Abstract
Unlike other bacteria that use FNR to regulate anaerobic respiration, Shewanella oneidensis MR-1 uses the cyclic AMP receptor protein (CRP) for this purpose. Three putative genes, cyaA, cyaB, and cyaC, predicted to encode class I, class IV, and class III adenylate cyclases, respectively, have been identified in the genome sequence of this bacterium. Functional validation through complementation of an Escherichia coli cya mutant confirmed that these genes encode proteins with adenylate cyclase activities. Chromosomal deletion of either cyaA or cyaB did not affect anaerobic respiration with fumarate, dimethyl sulfoxide (DMSO), or Fe(III), whereas deletion of cyaC caused deficiencies in respiration with DMSO and Fe(III) and, to a lesser extent, with fumarate. A phenotype similar to that of a crp mutant, which lacks the ability to grow anaerobically with DMSO, fumarate, and Fe(III), was obtained when both cyaA and cyaC were deleted. Microarray analysis of gene expression in the crp and cyaC mutants revealed the involvement of both genes in the regulation of key respiratory pathways, such as DMSO, fumarate, and Fe(III) reduction. Additionally, several genes associated with plasmid replication, flagellum biosynthesis, and electron transport were differentially expressed in the cyaC mutant but not in the crp mutant. Our results indicated that CyaC plays a major role in regulating anaerobic respiration and may contribute to additional signaling pathways independent of CRP.
Shewanella oneidensis MR-1 is a metal reducer that uses more than 14 terminal electron acceptors for respiration. These electron acceptors include oxygen, nitrate, fumarate, dimethyl sulfoxide (DMSO), Fe(III) oxides, uranium, and chromium (21, 25, 26). In Escherichia coli and other bacteria, the shift from aerobic respiration to anaerobic respiration requires activation of the global transcriptional regulator FNR (11, 34). FNR is an oxygen-sensing protein that is activated under anaerobic conditions by the formation of a [4Fe-4S] cluster (14). The S. oneidensis FNR homolog, EtrA, complements an E. coli FNR mutant (27) but does not appear to have the same role as the E. coli protein in S. oneidensis (17). Our previous findings demonstrated that instead of EtrA, the cyclic AMP (cAMP) receptor protein (CRP) controls anaerobic respiration in S. oneidensis MR-1 (28). crp mutants are deficient in anaerobic respiration of Fe(III), Mn(IV), fumarate, nitrate, and DMSO. Furthermore, fumarate, DMSO, and nitrate reductase activities are either severely decreased or undetectable in the crp mutants, suggesting that CRP regulates the expression of these anaerobic reductases (28).
Although genetic and phenotypic data clearly have implicated CRP in the activation of anaerobic reductase systems in S. oneidensis MR-1, the mechanisms of this regulation remain unclear. CRP lacks obvious redox-sensing domains and is not expected to respond to changes in oxygen concentrations like FNR. Complementation of the S. oneidensis crp mutants with E. coli crp indicates that CRP is activated similarly in these two organisms. Furthermore, addition of cAMP to aerobic cultures of S. oneidensis MR-1 leads to significant induction of the activity of the anaerobic fumarate reductase (28). Therefore, transcriptional regulation by CRP under anaerobic conditions is likely to be directly linked to adenylate cyclase activity and cAMP production. In this paper, we investigate the role of the S. oneidensis adenylate cyclases in the regulation of anaerobic respiration. Genetic, biochemical, and genome-wide transcriptome analyses indicated that although MR-1 has three adenylate cyclases, the membrane-bound class III enzyme, CyaC, appears to play a more significant role in CRP-dependent anaerobic gene expression.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
A list of the bacterial strains and plasmids used in this study is given in Table 1. S. oneidensis and E. coli strains were routinely cultured in Luria-Bertani (LB) medium at 30°C and 37°C, respectively. Antibiotics (20 μg/ml chloramphenicol, 25 μg/ml kanamycin, and 20 μg/ml gentamicin) were added as needed. Anaerobic growth of S. oneidensis strains was performed in Hungate tubes filled with minimal medium (28) supplemented with 50 mM lactate and 0.02% Casamino Acids. Disodium fumarate, DMSO, and ferric citrate were used as electron acceptors at a final concentration of 10 mM.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| S. oneidensis strains | ||
| MR-1 | Lake Oneida isolate | 23 |
| SR660 | MR-1 ΔcyaA | This study |
| SR945 | MR-1 ΔcyaB | This study |
| SR672 | MR-1 ΔcyaC | This study |
| SR665 | MR-1 ΔcyaA ΔcyaC | This study |
| SR946 | MR-1 ΔcyaA ΔcyaB ΔcyaC | This study |
| SR694 | MR-1 Δcrp | This study |
| SR956 | SR946 with cyaA in pJB3Cm6, Cmr | This study |
| SR957 | SR946 with cyaB in pJB3Cm6, Cmr | This study |
| SR958 | SR946 with cyaC in pJB3Cm6, Cmr | This study |
| E. coli strains | ||
| EC100 | mcrA Δ(mrr-hsdRMS-mcrBC) recA1 | Epicenter Technologies |
| EC100D+ | E. coli EC100 derivative, pir+ | Epicenter Technologies |
| β2155 | pir::RP4, Kmr | 5 |
| SP850 | ΔcyaA1400 (::kan) | 31 |
| E299 | E. coli SP850 with pJB3Cm6, Kmr Cmr | This study |
| E300 | E. coli SP850 with cyaA in pJB3Cm6, Kmr Cmr | This study |
| E301 | E. coli SP850 with cyaC in pJB3Cm6, Kmr Cmr | This study |
| E347 | E. coli SP850 with Po-cyaB in pJB3Cm6, Kmr Cmr | This study |
| E456 | E. coli SP850 with cyaB in pJB3Cm6, Kmr Cmr | This study |
| Plasmids | ||
| pJB3Cm6 | Cloning and sequencing vector, Cmr | 2 |
| pDS3.1 | R6K ori GmrsacB | 38 |
| pPR9TT | Promoter probe vector, lacZ Cmr Apr | 30 |
| pKB1 | cyaC in pJB3Cm6, Cmr | This study |
| pMC2 | cyaA in pJB3Cm6, Cmr | This study |
| pMC3 | cyaB fused to operon promoter (Po) in pJB3Cm6 | This study |
| pMC4 | cyaB and 300 bp of upstream DNA in pJB3Cm6 | This study |
| pDS10 | dms promoter fused to lacZ in pPR9TT | This study |
| pDS11 | fccA promoter fused to lacZ in pPR9TT | This study |
| pDS12 | cyaC promoter fused to lacZ in pPR9TT | This study |
| pDS13 | cyaA promoter fused to lacZ in pPR9TT | This study |
For transcriptome-profiling experiments, O2-limited cultivation was selected primarily because of the inability of Δcrp mutants to grow anaerobically with fumarate or DMSO. Wild-type S. oneidensis MR-1 and mutant strains were grown under O2-limited conditions in chemostats using 6-liter Bioflow 3000 bioreactors (New Brunswick Scientific, Edison, NJ) containing 3 liters of minimal medium (pH 7.0) supplemented with 90 mM dl-lactate and 10 ml of 10× Wolfe's vitamin solution (13). The bioreactors were each inoculated with 1 ml/liter of an overnight LB medium-grown culture and maintained in a batch mode until the late logarithmic stage. Continuous cultures were initiated and maintained at a dilution rate of 0.06/h. The bioreactors were constantly sparged with gas (60% pure N2, 40% air) at a rate of 4 liters/min. Agitation and temperature were automatically maintained at 150 rpm and 30°C, respectively. The pH was maintained at 7.0 by automatic addition of 2 M HCl. The dissolved oxygen tension was constantly monitored using an Ingold polarographic oxygen electrode. The onset of O2 limitation was defined by a decrease in the dissolved O2 tension to a level below the detection threshold (≤1%). To induce the expression of anaerobic respiratory genes, continuous addition of sodium fumarate (final concentration, 10 mM) to the bioreactors was initiated at the onset of steady-state O2-limited growth. Samples used for RNA isolation and transcriptome analysis were taken after at least five volume changes under steady-state conditions.
