia tsutsugamushi genome reveals massive proliferation of conjugative type IV secretion system and host-cell interaction genes. Proc. Natl. Acad. Sci. USA 1047981-7986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cockrell, D. C., P. A. Beare, E. R. Fischer, D. Howe, and R. A. Heinzen. 2008. A method for purifying obligate intracellular Coxiella burnetii that employs digitonin lysis of host cells. J. Microbiol. Methods 72321-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coleman, S. A., E. R. Fischer, D. Howe, D. J. Mead, and R. A. Heinzen. 2004. Temporal analysis of Coxiella burnetii morphological differentiation. J. Bacteriol. 1867344-7352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conover, G. M., I. Derre, J. P. Vogel, and R. R. Isberg. 2003. The Legionella pneumophila LidA protein: a translocated substrate of the Dot/Icm system associated with maintenance of bacterial integrity. Mol. Microbiol. 48305-321. [DOI] [PubMed] [Google Scholar]
- 14.de Felipe, K. S., R. T. Glover, X. Charpentier, O. R. Anderson, M. Reyes, C. D. Pericone, and H. A. Shuman. 2008. Legionella eukaryotic-like type IV substrates interfere with organelle trafficking. PLoS Pathog. 4e1000117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Felipe, K. S., S. Pampou, O. S. Jovanovic, C. D. Pericone, S. F. Ye, S. Kalachikov, and H. A. Shuman. 2005. Evidence for acquisition of Legionella type IV secretion substrates via interdomain horizontal gene transfer. J. Bacteriol. 1877716-7726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feldman, M., T. Zusman, S. Hagag, and G. Segal. 2005. Coevolution between nonhomologous but functionally similar proteins and their conserved partners in the Legionella pathogenesis system. Proc. Natl. Acad. Sci. USA 10212206-12211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Habyarimana, F., S. Al-Khodor, A. Kalia, J. E. Graham, C. T. Price, M. T. Garcia, and Y. A. Kwaik. 2008. Role for the ankyrin eukaryotic-like genes of Legionella pneumophila in parasitism of protozoan hosts and human macrophages. Environ. Microbiol. 101460-1474. [DOI] [PubMed] [Google Scholar]
- 18.Howe, D., and L. P. Mallavia. 2000. Coxiella burnetii exhibits morphological change and delays phagolysosomal fusion after internalization by J774A.1 cells. Infect. Immun. 683815-3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Howe, D., J. Melnicakova, I. Barak, and R. A. Heinzen. 2003. Maturation of the Coxiella burnetii parasitophorous vacuole requires bacterial protein synthesis but not replication. Cell. Microbiol. 5469-480. [DOI] [PubMed] [Google Scholar]
- 20.IJdo, J. W., A. C. Carlson, and E. L. Kennedy. 2007. Anaplasma phagocytophilum AnkA is tyrosine-phosphorylated at EPIYA motifs and recruits SHP-1 during early infection. Cell. Microbiol. 91284-1296. [DOI] [PubMed] [Google Scholar]
- 21.Iturbe-Ormaetxe, I., G. R. Burke, M. Riegler, and S. L. O'Neill. 2005. Distribution, expression, and motif variability of ankyrin domain genes in Wolbachia pipientis. J. Bacteriol. 1875136-5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kubori, T., A. Hyakutake, and H. Nagai. 2008. Legionella translocates an E3 ubiquitin ligase that has multiple U-boxes with distinct functions. Mol. Microbiol. 671307-1319. [DOI] [PubMed] [Google Scholar]
- 23.Leclerque, A., and R. G. Kleespies. 2008. Type IV secretion system components as phylogenetic markers of entomopathogenic bacteria of the genus Rickettsiella. FEMS Microbiol. Lett. 279167-173. [DOI] [PubMed] [Google Scholar]
- 24.Lin, M., A. den Dulk-Ras, P. J. Hooykaas, and Y. Rikihisa. 2007. Anaplasma phagocytophilum AnkA secreted by type IV secretion system is tyrosine phosphorylated by Abl-1 to facilitate infection. Cell. Microbiol. 92644-2657. [DOI] [PubMed] [Google Scholar]
