Abstract
In the food-borne pathogen Bacillus cereus F4430/73, the production of major virulence factors hemolysin BL (Hbl) and nonhemolytic enterotoxin (Nhe) is regulated through complex mechanisms. The two-component regulatory system ResDE is involved in the activation of hbl and nhe transcription. Here, the response regulator ResD and the sensor kinase ResE were overexpressed and purified, and autophosphorylation of ResE and transphosphorylation of ResD by ResE were demonstrated in vitro. ResD is mainly monomeric in solution, regardless of its phosphorylation state. ResD was shown to interact directly with promoter regions (p) of the enterotoxin regulator genes resDE, fnr, and plcR and the enterotoxin structural genes nhe and hbl, but with different affinities. Binding of ResD to pplcR, pnhe, and phbl was not dependent on the ResD phosphorylation status. In contrast, ResD phosphorylation significantly increased interactions between ResD and presDE and pfnr. Taken together, these results showed that phosphorylation of ResD results in a different target expression pattern. Furthermore, ResD and the redox activator Fnr were found to physically interact and simultaneously bind their target DNAs. We propose that unphosphorylated ResD acts as an antiactivator of Fnr, while phosphorylated ResD acts as a coactivator of Fnr. Finally, our findings represent the first molecular evidence of the role of ResDE as a sentinel system capable of sensing redox changes and coordinating a response that modulates B. cereus virulence.
Bacteria are often able to sense and respond to the surrounding environment via two-component systems (TCSs) (18, 22, 24, 34). In a typical TCS, a sensor kinase autophosphorylates in response to an extracellular and/or intracellular signal. Usually, a histidine residue in the sensor kinase receives a phosphoryl group from the low-molecular-weight donor ATP. This phosphoryl group is then transferred to an aspartate residue on a second protein, the response regulator (RR). Phosphorylation of the RR alters its ability to interact with either DNA or DNA and RNA polymerase and thus to activate or repress transcription in response to the signal received by the sensor histidine kinase (HK). Some HKs are bifunctional (19), not only acting as kinases but modulating the activity of their cognate RR proteins by acting as phosphatases that can remove the phosphoryl group from the RR.
ResD is an RR found in the opportunistic human pathogen Bacillus cereus (8). It has been proposed to classify ResD in the subfamily of RRs exemplified by OmpR and PhoB proteins from Escherichia coli (21). Members of this subfamily typically have two domains: an N-terminal receiver domain that acts as the phosphoryl acceptor and a C-terminal transactivation domain which contains a winged-helix-turn-helix DNA binding motif (10, 21). The linker connecting the two domains is variable in length (6 to 15 residues). Although all members of the OmpR/PhoB subfamily share a similar three-dimensional structure and appear to be activated by phosphorylation, they use different mechanisms to regulate their DNA-binding domains and modulate transcription (2, 11, 28). B. cereus ResD is encoded from the resDE operon that also encodes ResE, a prototypical HK. Because of their genomic context, ResD and ResE are thought to act as a TCS in B. cereus (8, 9). Previous in vivo studies have shown that ResDE is required for B. cereus growth under low oxydoreduction (ORP) conditions (8). Such conditions favor production of the PlcR-regulated HBL (hemolysin BL) enterotoxin and Nhe (nonhemolytic enterotoxin) (7), which are recognized as major virulence factors (33). Although it plays an important role in ORP-dependent regulation of enterotoxins, the ResDE system is not essential for enterotoxin production. In contrast, the redox regulator Fnr is essential for toxinogenesis, and its redox-dependent activity was clearly demonstrated (9). Previous data also suggest that both ResDE and Fnr could belong to the same redox regulatory pathway that may function at least partially independently of the pleiotropic virulence gene regulator PlcR (8).
In the study reported here, we purified and functionally characterized ResD and ResE His-tag-labeled variants in order to better understand the complex mechanisms employed by B. cereus to regulate enterotoxin gene expression. We demonstrate that both unphosphorylated and phosphorylated ResD directly interact with the promoter regions of the enterotoxin regulator genes fnr, resDE, and plcR (14) and the enterotoxin structural operons hbl and nhe. ResD is shown to exhibit multiple modes of binding, which may explain differences in the expressions of genes involved in enterotoxin expression. We evidenced a physical interaction between ResD and Fnr, and we propose that they thus potentially coregulate enterotoxin gene expression.
MATERIALS AND METHODS
Bacterial strains and growth conditions. Escherichia coli strain TOP10 (Invitrogen) [F− mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 deoR recA1 araD139 Δ(ara-leu)-7697 galU galK rpsL (Strr) endA1 nupG] was used as the general cloning host, and strain BL21-CodonPlus(DE3)-RIL (Stratagene) [F− ompT hsdS(rB− mB−) dcm+ Tetr gal λ (DE3) endA Hte (argU ileY leuW Cmr)] was used to overexpress resD and resE. Both E. coli strains were routinely grown in Luria broth, with vigorous agitation at 37°C. The B. cereus F4430/73 wild type (31) and resE and resDE mutants (8) were grown under microaerobiosis and N2-generated anaerobiosis, as previously described (8).
General molecular methods.
Restriction endonucleases and T4 DNA ligase were obtained from Promega. Genomic DNA of B. cereus was purified as described in reference 16. Plasmid DNA was purified using anion-exchange columns (Promega). PCR amplification of DNA was carried out with Taq polymerase (Roche Molecular Biochemicals).
Cloning and overexpression of resD and resE.