Generation of cya and crp chromosomal deletions.
Chromosomal deletions of crp and the predicted adenylate cyclase genes were performed using a previously published method (38), with minor modifications. Approximately 1-kb upstream and downstream fragments flanking the target genes were amplified by PCR with Phusion polymerase (New England Biolabs, Ipswich, MA) using the primers listed in Table S1 in the supplemental material. The fragments were fused by overlap extension PCR (12). The fusion fragments generated complete deletions of the cyaC and crp genes and removed the internal 1,860 bp (bases 435 to 2295) and 430 bp (bases 154 to 585) of cyaA and cyaB, respectively. The fusion amplicons were ligated into SmaI-digested pDS3.1 (38). Plasmids with the desired inserts were used to transform E. coli β2155 (5) or WM3063 (29) and subsequently transferred to the S. oneidensis strains by conjugation. The primary integrants were selected by plating preparations on LB agar supplemented with 25 μg/ml gentamicin. To isolate mutants with cya or crp chromosomal deletions, transconjugants were grown on 10% tryptone-5% yeast extract agar plates supplemented with 5% sucrose. Sucrose-resistant colonies were screened for deletion of the genes by PCR.
Complementation of E. coli adenylate cyclase mutant.
DNA fragments carrying the complete cyaA and cyaC genes with upstream flanking regions were amplified by PCR using Phusion polymerase and the primers listed in Table S1 in the supplemental material. The fragments were cloned into SmaI-digested broad-host-range vector pJB3Cm6 (2) to generate plasmids pMC2 and pKB1. To clone cyaB, which appears to be part of a three-gene operon, we first amplified this gene using primers cyaBFN1 and cyaBRE (see Table S1 in the supplemental material) to introduce an NdeI site at the 5′ end of the gene. A second DNA fragment, Po, which contains the promoter region that lies upstream of cydDC, cyaB, and an NdeI site at the 3′ end, was amplified using primers cyaBPROF and cyaBProR1. The two fragments (cyaB and Po) were digested with NdeI, ligated, and cloned into SmaI-digested pJB3Cm6 to generate pMC3. We also cloned cyaB and 300 bp of DNA directly upstream of this gene to generate pMC4. Plasmids pKB1, pMC2, pMC3, and pMC4 were transferred by electroporation into E. coli SP850 (obtained from the E. coli Genetic Stock Center, Yale University, New Haven, CT). This strain is deficient in lacZ expression because it lacks the adenylate cyclase needed for cAMP production and cannot activate CRP (31). To test for β-galactosidase activity, E. coli SP850 cells were grown in LB medium to early mid-log phase in the presence or absence of 3 mM cAMP. Isopropyl-β-d-thiogalactopyranoside (IPTG) was added at a concentration of 1 mM, and the cells were incubated for an additional hour. E. coli SP850 cells carrying the S. oneidensis cyaA, cyaB, or cyaC gene were grown in a similar manner but without addition of cAMP. β-Galactosidase activity was measured as described previously (19).
Localization of CyaA and CyaC.
Antibodies against the CyaA and CyaC peptides (N′-LDEENQLSHYVQSGSDMSELV and N′-ALAGRLDNPDSAYRLTNLGP, respectively) were generated by Protein Tech Group, Inc. (Chicago, IL). Membrane and soluble fractions were obtained from wild-type and mutant cells grown either aerobically or anaerobically as described previously (1). Proteins were separated on 10% or 4 to 20% sodium dodecyl sulfate-polyacrylamide gels (Pierce, Rockford, IL), transferred to a polyvinylidene difluoride membrane, and then incubated with CyaA or CyaC antibodies. Blots were developed using Supersignal West Pico chemiluminescent substrate (Pierce, Rockford, IL).
Enzyme activity and heme staining.
Wild-type S. oneidensis and mutant strains were grown anaerobically with fumarate or DMSO for 2 h. Cells were pelleted and then lysed using B-PER (Pierce, Rockford, IL). Total protein concentrations were measured using a BCA protein assay kit (Pierce, Rockford, IL). Enzyme activity was detected on native polyacrylamide gels as previously described using methyl viologen as the artificial electron donor and 10 mM DMSO as the electron acceptor (28). Heme staining of proteins in sodium dodecyl sulfate-polyacrylamide gels was performed using 3,3′,5,5′-tetramethylbenzidine dihydrochloride as described previously (37).
Generation of promoter-lacZ fusions.
DNA fragments directly upstream of the DMSO reductase operon (dmsEFAB; SO_1427 to SO_1430), the fumarate reductase gene (fccA; SO_0970), cyaA, and cyaC were amplified by PCR using the primers listed in Table S1 in the supplemental material and Phusion polymerase (New England Biolabs). The resulting 597-, 526-, 321-, and 207-bp fragments were digested with HindIII and BamHI and then cloned into pPR9TT (30) that carried a promoterless lacZ gene. Following transformation of E. coli β2155 (5), the plasmids were transferred into S. oneidensis wild-type and mutant strains by conjugation as described above. Recombinant strains were grown anaerobically for 2 h with either 10 mM fumarate or 10 mM DMSO in minimal medium supplemented with 50 mM lactate and 0.02% Casamino Acids. For aerobic growth, 10-ml cultures were grown in 500-ml flasks with vigorous shaking for 2 h. Cultures were assayed for β-galactosidase activity as described previously (19).
Microarray analysis.
Microarray expression profiling of S. oneidensis wild-type strain MR-1 and mutants was carried out using whole-genome Affymetrix custom expression arrays (Affymetrix, Inc., Santa Clara, CA) comprised of 103,797 11-μm synthesis features. The design used contains both antisense oligonucleotides and a complementary set of single-base-mismatched oligonucleotides to facilitate detection of background signals. Total cellular RNA from cultures grown in chemostats, as described above, was isolated using the TRIzol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. RNA samples were treated with RNase-free DNase I (Ambion, Inc., Austin, TX) to digest residual DNA and purified using a mini RNeasy kit (Qiagen, Inc., Valencia, CA). The concentrations and purity of the RNA samples were determined by spectrophotometric measurement at 260 and 280 nm, respectively. Generation of cDNA using reverse transcriptase, cDNA fragmentation, labeling, and hybridization were performed according to the instructions in the manufacturer's GeneChip expression analysis technical manual. Scanning of the hybridized microarrays was performed using an Affymetrix GeneChip 3000 scanner. Probe-level intensity values from Affymetrix CEL files were processed using the MAS5 and RMA algorithms to compute gene expression values as described previously (6, 7). Using the RMA approach, the mean expression for each gene was compared with a background mean for all other treatments. Using the difference of means, along with a pooled estimate of variance, the significance of up- or downregulation (expressed as a P value) was computed using a two-sample t test statistic (6). Genes that exhibited a change in expression in ΔcyaC or Δcrp mutants are listed in Tables S2 to S5 in the supplemental material.
