Abstract
Periplasmic membrane fusion proteins (MFPs) are essential components of multidrug efflux pumps and type I protein secretion systems of gram-negative bacteria. Located in the periplasm, MFPs function by creating a physical link between inner membrane transporters and outer membrane channels. The most conserved sequence of MFPs is located in their distal C-terminal domain. However, neither the structure nor the function of this domain is known. In this study, we investigated the structural and functional role of the C-terminal domain of Escherichia coli AcrA, a periplasmic component of the multidrug efflux pump AcrAB-TolC. Using trypsin proteolysis, we identified the proteolytically labile sites in the C-terminal domain (amino acid residues 315 to 397) of AcrA in vitro. We next used these sites as a map to evaluate the structural integrity of this domain of AcrA inside the periplasm. We found that the C-terminal domain of AcrA is protected from trypsin when the tripartite efflux pump AcrAB-TolC is assembled. In contrast, this domain remains proteolytically labile in cells producing only one of the AcrB or TolC components of the complex. Site-directed mutagenesis of 12 highly conserved amino acid residues of the C-terminal domain of AcrA showed that a single G363C substitution dramatically impairs the multidrug efflux activity of AcrAB-TolC. The G363C mutant interacts with both AcrB and TolC but fails to properly assemble into a functional complex. We conclude that the C-terminal domain of AcrA plays an important role in the assembly and function of AcrAB-TolC efflux pump.
AcrA, the multidrug efflux protein from Escherichia coli, is the best-characterized member of the membrane fusion protein (MFP) family (24). Periplasmic AcrA associates with the inner-membrane transporter AcrB, belonging to the RND superfamily of proteins, and the outer-membrane factor TolC (22, 23). Together, the three components form a transenvelope multidrug efflux pump responsible for the high levels of intrinsic as well as acquired antibiotic resistance of E. coli.
AcrA is anchored into the inner membrane by N-terminal lipid modification. However, genetic complementation studies showed that the presence of the lipid moiety is not required for AcrA function (14, 24). Structural studies of the proteolytically stable core of AcrA (amino acid [aa] residues 46 to 312) and of whole-length MexA, a homologous protein from Pseudomonas aeruginosa, showed that these proteins have modular structures (Fig. 1A). They comprise the α-helical hairpin, the lipoyl-binding domain, and the α-β-barrel domain (2, 9, 14). Mutagenesis and chemical cross-linking studies identified the α-helical hairpin of AcrA as a TolC-binding domain, whereas the α-β-barrel domain was proposed to bind AcrB (6, 11, 12, 16). Surprisingly, in isothermal calorimetry experiments, the core fragment of AcrA without its C-terminal domain (C-domain) was able to bind neither AcrB nor TolC (23). In contrast, the whole-length AcrA interacted with both components. This result suggested that the C-domain of AcrA might be important for these interactions. In crystal structures, however, the C-domains of AcrA and MexA were not resolved, and their structures remain unknown.
FIG. 1.
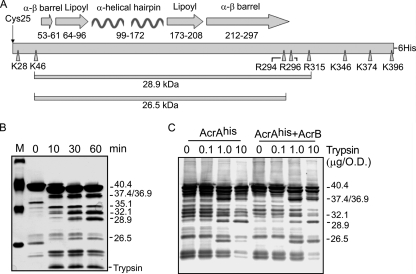
Proteolytic profiles of AcrAhis in vitro and in vivo. (A) Schematic representation of the secondary structure of AcrA. The unique N-terminal Cys25, which is lipid modified after processing in the periplasm, is shown with an arrow. Positions of amino acid residues that form the α-β-barrel, lipoyl-binding, and α-helical hairpin domains are indicated. AcrA residues cleaved by trypsin are indicated by arrowheads. The 28.9-kDa (K46-R315) core and the 26.5-kDa fragment (K46-R294) are also indicated. (B) Purified AcrAhis (final concentration, 1.95 μM) was digested with trypsin (final concentration, 0.10 μM) at 37°C. Aliquots (10 μl) were taken at different time points, and reactions were terminated by boiling in the SDS sample buffer for 5 min. Tryptic fragments were resolved by SDS-PAGE and analyzed by silver nitrate staining. Minor fragments in the untreated control (0 min) are contaminants that copurify with AcrAhis. Lane M, molecular marker. (C) Proteolytic profiles of AcrAhis in E. coli AG100AX cells carrying pAhis and pAhisB plasmids. After treatment with increasing concentrations of trypsin for 60 min at 37°C, the whole-cell proteins were resolved by SDS-PAGE and analyzed by immunoblotting with a polyclonal anti-AcrA antibody. Masses of tryptic fragments of the C-domain of AcrAhis identified by mass spectrometry and by mobility in SDS-PAGE are indicated. O.D., optical density as determined by absorbance at 600 nm.
The alignment of sequences of highly diverse MFPs from both gram-negative and gram-positive bacteria showed that amino acid sequences of the C-domains are conserved among members of the MFP family (4). In addition, several studies suggested that this region is important for the function of AcrA. The deletion mutant of AcrA lacking 85 C-terminal aa residues is poorly expressed and nonfunctional in multidrug efflux (14). The replacement of aa 290 to 357 of AcrA with an analogous region of YhiU disrupted AcrA function possibly because of the loss of interaction with the AcrB transporter (5). Random mutagenesis of MexA identified C-terminal amino acid residues as important for MexA oligomerization and interaction with MexB (16, 17).