The coding sequence for B. cereus resD was PCR amplified from B. cereus F4430/73 genomic DNA using the forward and reverse primers PET101F (5′-CACCATGGAAAATGAATCAAGAATTTTAATTGTAG-3′) and PET101R (5′-GTCGTTCACAACCTCAAATTTGTAACCTAC-3′). The nucleotide sequence of resE coding for the cytoplasmic domain (the amino acid residues 199 to 565) of ResE (38) was amplified from the F4430/73 genome using the forward and reverse primers PET100F (5′-CACCATGACAGCACCGCTTCGTAAAATGCGTGAG-3′) and PET100R (5′-CTAAATTATACGATTCGGTAAATATACAG-3′). Both amplicons were introduced as blunt-end PCR products into pET101/D-TOPO and pET100/D-TOPO (Invitrogen), yielding pET101resD and pET100resE, respectively. The integrity of both inserted sequences was confirmed by DNA sequencing. The resulting constructs were transformed into E. coli BL21-CodonPlus(DE3)-RIL (Stratagene) for expression. E. coli BL21-CodonPlus(DE3)-RIL (pET101resD) and E. coli BL21-CodonPlus(DE3)-RIL (pET100resE) were grown in a 1-liter Luria broth medium supplemented with 100 μg ml−1 ampicillin, at 37°C. When the optical density at 600 nm reached ∼1.0, expression was induced by adding 0.2 mM isopropyl-β-d-thiogalactopyranoside (IPTG), and cells were grown for another 16 h at 37°C before being harvested by centrifugation (10,000 × g for 15 min) and directly used for protein purification.
Purification of the ResD and ResE proteins.
E. coli BL21-CodonPlus(DE3)-RIL (pET101resD) cells were resuspended in 50 mM sodium phosphate buffer (pH 7.0), 300 mM NaCl (buffer A). E. coli BL21-CodonPlus(DE3)-RIL (pET100resE) cells were resuspended in 50 mM sodium phosphate; 6 M guanidine-HCl; 300 mM NaCl, pH 7.0 (denaturing buffer C). Cells were incubated with 0.5 mg/ml lysozyme for 30 min under gentle agitation and sonicated for 3 min at 80% of maximum amplitude using a Vibra cell ultrasonicator (Fisher Bioblock Scientific). Cell debris was removed by centrifugation at 12,000 × g for 20 min. Purification of C-terminal His-tagged ResD and N-terminal His-tagged ResEΔ(1-198) was carried out using a 2-ml Co2+ immobilized metal affinity chromatography column (Clontech) equilibrated with buffer A and buffer C, respectively. The columns were washed with 5 ml of buffer A, and the recombinant proteins were eluted with 5 ml of buffer B (50 mM sodium phosphate [pH 7.0], 300 mM NaCl, and 150 mM imidazole). As reported above, the N-terminal His-tagged ResEΔ(1-198) was purified in denaturing conditions and allowed to renature on the column during washing and elution. To verify that the purified protein represented intact, unaggregated protein, the elution fraction was analyzed using blue native polyacrylamide gel electrophoresis (BN-PAGE) (see Fig. S1C in the supplemental material).
Protein biochemical analyses.
Protein concentrations were determined by bicinchoninic acid assay (Interchim), with bovine serum albumin (BSA) used as standard. Overproductions of ResD and ResEΔ(1-198) in induced cultures and their purification were monitored by sodium dodecyl sulfate (SDS)-PAGE (23). Proteins were stained with Coomassie brilliant blue. ResD oligomeric states were determined using BN-PAGE in which the anionic dye Coomassie blue G-250 was added to the sample buffer to impose a net negative charge upon the protein (37). The quaternary structure of purified ResD was measured in solution by dynamic light scattering (DLS), as previously described (9). Briefly, the ResD sample was applied onto a 24-ml Superdex 200 column (HR10/30) equilibrated and run at a flow rate of 0.5 ml/min with 50 mM Tris-HCl (pH 8.3) containing 120 mM NaCl and 0.05% NaN3 filtered at 0.1 μm. The elution profile was monitored with a miniDawn Tristar multiangle laser static light scattering detector (three angles, 45°, 90°, and 135°) coupled to a DynaPro Titan light scattering instrument (Wyatt Technology) placed at 90° and an Optilab rEX differential refractometer (Wyatt Technology).
Phosphorylation assays.
Purified ResE (6 μM) was incubated in 50 mM Tris-HCl (pH 7.5), 50 mM NaCl, 1 mM EDTA, 10% glycerol, 4 mM dithiothreitol, and 4 mM MgCl2 supplemented with 5 μCi of [γ-32P]ATP (∼6,000 Ci/mmol; Amersham Biosciences). The ResE autophosphorylation reaction was carried out at 37°C until equilibrum (15 min). trans-phosphorylation of ResD (9.6 μM) by phosphorylated ResE (6 μM) was performed at 37°C, according to the method used by Zhang and Hulett (38). The reactions were terminated by adding an equal volume of 6× SDS sample buffer. The resulting products were resolved by SDS-PAGE on 12% polyacrylamide gels (23). Labeled proteins were detected and analyzed using a PhosphorImager (Molecular Dynamics). Purified ResD (32 μM) was subjected to phosphorylation by incubation with 32 mM acetyl phosphate (AcP, Sigma) as described in references 25 and 26. Phosphorylation of ResD was evaluated using two-dimensional gel electrophoresis (29), as well as SDS-PAGE and Pro-Q diamond dye staining (Interchim) (32). Tandem mass spectrometry identification of ResD after trypsin proteolysis was carried out with a LTQ Orbitrap hybrid mass spectrometer (Thermo) operated as described in reference 6. Tandem mass spectrometry spectra were assigned to the ResD sequence with the MASCOT search engine (see Fig. S2C in the supplemental material).
Protein cross-linking.
Purified ResD (5 μM) pretreated in the absence or presence of AcP was incubated with the protein cross-linking agents N-hydroxysulfosuccinimide (NHS) and 1-ethyl-3-[3-dimethylaminopropyl]carbodiimide hydrochloride (EDC) as previously described (9). The resulting products were detected by Western blotting using an anti-His antibody (Roche).
Western blot analysis.