Microarray accession number.
TheAffymetrix CEL files generated in our experiments have been deposited in the NCBI Gene Expression Ominbus microarray database (http://ncbi.nlm.nih.gov/geo/) under accession number GSE15657.
RESULTS
Functional analysis of S. oneidensis MR-1 adenylate cyclase genes.
The S. oneidensis genome (http://www.tigr.org) contains three putative adenylate cyclase genes. Two of these genes are designated cyaA and cyaB (SO_4312 and SO_3778, respectively), while the third is annotated as a putative adenylate cyclase gene (SO_1329) and is designated cyaC in this paper. CyaA is predicted to be a cytoplasmic protein consisting of 804 amino acids. It is similar to the bacterial class I adenylate cyclases and shares 30% identity and 49% similarity with E. coli CyaA. CyaB is predicted to be a soluble protein consisting of 194 amino acids with 69% identity to the class IV adenylate cyclase of Aeromonas hydrophila (33). The third adenylate cyclase, CyaC, consists of 335 amino acids and contains three putative membrane-spanning domains, suggesting that it is anchored to the cytoplasmic membrane. The C terminus of CyaC exhibits significant degrees of identity (33 to 39%) and similarity (54 to 65%) to the catalytic domain of the class III adenylate cyclase from Rhizobium etli (gi: 21616131; 35) and to putative class III adenylate cyclases from Psychroflexus torquis and Flavobacterium species. The predicted membrane-spanning domain of CyaC did not exhibit similarity to other sequences in the databases outside the genus Shewanella.
To confirm the predicted function of CyaA, CyaB, and CyaC, plasmids carrying the corresponding S. oneidensis MR-1 genes (Table 1) were used to complement E. coli SP850, which lacks the adenylate cyclase gene and cannot induce lacZ expression. This deficiency can be overcome by addition of cAMP to the growth medium or by introduction of a functional cya gene (35). Introduction of pKB1, which carries cyaC and 327 bp of upstream DNA, and pMC2, which carries cyaA and 200 bp of upstream DNA, into SP850 led to restoration of β-galactosidase expression in this strain. The levels of β-galactosidase activity in SP850 complemented with plasmids carrying cyaA and cyaC were ∼1,035 and 1,058 Miller units, respectively (Table 2). Similar levels of β-galactosidase activity were obtained when SP850 was complemented by addition of exogenous cAMP to the growth medium (Table 2), indicating that both genes encode adenylate cyclases, as predicted by sequence analysis.
TABLE 2.
β-Galactosidase activity of an E. coli cyaA mutant (strain SP850) complemented with S. oneidensis cya genes
| Strain | β-Galactosidase activity (Miller units)a |
|---|---|
| E. coli SP850 | 26.0 ± 10.5 |
| SP850 + pJB3Cm6 | 31.7 ± 4.2 |
| SP850 + cAMP | 1,284.6 ± 143.8 |
| SP850 + S. oneidensis cyaA | 1,034.9 ± 266.7 |
| SP850 + S. oneidensis cyaC | 1,057.8 ± 281.0 |
| SP850 + S. oneidensis Po-cyaB | 747.6 ± 80.6 |
| SP850 + cyaB | 10.7 ± 1.6 |
The data are the means ± standard deviations of three independent experiments performed in duplicate.
To determine the role of cyaB in cAMP production, two plasmids were generated. pMC4 carries cyaB and 300 bp of upstream DNA in cydC which is 25 bp upstream of cyaB, while pMC3 carries cyaB fused to 360 bp of DNA upstream of cydDC (designated Po as described in Materials and Methods). Since cyaB is located downstream of cydDC and is separated from this operon by 25 bp, generation of the two plasmids served to identify the promoter region of cyaB and determine its involvement in cAMP production. Introduction of pMC3 into E. coli SP850 restored β-galactosidase activity (Table 2), indicating that cyaB encodes an adenylate cyclase, as predicted. Introduction of pMC4, however, failed to complement SP850, suggesting that the DNA directly upstream of cyaB lacks a promoter and that cyaB is part of the cydDC-cyaB operon.
Cellular localization of CyaA and CyaC.
Antibodies against CyaA and CyaC peptides were used to determine the locations of these proteins in S. oneidensis MR-1 cellular subfractions. A band that reacted with the CyaA antiserum and corresponded to the predicted size of CyaA (∼ 90 kDa) was detected in the soluble fraction of wild-type strain MR-1 but not in the soluble fraction of the cyaAC mutant (Fig. 1). When antibodies raised against the CyaC peptide were used, a band at ∼37 kDa, which corresponded to the predicted size of CyaC, was detected in the membrane fraction of the wild type but not in the cyaAC mutant fraction (Fig. 1). These results are in agreement with the initial sequence analysis that predicted cytoplasmic and membrane locations of CyaA and CyaC, respectively.
FIG. 1.
Localization of CyaA (top blot) and CyaC (bottom blot) by Western blot analysis of soluble (lanes S) and membrane (lanes M) fractions of wild-type strain MR-1 and the cyaAC double mutant (A−C−). A band that reacted with CyaA antibodies was detected in the soluble fraction of the wild type but not in the mutant fractions. A band that reacted with the CyaC antibodies was detected in the membrane fraction of the wild type but not in the mutant fractions.
Effect of adenylate cyclases on anaerobic growth.
To further elucidate the role of adenylate cyclases in anaerobic respiration of S. oneidensis MR-1, we generated mutants with single, double, and triple chromosomal deletions of cyaA, cyaB, and cyaC. The mutants were tested for anaerobic respiration with fumarate, DMSO, and Fe(III) citrate as electron acceptors. Loss of either cyaA or cyaB did not affect the ability of the mutants to use any of these electron acceptors (data not shown). However, significant deficiencies were observed for the ΔcyaC mutant that lacked the membrane-bound class III adenylate cyclase. When DMSO was used as the electron acceptor, a lag phase of 10 to 12 h was routinely observed for the ΔcyaC mutant, compared to a ∼5 h-lag for the wild type (Fig. 2A). To determine if growth of this mutant with DMSO after the long lag phase was due to suppressor mutations, stationary-phase cells from the cultures were grown aerobically in LB medium and then retested for anaerobic growth in minimal medium with DMSO as described above. Significant changes in the duration of the lag phase were not observed (data not shown), indicating that growth of the ΔcyaC mutant after a long lag phase was not due to accumulation of suppressor mutations.
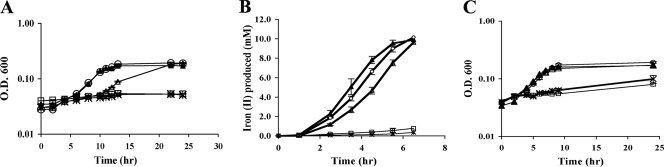
FIG. 2.