In this study, we identified proteolytically labile sites in the C-domain (aa 315 to 397) of the purified AcrA and compared the accessibility of these sites to that in free AcrA and when engaged in the bipartite and tripartite AcrA, AcrB, and TolC interactions in vivo. We found that the assembly of the AcrAB-TolC complex, but not bipartite AcrA-AcrB and AcrA-TolC interactions, protects the C-domain of AcrA from proteolytic digestion. This result suggested that this domain of AcrA interacts with AcrB, TolC, or both. The functional significance of the C-domain was confirmed by site-directed mutagenesis. A single G363C substitution significantly impairs the multidrug efflux activity of AcrAB-TolC.
MATERIALS AND METHODS
Bacterial growth conditions and media.
E. coli strains W4680 (K-12), W4680AE (K-12 but ΔacrAB ΔacrEF), AG100AX (AG100 ΔacrAB ΔacrEF), AG102MB (AG100 ΔacrB), ZK4 (MC4100), ZK796 (MC4100 ΔtolC), and ECM2112 (MC4100 ΔacrAB ΔtolC) were reported previously (7, 8, 15). The native AcrA, AcrB, and TolC proteins or their six-His-tagged versions were expressed from the previously described plasmids pUC151A (acrAB), pAhis (six-His-tagged acrA), pAhisB (six-His-tagged acrA, acrB), pBhis (six-His-tagged acrB) and pTolChis (six-His-tagged tolC) (13, 22). E. coli cells were grown at 37°C in Luria-Bertani (LB) broth (10 g of Bacto tryptone, 5 g of yeast extract, and 5 g of NaCl per liter). Antibiotics were added when needed to the following final concentrations: ampicillin, 100 μg/ml; kanamycin, 34 μg/ml; tetracycline, 25 μg/ml; and spectinomycin, 50 μg/ml.
Protein purification.
AG100AX E. coli cells were transformed with pAHisB plasmid producing six-His-tagged AcrA (AcrAhis) and native AcrB (22). Cells were grown overnight and reinoculated into 500 ml of LB medium supplemented with ampicillin. At an A600 of ∼0.5 to 0.7, the expression of AcrA was induced by the addition of isopropyl-β-d-1-thiogalactopyranoside (IPTG) to the final concentration, 1 mM. Three hours after induction, cells were collected by low-speed centrifugation and washed in a binding buffer containing 5 mM imidazole, 500 mM NaCl, 1 mM phenylmethylsulfonyl fluoride, and 20 mM Tris-HCl (pH 8.0). Cells were lysed by EDTA-lysozyme treatment followed by sonication (22). Membrane fractions isolated by ultracentrifugation were solubilized in the binding buffer supplemented with 80 mM Triton X-100 (TX). The detergent-soluble proteins were loaded onto a HisBind column (Novagen). The column was washed twice, first with the binding buffer containing 5 mM imidazole and 3.2 mM TX and then with the same buffer but containing 60 mM imidazole. Bound AcrAhis was eluted with the binding buffer containing 500 mM imidazole and 3.2 mM TX. After purification, protein was dialyzed against a buffer containing 20 mM Tris-HCl (pH 7.5), 200 mM NaCl, 3.2 mM TX, and 1 mM EDTA and stored at 4°C until needed. For prolonged storage, dialysis buffer was supplemented with 50% (vol/vol) glycerol, and protein was stored at −20°C.
Limited proteolysis.
For the whole-cell proteolysis, exponentially growing cells (A600 of ∼1.0 to 1.2) were harvested in 0.3- to 0.6-ml aliquots by centrifugation and washed with buffer containing 20 mM Tris-HCl (pH 7.5) and 100 mM NaCl. Cells were resuspended in 90 μl of buffer containing 580 mM sucrose, 20 mM Tris-HCl (pH 7.5), and 5 mM EDTA and incubated on ice for 5 min. Trypsin in the indicated concentrations was added (Fig. 1 and 2), and reactions were carried out at 37°C. Aliquots were withdrawn at 5 and 60 min, and reactions were terminated by the addition of sodium dodecyl sulfate (SDS) sample buffer and by boiling for 5 min. Whole-cell proteins were resolved by SDS-polyacrylamide gel electrophoresis (PAGE) (12% [wt/vol] acrylamide) and analyzed by immunoblotting with polyclonal anti-AcrA antibody. A cell aliquot removed just before the addition of trypsin was used for the zero time point.
FIG. 2.
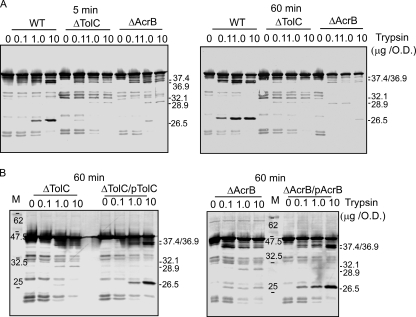
Proteolytic profile of AcrA assembled into AcrAB-TolC complex differs from that of the purified or partially assembled AcrA. (A) E. coli ZK4 (wild type [WT]), ZK796 (ΔTolC), and AG102MB (ΔAcrB) were grown to the mid-exponential phase (A600, ∼1.0). Cells were collected and digested with increasing concentrations of trypsin for 5 min (left panel) and 60 min (right panel) at 37°C. Total proteins were resolved by 12% SDS-PAGE, and AcrA fragments were visualized by immunoblotting with anti-AcrA antibody. (B) ZK796 (ΔTolC) and AG102MB (ΔAcrB) cells were transformed with pTolChis and pBhis plasmids producing TolC and AcrB, respectively, or with pUC18 vector alone. Cells were processed and analyzed as in panel A. Tryptic fragments of AcrA identified by comparison to the proteolytic profiles of the purified AcrA are indicated. Lanes M, molecular marker.