Samples were resolved by SDS-PAGE and electrotransferred onto nitrocellulose membranes (Amersham Bioscience) for Western blotting according to standard procedures (Bio-Rad). Membranes were probed with either anti-His antibodies or with a 1:1,000 dilution of polyclonal antibodies raised against ResD (anti-ResD) generated in-house as follows: rabbits were immunized with a total of 2 mg of purified ResD administered in four equal doses over a 90-day period and bled on day 20. The blotted membranes were developed with a 1:2,000 dilution of peroxidase-conjugated goat anti-rabbit immunoglobulin G (Sigma) and an enhanced chemiluminescence substrate (Immobilon Western; Millipore). Quantitative analysis of the signals was carried out using ImageJ (version 1.3, NIH).
Far-Western assays.
Biotinylated ResD was prepared with a biotin labeling kit-NH2 (Dojindo Laboratories). Purified six-His-tagged proteins were spotted onto two nitrocellulose membranes (Amersham Bioscience). One membrane was stained with Ponceau S solution (Sigma) to control the amount of proteins used for the protein-protein interaction studies. The other membrane was blocked with dilution buffer (Profound far-Western biotinylated protein-protein interaction kit; Pierce) for 1 h at room temperature and washed with phosphate buffer saline containing 0.05% Tween 20 (T-PBS). The membrane was then incubated for 1 h with 1 μg/ml biotinylated ResD. After the membrane was washed with phosphate buffer saline containing 0.05% Tween 20, it was incubated with anti-streptavidin-horseradish peroxidase (HRP; 0,1 mg/ml) and ResD was visualized by chemiluminescence (Pierce). Purified ResE was blotted as a positive control. BSA and blotting buffer alone were used as negative controls.
EMSAs.
Fragments containing the promoter regions of fnr, resDE, plcR, hbl, and nhe (see Fig. S3 in the supplemental material) were amplified and end labeled as described previously (9). Electrophoretic mobility shift assays (EMSAs) were performed by incubating the labeled fragments (1,000 cpm) with the specified amount of purified ResD in the presence or absence of ResE (6 μM) at 37°C for 30 min in 20 μl binding buffer (50 mM Tris-HCl, pH 7.5; 50 mM NaCl; 1 mM EDTA; 10% glycerol; 4 mM dithiothreitol; and 4 mM MgCl2) containing 200 μM ATP. The samples were loaded onto a 6% native polyacrylamide gel run with Tris-borate-EDTA buffer at 4°C and 200 V. Gels were dried and analyzed using a Molecular Dynamics PhosphorImager.
RESULTS
Autophosphorylation of ResE and trans-phosphorylation of ResD by ResE.
To demonstrate the phosphorylation activities of B. cereus ResDE, the cytoplasmic domain of ResE [ResEΔ(1-198)] (38) and the full-length wild-type ResD were expressed as six-histidine-tagged fusion proteins in E. coli and purified to homogeneity (see Fig. S1A and S1B in the supplemental material). Given that in vitro autophosphorylation of HK signaling domains is a common property of TCS histidine kinases that occurs in the absence of any signaling input, ResEΔ(1-198) was subjected to autophosphorylation in the presence of [γ-32P]ATP (see Fig. S1D in the supplemental material). Peak autophosphorylation was observed within 15 min (data not shown). Unlike ResEΔ(1-198), ResD was not phosphorylated by ATP. Adding ResD to phosphorylated ResEΔ(1-198) resulted in dephosphorylation of ResEΔ(1-198) and transphosphorylation of ResD (see Fig. S1D in the supplemental material). A sequence alignment of ResE and ResD from B. cereus with their orthologs from Bacillus subtilis, E. coli, and Salmonella enterica serovar Typhimurium showed strong sequence conservation in the phosphorylatable regions of both proteins (data not shown). Consequently, autophosphorylation of ResE may occur at histidine 374 and phosphorylation of ResD should occur on aspartate 55.
DNA-binding properties of ResD.
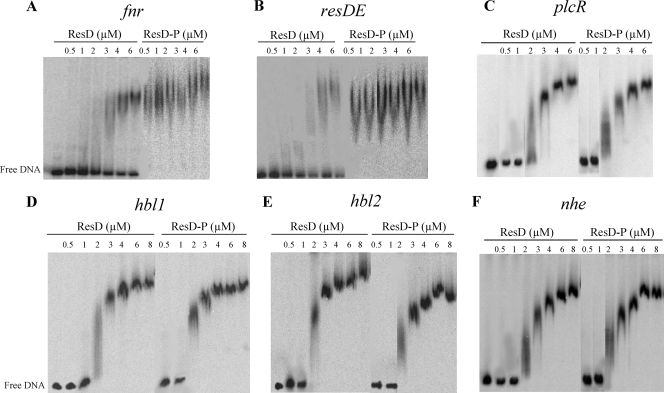
Figure 1 shows the EMSAs that were performed with both ResD and phosphorylated ResD (ResD∼P) (generated after incubation with ResE and ATP) and DNA fragments (1,000 cpm) containing the promoter regions of resDE, fnr, plcR, hbl, and nhe (presDE, pfnr, pplcR, phbl, and pnhe). In view of its size (1,157 bp), the promoter region of hbl was divided into two overlapping fragments of 636 bp (hbl1) and 610 bp (hbl2), as defined in Fig. S3 in the supplemental material. Both ResD and ResD∼P caused mobility shifts in the six tested DNA fragments. The specificity of the binding was evidenced by (i) the disappearance of complexes in competition assays using 50-fold excess of homologous unlabeled promoter regions and the absence of any competition when unlabeled heterologous DNA was used, (ii) the absence of ResD and ResD∼P binding above 8 μM with the negative control derived from the ssu promoter region (20), and (iii) the inability of ResE to interact with any of the six DNA fragments (see Fig. S4 in the supplemental material). Interestingly, the concentrations of ResD that were required for a mobility shift to occur with pplcR, phbl, and pnhe were lower than those required for a shift with presDE and pfnr, which suggests that ResD had greater affinity for pplcR, phbl, and pnhe. In addition, the concentration of ResD∼P that was needed to form complexes with pfnr and presDE was lower than the concentration of unphosphorylated ResD. In contrast, ResD∼P behaved like unphosphorylated ResD when binding to pplcR, phbl, and pnhe. Taken together, these data indicate that phosphorylation of ResD is not essential for DNA binding and that ResD phosphorylation affects the DNA binding affinity of some but not all target promoters. Moreover, whatever the promoter used for these assays, we observed that the gel shift not only required a high ResD or ResD∼P concentration but also exhibited a smear instead of a discrete band. This kind of smearing is usually observed when a DNA-protein complex tends to dissociate during its migration during electrophoresis. These data suggest the existence of a weak association between ResD and its nucleic targets. Hence, the involvement of the regulator acting together with ResD is a possible means of stabilizing this interaction.