Anaerobic respiration with DMSO (A), Fe+3 (B), and fumarate (C) by wild-type strain MR-1 (○), the ΔcyaC mutant (⧫), the ΔcyaA ΔcyaC mutant (▵), and the Δcrp mutant (▪). The cyaC mutant was deficient in anaerobic respiration with DMSO and Fe(III). The cyaAC double mutant was deficient in anaerobic respiration with the three electron acceptors, and its phenotype mimicked the phenotype of the crp mutant. The data are the means and standard deviations of three independent experiments. O.D. 600, optical density at 600 nm.
In addition to DMSO growth deficiencies, the cyaC mutant was deficient in Fe(III) reduction (Fig. 2B). At 4 h, ∼6.7 mM Fe(II) and 3.2 mM Fe(II) were produced by the wild type and the ΔcyaC mutant, respectively. When fumarate was used as the electron acceptor, the ΔcyaC mutant exhibited a slight deficiency in growth (Fig. 2C), but this deficiency was not as significant as the deficiency observed when DMSO or Fe(III) was used as the electron acceptor.
More pronounced differences in anaerobic growth with DMSO, fumarate, and Fe(III) were observed for the cyaAC double and cyaABC triple mutants. These two mutants exhibited similar deficiencies in anaerobic respiration with DMSO, fumarate, and Fe(III) (Fig. 2 and Fig. 3). The phenotype of the cyaAC double mutant was essentially identical to the phenotype of the Δcrp mutant (Fig. 2) (28). This suggests that CyaA and CyaC are both involved in CRP activation under anaerobic conditions, while CyaB does not appear to contribute significantly to this process.
FIG. 3.
Complementation of the cya triple mutant with cyaA, cyaB, or cyaC. The mutant (□) was deficient in anaerobic respiration with DMSO (A), Fe(III) citrate (B), and fumarate (C). Introduction of cyaC (○) restored the ability of the triple mutant to grow with these electron acceptors. Introduction of cyaA (▵) led to wild-type levels of growth only when fumarate was used as the electron acceptor. Introduction of cyaB (*) into the mutant did not restore anaerobic respiration with any of the electron acceptors. The data are the means and standard deviations of three independent experiments. O.D. 600, optical density at 600 nm.
Complementation of cya triple mutant with cyaC.
To determine if CyaC is sufficient for CRP activation under anaerobic conditions or if other adenylate cyclases are essential for this process, we introduced cyaA, cyaB, and cyaC on plasmids into the cya triple mutant. Introduction of cyaB did not restore anaerobic respiration with any of the electron acceptors tested (Fig. 3), further confirming that this gene does not have a significant role in anaerobic respiration. Interestingly, the expression of cyaA alone restored anaerobic growth with fumarate to levels similar to the wild-type levels, although the growth with DMSO and the reduction of Fe(III) were slow (Fig. 3) and similar to those of the ΔcyaC mutant (Fig. 2). In contrast, expression of cyaC in the cya triple mutant restored the ability to grow with fumarate and DMSO and the ability to reduce Fe(III) like the wild type (Fig. 3). These results clearly demonstrate that the class III membrane-bound CyaC protein was sufficient for anaerobic reductase gene expression in the absence of other cAMP-producing enzymes.
To begin to understand the reason for the partial redundancy of CyaA and CyaC, we used cyaA and cyaC promoter-lacZ fusions to measure the relative activities of the promoters in wild-type S. oneidensis under different growth conditions. The presence of the cyaA and cyaC promoters resulted in ∼23 to 25 Miller units of β-galactosidase activity when the cells were grown aerobically. Under anaerobic conditions with fumarate, the β-galactosidase activity increased almost threefold (62.4 ± 5 Miller units) in cells carrying the cyaC promoter-lacZ fusion. Surprisingly, the β-galactosidase activity decreased twofold (9.7 ± 1.1 Miller units) in fumarate-grown cells carrying the cyaA promoter-lacZ fusion plasmid. These results suggest that CyaC may be the dominant adenylate cyclase under anaerobic conditions, which may explain the observation that its presence is sufficient for anaerobic respiration with several electron acceptors.
Microarray analysis of genes regulated by CyaC and CRP.
The results described above indicate that the phenotype of the crp mutant is reproduced when both cyaA and cyaC are absent, while loss of cyaC alone results in a partial deficiency in anaerobic respiration. To further explore the roles of CyaC and CRP in the regulation of anaerobic respiration, we compared the genome-wide transcriptome profiles of ΔcyaC and Δcrp mutants to those of wild-type strain MR-1. Given the growth deficiencies displayed by the Δcrp and ΔcyaC mutants under anaerobic conditions, all strains were cultured in chemostats under O2-limited conditions. Since both the Δcrp and ΔcyaC mutants are not impaired in aerobic respiration, oxygen-limited conditions provided the means to maintain equal growth rates (0.06 h−1) for the mutant and wild-type strains, thus eliminating the effect of growth on the overall transcriptome profiles. At the same time, while growing aerobically, the cells decreased the dissolved O2 concentration in the medium to levels below the detection threshold (dissolved O2 tension, ≤1%), allowing expression of anaerobic respiratory genes. The expression of these genes was confirmed by the reduction of fumarate by the wild-type culture, which produced stoichiometric amounts of succinate under O2-limited conditions. The optical densities at 600 nm of the wild-type, ΔcyaC, and Δcrp strains in chemostat cultures were 0.50, 0.45, and 0.40, respectively.
Complete lists of genes that exhibited significant changes in expression in the Δcrp or ΔcyaC mutant are shown in Tables S2 to S5 in the supplemental material. Genes primarily predicted or known to be involved in electron transport and respiration are listed in Tables 3, 4, and 5. Of 5,066 genes represented on the microarray, 280 exhibited an increase or decrease in the expression level in the Δcrp mutant but not in the ΔcyaC mutant. These genes included genes involved in heme biosynthesis (hemH-1, hemB, hemG-3, and hemB-2), whose expression was significantly reduced in the Δcrp mutant (Table 3). The expression data for these genes were consistent with the observed lack of pigmentation of Δcrp colonies compared to the orange wild-type colonies (data not shown). Orange pigmentation is attributed to the large amount of c cytochromes produced by S. oneidensis wild-type strain MR-1 (3). Heme staining further confirmed the absence of a large number of c cytochromes in the Δcrp mutant (Fig. 4A). It should be noted that the genome sequence of S. oneidensis contains redundant homologues of several heme biosynthesis genes, and the crp mutant does not appear to be completely deficient in heme production. Therefore, it is likely that the significant loss of c cytochromes in the Δcrp mutant is due to a combination of reduced heme synthesis and downregulation of the structural c-type cytochrome genes.
TABLE 3.
