Abstract
The biofilm matrix contributes to the chemistry, structure, and function of biofilms. Biofilm-derived membrane vesicles (MVs) and DNA, both matrix components, demonstrated concentration-, pH-, and cation-dependent interactions. Furthermore, MV-DNA association influenced MV surface properties. This bears consequences for the reactivity and availability for interaction of matrix polymers and other constituents.
The biofilm matrix contributes to the chemistry, structure, and function of biofilms and is crucial for the development of fundamental biofilm properties (46, 47). Early studies defined polysaccharides as the matrix component, but proteins, lipids, and nucleic acids are all now acknowledged as important contributors (7, 15). Indeed, DNA has emerged as a vital participant, fulfilling structural and functional roles (1, 5, 6, 19, 31, 34, 36, 41, 43, 44). The phosphodiester bond of DNA renders this polyanionic at a physiological pH, undoubtedly contributing to interactions with cations, humic substances, fine-dispersed minerals, and matrix entities (25, 41, 49).
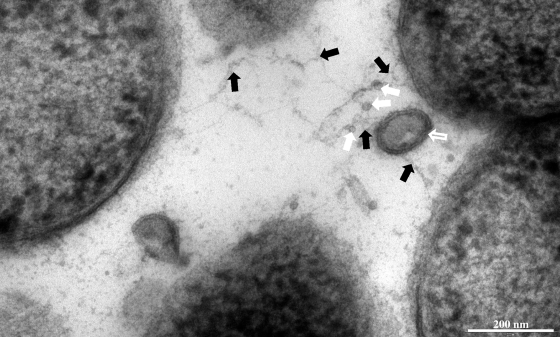
In addition to particulates such as flagella and pili, membrane vesicles (MVs) are also found within the matrices of gram-negative and mixed biofilms (3, 16, 40). MVs are multifunctional bilayered structures that bleb from the outer membranes of gram-negative bacteria (reviewed in references 4, 24, 27, 28, and 30) and are chemically heterogeneous, combining the known chemistries of the biofilm matrix. Examination of biofilm samples by transmission electron microscopy (TEM) has suggested that matrix material interacts with MVs (Fig. 1). Since MVs produced in planktonic culture have associated DNA (11, 12, 13, 20, 21, 30, 39, 48), could biofilm-derived MVs incorporate DNA (1, 39, 40, 44)?
FIG. 1.
Possible interactions between matrix polymers and particulate structures. Shown is an electron micrograph of a thin section through a P. aeruginosa PAO1 biofilm. During processing, some dehydration occurred, resulting in collapse of matrix material into fibrillate arrangements (black filled arrows). There is a suggestion of interactions occurring with particulate structures such as MVs (hollow white arrow) and flagella (filled white arrows) (identified by the appearance and cross-dimension of these highly ordered structures when viewed at high magnification), which was consistently observed with other embedded samples and also with whole-mount preparations of gently disrupted biofilms (data not shown). The scale bar represents 200 nm.
MATERIALS AND METHODS
Biofilm growth and isolation and purification of matrix and MVs.
Pseudomonas aeruginosa PAO1 and green fluorescent protein-tagged PAO1 (17) biofilms were grown using the agar plate model (40). Matrix was isolated (40) and sequentially filtered through 0.22-, 0.45-, and 1.2-μm cellulose acetate filters and the filtrate collected. Absence of cells was confirmed by plating 100-μl aliquots (18 h at 37°C; trypticase soy agar; n = 3) and TEM of whole-mount preparations (see below). Matrix for characterization was dialyzed (24 h at 4°C; Spectrapor regenerated cellulose dialysis membranes [molecular mass cutoff, 3,000 Da]; Fisher). Particulate components were harvested by ultracentrifugation (125,000 × g for 1.5 h at 5°C; Beckman Ti45 rotor), the pellets resuspended in 30% (vol/vol) Optiprep (Sigma), and the components separated on Optiprep density gradients (2) at 0% (1 ml), 18% (1 ml), 20% (1 ml), 22.5% (3 ml), 25% (3 ml), 27.5% (3 ml), 30% (3 ml), and 50% (1 ml) (vol/vol). All Optiprep solutions contained 10 mM HEPES, 0.85% (wt/vol) NaCl (pH 7.4). Samples were centrifuged to equilibrium (100,000 × g for 16 h at 5°C; Beckman SW28.1 rotor) and fractionated (200-μl aliquots) and whole mounts assessed by TEM (see below). MV-containing fractions were combined, washed twice in water (125,000 × g for 1.5 h at 5°C; Beckman Ti45 rotor), resuspended in water, and frozen.
Quantitative and biochemical characterization.
Dry samples were obtained by freeze drying.