For proteolysis in vitro, purified AcrAhis (final concentration, 1.95 μM) was mixed with trypsin (final concentration, 0.10 μM) and incubated at 37°C. Aliquots were withdrawn at different time points, the reaction was terminated by boiling in the SDS sample buffer, and the samples were analyzed by SDS-PAGE followed by silver nitrate staining (3) or immunoblotting with the anti-AcrA polyclonal antibody.
Immunoblotting analyses.
Whole-cell extracts or purified proteins were separated by SDS-PAGE (12% [wt/vol] acrylamide) and transferred onto polyvinylidene difluoride membranes using standard protocols. AcrA, AcrB, and TolC were detected after incubation with primary polyclonal rabbit antibodies as described previously (22).
Mass spectrometry.
AcrAhis (final concentration, 1.95 μM) was digested with trypsin (final concentration, 0.10 μM) at 37°C for 90 min. The reaction was terminated by the addition of acetic acid to a final concentration of 2% (vol/vol). C18 and C4 ZipTips (Millipore) were used to desalt and concentrate tryptic fragments of AcrAhis. AcrA fragments were eluted using an aqueous solution containing 50% (vol/vol) acetonitrile and 0.1% (vol/vol) trifluoroacetic acid. Matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) analysis was carried out at the Molecular Biology-Proteomics Facility, University of Oklahoma Health Sciences Center. Samples were mixed with sinapinic acid (3,5-dimethoxy-4-hydroxy-cinnamic acid) and then spotted and dried on specimen grids. MALDI-TOF mass spectra of peptide fragments were collected by using the linear mode of a Voyager-DE Pro mass spectrometer (Applied Biosystems) equipped with a delayed-extraction device.
Site-directed mutagenesis.
All point mutations were introduced using a QuikChange XL site-directed mutagenesis kit (Stratagene) as recommended by the manufacturer. The pAhisB plasmid was used as a template in PCRs. After mutagenesis, all plasmids were resequenced to confirm the presence of the desired mutation and the lack of unwanted substitutions (Oklahoma Medical Research Foundation Sequencing Facility). Functionalities of AcrA mutants were judged by their abilities to complement the drug-susceptible phenotype of W4680AE cells. MICs of various antimicrobial agents were measured by the twofold dilution method as described previously (21).
Copurification of DSP cross-linked AcrAB-TolC complexes.
Bacterial cells were grown to an A600 of 0.8 to 1.0 in 250 ml of LB broth supplemented with ampicillin, which was collected by centrifugation and washed with 25 ml of 0.1 M sodium phosphate buffer (pH 7.5). Cross-linking with dithiobissuccinimidyl propionate (DSP; Pierce) and protein copurification by metal affinity chromatography were carried out as described previously (22).
RESULTS
Identification of the proteolytically labile sites in the C-domain of AcrA.
Since the structural role of the N-terminal lipid modification of AcrA remains unclear, for this study we purified the native AcrA, which is lipid modified in the periplasm. For purification purposes, the protein was modified with the C-terminal six-histidine tag (AcrAhis). MALDI-TOF analysis of the purified AcrAhis showed a single peak with a molecular mass of 41,696 Da. This value is in good agreement with the expected 41,624 Da for AcrA modified with N-acyl-S-diacylglycerol containing two palmitoyl residues and one oleoyl residue (Table 1). No additional peaks were detected in this preparation, suggesting that most of the purified AcrAhis is lipid modified.
TABLE 1.
Peptide masses of whole-length AcrAhis and its major tryptic fragments
| Peptide | Mass of AcrAhis and its peptides (Da) based on:
|
|
|---|---|---|
| MALDI-TOF | aa sequence | |
| Whole length | 41,696 | 41,622 |
| N-K396 | 40,411 | 40,471 |
| K28-K396 | 39,362 | 39,303 |
| K46-K396 | 37,308 | 37,495 |
| K28-K374 | NDa | 36,920 |
| K46-K374 | 34,909 | 35,113 |
| K46-K346 | 32,056 | 32,097 |
| K46-R315 | 28,886 | 28,935 |
ND, not detected.
We next used limited proteolysis to evaluate the structure of the purified AcrAhis. After 60 min of tryptic digestion, AcrAhis was completely cleaved into five major fragments (Fig. 1B). The molecular masses of these AcrAhis-derived peptides were determined to be 40.4, 36.9, 35.1, 32.1, and 28.9 kDa (Table 1 and Fig. 1B). In addition, several minor fragments are also seen in the tryptic profile of AcrAhis. Based on the sequence analysis and extant literature, we identified the 36.9-kDa fragment as a product of cleavage of the distal N- and C-terminal ends of AcrA (28 to 374 aa) (Table 1 and Fig. 1A) (14, 23). The 32.1- and 35.1-kDa fragments are generated by further digestion from the N and C termini of AcrA. Finally, the 28.9-kDa band is the previously characterized 46- to 315-aa proteolytically stable core of AcrA (14, 23).