- 25.Luhrmann, A., and C. R. Roy. 2007. Coxiella burnetii inhibits activation of host cell apoptosis through a mechanism that involves preventing cytochrome c release from mitochondria. Infect. Immun. 755282-5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moos, A., and T. Hackstadt. 1987. Comparative virulence of intra- and interstrain lipopolysaccharide variants of Coxiella burnetii in the guinea pig model. Infect. Immun. 551144-1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mosavi, L. K., T. J. Cammett, D. C. Desrosiers, and Z. Y. Peng. 2004. The ankyrin repeat as molecular architecture for protein recognition. Protein Sci. 131435-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nagai, H., E. D. Cambronne, J. C. Kagan, J. C. Amor, R. A. Kahn, and C. R. Roy. 2005. A C-terminal translocation signal required for Dot/Icm-dependent delivery of the Legionella RalF protein to host cells. Proc. Natl. Acad. Sci. USA 102826-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagai, H., J. C. Kagan, X. Zhu, R. A. Kahn, and C. R. Roy. 2002. A bacterial guanine nucleotide exchange factor activates ARF on Legionella phagosomes. Science 295679-682. [DOI] [PubMed] [Google Scholar]
- 30.Ninio, S., and C. R. Roy. 2007. Effector proteins translocated by Legionella pneumophila: strength in numbers. Trends Microbiol. 15372-380. [DOI] [PubMed] [Google Scholar]
- 31.Ninio, S., D. M. Zuckman-Cholon, E. D. Cambronne, and C. R. Roy. 2005. The Legionella IcmS-IcmW protein complex is important for Dot/Icm-mediated protein translocation. Mol. Microbiol. 55912-926. [DOI] [PubMed] [Google Scholar]
- 32.Ogata, H., P. Renesto, S. Audic, C. Robert, G. Blanc, P. E. Fournier, H. Parinello, J. M. Claverie, and D. Raoult. 2005. The genome sequence of Rickettsia felis identifies the first putative conjugative plasmid in an obligate intracellular parasite. PLoS Biol. 3e248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pan, X., A. Luhrmann, A. Satoh, M. A. Laskowski-Arce, and C. R. Roy. 2008. Ankyrin repeat proteins comprise a diverse family of bacterial type IV effectors. Science 3201651-1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park, J., K. J. Kim, K. S. Choi, D. J. Grab, and J. S. Dumler. 2004. Anaplasma phagocytophilum AnkA binds to granulocyte DNA and nuclear proteins. Cell. Microbiol. 6743-751. [DOI] [PubMed] [Google Scholar]
- 35.Ragaz, C., H. Pietsch, S. Urwyler, A. Tiaden, S. S. Weber, and H. Hilbi. 2008. The Legionella pneumophila PtdIns(4)P-binding Icm/Dot substrate SidC recruits ER vesicles to a replication-permissive vacuole. Cell. Microbiol. 102416-2433. [DOI] [PubMed] [Google Scholar]
- 36.Raoult, D., T. Marrie, and J. Mege. 2005. Natural history and pathophysiology of Q fever. Lancet Infect. Dis. 5219-226. [DOI] [PubMed] [Google Scholar]
- 37.Romano, P. S., M. G. Gutierrez, W. Beron, M. Rabinovitch, and M. I. Colombo. 2006. The autophagic pathway is actively modulated by phase II Coxiella burnetii to efficiently replicate in the host cell. Cell. Microbiol. 9891-909. [DOI] [PubMed] [Google Scholar]
- 38.Roux, V., M. Bergoin, N. Lamaze, and D. Raoult. 1997. Reassessment of the taxonomic position of Rickettsiella grylli. Int. J. Syst. Bacteriol. 471255-1257. [DOI] [PubMed] [Google Scholar]
- 39.Roy, C. R., K. H. Berger, and R. R. Isberg. 1998. Legionella pneumophila DotA protein is required for early phagosome trafficking decisions that occur within minutes of bacterial uptake. Mol. Microbiol. 28663-674. [DOI] [PubMed] [Google Scholar]
- 40.Roy, C. R., and R. R. Isberg. 1997. Topology of Legionella pneumophila DotA: an inner membrane protein required for replication in macrophages. Infect. Immun. 65571-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sadosky, A. B., L. A. Wiater, and H. A. Shuman. 1993. Identification of Legionella pneumophila genes required for growth within and killing of human macrophages. Infect. Immun. 615361-5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sauer, J. D., J. G. Shannon, D. Howe, S. F. Hayes, M. S. Swanson, and R. A. Heinzen. 2005. Specificity of Legionella pneumophila and Coxiella burnetii vacuoles and versatility of Legionella pneumophila revealed by coinfection. Infect. Immun. 734494-4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Segal, G., J. J. Russo, and H. A. Shuman. 1999. Relationships between a new type IV secretion system and the icm/dot virulence system of Legionella pneumophila. Mol. Microbiol. 34799-809. [DOI] [PubMed] [Google Scholar]