Protein was quantified using a micro-bicinchoninic acid protein assay kit (Pierce Bioassay; n = 3).
DNA was quantified by a modified PicoGreen assay (Molecular Probes, Invitrogen) (23); 1:100 GES (5 M guanidinium thiocyanate, 100 mM EDTA, 0.5% [vol/vol] Sarkosyl) yielded optimal results. Independent samples were assayed using a Spex fluorescence spectrophotometer (excitation λ, 480 nm; emission λ, 523 nm); the slit widths were 4 and 8 nm, respectively. MVs were treated with DNase I (Roche) to degrade externally associated DNA which was susceptible, establishing internal/external DNA content (12, 13, 20, 23, 39). Fifty micrograms of MV protein (10 ml 50 mM HEPES buffer, pH 6.8, 10 mM MgCl2) was digested with 400 μg DNase·ml−1 (2 h at 37°C and 125 rpm), and MVs were harvested by ultracentrifugation. TEM and sodium dodecyl sulfate-polyacrylamide gel electrophoresis indicated no gross changes (data not shown). Henceforth, DNase-treated MVs are referred to as “DNase-treated MVs” and untreated MVs as “native MVs.”
Zeta potential analysis.
The relative surface charges of MVs were estimated using a Zetasizer Nano ZS particle analyzer (Malvern Scientific). The microelectrophoresis cell was filled with 8 μg of MV protein·ml−1 HEPES buffer (10, 50, or 100 mM; 4.4 to 9.8 pH units). A voltage difference of 150 V was applied across the chamber. Five readings were done on each of three independent samples.
Titration analysis.
Available surface ligands were estimated by titration analysis (37). Three independent wet samples, each equivalent to 40 mg (dry weight), were titrated.
Isolation of DNA.
Three grams of biofilm (wet weight) was resuspended in 40 ml of lysis buffer (10 mM Tris, 10 mM EDTA, 2% sodium dodecyl sulfate, 150 mM NaCl), an equal volume of phenol-chloroform-isoamyl alcohol (25:24:1, vol/vol/vol) was added, and the phases were mixed gently (10 min) and then centrifuged (12,000 × g for 20 min) (32, 38, 45). A 1/100 volume of 5 M NaCl was added to the collected upper aqueous phase, extracted with an equal volume of chloroform-isoamyl alcohol (24:1, vol/vol), and centrifuged and the aqueous phase collected. Chloroform-isoamyl alcohol extraction was repeated if material was present at the interface. DNA was precipitated by adding 2 volumes of ice-cold 100% ethanol, the phases were gently mixed, and DNA was spooled onto a glass rod and dissolved in and dialyzed against nanopure water (24 h at 4°C; Spectrapor regenerated cellulose dialysis membranes [molecular mass cutoff, 3,000 Da; Fisher). Solutions having absorbance maximum ratios for nucleic acids/proteins (A260/A280) of ≥1.8 were considered of adequate purity. Concentration was estimated by the PicoGreen assay.
DNA binding studies.
MV and exogenous DNA interactions were assessed by incubating aliquots of MVs and DNA together. Difficulties were encountered isolating DNA from matrix (see Results and Discussion). Since extracellular DNA is similar to whole-genome DNA (1), DNA was isolated from biofilms. MVs were incubated with DNA (1 h at 37°C and 50 rpm) in HEPES buffer (50 mM; pH 5.6, 6.0, 6.4, 6.8, 7.2, or 7.6, adjusted with HCl or NaOH), supplemented with NaCl (0.1 to 100 mM) or MgCl2 (0.1 to 100 mM) where indicated. After incubation, samples were harvested by ultracentrifugation (see above) and resuspended. Negative controls contained MVs or DNA.
Fluorescence microscopy.
Biofilms were carefully excised and stained with the DNA indicator dye 7-hydroxy-9H-(1,3-dichloro-9,9-dimethylacridin-2-one) (DDAO [1]; 1 mM solution in tetrahydrofuran, diluted 1:1,000 in 50 mM HEPES buffer, pH 7.0, for 15 min; Molecular Probes, Invitrogen). All images were collected using a Leica TCS SP2 confocal-scanning-laser microscope (Leica) equipped with Ar-Kr 488-nm and He-Ne 633-nm lasers and a 43× phase contrast lens. Stained and unstained biofilms were excited (488 and 633 nm, respectively) and emission data collected (510 and 659 nm, respectively).
TEM.
Negatively stained whole mounts were prepared as detailed elsewhere (40).