FIG. 1.
Binding of ResD and ResD∼P to the regulatory regions of the fnr, resDE, plcR, hbl, and nhe genes determined by EMSA. DNA (1,000 cpm) corresponding to fnr (A), resDE (B), plcR (C), hbl1 (D), hbl2 (E), and nhe (F) was incubated with increasing concentrations of ResD and ResD∼P obtained after incubation with purified ResE and ATP, as indicated. The results presented are representative examples of experiments performed in triplicate. The concentrations of ResD and ResD∼P used in each reaction are, from left to right, 0.5, 1, 2, 3, 4, 6 and eventually 8 μM. The first lane of each panel corresponds to the labeled DNA fragment alone.
Phosphorylation or DNA binding does not influence the oligomerization state of ResD.
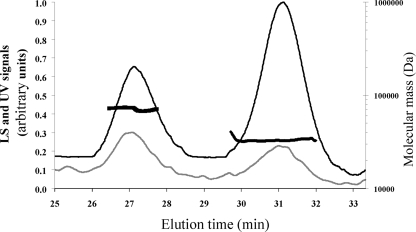
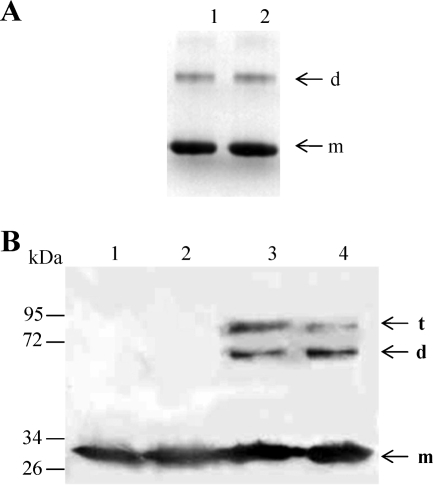
The oligomerization state of ResD in solution was first analyzed by DLS (9). Figure 2 shows the elution profiles obtained after resolution of the ResD entities on Superdex 200 gel filtration. The molecular mass estimates were derived from the light scattering signal measured at different angles. ResD was resolved into two peaks eluting at 31.0 and 27.4 min, as detected on the UV and DLS traces (Fig. 2). The distribution of molar masses across these two peaks was constant, indicating a monodisperse distribution for each peak with molecular masses of 28 and 67 kDa (± 10%), respectively. These values indicate that ResD exists as a mixture of monomer and dimer in solution. Considering the relative mass ratio that can be estimated from the UV trace (2/3 and 1/3, respectively), the predominant form was the monomer. Analysis of purified ResD in BN-PAGE (Fig. 3A, lane 1) showed a two-band pattern, revealing the presence of a mixed population of monomeric and dimeric forms. Like in the DLS experiment (Fig. 2), the predominant form is the monomer. To test the effect of phosphorylation on the ResD oligomerization state, purified ResD was phosphorylated using AcP (see Fig. S2 in the supplemental material). Figure 4A shows that ResD∼P (lane 2) exhibits the same band pattern as unphosphorylated ResD (lane 1). This indicates that phosphorylation did not promote oligomerization of ResD in solution and that both ResD and ResD∼P are mainly monomeric in solution. We then quantified and compared cross-linking efficiency of ResD and ResD∼P upon NHS/EDC chemical cross-linking from two independent experiments. Figure 3B showes that under the conditions examined, the two ResD forms are equally effective in forming stable cross-linked oligomers (36% ± 3%). However, there was a slight but noticeable reduction in trimer formation (9% ± 1% instead of 15% ± 1%) and a concomitant increase in dimer formation (27% ± 2% instead of 21% ± 2%) in the presence of ResD∼P. This suggests that there is a phosphorylation-induced conformational change that could affect transiently the oligomerization state of ResD. We concluded that phosphorylation does not modify the equilibrium between monomer and dimer.
FIG. 2.
Gel filtration and DLS chromatograms of purified ResD. ResD protein was desalted on a G25 gel filtration unit, concentrated on an Amicon 5,000-Da-cut-off Ultra device (Amicon), and injected (∼80 μg in 70 μl) into a Superdex 200 column (HR 10/30) with 50 mM Tris-HCl (pH 8.3) and with 120 mM NaCl as the eluent, at a flow rate of 0.5 ml/min. The gray and black lines correspond to the light scattering (LS) signal and the UV signal recorded at 280 nm, respectively (y axis). LS signal noise was removed by moving average smoothing. The molecular mass estimates of the two major peaks are indicated by thick black broken lines (y axis).
FIG. 3.