Proteolytic profile of the overproduced AcrAhis in intact cells is similar to that of the purified protein.
We next used tryptic digestion to compare conformations of the C-domain of AcrAhis in vitro and in intact cells. For this purpose, AG100AX cells lacking both AcrAB and AcrEF multidrug efflux pumps were transformed with the pAhis plasmid. We took advantage of the fact that AcrA is located in the periplasm and made it accessible to trypsin by means of osmotic shock. Even before treatment with trypsin, the overproduced AcrAhis was notably degraded by periplasmic proteases (Fig. 1C). Incubation with increasing concentrations of trypsin produced a limited number of the specific bands. By comparison to the immune- and silver-stained proteolytic profiles of the in vitro-digested AcrAhis, these bands were identified as 40.4-, 37.4-, 36.9-, 32.1-, 28.9-, and 26.5-kDa fragments. Overall, immunoblots of the proteolytic profile of the in vivo-overproduced AcrAhis were surprisingly similar to those of the purified protein. The 26.5-kDa fragment (this mass is estimated from the mobility in SDS-PAGE), which is a minor band in the proteolytic profile of the purified AcrAhis (Fig. 1B), could be clearly detected by immunoblotting in the AcrAhis digests in vivo. The proteolytic profile of AcrAhis overproduced in ECM2112 cells lacking all three genes, acrA, acrB, and tolC, was similar to that in AG100AX cells (data not shown). We conclude that in intact cells, the overall conformation of the overproduced AcrAhis closely resembles that of the purified protein.
The coexpression of AcrAhis with AcrB under the native acrAB promoter (pAhisB plasmid) did not significantly affect the proteolytic profile of AcrAhis. However, we noticed that at high trypsin concentrations, amounts of the 26.5-kDa fragment were higher when AcrB was overproduced together with AcrA, suggesting that the interaction with AcrB protects this fragment from trypsin degradation (see also below).
Assembly of the tripartite AcrAB-TolC complex protects the C-domain of AcrA from trypsin.
Overproduction of AcrAhis from pAhis and pAhisB plasmids results in a significant excess of AcrA compared to levels of the two other components of the multidrug efflux complex, AcrB and TolC. Thus, the tryptic digestion profile of the overproduced AcrAhis reflects mainly the conformation of the free AcrAhis. To investigate whether association with AcrB, TolC, or both brings any changes into AcrA conformation, we compared proteolytic profiles of AcrA in wild-type E. coli cells producing all three components of the complex from the chromosome and in mutant cells lacking either AcrB or TolC.
Surprisingly, we found that the tryptic digestion profiles of the chromosomally produced AcrA differ significantly from those of the purified/overproduced protein (Fig. 2). In the wild-type E. coli cells producing all three proteins, AcrA, AcrB, and TolC, only three tryptic fragments of AcrA are accumulated; by comparison to the tryptic digestion of the purified AcrAhis, these were identified as 37.4-, 36.9-, and 26.5-kDa fragments (Fig. 2A). Not even traces of the core 28.9-kDa fragment of AcrA could be detected in the wild-type E. coli cells. Thus, the 28.9-kDa fragment, which is invariably present in tryptic digests of free AcrA, is not accumulated when AcrA is assembled into a complex. In contrast, large amounts of the 26.5-kDa fragment of AcrA could be detected already after a 5-min digestion with trypsin, and this fragment resists further cleavage even after 60 min of incubation with large amounts of trypsin (Fig. 2A).
The proteolytic profile of AcrA changed dramatically in cells lacking either AcrB or TolC. Only traces of the 26.5-kDa fragment of AcrA could be seen on immunoblots of cells treated with trypsin for 5 min, and none could be seen after 60-min digests (Fig. 2A). When ΔacrB and ΔtolC strains were transformed with plasmids producing AcrB or TolC, respectively, the 26.5-kDa fragment of AcrA became highly abundant again (Fig. 2B). We conclude that the increase in the amounts of the 26.5-kDa fragment is specific for the assembly of the functional tripartite AcrAB-TolC complex.
Previous genetic and biochemical studies demonstrated that AcrA forms bipartite complexes with AcrB and TolC even in the absence of the respective third component (22, 23). Thus, in ΔacrB and ΔtolC strains, AcrA is engaged in interactions with TolC and AcrB, respectively. Yet the proteolytic profiles of AcrA are very similar in these two genetic backgrounds, with small amounts of the 26.5-kDa fragment and the 28.9-kDa core present at low concentrations of trypsin and short digestion times (Fig. 2). To determine whether such small amounts of fragments reflect the increased stability of the whole-length AcrA or, alternatively, indicate the rapid degradation of fragments, we quantified relative amounts of the whole-length AcrA on immunoblots of mutants and wild-type cells. We found that the decreases in the amounts of AcrA with increasing concentrations of trypsin and times of digestion were similar for all three strains (data not shown). Thus, the C-domain of AcrA is accessible to trypsin and rapidly degraded when one of the components of the AcrAB-TolC complex is missing.