Thin-sectioned biofilm samples were prepared by freeze substitution (19). On completion, the medium was replaced with 100% ethanol (30 min) and two changes of LR White-ethanol (1:1; London Resin Company) over a period of 45 min and 100% LR-White (30 min). The sample was polymerized in fresh LR White (60°C for 1 h), and sections were prepared and poststained with 2% (wt/vol) uranyl acetate and Reynold's lead citrate (42).
All samples were examined under standard operating conditions, using a Philips CM10 TEM (acceleration voltage, 80 kV). Images were archived using the iTEM program (version 5.0; Soft Imaging Systems).
RESULTS AND DISCUSSION
DNA is associated with MVs isolated from the matrices of biofilms. The selective dye DDAO established the presence and locality of matrix DNA in biofilms (Fig. 2) (1). In agreement with reported double-stranded-DNA (dsDNA) contributions to P. aeruginosa matrix polymers (29, 33, 34), a biochemical assay indicated that 6.1% (wt/wt) of the matrix was dsDNA (Table 1).
FIG. 2.

DDAO staining of P. aeruginosa PAO1 biofilm revealed the presence of extracellular DNA. Areas where the DDAO stain indicated the presence of dsDNA (shown in red) were seen in between green fluorescent protein-tagged cells (shown in green) within the biofilm matrix. Note how the stain was not uniform. The presence of dsDNA was supported by biochemical assays with isolated matrix material (Table 1). The scale bar represents 4.69 μm.
TABLE 1.
DNA and its contribution to the matrix and matrix-derived MVsa
| Isolated fraction | Dry wt (mg) | Amt of DNA (mg) | % DNA/dry wt |
|---|---|---|---|
| Matrix | 56.6 ± 11.1 | 3.46 ± 0.52 | 6.11 |
| MV | 24.5 ± 3.9 | 0.012 ± 0.002 | 0.05 |
MV DNA represents 0.35% (wt/wt) of the matrix DNA. Dry weight values are averages ± standard deviations of results from three independent samples. DNA values are averages ± standard errors of the means of results from three independent samples assayed in triplicate (n = 180).
Biofilm MVs (b-MVs) contained 91 ± 1 ng DNA/20 μg MV protein (mean ± standard error of the mean [SEM]; n = 180), much greater than planktonic MVs (p-MVs) (20, 39), possibly due to higher local concentrations and physical and temporal proximity (31) or the strikingly different chemistry of b-MVs (40). b-MV-associated DNA, however, represented a minute proportion of matrix DNA (Table 1). DNA is notorious for its fragility; our values reflect the DNA remaining associated after isolation. Attempted DNA isolation from matrix yielded no matter, suggesting that the DNA was sheared or short stranded; anecdotally, the matrix was more viscous prior to filtering.
DNA is associated with the lumen and external face of MVs.
p-MVs have DNA associated with their lumen, external surface, or both (11, 12, 13, 39). Mechanisms for this have been described (12, 13, 20, 39). The distribution is relevant: location and arrangement affect availability and interactions. Forty-one percent of b-MV DNA was located at their outer surfaces (PAO1 p-MVs; 25%) (39). The lumen contained 54 ± <1 ng DNA/20 μg MV protein (mean ± SEM; n = 180), greater than the level reported for p-MVs (39), supporting b-MVs as distinct from p-MVs (40).
Exogenous DNA interacts and associates with MVs.
b-MVs were incubated with DNA at the ratio at which these occurred within the matrix. In agreement with the DNA-retaining properties of p-MVs (11, 12, 21, 39), b-MVs showed a substantial increase in associated DNA (281 ± <1 ng DNA/20 μg MV protein [mean ± SEM; n = 60]). DNase-treated MVs showed a propensity similar to that for native MVs (269 ± 1 ng DNA/20 μg MV protein [mean ± SEM; n = 60]), suggesting that, for a given amount of DNA, there is a maximum DNA load per unit MV protein.
MV-DNA interaction is dependent upon physical conditions.