Effects of phosphorylation on the oligomeric state of ResD. (A) BN-PAGE analysis of ResD and ResD∼P. ResD (2 μg) was subjected to phosphorylation by acetyl phosphate as described in Materials and Methods. Lane 1, ResD; lane 2, ResD∼P. (B) SDS-PAGE profile of ResD cross-linked with NHS/EDC. Cross-linking was performed as described in Materials and Methods. Proteins were visualized by immunoblotting with anti-His antibodies. Lane 1, untreated ResD; lane 2, phosphorylated ResD; lane 3, cross-linked ResD; lane 4, cross-linked ResD∼P (generated with acetyl phosphate). The arrows indicate monomers (m), dimers (d), and trimers (t). The sizes of the molecular mass markers (in thousands) are indicated on the left.
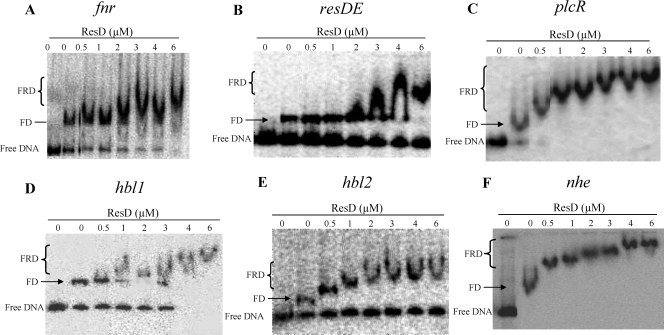
FIG. 4.
Cooperative binding between ResD and Fnr on the regulatory regions of the fnr, resDE, plcR, hbl, and nhe genes. Cooperativity between ResD and Fnr was analyzed by EMSA. Increasing amounts of ResD were incubated with labeled DNA fragments (1,000 cpm) corresponding to the fnr (A), resDE (B), plcR (C), hbl1 (D), hbl2 (E), and nhe (F) regulatory regions in the presence of 0.6 μM of Fnr. FD indicates the Fnr-DNA complex, and FRD indicates the Fnr-ResD-DNA ternary complex.
ResD and Fnr interaction when binding their target promoters.
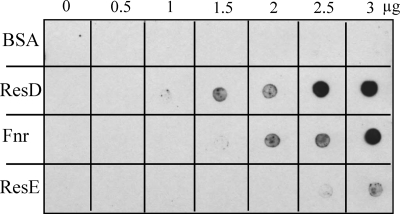
Like ResD, the redox regulator Fnr binds to presDE, pfnr, pplcR, phbl, and pnhe (9). To determine whether ResD and Fnr can bind simultaneously to these DNAs, we performed a competitive EMSA. Figure 4 shows the results obtained after incubating purified Fnr and ResD with presDE, pfnr, pplcR, phbl, or pnhe. At a constant Fnr concentration (0.6 μM), increasing the ResD concentration resulted in the appearance of a high-molecular-mass complex, whose mobility corresponds to an Fnr-ResD-DNA ternary complex and the disappearance of the complex containing only Fnr and DNA. This was detected with whichever of the six promoters tested. Moreover, this ternary complex is formed at a lower ResD concentration than that of the ResD-DNA binary complex at pnhe, phbl, and pplcR but not at pfnr and presDE (Fig. 1). The same results were obtained using phosphorylated ResD (generated after incubation with ResE and ATP). We also evidenced that for a constant amount of ResD or ResD∼P, titration by increasing amounts of Fnr resulted in the formation of the Fnr-ResD-DNA complex (data not shown). To determine whether the formation of the Fnr-ResD-DNA complex may involve a direct protein-protein interaction, far-Western analysis was conducted with purified monomeric Fnr adsorbed onto a dot blot. Figure 5 shows the nitrocellulose membrane that was incubated with biotinylated ResD and then revealed after incubation with HRP-conjugated streptavidin. The direct application of proteins to nitrocellulose made it possible to test interactions under native conditions, i.e., without denaturation of the proteins. The data indicate that ResD binds directly to Fnr with a lower affinity than with itself but with higher affinity than with its cognate partner, ResE. In the control experiment, BSA did not show any binding to ResD. To examine whether phosphorylation influences protein-protein interactions, equal molar amounts of ResD∼P generated with AcP (see Fig. S2 in the supplemental material) were tested by following the same protocol. Results with ResD∼P were essentially identical to those obtained with ResD (data not shown), suggesting that the interaction between Fnr and ResD was not affected by the phosphorylation status of ResD. In conclusion, we evidenced a direct and specific interaction between the ResD and Fnr proteins, independently of the presence of their nucleic targets.
FIG. 5.
Far-Western analysis of ResD-Fnr interaction. Increased amounts of purified ResD, Fnr, and ResE were spotted onto nitrocellulose membranes and incubated with biotinylated ResD, as indicated in Materials and Methods. ResD binding to proteins was detected using the streptavidin-HRP complex and visualized by chemiluminescence, with BSA used as negative control.
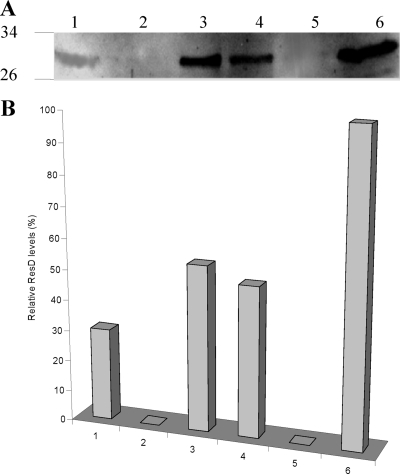
Endogeneous ResD protein levels in the wild type and resE and resDE mutants.