Genes with altered expression profiles in the Δcrp mutant compared to the wild type for cells grown under O2-limited conditions
| Locus | Gene | Function or description | Δcrp mutant/wild type ratioa |
|---|---|---|---|
| Upregulated genes | |||
| Heme biosynthesis | |||
| SO_0027 | hemG-2 | Protoporphyrinogen oxidase | 0.05 |
| SO_0435 | hemE | Uroporphyrinogen decarboxylase | 0.22 |
| SO_2109 | hemH-1 | Ferrochelatase | 0.34 |
| SO_2587 | hemB-1 | Delta-aminolevulinic acid dehydratase | 0.20 |
| SO_3720 | hemG-3 | Protoporphyrinogen oxidase | 0.30 |
| SO_4208 | hemB-2 | Delta-aminolevulinic acid dehydratase | 0.25 |
| Respiratory functions | |||
| SO_0396 | frdC | Quinol:fumarate reductase | 0.13 |
| SO_0848 | napA | Periplasmic nitrate reductase | 0.30 |
| SO_0902 | nqrA | Na-translocating NADH-quinone reductase subunit A | 0.21 |
| SO_0903 | nqrB | Na-translocating NADH-quinone reductase subunit B | 0.14 |
| SO_0904 | nqrC | Na-translocating NADH-quinone reductase subunit C | 0.11 |
| SO_0970 | fccA | Periplasmic fumarate reductase | 0.14 |
| SO_2912 | pflB | Pyruvate formate lyase | 0.17 |
| SO_2913 | pflA | Pyruvate formate lyase 1 activating subunit | 0.20 |
| SO_3922 | fdh | Formate dehydrogenase, cytochrome b | 0.32 |
| SO_4591 | cymA | Membrane-anchored tetraheme c cytochrome | 0.22 |
| Regulatory functions | |||
| SO_0490 | Transmembrane transcriptional regulator | 0.19 | |
| SO_0940 | Transmembrane transcriptional regulator | 0.07 | |
| SO_1415 | Transcriptional regulator, TetR family | 0.12 | |
| SO_3059 | Sigma 54 transcriptional regulator, Fis family | 0.28 | |
| SO_3538 | hlyU | Transcriptional regulator | 0.06 |
| SO_3627 | Transcriptional regulator, TetR family | 0.14 | |
| Downregulated genes | |||
| SO_0852 | Type IV pilin, putative | 4.76 | |
| SO_1329 | cyaC | Adenylate cyclase | 2.08 |
| SO_4312 | cyaA | Adenylate cyclase | 3.97 |
| SO_4609 | coxC | aa3-type cytochrome c oxidase, subunit III | 2.68 |
Ratio of the mean hybridization signal intensity for the crp mutant to the mean hybridization signal intensity for the wild type. Values greater than 1 indicate an increase in the mRNA level in the mutant; values less than 1 indicate a decrease.
TABLE 4.
Genes that exhibited decreased or increased expression in Δcrp and ΔcyaC mutants compared to the wild type for cells grown under O2-limited conditions
| Locus | Gene | Function or description | Δcrp mutant/wild type ratioa | ΔcyaC mutant/wild type ratioa |
|---|---|---|---|---|
| CRP- and CyaC-upregulated genes | ||||
| SO_0827 | Lactate transport protein | 0.03 | 0.23 | |
| SO_1422 | ifcR | Fe(III)-induced transcriptional regulator | 0.12 | 0.29 |
| SO_1427 | dmsE | Cytochrome c component of DMSO reductase | 0.08 | 0.38 |
| SO_1428 | dmsF | Outer membrane protein | 0.07 | 0.25 |
| SO_1429 | dmsA | Outer membrane DMSO reductase | 0.09 | 0.23 |
| SO_1430 | dmsB | FeS subunit of DMSO reductase | 0.13 | 0.12 |
| SO_1431 | dmsG | DMSO reductase chaperone | 0.14 | 0.16 |
| SO_1432 | dmsH | Conserved hypothetical protein | 0.24 | 0.22 |
| SO_1776 | mtrB | Outer membrane protein, metal reduction | 0.06 | 0.20 |
| SO_1777 | mtrA | Decaheme c cytochrome, metal reduction | 0.05 | 0.31 |
| SO_1778 | mtrC | Outer membrane c cytochrome, metal reduction | 0.04 | 0.31 |
| SO_1779 | omcA | Outer membrane c cytochrome, metal reduction | 0.09 | 0.24 |
| SO_3285 | cydB | Cytochrome d ubiquinol oxidase | 0.14 | 0.20 |
| SO_3286 | cydA | Cytochrome d ubiquinol oxidase | 0.12 | 0.19 |
| CRP- and CyaC-downregulated genes | ||||
| SO_3503 | nagP | N-Acetylglucosamine transporter | 6.50 | 2.58 |
| SO_3504 | nagX | N-Acetylglucosamine-related transporter | 10.06 | 8.00 |
| SO_3505 | nagA | N-Acetylglucosamine-6-phosphate deacetylase | 14.42 | 8.28 |
| SO_3506 | nagB | Glucosamine-6-phosphate deaminase | 27.47 | 17.15 |
| SO_3507 | nagK | N-Acetylglucosamine kinase | 23.75 | 14.83 |
| SO_3509 | hexB | Beta-N-acetylhexosaminidase | 1.00 | 1.00 |
| SO_3510 | Methyl-accepting chemotaxis protein | 10.27 | 6.06 | |
| SO_3513 | Tryptophan halogenase | 9.71 | 5.06 |
Ratio of the mean hybridization signal intensity for the crp or cyaC mutant to the mean hybridization signal intensity for the wild type. Values greater than 1 indicate an increase in the mRNA level in the mutant; values less than 1 indicate a decrease.
TABLE 5.
Genes with altered expression profiles in the ΔcyaC mutant compared to the wild type for cells grown under O2-limited conditions
| Locus | Gene | Function or description | ΔcyaC mutant/wild type ratioa |
|---|---|---|---|
| Genes upregulated by CyaC | |||
| SO_0026 | Transcriptional regulator | 0.34 | |
| SO_0268 | ccmH-1 | Cytochrome c biogenesis protein | 0.33 |
| SO_0269 | Cytochrome c biogenesis thioredoxin | 0.28 | |
| SO_0282 | pilN | Type IV pilus biogenesis protein | 0.26 |
| SO_0521 | Monooxygenase domain protein | 0.28 | |
| SO_1215 | ompK | Outer membrane protein | 0.33 |
| SO_1490 | adhB | Alcohol dehydrogenase II | 0.35 |
| SO_1824 | TonB system biopolymer transport component | 0.34 | |
| SO_2361 | ccoP | cbb3-type cytochrome c oxidase, subunit III, CcoP | 0.26 |
| SO_2362 | ccoQ | cbb3-type cytochrome c oxidase, subunit IV, CcoQ | 0.21 |
| SO_2363 | ccoO | cbb3-type cytochrome c oxidase, subunit II, CcoO | 0.15 |
| SO_2364 | ccoN | cbb3-type cytochrome c oxidase, subunit I, CcoN | 0.21 |
| SO_2942 | Prophage lambda So transcriptional regulator | 0.31 | |
| SO_2993 | Prophage lambda type II DNA methyltransferase | 0.16 | |
| SO_3146 | DNA-binding protein, H-NS family | 0.29 | |
| SO_3211 | flhG | Flagellar biosynthesis protein | 0.21 |
| SO_3212 | flhF | Flagellar biosynthesis protein | 0.31 |
| SO_3213 | flhA | Flagellar biosynthesis protein | 0.29 |
| SO_3221 | fliM | Flagellar motor switching | 0.30 |
| SO_3223 | fliK | Flagellar hook length control protein | 0.18 |
| SOA_0001 | repA | Plasmid replication protein | 0.10 |
| Genes downregulated by CyaC | |||
| SO_0312 | Outer membrane porin | 3.06 | |
| SO_0630 | nosA | TonB-dependent outer membrane protein | 6.87 |
| SO_2318 | cheY | Chemotaxis response regulator | 5.97 |
| SO_3687 | csgE | Curli secretion protein | 6.67 |
Ratio of the mean hybridization signal intensity for the cyaC mutant to the mean hybridization signal intensity for the wild type. Values greater than 1 indicate an increase in the mRNA level in the mutant; values less than 1 indicate a decrease.