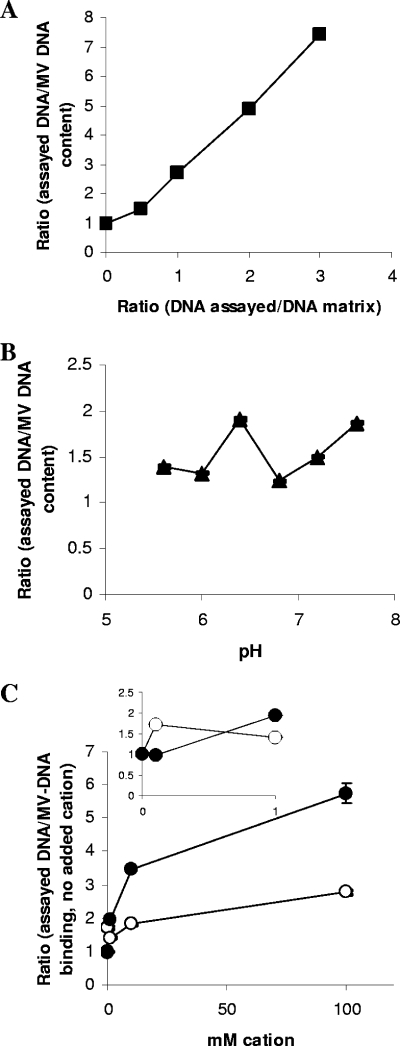
Microenvironments throughout biofilms mean that diverse environmental conditions, such as concentration and pH, are at play. DNA and MVs were incubated together at different ratios; a ratio value of 1 (Fig. 3A, x axis) reflects the relative amount of DNA to MV protein as it occurred in the matrix. MV protein was kept constant and DNA concentration altered. A very strong positive linear correlation relationship was found between DNA concentration and DNA association with MVs (y = 2.2084x + 0.6319; R2 = 0.99). Even when the DNA concentration was increased threefold, the system remained unsaturated.
FIG. 3.
Assessment of the effect of environmental conditions upon the interaction between b-MVs and exogenous DNA. MVs were incubated with DNA (see Materials and Methods for details), and the effects of concentration of DNA (▪) (A), altered pH (▴) (B), and concentration of Na+ (○) or Mg2+ (•) cations, respectively (C), upon MV-DNA interactions were assessed. Note that for panel A, the ratio on the x axis is equal to the concentration of the assayed DNA/concentration of DNA in the matrix. Quantities of DNA (ng)/20 μg MV protein for R values of 1 were 116 ± 3 (DNA concentration; MVs only), 86 ± 3 (pH; MVs only), 275 ± 3 (Na+; MVs plus DNA only), and 236 ± 6 (Mg2+; MVs plus DNA only) (means ± standard deviations; n = 60).
pH is another well-established variable within biofilms (8, 17). MVs were incubated with DNA at the same ratio as they were within the matrix, and pH was altered (Fig. 3B). The baseline R value of 1 describes the amount of DNA per 20 μg MV protein. Retained DNA increased from pH 5.6 to pH 6.4 and dropped at pH 6.8, reaching a maximum at pH 7.6. Deprotonation of the main surface groups increases with pH (carboxyl > phosphate > amine). As available carboxyl groups interact with DNA, diminished electrostatic repulsion promotes further binding. As pH further increases, deprotonation of phosphate groups (titration of MVs [see below], pH 6.8 to 7.1) renders the surface more negatively charged, repulsing MV-DNA interactions. The mechanistics from pH 6.8 to 7.6 are not clear, although it mirrors the lowering of net negative surface charge (Fig. 4). pH change may also cause reorganization of surface components or electrostatic destabilization, exposing previously inaccessible functional groups (9).
FIG. 4.
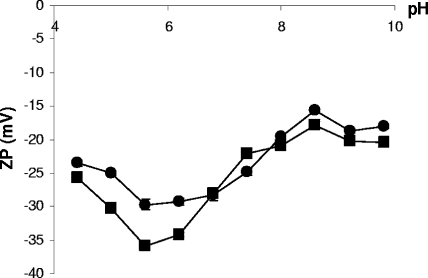
Relative surface charges of native and DNase-treated MVs. Suspensions of native MVs (▪) or DNase-treated MVs (•; 8 μg of MV protein·ml−1) were assessed by zeta potentiometry. Data for 10 and 100 mM buffer (not shown) yielded the expected concentration-dependent effect whereby an increase in buffer concentration causes a subsequent lessening of the apparent negative surface charge (mean ± SEM; n = 15). A two-tailed Mann-Whitney test was performed using Analyze-It for Microsoft Excel (version 2.11) and indicated that all data, with the exception of that collected at pH 6.8, showed highly significant differences (P < 0.01).
The third assessed parameter was the presence of monovalent and divalent cations. MVs were incubated with DNA at the same ratio as they were within the matrix, and cation concentration was altered (Fig. 3C). The baseline R value of 1 describes the amount of DNA per 20 μg MV protein following incubation with DNA. As the Na+ concentration increased, this increase was accompanied by a concomitant gradual increase in associated MV-DNA. As the concentration of Na+ increases, electrostatic repulsion is reduced, permitting positively charged surface groups, e.g., amines, to interact more readily with DNA. The presence of Mg2+ ions displayed behavior similar to that of Na+ ions; however, the association was greater (Fig. 3C). Polyvalent cations screen negative charges more effectively and also facilitate salt bridging of the phosphate groups on DNA and negatively charged groups on the MV surface.