Although the EMSA results described above indicated that ResE-dependent phosphorylation of ResD did not affect its binding to phbl and pnhe, our previous study showed that hbl and nhe transcription was dramatically downregulated in ResE-deficient cells (resE mutant cells) (37). To determine whether the defective hbl and nhe transcription was due to the lack of the kinase activity of ResE and not to a strongly reduced level of the ResD protein, ResD protein levels were measured in resE mutant cells by Western blotting using anti-ResD polyclonal antibodies. A protein band corresponding to the molecular mass of ResD (∼30 kDa) was detected in both microaerobically and anaerobically grown resE mutant and wild-type cells (Fig. 6A). This band was absent when protein extracts from resDE mutant cells (lacking both ResE and ResD) were studied, confirming that it was indeed ResD. Densitometric analysis of the ResD bands on two independent Western blots (Fig. 6B) revealed the relative intensities of the ResD band from anaerobically grown wild-type cells (100%) and resE mutant cells (49% ± 5%) and from microaerobically grown wild-type cells (54% ± 5%) and resE mutant cells (30% ± 3%). These results indicated that (i) anaerobically grown cells contained higher levels of ResD than microaerobically grown cells, (ii) a strain lacking ResE synthesized only half as much ResD as the wild-type strain when grown in the same conditions, and (iii) anaerobically grown resE mutant cells exhibit levels of ResD similar to those of microaerobically grown wild-type cells. Our previous data showed that microaerobically and anaerobically grown wild-type cells exhibited the same nhe and hbl mRNA levels and that these levels were approximately 50-fold reduced in the resE background under both conditions (8). Taken together, these results indicate that the level of enterotoxin gene transcription is not correlated with the intracellular ResD level. We thus conclude that ResD-dependent hbl and nhe transcription is more sensitive to the ResE-dependent ResD phosphorylation status than to the ResD level.
FIG. 6.
Quantification of ResD levels in B. cereus cells by densitometric analysis of the ResD bands on the Western blot. (A) Cytoplasmic proteins (5 μg) of the B. cereus F4430/73 resE mutant, resDE mutant, and wild type grown under microaerobiosis (lanes 1, 2, and 3, respectively) and under anaerobiosis (lanes 4, 5, and 6, respectively) were separated using SDS-PAGE and probed with anti-ResD polyclonal antibodies. (B) Quantitative analysis of the ResD bands was carried out using Image J (version 1.3, NIH). Relative levels can be compared to the 100% value that was arbitrarily fixed for anaerobically grown wild-type cells. High reproducibility was reached (<10%).
DISCUSSION
Although the complete set of TCSs in different members of the B. cereus group has been identified and confirmed as playing important roles in gene regulation (3-5), only the biological role of ResDE has thus far been studied (8, 35, 36). The molecular mechanisms responsible for signal transduction remained uncharacterized. The present study reports the biochemical properties of B. cereus ResDE and its link with virulence gene expression.
The signaling-transduction process is mediated by two phosphorylation events. In the first phosphorylation event, the HK is autophosphorylated at the conserved His residue in the so-called activation loop. Our study shows that B. cereus ResE is able to autophosphorylate in vitro in the presence of ATP. Although in vitro experiments are carried out with the cytosolic portion of ResE lacking the transmembrane region, data reported from B. subtilis have shown that this fragment has enzymatic activity similar to the full-length HK (13). In the second phosphorylation event, the phosphoryl group is transferred from the HK to the cognate RR, in a response regulator-catalyzed reaction. Our experiments show that the ResD-dependent phosphoryl group transfer occurs in vitro. These biochemical data suggest that the ResDE system is able to transduce cellular stress signals in vivo, in agreement with previous in vivo characterization studies (8). The genomic context and previous in vivo studies suggest that ResE may be the only biologically relevant kinase able to phosphorylate ResD. However, we cannot exclude the possibility that phosphorylation by acetyl phosphate could play an important role in modulating ResD activity in vivo, linking its phosphorylation state to the metabolic status of B. cereus (15).
Since putative ResD binding consensus motifs could be predicted in the enterotoxin gene promoter regions (12) (see Fig. S3 in the supplemental material), we expected to see ResD binding to these fragments. However, the ResD binding pattern within the promoter regions of the plcR-regulated genes nhe, hbl, and plcR was significantly different from that of resDE and fnr promoters. First, the DNA binding affinity of unphosphorylated ResD was higher for pnhe, phbl, and pplcR than for presDE and pfnr. Second, phosphorylation of ResD enhanced its ability to bind to presDE and pfnr but not to pnhe, phbl, and pplcR. Third, a Fnr-ResD-DNA ternary complex was formed at a lower concentration than that of the ResD-DNA binary complex at pnhe, phbl, and pplcR but not at presDE and pfnr. The existence of different patterns of recognition by ResD and ResD∼P suggests that differences may exist in the ResD-binding affinities, depending on the different ResD binding sites in the studied promoters. We planned new specific studies to understand how differences in promoter organizations may affect ResD binding.
Previous data have established the following points. First, the enterotoxin gene promoters (pnhe and phbl) require Fnr for activation (39). Such activation probably involves direct protein-protein contact between Fnr and RNA polymerase (9). Second, ResD was also required for pnhe and phbl activation but only when phosphorylated by ResE. Third, Fnr cannot activate pnhe and phbl without ResD∼P but can activate pnhe and phbl without ResD (8). The experiments presented here showed that (i) phosphorylation of ResD affects neither its oligomeric state nor its ability to interact with phbl and pnhe, (ii) ResD and ResD∼P can bind to pnhe and phbl in concert with Fnr, and (iii) both ResD and ResD∼P physically interact with Fnr. Taken together, these results suggest that ResD∼P could interact with Fnr to form transcriptionally active ResD∼P-Fnr-pnhe-phbl complexes and ResD could interact with Fnr to form transcriptionally inactive ResD-Fnr-pnhe/phbl complexes. Therefore, ResD∼P may act as an Fnr coactivator and ResD as an Fnr antiactivator. However, this finding needs to be confirmed by analyzing more particularly the ResD-Fnr interaction using the holoform of Fnr instead of the apoform, as was the case in this study. The enterotoxin gene expression level depends on environmental redox conditions. In particular, low ORP conditions favored higher levels of hbl and nhe expression than high ORP conditions (8). Low ORP conditions could thus favor the formation of high levels of the ResD∼P-Fnr-pnhe-phbl activation complex, ensuring high levels of enterotoxin gene expression. This possibility is sustained by previous and present results showing that ResD and ResE synthesis, together with Fnr synthesis, is enhanced in response to low ORP conditions (8).