The C-domain of AcrA is important for multidrug efflux function.
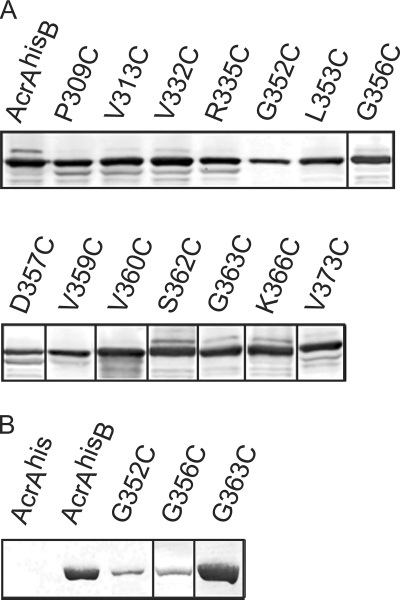
The experiments described above showed that the C-domain of AcrA is protected from tryptic digestion in AcrAB-TolC, suggesting that this domain might be involved in the assembly of the complex. To test the functional significance of the C-domain, we identified 12 highly conserved amino acid residues of AcrA, P309, V313, V332, R335, G352, L353, G356, D357, V359, V360, G363, and V373, and replaced them with cysteines by site-directed mutagenesis. In addition, cysteine substitutions were introduced into the variable S362 and K366 positions. Plasmids were transformed into E. coli cells deficient in acrAB and acrEF, and the expression of AcrAhis and its derivatives was determined by immunoblotting (Fig. 3A). The expression levels of all AcrAhis mutants were similar to that of wild-type AcrAhis, with the exception of that of the G352C mutant, which was reproducibly expressed at levels two- to threefold below that of the wild-type protein.
FIG. 3.
Expression of AcrAhis and AcrB is not affected by mutations in the C-domain of AcrAhis. (A) Total E. coli W4680AE cells harboring plasmids producing AcrB and either wild-type or mutant AcrAhis were boiled in the SDS sample buffer for 5 min, separated by 10% SDS-PAGE, and analyzed by immunoblotting with the polyclonal anti-AcrA antibody. (B) Membrane fractions isolated from E. coli W4680AE harboring pAhis, pAhisB, pAhisG352CB, pAhisG356CB, and pAhisG363CB plasmids were separated by 10% SDS-PAGE and analyzed by immunoblotting with the polyclonal anti-AcrB antibody.
To determine the functional competence of the AcrAhis mutants, we measured MICs of the selected known substrates of AcrAB-TolC, including erythromycin, norfloxacin, novobiocin, ethidium bromide, and SDS (Table 2). Most of the AcrAhis mutants fully complemented the function of the wild-type AcrAhis. However, E. coli W4680AE cells carrying plasmids with AcrAhis G352C, G356C, and G363C mutants were more susceptible to multiple drugs. The effect of G356C substitution was modest, with only a twofold decrease in the MICs of the tested compounds. The G352C mutation in AcrAhis resulted in a two- to eightfold decrease in MICs of the tested drugs, with the exception of SDS. Surprisingly, cells carrying the AcrAhis G363C mutant were highly susceptible (up to 32-fold) to all tested compounds.
TABLE 2.
Antibiotic susceptibility of E. coli W4680AE cells carrying plasmids that produce the wild-type and mutant derivatives of AcrAhis
| Plasmid | MIC (μg/ml)a
|
||||
|---|---|---|---|---|---|
| EM | NFLX | NB | EtBr | SDS | |
| pUC18 | 2 | 0.016 | 2 | 3.125 | 32 |
| pAhisB | 128 | 0.250 | 256 | 800 | >65,540 |
| pAhisG352CB | 64 | 0.125 | 64 | 100 | >65,540 |
| pAhisG356CB | 64 | 0.125 | 128 | 400 | >65,540 |
| pAhisG363CB | 4 | 0.016 | 16 | 50 | 16,380 |
All MIC measurements were done in triplicate. EM, erythromycin; NFLX, norfloxacin; NB, novobiocin; EtBr, ethidium bromide. Plasmids pAhisP309CB, pAhisV313CB, pAhisV332CB, pAhisR335CB, pAhisL353CB, pAhisD357CB, pAhisV359CB, pAhisV360CB, pAhisS362CB, pAhisK366CB, and pAhisV373CB fully complemented the drug-susceptible phenotype of W4680AE cells.
As shown in Fig. 3A, all three antibiotic-susceptible mutants were produced at the levels similar to those of the wild-type AcrAhis. Thus, changes in protein expression cannot account for the decrease in multidrug efflux activity. In agreement, we found no undesired mutations in the sequences of the upstream noncoding region of plasmids producing these three AcrAhis mutants (data not shown). Also, no mutations were found in the acrB sequence of the pAhisG363CB plasmid. Furthermore, the immunoblotting analysis showed that the amounts of AcrB transporter were similar in E. coli W4680AE cells carrying pAhisG363CB and pAhisB plasmids (Fig. 3B). We conclude that the decreased antibiotic resistance is caused by defects in the C-domain of AcrA. Cells carrying pAhisG352CB and pAhisG356CB plasmids produced notably smaller amounts of AcrB (Fig. 3B). Thus, in these cells, the low expression levels of AcrB could contribute to the decreased resistance to antibiotics.