Collectively, the data indicated that conditions mimicking variation within biofilm microenvironments altered interactions between particulate components (MVs) and matrix polymers (DNA).
DNA associated with the external face of MVs influences MV surface properties.
Biofilm-derived MVs and DNA interact, but does this alter the apparent surface properties of MVs? Native and DNase-treated MVs were compared by zeta potentiometry. DNase-treated MVs were generally less negatively charged (Fig. 4). Readings within the pH range of 5.2 to 7.8, conditions under which phosphate groups deprotonate (pKa, 6.4 to 7.4) (14) and contribute to surface charge, indicated that native MVs had a more negative charge. Sources for this phosphate group include lipopolysaccharide and terminal phosphate groups of DNA; the phosphodiester bond (pKa, ∼3 units) of DNA or other exposed molecules accounts for some of the negative charge at pH values of just over 4 units (Fig. 4).
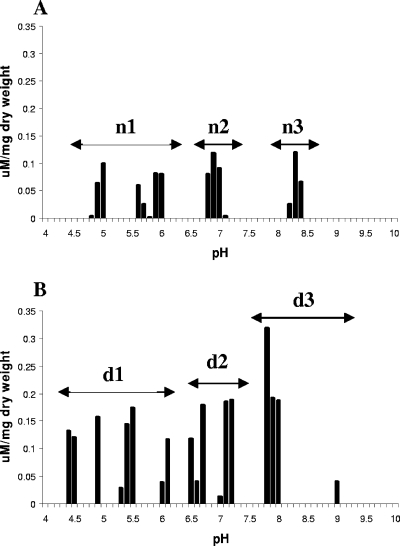
Titration analysis confirmed the data and yielded information on the available surface groups. Differences in the abundance and distribution of ligands of native or DNase-treated MVs were obvious (Fig. 5). Analysis of the data (Table 2) showed that DNase-treated MVs had a total ligand concentration 2.5 times (wt/wt) greater than that of the native MVs, i.e., extravesicular DNA interacted with surface groups masking them. The assignment of functional group identities (Table 2) (10, 14, 26, 35, 37) revealed increases of ∼2.5-fold in the designated carboxyl and phosphate groups of DNase-treated MVs but 3.5-fold for amine groups. The data supported that interactions between MVs and DNA were occurring via both salt-bridging mechanisms and electrostatic interactions. The greater increases in the amine groups following DNA digestion indicated that the latter had a greater impact on binding. The data also indicated that despite the extremely low-level contribution of DNA to MVs (Table 1), this altered the surface chemistry and clearly has broad implications in terms of the functionality and reactivity of MVs.
FIG. 5.
Titration analysis of the available functional groups present on the native and DNase-treated MVs. pKa spectra for native MVs (A) and DNase-treated MVs (B) composite plot of three spectra. The site density (concentration of ligand) is normalized to the dry weight of the sample used. The arrows (A, n1 to n3; B, d1 to d3) indicate the groups that the data cluster in. Please refer to Table 2 for details of these.
TABLE 2.
Composite titration data for native and DNase-treated MVsa
| Ligand classb | Mean pKac | Mean LT (μM/mg [dry wt])d | Proposed functional group |
|---|---|---|---|
| n1 | 5.48 (4.8-6.0) | 0.138 (45) | Carboxyl |
| n2 | 6.91 (6.8-7.1) | 0.098 (32) | Phosphate |
| n3 | 8.32 (8.2-8.4) | 0.070 (23) | Amine |
| d1 | 5.18 (4.4-6.1) | 0.305 (38.5) | Carboxyl |
| d2 | 6.90 (6.5-7.2) | 0.242 (30.5) | Phosphate |
| d3 | 7.94 (7.8-9.0) | 0.246 (31) | Amine |
The total available surface ligand concentrations for native and DNase-treated MVs were 0.306 and 0.793 μM/mg (dry weight), respectively (DNase/native ratio of 2.5). A ratiometric comparison of DNase-treated to native MVs indicated values of 2.2 (d1/n1), 2.5 (d2/n2), and 3.5 (d3/n3). Native MVs contributed 8% (wt/wt) of the total ligand concentration of the matrix material. Data were obtained from titration analysis of three independent samples.
Based on pKa clusters in Fig. 5A and B.
Values in parentheses represent ranges for given pKa clusters. Mean pKa values were estimated using a weighted mean calculation, with the corresponding available surface ligand concentration as the weighting factor.
LT refers to site density (μM/mg [dry weight]). Values in parentheses represent the percent contribution of any functional group to the sample's total ligand concentration.
An additional facet to the MV paradigm.