Although plcR belongs to the same regulon as nhe and hbl (the plcR regulon) (14), plcR activation did not require ResD phosphorylation (8). This indicates that ResD phosphorylation is not a prerequisite for the activation of pplcR (unlike pnhe and phbl) in response to environmental stress. PlcR is known to autoactivate and activate enterotoxin gene expression by binding specific sequences in pplcR, pnhe, and phbl (1). The possibility that PlcR might form with Fnr, ResD, and DNA a quaternary complex is thus not excluded. In conclusion, the mechanism of ResD-dependent regulation of enterotoxin gene expression in response to redox conditions is undoubtedly complex, and further studies are required. However, deciphering the complexities of this regulation under low ORP conditions, such as those encountered in the human small intestine (17, 27, 30), represents the most critical step toward understanding the mechanisms employed by B. cereus to unleash optimal virulence gene expression. Finally, this study is one of the first examples of in vitro TCS reconstitution to report the characterization of important signaling and DNA binding events for B. cereus virulence.
Supplementary Material
Acknowledgments
J.E. is supported by a fellowship from the Ministère de la Recherche et de l'Enseignement supérieur.
We thank our colleagues from CEA-Marcoule: Bernard Fernandez for help with the DLS experiments, Philippe Guérin for performing two-dimensional gel electrophoresis and mass spectrometry experiments, and Valérie Tanchou for her kind help with the production of anti-ResD polyclonal antibodies.
Footnotes
Published ahead of print on 24 April 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Agaisse, H., M. Gominet, O. A. Okstad, A. B. Kolsto, and D. Lereclus. 1999. PlcR is a pleiotropic regulator of extracellular virulence factor gene expression in Bacillus thuringiensis. Mol. Microbiol. 321043-1053. [DOI] [PubMed] [Google Scholar]
- 2.Allen, M. P., K. B. Zumbrennen, and W. R. McCleary. 2001. Genetic evidence that the α5 helix of the receiver domain of PhoB is involved in interdomain interactions. J. Bacteriol. 1832204-2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brillard, J., K. Susanna, C. Michaud, C. Dargaignaratz, M. Gohar, C. Nielsen-Leroux, N. Ramarao, A. B. Kolsto, C. Nguyen-The, D. Lereclus, and V. Broussolle. 2008. The YvfTU two-component system is involved in plcR expression in Bacillus cereus. BMC Microbiol. 8183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Been, M., M. J. Bart, T. Abee, R. J. Siezen, and C. Francke. 2008. The identification of response regulator-specific binding sites reveals new roles of two-component systems in Bacillus cereus and closely related low-GC Gram-positives. Environ. Microbiol. 102796-2809. [DOI] [PubMed] [Google Scholar]
- 5.de Been, M., C. Francke, R. Moezelaar, T. Abee, and R. J. Siezen. 2006. Comparative analysis of two-component signal transduction systems of Bacillus cereus, Bacillus thuringiensis and Bacillus anthracis. Microbiology 1523035-3048. [DOI] [PubMed] [Google Scholar]
- 6.de Groot, A., R. Dulermo, P. Ortet, L. Blanchard, P. Guérin, B. Fernandez, B. Vacherie, C. Dossat, E. Jolivet, P. Siguier, M. Chandler, M. Barakat, A. Dedieu, V. Barbe, T. Heulin, S. Sommer, W. Achouak, and J. Armengaud. 2009. Alliance of proteomics and genomics to unravel the specificities of Sahara bacterium Deinococcus deserti. PLoS Genet. 5e1000434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duport, C., S. Thomassin, G. Bourel, and P. Schmitt. 2004. Anaerobiosis and low specific growth rates enhance hemolysin BL production by Bacillus cereus F4430/73. Arch. Microbiol. 18290-95. [DOI] [PubMed] [Google Scholar]
- 8.Duport, C., A. Zigha, E. Rosenfeld, and P. Schmitt. 2006. Control of enterotoxin gene expression in Bacillus cereus F4430/73 involves the redox-sensitive ResDE signal transduction system. J. Bacteriol. 1886640-6651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Esbelin, J., Y. Jouanneau, J. Armengaud, and C. Duport. 2008. ApoFnr binds as a monomer to promoters regulating the expression of enterotoxin genes of Bacillus cereus. J. Bacteriol. 1904242-4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Galperin, M. Y. 2006. Structural classification of bacterial response regulators: diversity of output domains and domain combinations. J. Bacteriol. 1884169-4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao, R., Y. Tao, and A. M. Stock. 2008. System-level mapping of Escherichia coli response regulator dimerization with FRET hybrids. Mol. Microbiol. 691358-1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geng, H., Y. Zhu, K. Mullen, C. S. Zuber, and M. M. Nakano. 2007. Characterization of ResDE-dependent fnr transcription in Bacillus subtilis. J. Bacteriol. 1891745-1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geng, H., P. Zuber, and M. M. Nakano. 2007. Regulation of respiratory genes by ResD-ResE signal transduction system in Bacillus subtilis. Methods Enzymol. 422448-464. [DOI] [PubMed] [Google Scholar]
- 14.Gohar, M., K. Faegri, S. Perchat, S. Ravnum, O. A. Okstad, M. Gominet, A. B. Kolsto, and D. Lereclus. 2008. The PlcR virulence regulon of Bacillus cereus. PLoS ONE 3e2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gueriri, I., S. Bay, S. Dubrac, C. Cyncynatus, and T. Msadek. 2008. The Pta-AckA pathway controlling acetyl phosphate levels and the phosphorylation state of the DegU orphan response regulator both play a role in regulating Listeria monocytogenes motility and chemotaxis. Mol. Microbiol. 701342-1357. [DOI] [PubMed] [Google Scholar]
- 16.Guinebretiere, M. H., and C. Nguyen-The. 2003. Sources of Bacillus cereus contamination in a pasteurized zucchini purée processing line, differentiated by two PCR-based methods. FEMS Microbiol. Ecol. 43207-215. [DOI] [PubMed] [Google Scholar]
- 17.Guyton, A. C. 1977. Basic human physiology: normal function and mechanisms of disease, 2nd ed. Elsevier Health Sciences, Philadelphia, PA.