The G363C mutant fails to properly assemble into the functional AcrAB-TolC complex.
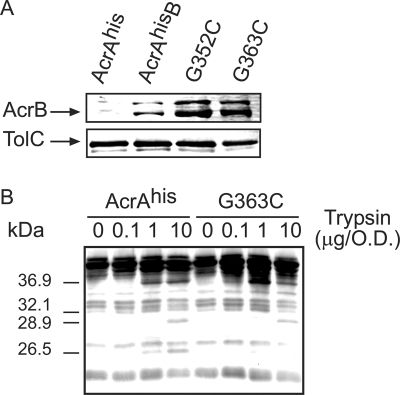
One possible reason why G352C and G363C substitutions inactivate AcrAhis is that these mutants have lost the ability to interact properly with AcrB, TolC, or both. Therefore, we next compared AcrAhis and its G352C and G363C variants in their ability to copurify with AcrB and TolC. For this purpose, all E. coli cells carrying pAHis (produces AcrAhis alone), pAhisB, pAhisG352CB, or pAhisG363CB plasmids were treated with the amine-reactive cross-linker DSP (spacer arm, 12 Å), and protein complexes were purified using metal affinity chromatography. AcrB and TolC copurified with AcrAhis, and its G352C and G363C mutants were detected by immunoblotting. As shown in Fig. 4A, in all three complexes, the amounts of AcrB and TolC were similar. Thus, G352C and G363C substitutions do not disrupt formation of the tripartite AcrAB-TolC complex.
FIG. 4.
The AcrAhis G363C mutant interacts with AcrB and TolC but fails to assemble into the functional AcrAB-TolC complex. (A) AcrB (top panel) and TolC (bottom panel) are copurified with the AcrAhis G363C mutant. E. coli W4680AE cells carrying pAhis, pAhisB, pAhisG352CB, and pAhisG363CB plasmids were collected at the mid-exponential phase and treated with the amino-reactive cross-linker DSP. Equal amounts of purified AcrAhis were separated by 10% SDS-PAGE and analyzed by immunoblotting with anti-AcrB and anti-TolC antibodies. (B) Trypsin digestion of E. coli W4680AE cells carrying pAhisB and its pAhisG363CB derivative. Trypsin digestion and analysis were carried out as described for Fig. 1C. O.D., optical density as determined by absorbance at 600 nm.
In vivo tryptic digestion was used to compare the structural features of the overproduced AcrAhis and its G363C derivative when these two proteins are coexpressed with AcrB. Overall, the proteolytic profile of G363C was similar to that of AcrAhis, with all major fragments in place (Fig. 4B). This result is consistent with the above finding that G363C substitution does not alter the structure of AcrA and that this mutant can assemble into the AcrAB-TolC complex. However, the G363C profile was more similar to that of the AcrAhis protein overproduced in the absence of AcrB (Fig. 1C). The 26.5-kDa fragment of G363C was notably more susceptible to trypsin than the corresponding fragment of AcrAhis (Fig. 4B). This result suggested that despite interactions with AcrB and TolC, the G363C mutant fails to properly assemble into the AcrAB-TolC complex.
DISCUSSION
Several models of AcrAB-TolC complex were recently proposed, all based on the structure of the 28.9-kDa AcrA core (6, 9, 18). The stoichiometry of the complex remains unclear, but a 1:1:1 AcrA/AcrB/TolC ratio is favored because of the trimeric structures of AcrB and TolC (6). The interaction between AcrA and TolC is proposed to involve the α-helical hairpin of AcrA and the periplasmic helices of TolC (12). The lipoyl- and α-β-barrel domains of AcrA are placed on the TolC-docking domain of AcrB (Fig. 5). Mutagenesis studies support this interaction: the suppressor mutations in MexA that either restore the activity of the defective MexB or mutations in AcrA that enable the functional complex with MexB all map to the α-β-barrel region of MFPs (11, 17). However, in all these models, the C-domain of AcrA does not contribute to interactions with TolC and AcrB, and the assembly of the AcrAB-TolC complex does not require any significant changes in AcrA structure.
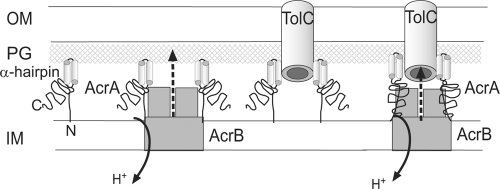
FIG. 5.
Schematic representation of the mechanism of assembly of AcrAB-TolC complex. AcrA forms bipartite complexes with AcrB and TolC. The N terminus of AcrA (N) is lipid modified and inserted into the inner membrane. The lipoyl and α-β-barrel domains of AcrA interact with AcrB, whereas the α-helical hairpin domain (α-hairpin) docks with TolC. In free AcrA and bipartite complexes, the C-domain (C) is unstructured and readily cleaved by trypsin. However, upon assembly of the functional tripartite AcrAB-TolC complex, this domain is protected from trypsin attack. The G363C mutation possibly induces a structural misfit between the C-terminal domain of AcrA and AcrB during assembly of the tripartite complex. OM, outer membrane; PG, peptidoglycan; IM, inner membrane.