Assessment of biofilms by TEM has long hinted at a relationship between particulates and matrix polymers (Fig. 1). MVs, in addition to their diverse known functionality, are capable of interacting with matrix polymers. This interaction will be modulated by regions of variable polymer and MV content within biofilms (1, 18, 40) and qualitative differences in MVs that will intuitively occur (22). The enmeshment, entanglement, and stiffness of matrix polymers will be influenced by the locality, chemical composition, and relative quantities of participant particulates and polymers. MVs are also trafficked out of biofilms (40). Surfaces may be preconditioned with a DNA-MV complex altering subsequent interactions, e.g., cell adhesion, and mobile MV-DNA complexes may transfer genetic material between otherwise immobile cell populations. However, MV-DNA transfer of genetic information between cells remains controversial (39). Yet, whether the internalized DNA differs from that on the MV surface, or even within the matrix, begs to be asked.
In summary, the matrix is central to the chemistry, structure, and function of biofilms. Its very nature is defined by the dynamic interchange between chemical constituents, environmental conditions, and cell members of the consortium. We must remember that interactions also occur between polymers and particulates. This work confirms the presence of MV-associated DNA within biofilms, the association and interaction of b-MVs with matrix components, and the impact of these processes on interacting moieties. We reemphasize the matrix as a complex amalgam and not a simple collection of chemistries. The matrix is a constantly changing entity, in many ways as responsive and reactive as the cells that it enshrouds.
Acknowledgments
This work was supported by funds to T.J.B. from NSERC and AFMNet-NCE. T.J.B. held a Canada Research Chair. TEM was performed in the NSERC Guelph Regional Integrated Imaging Facility (GRIIF), which is partially funded by an NSERC-MFA grant.
We thank Frances Sharom and Milena Corredig (University of Guelph) for use of the Spex fluorescence spectrophotometer and the Zetasizer Nano, respectively; Anuradha Saxena (University of Guelph) for running titrations; Ryan Hunter (Massachusetts Institute of Technology) and Michaela Strüder-Kypke (University of Guelph) for assistance with freeze substitution and confocal work, respectively; Vernon Phoenix (University of Glasgow) for insightful discussion; and Chris Whitfield, Cecil Forsberg, and Anton Korenevski (University of Guelph) as well as the editor and reviewers for reading the manuscript and providing useful criticism.
Footnotes
Published ahead of print on 8 May 2009.
This paper is dedicated to the late Terry Beveridge, friend and mentor, and a belief of science—“isn't this fun?”
REFERENCES
- 1.Allesen-Holm, M., K. B. Barken, L. Yang, M. Klausen, J. S. Webb, S. Kjelleberg, S. Molin, M. Givskov, and T. Tolker-Nielsen. 2006. A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol. Microbiol. 591114-1128. [DOI] [PubMed] [Google Scholar]
- 2.Bauman, S. J., and M. J. Kuehn. 2006. Purification of outer membrane vesicles from Pseudomonas aeruginosa and their activation of an IL-8 response. Microbes Infect. 82400-2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beveridge, T. J., S. A. Makin, J. L. Kadurugamuwa, and Z. Li. 1997. Interactions between biofilms and the environment. FEMS Microbiol. Rev. 20291-303. [DOI] [PubMed] [Google Scholar]
- 4.Beveridge, T. J. 1999. Structures of gram-negative cell walls and their derived membrane vesicles. J. Bacteriol. 1814725-4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Böckelmann, U., A. Janke, R. Kuhn, T. R. Neu, J. Wecke, J. R. Lawrence, and U. Szewzyk. 2006. Bacterial extracellular DNA forming a defined network-like structure. FEMS Microbiol. Lett. 26231-38. [DOI] [PubMed] [Google Scholar]
- 6.Böckelmann, U., H. Lünsdorf, and U. Szewzyk. 2007. Ultrastructural and electron energy-loss spectroscopic analysis of an extracellular filamentous matrix of an environmental bacterial isolate. Environ. Microbiol. 92137-2144. [DOI] [PubMed] [Google Scholar]
- 7.Branda, S. S., Å. Vik, L. Friedman, and R. Kolter. 2005. Biofilms: the matrix revisited. Trends Microbiol. 1320-26. [DOI] [PubMed] [Google Scholar]
- 8.Caldwell, D. E., D. R. Korber, and J. R. Lawrence. 1992. Confocal laser microscopy and digital image analysis in microbial ecology. Adv. Microb. Ecol. 121-67. [Google Scholar]
- 9.Claessens, J., T. Behrends, and P. V. Cappallen. 2004. What do acid-base titrations of live bacteria tell us? A preliminary assessment. Aquat. Sci. 6619-26. [Google Scholar]
- 10.Cox, J. S., D. S. Smith, L. A. Warren, and F. G. Ferris. 1999. Characterizing heterogeneous bacterial surface functional groups using discrete affinity spectra for proton binding. Environ. Sci. Technol. 334514-4521. [Google Scholar]