- 18.Hoch, J. A. 2000. Two-component and phosphorelay signal transduction. Curr. Opin. Microbiol. 3165-170. [DOI] [PubMed] [Google Scholar]
- 19.Igoshin, O. A., R. Alves, and M. A. Savageau. 2008. Hysteretic and graded responses in bacterial two-component signal transduction. Mol. Microbiol. 681196-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ivanova, N., A. Sorokin, I. Anderson, N. Galleron, B. Candelon, V. Kapatral, A. Bhattacharyya, G. Reznik, N. Mikhailova, A. Lapidus, L. Chu, M. Mazur, E. Goltsman, N. Larsen, M. D'Souza, T. Walunas, Y. Grechkin, G. Pusch, R. Haselkorn, M. Fonstein, S. D. Ehrlich, R. Overbeek, and N. Kyrpides. 2003. Genome sequence of Bacillus cereus and comparative analysis with Bacillus anthracis. Nature 42387-91. [DOI] [PubMed] [Google Scholar]
- 21.Kenney, L. J. 2002. Structure/function relationships in OmpR and other winged-helix transcription factors. Curr. Opin. Microbiol. 5135-141. [DOI] [PubMed] [Google Scholar]
- 22.Khorchid, A., and M. Ikura. 2006. Bacterial histidine kinase as signal sensor and transducer. Int. J. Biochem. Cell Biol. 38307-312. [DOI] [PubMed] [Google Scholar]
- 23.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227680-685. [DOI] [PubMed] [Google Scholar]
- 24.Mascher, T., J. D. Helmann, and G. Unden. 2006. Stimulus perception in bacterial signal-transducing histidine kinases. Microbiol. Mol. Biol. Rev. 70910-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCleary, W. R., and J. B. Stock. 1994. Acetyl phosphate and the activation of two-component response regulators. J. Biol. Chem. 26931567-31572. [PubMed] [Google Scholar]
- 26.Miller, A. A., N. C. Engleberg, and V. J. DiRita. 2001. Repression of virulence genes by phosphorylation-dependent oligomerization of CsrR at target promoters in S. pyogenes. Mol. Microbiol. 40976-990. [DOI] [PubMed] [Google Scholar]
- 27.Moriarty-Craige, S. E., and D. P. Jones. 2004. Extracellular thiols and thiol/disulfide redox in metabolism. Annu. Rev. Nutr. 24481-509. [DOI] [PubMed] [Google Scholar]
- 28.Rhee, J. E., W. Sheng, L. K. Morgan, R. Nolet, X. Liao, and L. J. Kenney. 2008. Amino acids important for DNA recognition by the response regulator OmpR. J. Biol. Chem. 2838664-8677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ruiz, D., P. Salinas, M. L. Lopez-Redondo, M. L. Cayuela, A. Marina, and A. Contreras. 2008. Phosphorylation-independent activation of the atypical response regulator NblR. Microbiology 1543002-3015. [DOI] [PubMed] [Google Scholar]
- 30.Skinner, F. A., and J. G. Carr. 1995. The normal microbial flora of man. Academic, London, United Kingdom.
- 31.Spira, W. M., and J. M. Goepfert. 1975. Biological characteristics of an enterotoxin produced by Bacillus cereus. Can. J. Microbiol. 211236-1246. [DOI] [PubMed] [Google Scholar]
- 32.Steinberg, T. H., B. J. Agnew, K. R. Gee, W. Y. Leung, T. Goodman, B. Schulenberg, J. Hendrickson, J. M. Beechem, R. P. Haugland, and W. F. Patton. 2003. Global quantitative phosphoprotein analysis using multiplexed proteomics technology. Proteomics 31128-1144. [DOI] [PubMed] [Google Scholar]
- 33.Stenfors Arnesen, L. P., A. Fagerlund, and P. E. Granum. 2008. From soil to gut: Bacillus cereus and its food poisoning toxins. FEMS Microbiol. Rev. 32579-606. [DOI] [PubMed] [Google Scholar]
- 34.Stock, J., and S. Da Re. 2000. Signal transduction: response regulators on and off. Curr. Biol. 10420-424. [DOI] [PubMed] [Google Scholar]
- 35.Vetter, S. M., and P. M. Schlievert. 2007. The two-component system Bacillus respiratory response A and B (BrrA-BrrB) is a virulence factor regulator in Bacillus anthracis. Biochemistry 467343-7352. [DOI] [PubMed] [Google Scholar]
- 36.Wilson, A. C., J. A. Hoch, and M. Perego. 2008. Virulence gene expression is independent of ResDE-regulated respiration control in Bacillus anthracis. J. Bacteriol. 1905522-5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wittig, I., H. P. Braun, and H. Schagger. 2006. Blue native PAGE. Nat. Protoc. 1418-428. [DOI] [PubMed] [Google Scholar]
- 38.Zhang, X., and F. M. Hulett. 2000. ResD signal transduction regulator of aerobic respiration in Bacillus subtilis: ctaA promoter regulation. Mol. Microbiol. 371208-1219. [DOI] [PubMed] [Google Scholar]
- 39.Zigha, A., E. Rosenfeld, P. Schmitt, and C. Duport. 2007. The redox regulator Fnr is required for fermentative growth and enterotoxin synthesis in Bacillus cereus F4430/73. J. Bacteriol. 1892813-2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.