FIG. 4.
(A and B) Heme (A) and Coomassie blue staining (B) of polyacrylamide gels with cell extracts from wild-type and mutant strains. The arrow indicates a heme-stained band that was overexpressed in the cyaC mutant. Twenty micrograms of protein was loaded in each lane. (C) DMSO reductase activity staining using methyl viologen as the electron donor. Similar levels of DMSO reductase activity (arrow) were detected in the wild type and the cyaA mutant. The levels were significantly reduced in the cyaC, cyaAC, and crp mutants. Twenty micrograms of protein was loaded in each lane.
Other genes that appeared to be positively regulated by CRP include SO_0970 and SO_0848, which encode the terminal fumarate and nitrate reductases, respectively, as well as SO_4591, which encodes the c cytochrome CymA (Table 3). CymA is needed for electron transfer to the fumarate, nitrate, and metal terminal reductases (24). Since loss of CymA affects anaerobic growth with fumarate, we tested the expression levels of the fumarate reductase gene (fccA; 28) in the wild-type, Δcya, and Δcrp strains using plasmids that carry the fccA promoter fused to a promoterless lacZ gene. The levels of β-galactosidase activity were reduced in the Δcrp mutant compared to the wild type (Table 6), in agreement with the microarray data that showed a lower level of fccA expression in the mutant. These results confirmed that the deficiency in anaerobic growth with fumarate is due to loss of the terminal reductase and not simply due to a lack of required intermediate electron transport chain components. We have previously shown that crp mutants of S. oneidensis lack fumarate and nitrate reductase activities, using methyl viologen as an electron donor (28). The results presented here are in agreement with these findings. CRP also appears to positively control the expression of genes that encode regulatory proteins, such as SO0490, SO0940, and SO3538. These proteins were shown to be overexpressed in response to cold shock (9), but their roles are not known.
TABLE 6.
β-Galactosidase activities for dms and fccA promoter-lacZ fusions under anaerobic conditions in wild-type and mutant strains of S. oneidensis MR-1
| Strain | β-Galactosidase activity (Miller units)a
|
|
|---|---|---|
| dms promoterb | fccA promoterc | |
| MR-1 | 237.1 ± 23.9 | 341 ± 6.2 |
| ΔcyaC mutant | 61.9 ± 1.7 | 242.8 ± 5.2 |
| ΔcyaAC mutant | 19.9 ± 0.7 | 55.9 ± 1.4 |
| Δcrp mutant | 25.0 ± 5.3 | 71.4 ± 1.8 |
The β-galactosidase activity in MR-1 carrying pPR9TT without an insert was 10 Miller units. The data are means ± standard deviations of three independent experiments.
Cultures were incubated anaerobically with DMSO for 2 h.
Cultures were incubated anaerobically with fumarate for 2 h.
In addition to positive regulation, microarray data suggested that CRP negatively regulates the expression of some genes, such as cyaA and cyaC (Table 3; see Table S3 in the supplemental material). The expression levels of cyaA and cyaC were increased in the Δcrp mutant compared to the wild type. These results were confirmed by β-galactosidase activity measurements for lacZ-promoter fusions. The β-galactosidase activity in wild-type cells carrying the cyaC promoter-lacZ fusion plasmid and incubated anaerobically with fumarate was 62.4 ± 5.0 Miller units. The β-galactosidase activity in the Δcrp mutant was 101 ± 2.1 Miller units. A more significant increase was observed for the Δcrp mutant when the cyaA promoter was fused to lacZ. The β-galactosidase activity in wild-type strain MR-1 cells carrying the fusion plasmid and incubated anaerobically with fumarate was 9.7 ± 1.1 Miller units, and the activity was 95.9 ± 0.7 Miller units in the Δcrp mutant under the same conditions. These data confirm the role of CRP in the negative regulation of cyaA and cyaC expression. This situation is similar to the situation in E. coli, where CRP negatively regulates the expression of cyaA (20).
Analysis of microarray data identified 77 genes that appeared to depend on both CRP and CyaC for proper expression. The transcript levels of the DMSO and metal reductase genes (SO_1427 to SO_1432, which encode DmsEFABGH, and SO_1776 to SO_1779, which encode MtrB, MtrA, MtrC, and OmcA) were reduced in the Δcrp and ΔcyaC mutants (Table 4). These data are in agreement with the results of a phenotypic analysis of these mutants. The downregulation of dms genes was further tested using a promoterless lacZ gene fused to the dms promoter. The β-galactosidase activity in these strains confirmed that the expression of the DMSO reductase operon was significantly reduced in both mutants (Table 6). We also tested the levels of DMSO reductase activity in the wild-type and mutant strains using methyl viologen as the electron donor. DMSO reductase activity was either severely diminished or absent in ΔcyaC, ΔcyaAC, and Δcrp mutant cell extracts (Fig. 4C), while wild-type activity levels were present in ΔcyaA mutant cell extracts. These results confirm the major role of CyaC in addition to CRP in regulation of the expression of the DMSO reductase.
Comparison of gene expression profiles in the crp and cyaC mutants identified 557 genes that appeared to be regulated by CyaC but not by CRP. Genes that encode cbb3-type cytochrome c oxidase subunits and flagellum biosynthesis proteins, as well as genes that encode phage-related proteins (Table 5), were downregulated in the ΔcyaC mutant. Similarly, genes involved in cytochrome c biosynthesis, such as ccmH-1, exhibited lower levels of expression in the ΔcyaC mutant than in wild-type strain MR-1 and the Δcrp mutant (Table 5). Heme staining of ΔcyaC mutant cell extracts indicated that there was a reduction in the number of proteins that possess peroxidase activity compared to the number in the wild type, suggesting that there were reduced levels of c cytochromes in the mutant (Fig. 4A). As observed for the heme biosynthesis genes described above, there are redundant copies of the cytochrome c maturation genes in the S. oneidensis genome, which may explain the incomplete loss of c cytochromes in the ΔcyaC mutant. Transcriptome analysis and heme staining of cell extracts suggested that CyaC may negatively regulate the expression of some genes (Table 5 and Fig. 4A). A band with increased peroxidase activity was detected in cell extracts of the ΔcyaC mutant but not in extracts of the wild type or the Δcrp mutant. These results strongly imply that CyaC may have other roles in addition to CRP activation.
DISCUSSION
The role of proteins that belong to the FNR-CRP family of global regulators has been extensively studied in many bacteria. Generally, the oxygen-sensing members of this family, such as E. coli FNR, are activated by anaerobiosis and play a major role in regulating anaerobic respiration. Contrary to expectations, S. oneidensis uses CRP instead of FNR to regulate anaerobic respiration (28). This finding, however, does not explain how cells sense redox changes in their environment. The S. oneidensis CRP lacks redox-sensing domains and appears to be activated solely by cAMP (22, 28). This conclusion is further supported by the observation that addition of cAMP to aerobically growing S. oneidensis cells results in anaerobic levels of fumarate reductase activity (28). This suggests that although CRP is a major regulator of anaerobic respiration in S. oneidensis, additional oxygen-sensing proteins must be involved. These proteins, such as ArcA (10), may function downstream of CRP and participate in activation of anaerobic gene expression, or they may function upstream of CRP and play a role in its activation. Since cAMP is needed for CRP activation, we investigated the role of three S. oneidensis adenylate cyclases in CRP activation and anaerobiosis. Although these three cyclases were capable of activating CRP in E. coli, only two enzymes, CyaA and CyaC, were needed for CRP activation under anaerobic conditions in S. oneidensis. The membrane-bound class III cyclase, CyaC, had a prominent role in this process since it was able to fully restore anaerobic respiration to a cya triple mutant.