- 44.Segal, G., and H. A. Shuman. 1997. Characterization of a new region required for macrophage killing by Legionella pneumophila. Infect. Immun. 655057-5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seshadri, R., I. T. Paulsen, J. A. Eisen, T. D. Read, K. E. Nelson, W. C. Nelson, N. L. Ward, H. Tettelin, T. M. Davidsen, M. J. Beanan, R. T. Deboy, S. C. Daugherty, L. M. Brinkac, R. Madupu, R. J. Dodson, H. M. Khouri, K. H. Lee, H. A. Carty, D. Scanlan, R. A. Heinzen, H. A. Thompson, J. E. Samuel, C. M. Fraser, and J. F. Heidelberg. 2003. Complete genome sequence of the Q-fever pathogen Coxiella burnetii. Proc. Natl. Acad. Sci. USA 1005455-5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shannon, J. G., and R. A. Heinzen. 2008. Infection of human monocyte-derived macrophages with Coxiella burnetii. Methods Mol. Biol. 431189-200. [DOI] [PubMed] [Google Scholar]
- 47.Shin, S., and C. R. Roy. 2008. Host cell processes that influence the intracellular survival of Legionella pneumophila. Cell. Microbiol. 101209-1220. [DOI] [PubMed] [Google Scholar]
- 48.Sory, M. P., and G. R. Cornelis. 1994. Translocation of a hybrid YopE-adenylate cyclase from Yersinia enterocolitica into HeLa cells. Mol. Microbiol. 14583-594. [DOI] [PubMed] [Google Scholar]
- 49.Steele-Mortimer, O., L. A. Knodler, S. L. Marcus, M. P. Scheid, B. Goh, C. G. Pfeifer, V. Duronio, and B. B. Finlay. 2000. Activation of Akt/protein kinase B in epithelial cells by the Salmonella typhimurium effector SigD. J. Biol. Chem. 27537718-37724. [DOI] [PubMed] [Google Scholar]
- 50.Stein, A., C. Louveau, H. Lepidi, F. Ricci, P. Baylac, B. Davoust, and D. Raoult. 2005. Q fever pneumonia: virulence of Coxiella burnetii pathovars in a murine model of aerosol infection. Infect. Immun. 732469-2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stoenner, H. G., and D. B. Lackman. 1960. The biologic properties of Coxiella burnetii isolated from rodents collected in Utah. Am. J. Hyg. 7145-51. [DOI] [PubMed] [Google Scholar]
- 52.Tissot-Dupont, H., and D. Raoult. 2008. Q fever. Infect. Dis. Clin. N. Am. 22505-514. [DOI] [PubMed] [Google Scholar]
- 53.Vogel, J. P. 2004. Turning a tiger into a house cat: using Legionella pneumophila to study Coxiella burnetii. Trends Microbiol. 12103-105. [DOI] [PubMed] [Google Scholar]
- 54.Voth, D. E., and R. A. Heinzen. 2007. Lounging in a lysosome: the intracellular lifestyle of Coxiella burnetii. Cell. Microbiol. 9829-840. [DOI] [PubMed] [Google Scholar]
- 55.Voth, D. E., D. Howe, and R. A. Heinzen. 2007. Coxiella burnetii inhibits apoptosis in human THP-1 cells and monkey primary alveolar macrophages. Infect. Immun. 754263-4271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weber, S. S., C. Ragaz, K. Reus, Y. Nyfeler, and H. Hilbi. 2006. Legionella pneumophila exploits PI(4)P to anchor secreted effector proteins to the replicative vacuole. PLoS Pathog. 2e46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zamboni, D. S., S. McGrath, M. Rabinovitch, and C. R. Roy. 2003. Coxiella burnetii express type IV secretion system proteins that function similarly to components of the Legionella pneumophila Dot/Icm system. Mol. Microbiol. 49965-976. [DOI] [PubMed] [Google Scholar]
- 58.Zusman, T., G. Yerushalmi, and G. Segal. 2003. Functional similarities between the icm/dot pathogenesis systems of Coxiella burnetii and Legionella pneumophila. Infect. Immun. 713714-3723. [DOI] [PMC free article] [PubMed] [Google Scholar]