- 11.Deich, R. A., and L. C. Hoyer. 1982. Generation and release of DNA-binding vesicles by Haemophilus influenzae during induction and loss of competence. J. Bacteriol. 152855-864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dorward, D. E., C. F. Garon, and R. C. Judd. 1989. Export and intercellular transfer of DNA via membrane blebs of Neisseria gonorrhoeae. J. Bacteriol. 1712499-2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dorward, D. E., and C. F. Garon. 1990. DNA is packaged within membrane-derived vesicles of gram-negative but not gram-positive bacteria. Appl. Environ. Microbiol. 561960-1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fein, J. B., C. J. Daughney, N. Yee, and T. A. Davis. 1997. A chemical equilibrium model for metal adsorption onto bacterial surfaces. Geochim. Cosmochim. Acta 613319-3328. [Google Scholar]
- 15.Flemming, H.-C., and J. Wingender. 1999. Relevance of microbial extracellular polymeric substances (EPSs)—part I: structural and ecological aspects. Water Sci. Technol. 43(6)1-8. [PubMed] [Google Scholar]
- 16.Halhoul, N., and J. R. Colvin. 1975. The ultrastructure of bacterial plaque attached to the gingiva of man. Arch. Oral Biol. 20115-118. [DOI] [PubMed] [Google Scholar]
- 17.Hunter, R. C., and T. J. Beveridge. 2005. Application of a pH-sensitive fluoroprobe (C-SNARF-4) for pH microenvironment analysis in Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 712501-2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hunter, R. C., and T. J. Beveridge. 2005. High-resolution visualization of Pseudomonas aeruginosa PAO1 biofilms by freeze-substitution transmission electron microscopy. J. Bacteriol. 1877619-7630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jurcisek, J. A., and L. O. Bakaletz. 2007. Biofilms formed by nontypeable Haemophilus influenzae in vivo contain both double-stranded DNA and type IV pilin protein. J. Bacteriol. 1893868-3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kadurugamuwa, J. L., and T. J. Beveridge. 1995. Virulence factors are released from Pseudomonas aeruginosa in association with membrane vesicles during normal growth and exposure to gentamicin: a novel mechanism of enzyme secretion. J. Bacteriol. 1773998-4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kahn, M., M. Concino, R. Gromkova, and S. Goodgal. 1979. DNA binding activity of vesicles produced by competence deficient mutants of Haemophilus. Biochem. Biophys. Res. Commun. 87764-772. [DOI] [PubMed] [Google Scholar]
- 22.Keenan, J. L., and R. A. Allardyce. 2000. Iron influences the expression of Helicobacter pylori outer membrane vesicle-associated virulence factors. Eur. J. Gastroenterol. Hepatol. 121267-1273. [DOI] [PubMed] [Google Scholar]
- 23.Kolling, G. L., and K. R. Matthews. 1999. Export of virulence genes and Shiga toxin by membrane vesicles of Escherichia coli O157:H7. Appl. Environ. Microbiol. 651843-1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuehn, M. J., and N. C. Kesty. 2005. Bacterial outer membrane vesicles and the host-pathogen interaction. Genes Dev. 192645-2655. [DOI] [PubMed] [Google Scholar]
- 25.Lorenz, M. G., and W. Wackernagel. 1994. Bacterial gene transfer by natural genetic transformation in the environment. Microbiol. Rev. 58563-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martinez, R. E., D. S. Smith, E. Kulczycki, and F. G. Ferris. 2002. Determination of intrinsic bacterial surface acidity constants using a Donnan shell model and a continuous pKa distribution method. J. Colloid Interface Sci. 253130-139. [DOI] [PubMed] [Google Scholar]
- 27.Mashburn-Warren, L., R. J. Mclean, and M. Whiteley. 2008. Gram-negative outer membrane vesicles: beyond the cell surface. Geobiology 6214-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mashburn-Warren, L. M., and M. Whiteley. 2006. Special delivery: vesicle trafficking in prokaryotes. Mol. Microbiol. 61839-846. [DOI] [PubMed] [Google Scholar]
- 29.Matsukawa, M., and E. P. Greenberg. 2004. Putative exopolysaccharide synthesis genes influence Pseudomonas aeruginosa biofilm development. J. Bacteriol. 1864449-4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mayrand, D., and D. Grenier. 1989. Biological activities of outer membrane vesicles. Can. J. Microbiol. 35607-613. [DOI] [PubMed] [Google Scholar]
- 31.Molin, S., and T. Tolker-Nielsen. 2003. Gene transfer occurs with enhanced efficiency in biofilms and induces enhanced stabilisation of the biofilm structure. Curr. Opin. Biotechnol. 14255-261. [DOI] [PubMed] [Google Scholar]
- 32.Moore, D., and D. Dowhan. 2002. Preparation and analysis of DNA, p. 2.1.1-2.1.10. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. John Wiley & Sons, Inc., New York, NY.