We used gene expression profiling to determine the connection between CyaC and CRP activation. As expected, several genes had similar changes in their expression patterns in the ΔcyaC and Δcrp mutants, including the DMSO reductase genes, whose expression was reduced in both mutants. The expression of more than 200 genes, however, was altered only in the Δcrp mutant. One example of these genes is fccA, which encodes the terminal fumarate reductase (28). Anaerobic growth of the Δcrp mutant with fumarate was severely impaired, while that of the ΔcyaC and ΔcyaA mutants was barely affected. Similarly, fccA promoter activity was greatly diminished only in the Δcrp mutant. These observations indicate that although CyaC and CyaA contribute to cAMP production and CRP activation, they are not completely redundant. This may be due to the relative abundance and activity of the two enzymes, as well as nature of the cAMP-CRP-regulated promoters. Assays of promoter-lacZ fusions in wild-type strain MR-1 indicated that cyaC was expressed at higher levels than cyaA in anaerobic cells. Based on these results, we expect cAMP production to be higher in the ΔcyaA mutant than in the ΔcyaC mutant, which may lead to increased levels of cAMP-CRP. The long lag phase observed for the cyaC mutant when it was grown with DMSO may be a reflection of the time required to reach optimum cAMP levels in the absence of CyaC.
One of the most surprising results of this study was the finding that a large number of genes appeared to be regulated by CyaC but not by CRP. These genes included a large number of prophage and plasmid-related genes, as well as genes involved in cytochrome c maturation, regulation, and electron transfer. These results point to additional roles for CyaC in the regulation of gene expression that may be independent of CRP activation. It is not clear at present what these roles are, but it is possible that additional cAMP-binding proteins, yet to be identified, may rely on CyaC for activation.
Although class III adenylate cyclases have been studied mostly in eukaryotes, they are beginning to receive considerable attention in prokaryotes. Several bacteria use class III adenylate cyclases to respond to osmotic stress, pH, light, and other environmental stimuli (16, 32). In Pseudomonas aeruginosa, the membrane-bound CyaB protein is involved in regulation of virulence genes and appears to respond to calcium limitation (39), and in Myxococcus xanthus cyaA plays a role in signal transduction during osmotic stress (15). Analysis of bacterial genomes has identified several membrane-bound class III adenylate cyclases in Bradyrhizobium japonicum, Sinorhizobium meliloti, and Mycobacterium tuberculosis (18, 32). One M. tuberculosis adenylate cyclase, Rv1264, is a pH-sensing enzyme (8, 36), while another, Rv1319, is regulated by bicarbonate ions (4). In S. oneidensis MR-1 and Shewanella sp. strain ANA-3 (22), the class III adenylate cyclases are involved in the regulation of anaerobic respiration, and a shift from aerobic to anaerobic conditions resulted in increased expression of cyclase genes in both organisms. It is interesting that class III adenylate cyclases in bacteria are proving to be versatile enzymes that respond to different stimuli based on the lifestyle of the organism (32). The results presented in this paper and the recently published reports of Murphy et al. (22) suggest that the Shewanella class III enzymes may represent a novel group of adenylate cyclases that may, directly or indirectly, sense and respond to anaerobiosis.
Supplementary Material
Acknowledgments
We gratefully acknowledge Michael Driscoll, Elissa Cosgrove, and Timothy Gardner for help with the microarray expression analysis. We thank Mark McBride and Patrick Trewitt for valuable discussions and critical reading of the manuscript.
This work was supported by a grant from the National Science Foundation (grant MCB 0543501), by the Office of Science (BER), U.S. Department of Energy (DOE) grant DE-FG02-07ER643820, and by the DOE Genomics:GTL Program via the Shewanella Federation consortium. Pacific Northwest National Laboratory is operated for the DOE by Battelle Memorial Institute under contract DE-AC05-76RLO 1830.
Footnotes
Published ahead of print on 24 April 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Beliaev, A., D. Saffarini, J. McLaughlin, and D. Hunnicut. 2001. MtrC, an outer membrane decaheme c cytochrome required for metal reduction in Shewanella putrefaciens MR-1. Mol. Microbiol. 39722-730. [DOI] [PubMed] [Google Scholar]
- 2.Blatny, J. M., T. Brautaset, H. C. Winther-Larsen, K. Haugan, and S. Valla. 1997. Construction and use of a versatile set of broad-host-range cloning and expression vectors based on the RK2 replicon. Appl. Environ. Microbiol. 63370-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bouhenni, R., A. Gehrke, and D. Saffarini. 2005. Identification of genes involved in cytochrome c biogenesis in Shewanella oneidensis, using a modified mariner transposon. Appl. Environ. Microbiol. 714935-4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cann, M., A. Hammer, J. Zhou, and T. Kanacher. 2003. A defined subset of adenylyl cyclases is regulated by bicarbonate ion. J. Biol. Chem. 27835033-35038. [DOI] [PubMed] [Google Scholar]
- 5.Dehio, C., and M. Meyer. 1997. Maintenance of broad-host-range incompatibility group P and group Q plasmids and transposition of Tn5 in Bartonella henselae following conjugal plasmid transfer from Escherichia coli. J. Bacteriol. 179538-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Driscoll, M., M. Romine, F. Juhn, M. Serres, L. McCue, A. Beliaev, J. Fredrickson, and T. S. Gardner. 2007. Identification of diverse carbon utilization pathways in Shewanella oneidensis MR-1 via expression profiling. Genome Inform. 18287-298. [PubMed] [Google Scholar]
- 7.Faith, J., B. Hayete, J. Thaden, I. Mogno, J. Wierzbowski, G. Cottarel, S. Kasif, J. Collins, and T. S. Gardner. 2007. Large-scale mapping and validation of Escherichia coli transcriptional regulation from a compendium of expression profiles. PLoS Biol. 5e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Findeisen, F., J. Linder, A. Schultz, J. Schultz, B. Brugger, F. Wieland, I. Sinnin, and I. Tews. 2007. The structure of the regulatory domain of the adenylyl cyclase Rv1264 from Mycobacterium tuberculosis with bound oleic acid. J. Mol. Biol. 3691282-1295. [DOI] [PubMed] [Google Scholar]
- 9.Gao, H., Z. Yang, L. Wu, D. Thompson, and J. Zhou. 2006. Global transcriptome analysis of the cold shock response of Shewanella oneidensis MR-1 and mutational analysis of its classical cold shock proteins. J. Bacteriol. 1884560-4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gralnick, J. A., H. Vali, D. P. Lies, and D. K. Newman. 2006. Extracellular respiration of dimethyl sulfoxide by Shewanella oneidensis strain MR-1. Proc. Natl. Acad. Sci. USA 1034669-4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guest, J., J. Green, A. Irvine, and S. Spiro. 1996. The FNR modulon and FNR-regulated gene expression, p. 317-342. In E. C. Lin and S. Lynch (ed.), Regulation of gene expression in Escherichia coli. Chapman & Hall, New York, NY.