In contrast, our results strongly suggest that the C-domain of AcrA is directly involved and required for the functional assembly of the multidrug efflux complex AcrAB-TolC. We found that the tryptic profile of AcrA within the fully assembled AcrAB-TolC pump differs significantly from that in the bipartite AcrA-AcrB and AcrA-TolC complexes. In wild-type cells, when the complex is assembled, the 28.9-kDa core characteristic for the free AcrA is undetectable, whereas the 26.5-kDa fragment is rapidly accumulated and resists further digestion by trypsin. In contrast, when only one of the components, AcrB or TolC, is present, both fragments are produced but rapidly reduced to trace amounts. Hence, the assembly of the AcrAB-TolC pump is not limited to the interactions of two nonoverlapping surfaces of AcrA with AcrB and TolC and likely engages new sites on the AcrA protein.
Previously, changes in proteolytic profiles were also reported for HlyD, the MFP components of hemolysin secretion complex HlyBD-TolC, and CvaA, the MFP component of the colicin V secretion complex CvaAB-TolC (10, 19). Both proteins displayed altered protease sensitivities in the functional tripartite complexes. Furthermore, CvaA was reported to be proteolytically unstable in cells lacking TolC. Thus, assembly of all tripartite complexes could involve rearrangement of MFPs.
We did not identify new tryptic fragments that would be specific for AcrA assembled into a complex. This result argues against significant conformational changes in AcrA during assembly of the AcrAB-TolC complex. Thus, the simplest explanation for the lack of 35.1-, 33.0-, 32.1-, and 28.9-kDa tryptic fragments in wild-type cells is that the amino acid residues K374, K346, and R315 of AcrA are protected from trypsin in AcrAB-TolC complex but readily cleaved in the free AcrA and in AcrA-AcrB and AcrA-TolC complexes. In addition, large amounts of the 26.5-kDa fragment in wild-type cells indicate that amino acid residues of AcrA responsible for this tryptic fragment remain accessible to trypsin in the AcrAB-TolC complex. However, tripartite interaction with AcrB and TolC protects this fragment from further degradation by trypsin and periplasmic proteases (Fig. 5). Analysis of the AcrA sequence suggested that the 26.5-kDa fragment could be generated by cleavage of the proteolytically stable core of AcrA K46-R315 at the carboxyl side of either residue R296 or residue R294. These residues terminate the α-β-barrel domain of AcrA (Fig. 1A).
The functional significance of the C-domain of AcrA was further confirmed by site-directed mutagenesis. A single substitution, G363C, reduced the activity of AcrAB-TolC to almost the null mutant level. However, the reduced level of multidrug efflux activity of the G363C mutant is still seen against SDS, novobiocin, and ethidium bromide (Table 2). This result is consistent with the finding that both AcrB and TolC can be copurified with the G363C mutant, indicating that the complex is assembled but cannot function properly.
It remains unclear why G363C fails to enable the full activity of the AcrAB-TolC multidrug efflux pump. One possibility is that the assembled G363C AcrAB-TolC complex is structurally defective. This notion is supported by the decreased stability of the 26.5-kDa fragment in the tryptic profile of periplasmic G363C (Fig. 4B). Our previous studies suggested that a structural misfit between components of the tripartite complex can significantly impact its multidrug efflux activity (11). Perhaps G363C mutation causes a similar structural misfit between AcrA, AcrB, and TolC. On the other hand, the defect in the G363C mutant could be functional. Several studies revealed that the role of MFPs in transport across two membranes is not limited to providing a structural link between transporters and outer-membrane channels. In addition, these proteins are functional subunits of transporters and stimulate their activities (1, 20, 25). AcrA was shown to stimulate substrate and proton transport activities of the reconstituted AcrB and AcrD. Similarly, MacA stimulated the ATPase activity of the reconstituted MacB, the macrolide transporter from E. coli. Thus, the G363C mutant could be defective because this protein has lost its ability to stimulate the activity of AcrB in the assembled AcrAB-TolC complex.
Footnotes
Published ahead of print on 1 May 2009.