- 33.Murakawa, T. 1973. Slime production by Pseudomonas aeruginosa. III. Purification of slime and its physicochemical properties. Jpn. J. Microbiol. 17273-281. [PubMed] [Google Scholar]
- 34.Nemoto, K., K. Hirota, K. Murakami, K. Taniguti, H. Murata, D. Viduvic, and Y. Miyake. 2003. Effect of varidase (streptodornase) on biofilm formed by Pseudomonas aeruginosa. Chemotherapy 49121-125. [DOI] [PubMed] [Google Scholar]
- 35.Ngwenya, B. T., I. W. Sutherland, and L. Kennedy. 2003. Comparison of the acid-base behaviour and metal adsorption characteristics of a gram-negative bacterium with other strains. Appl. Geochem. 18527-538. [Google Scholar]
- 36.Petersen, F. C., L. Tao, and A. A. Scheie. 2005. DNA binding-uptake system: a link between cell-to-cell communication and biofilm formation. J. Bacteriol. 1874392-4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Phoenix, V. R., R. E. Martinez, K. Konhauser, and F. G. Ferris. 2002. Characterization and implications of the cell surface reactivity of Calothrix sp. strain KC97. Appl. Environ. Microbiol. 684827-4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pink, J., T. Smith-Palmer, D. Chisholm, T. J. Beveridge, and D. A. Pink. 2005. An FTIR study of Pseudomonas aeruginosa PAO1 biofilm development: interpretation of ATR-FTIR data in the 1500-1180 cm−1 region. Biofilms 2165-175. [Google Scholar]
- 39.Renelli, M., V. Matias, R. Lo, and T. J. Beveridge. 2004. DNA-containing membrane vesicles of Pseudomonas aeruginosa PAO1 and their genetic information potential. Microbiology 1502161-2169. [DOI] [PubMed] [Google Scholar]
- 40.Schooling, S. R., and T. J. Beveridge. 2006. Membrane vesicles: an overlooked component of the matrices of biofilms. J. Bacteriol. 1885945-5957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Schaik, E. J., C. L. Giltner, G. F. Audette, D. W. Keizer, D. L. Bautista, C. M. Slupsky, B. D. Sykes, and R. T. Irvin. 2005. DNA binding: a novel function of Pseudomonas aeruginosa type IV pili. J. Bacteriol. 1871455-1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Venable, J. H., and R. Coggeshall. 1965. A simplified lead citrate stain for use in electron microscopy. J. Cell Biol. 25407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walker, T. S., K. L. Tomlin, G. S. Worthen, K. R. Poch, J. G. Lieber, M. T. Saavedra, M. B. Fessler, K. C. Malcolm, M. L. Vasil, and J. A. Nick. 2005. Enhanced Pseudomonas aeruginosa biofilm development mediated by human neutrophils. Infect. Immun. 733693-3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Whitchurch, C. B., T. Tolker-Nielsen, P. C. Ragas, and J. S. Mattick. 2002. Extracellular DNA required for biofilm formation. Science 2951487. [DOI] [PubMed] [Google Scholar]
- 45.Wilson, K. 1997. Preparation and analysis of DNA, p. 2.4.1-2.4.5. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. John Wiley & Sons, Inc., New York, NY.
- 46.Wingender, J., T. R. Neu, and H.-C. Flemming (ed.). 1999. Microbial extracellular polymeric substances: characterization, structure and function, p. 1-19. Springer-Verlag, Berlin, Germany.
- 47.Wolfaardt, G. M., J. R. Lawrence, and D. R. Korber. 1999. Function of EPS, p. 171-200. In J. Wingender, T. R. Neu, and H.-C. Flemming (ed.), Microbial extracellular polymeric substances: characterization, structure and function. Springer-Verlag, Berlin, Germany.
- 48.Yaron, S., G. L. Kolling, L. Simon, and K. R. Matthews. 2000. Vesicle-mediated transfer of virulence genes from Escherichia coli O157:H7 to other enteric bacteria. Appl. Environ. Microbiol. 664414-4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou, S., D. Liang, C. Burger, F. Yeh, and B. Chu. 2004. Nanostructures of complexes formed by calf thymus DNA interacting with cationic substances. Biomacromolecules 51256-1261. [DOI] [PubMed] [Google Scholar]