- 12.Ho, S. N., H. Hunt, H. Horton, J. Pullen, and L. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 7751-59. [DOI] [PubMed] [Google Scholar]
- 13.Kieft, T. L., J. K. Fredrickson, T. C. Onstott, Y. A. Gorby, H. M. Kostandarithes, T. J. Bailey, D. W. Kennedy, S. W. Li, A. E. Plymale, C. M. Spadoni, and M. S. Gray. 1999. Dissimilatory reduction of Fe(III) and other electron acceptors by a Thermus isolate. Appl. Environ. Microbiol. 651214-1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kiley, P. J., and H. Beinert. 1998. Oxygen sensing by the global regulator, FNR: the role of the iron-sulfur cluster. FEMS Microbiol. Rev. 22341-352. [DOI] [PubMed] [Google Scholar]
- 15.Kimura, Y., Y. Mishima, H. Nakano, and K. Takegawa. 2002. An adenylyl cyclase, CyaA, of Myxococcus xanthus functions in signal transduction during osmotic stress. J. Bacteriol. 1843578-3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Linder, J., and A. Schultz. 2003. The class III adenylyl cyclases: multi-purpose signalling modules. Cell. Signal. 151081-1089. [DOI] [PubMed] [Google Scholar]
- 17.Maier, T. M., and C. R. Myers. 2001. Isolation and characterization of a Shewanella putrefaciens MR-1 electron transport regulator etrA mutant: reassessment of the role of EtrA. J. Bacteriol. 1834918-4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCue, L. A., K. McDonough, and C. E. Lawrence. 2000. Functional classification of cNMP-binding proteins and nucleotide cyclases with implications for novel regulatory pathways in Mycobacterium tuberculosis. Genome Res. 10204-219. [DOI] [PubMed] [Google Scholar]
- 19.Miller, J. 1972. Experiments in molecular genetics, p. 352-355. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 20.Mori, K., and H. Aiba. 1985. Evidence for negative control of cya transcription by cAMP and cAMP receptor protein in intact Escherichia coli cells. J. Biol. Chem. 26014838-14843. [PubMed] [Google Scholar]
- 21.Moser, D., and K. Nealson. 1996. Growth of the facultative anaerobe Shewanella putrefaciens by elemental sulfur reduction. Appl. Environ. Microbiol. 622100-2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murphy, J., K. Durbin, and C. Saltikov. 2009. The functional roles of arcA, etrA, cyclic AMP (cAMP)-cAMP receptor protein, and cya in the arsenate respiration pathway in Shewanella sp. strain ANA-3. J. Bacteriol. 1911035-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Myers, C., and K. Nealson. 1988. Microbial reduction of manganese oxides: interactions with iron and sulfur. Geochim. Cosmochim. Acta 522727-2732. [Google Scholar]
- 24.Myers, C. R., and J. M. Myers. 1997. Cloning and sequence of cymA, a gene encoding tetraheme cytochrome c required for reduction of iron(III), fumarate, and nitrate by Shewanella putrefaciens MR-1. J. Bacteriol. 1791143-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Myers, J., and C. Myers. 2000. Chromium(VI) reductase activity is associated with the cytoplasmic membrane of anaerobically grown Shewanella putrefaciens MR-1. J. Appl. Microbiol. 8898-106. [DOI] [PubMed] [Google Scholar]
- 26.Nealson, K., and D. Saffarini. 1994. Iron and manganese in anaerobic respiration: environmental significance, physiology, and regulation. Annu. Rev. Microbiol. 48311-343. [DOI] [PubMed] [Google Scholar]
- 27.Saffarini, D., and K. Nealson. 1993. Sequence and genetic characterization of etrA, an fnr analog that regulates anaerobic respiration in Shewanella putrefaciens MR-1. J. Bacteriol. 1757938-7944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saffarini, D. A., R. Schultz, and A. Beliaev. 2003. Involvement of cyclic AMP (cAMP) and cAMP receptor protein in anaerobic respiration of Shewanella oneidensis. J. Bacteriol. 1853668-3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saltikov, C. W., A. Cifuentes, K. Venkateswaran, and D. K. Newman. 2003. The ars detoxification system is advantageous but not required for As(V) respiration by the genetically tractable Shewanella species strain ANA-3. Appl. Environ. Microbiol. 692800-2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Santos, P., J. M. Blatny, I. Di Bartolo, S. Valla, and E. Zennaro. 2000. Physiological analysis of the expression of the styrene degradation gene cluster in Pseudomonas fluorescens ST. Appl. Environ. Microbiol. 661305-1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shah, S., and A. Peterkofsky. 1991. Characterization and generation of Escherichia coli adenylate cyclase deletion mutants. J. Bacteriol. 1733238-3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shenroy, A., and S. Visweswariah. 2004. Class III nucleotide cyclases in bacteria and archaebacteria: lineage-specific expansion of adenylyl cyclases and a dearth of guanylyl cyclases. FEBS Lett. 56111-21. [DOI] [PubMed] [Google Scholar]
- 33.Sismeiro, O., P. Trotot, F. Biville, C. Vivares, and A. Danchin. 1998. Aeromonas hydrophila adenylyl cyclase 2: a new class of adenylyl cyclases with thermophilic properties and sequence similarities to proteins from hyperthermophilic archaebacteria. J. Bacteriol. 1803339-3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spiro, S., and J. Guest. 1990. FNR and its role in oxygen-regulated gene expression in Escherichia coli. FEMS Microbiol. Rev. 75399-428. [DOI] [PubMed] [Google Scholar]
- 35.Tellez-Sosa, J., N. Soberon, A. Vega-Segura, M. Torres-Marquez, and M. Cevallos. 2002. The Rhizobium etli cyaC product: characterization of a novel adenylate cyclase class. J. Bacteriol. 1843560-3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tews, I., F. Findeisen, I. Sinning, A. Schultz, J. Schultz, and J. Linder. 2005. The structure of a pH-sensing mycobacterial adenylyl cyclase holoenzyme. Science 3081020-1023. [DOI] [PubMed] [Google Scholar]
- 37.Thomas, P. E., D. Ryan, and W. Levin. 1976. An improved staining procedure for the detection of the peroxidase activity of cytochrome P-450 on sodium dodecyl sulfate polyacrylamide gels. Anal. Biochem. 75168-176. [DOI] [PubMed] [Google Scholar]
- 38.Wan, X., N. VerBerkmoes, L. A. McCue, D. Stanek, H. Connelly, L. Hauser, L. Wu, X. Liu, T. Yan, A. Leaphart, R. Hettich, J. Zhou, and D. Thompson. 2004. Transcriptomic and proteomic characterization of the Fur modulon in the metal-reducing bacterium Shewanella oneidensis. J. Bacteriol. 1868385-8400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wolfgang, M. C., V. T. Lee, M. E. Gilmore, and S. Lory. 2003. Coordinate regulation of bacterial virulence genes by a novel adenylate cyclase-dependent signaling pathway. Dev. Cell 4253-263. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.