REFERENCES
- 1.Aires, J. R., and H. Nikaido. 2005. Aminoglycosides are captured from both periplasm and cytoplasm by the AcrD multidrug efflux transporter of Escherichia coli. J. Bacteriol. 1871923-1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akama, H., T. Matsuura, S. Kashiwagi, H. Yoneyama, S. Narita, T. Tsukihara, A. Nakagawa, and T. Nakae. 2004. Crystal structure of the membrane fusion protein, MexA, of the multidrug transporter in Pseudomonas aeruginosa. J. Biol. Chem. 27925939-25942. [DOI] [PubMed] [Google Scholar]
- 3.Blum, H., H. Beier, and H. J. Gross. 1987. Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis 893-99. [Google Scholar]
- 4.Dinh, T., I. T. Paulsen, and M. H. Saier, Jr. 1994. A family of extracytoplasmic proteins that allow transport of large molecules across the outer membranes of gram-negative bacteria. J. Bacteriol. 1763825-3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elkins, C. A., and H. Nikaido. 2003. Chimeric analysis of AcrA function reveals the importance of its C-terminal domain in its interaction with the AcrB multidrug efflux pump. J. Bacteriol. 1855349-5356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fernandez-Recio, J., F. Walas, L. Federici, J. Venkatesh Pratap, V. N. Bavro, R. N. Miguel, K. Mizuguchi, and B. Luisi. 2004. A model of a transmembrane drug-efflux pump from Gram-negative bacteria. FEBS Lett. 5785-9. [DOI] [PubMed] [Google Scholar]
- 7.George, A. M., and S. B. Levy. 1983. Gene in the major cotransduction gap of the Escherichia coli K-12 linkage map required for the expression of chromosomal resistance to tetracycline and other antibiotics. J. Bacteriol. 155541-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gilson, L., H. K. Mahanty, and R. Kolter. 1990. Genetic analysis of an MDR-like export system: the secretion of colicin V. EMBO J. 93875-3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Higgins, M. K., E. Bokma, E. Koronakis, C. Hughes, and V. Koronakis. 2004. Structure of the periplasmic component of a bacterial drug efflux pump. Proc. Natl. Acad. Sci. USA 1019994-9999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hwang, J., X. Zhong, and P. C. Tai. 1997. Interactions of dedicated export membrane proteins of the colicin V secretion system: CvaA, a member of the membrane fusion protein family, interacts with CvaB and TolC. J. Bacteriol. 1796264-6270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krishnamoorthy, G., E. B. Tikhonova, and H. I. Zgurskaya. 2008. Fitting periplasmic membrane fusion proteins to inner membrane transporters: mutations that enable Escherichia coli AcrA to function with Pseudomonas aeruginosa MexB. J. Bacteriol. 190691-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lobedanz, S., E. Bokma, M. F. Symmons, E. Koronakis, C. Hughes, and V. Koronakis. 2007. A periplasmic coiled-coil interface underlying TolC recruitment and the assembly of bacterial drug efflux pumps. Proc. Natl. Acad. Sci. USA 1044612-4617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma, D., D. N. Cook, M. Alberti, N. G. Pon, H. Nikaido, and J. E. Hearst. 1993. Molecular cloning and characterization of acrA and acrE genes of Escherichia coli. J. Bacteriol. 1756299-6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mikolosko, J., K. Bobyk, H. I. Zgurskaya, and P. Ghosh. 2006. Conformational flexibility in the multidrug efflux system protein AcrA. Structure 14577-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miyamae, S., O. Ueda, F. Yoshimura, J. Hwang, Y. Tanaka, and H. Nikaido. 2001. A MATE family multidrug efflux transporter pumps out fluoroquinolones in Bacteroides thetaiotaomicron. Antimicrob. Agents Chemother. 453341-3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nehme, D., X.-Z. Li, R. Elliot, and K. Poole. 2004. Assembly of the MexAB-OprM multidrug efflux system of Pseudomonas aeruginosa: identification and characterization of mutations in mexA compromising MexA multimerization and interaction with MexB. J. Bacteriol. 1862973-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nehme, D., and K. Poole. 2007. Assembly of the MexAB-OprM multidrug pump of Pseudomonas aeruginosa: component interactions defined by the study of pump mutant suppressors. J. Bacteriol. 1896118-6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stegmeier, J. F., G. Polleichtner, N. Brandes, C. Hotz, and C. Andersen. 2006. Importance of the adaptor (membrane fusion) protein hairpin domain for the functionality of multidrug efflux pumps. Biochemistry 4510303-10312. [DOI] [PubMed] [Google Scholar]
- 19.Thanabalu, T., E. Koronakis, C. Hughes, and V. Koronakis. 1998. Substrate-induced assembly of a contiguous channel for protein export from E. coli: reversible bridging of an inner-membrane translocase to an outer membrane exit pore. EMBO J. 176487-6496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tikhonova, E. B., V. K. Devroy, S. Y. Lau, and H. I. Zgurskaya. 2007. Reconstitution of the Escherichia coli macrolide transporter: the periplasmic membrane fusion protein MacA stimulates the ATPase activity of MacB. Mol. Microbiol. 63895-910. [DOI] [PubMed] [Google Scholar]
- 21.Tikhonova, E. B., Q. Wang, and H. I. Zgurskaya. 2002. Chimeric analysis of the multicomponent multidrug efflux transporters from gram-negative bacteria. J. Bacteriol. 1846499-6507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tikhonova, E. B., and H. I. Zgurskaya. 2004. AcrA, AcrB, and TolC of Escherichia coli form a stable intermembrane multidrug efflux complex. J. Biol. Chem. 27932116-32124. [DOI] [PubMed] [Google Scholar]
- 23.Touzé, T., J. Eswaran, E. Bokma, E. Koronakis, C. Hughes, and V. Koronakis. 2004. Interactions underlying assembly of the Escherichia coli AcrAB-TolC multidrug efflux system. Mol. Microbiol. 53697-706. [DOI] [PubMed] [Google Scholar]
- 24.Zgurskaya, H. I., and H. Nikaido. 1999. AcrA is a highly asymmetric protein capable of spanning the periplasm. J. Mol. Biol. 285409-420. [DOI] [PubMed] [Google Scholar]
- 25.Zgurskaya, H. I., and H. Nikaido. 1999. Bypassing the periplasm: reconstitution of the AcrAB multidrug efflux pump of Escherichia coli. Proc. Natl. Acad. Sci. USA 967190-7195. [DOI] [PMC free article] [PubMed] [Google Scholar]