Abstract
Although glycerol is the primary carbon source available to halophilic heterotrophic communities, little is known regarding haloarchaeal glycerol metabolism. In this study, a gene encoding a glycerol kinase homolog (glpK; HVO_1541) was deleted from the genome of the haloarchaeon Haloferax volcanii by a markerless knockout strategy. The glpK mutant, KS4, readily grew on yeast extract-peptone complex medium and glucose minimal medium but was incapable of growth on glycerol. Glycerol kinase activity was dependent on the glpK gene and readily detected in cells grown on glucose and/or glycerol, with the activity level higher in medium supplemented with glycerol (with or without glucose) than in medium with glucose alone. An analysis of carbon utilization revealed that glycerol suppressed the metabolism of glucose in both the parent H26 and glpK mutant strains, with catabolite repression more pronounced in the glycerol kinase mutant. Transcripts specific for glpK and an upstream gene, gpdA, encoding a homolog of glycerol-3-phosphate dehydrogenase subunit A, were upregulated (8- and 74-fold, respectively) in the presence of glycerol and glucose compared to those in the presence of glucose alone. Furthermore, glpK was transcriptionally linked to the gpdC gene of the putative glycerol-3-phosphate dehydrogenase operon (gpdABC), based on the findings of reverse transcriptase PCR analysis. The results presented here provide genetic and biochemical evidence that glycerol metabolism proceeds through a glycerol kinase encoded by glpK and suggest that a glycerol-3-phosphate dehydrogenase encoded by the upstream gpdABC operon is also involved in this pathway. Furthermore, our findings reveal a unique example of glycerol-induced repression of glucose metabolism in H. volcanii.
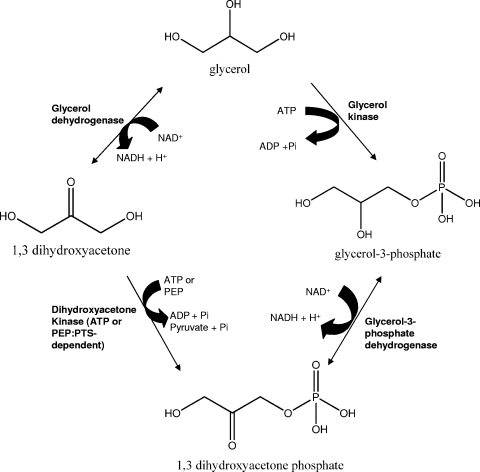
Halophilic and halotolerant microorganisms have adapted different methods for withstanding the high osmotic pressure exerted by their surrounding hypersaline environment. Halophilic archaea (7, 13), as well as the halophilic bacterium Salinibacter ruber (19), maintain a high intracellular salt concentration by accumulating K+ and Cl− ions and excluding Na+ ions, thus requiring intracellular proteins to be active under high-salt conditions. Many halophilic bacteria (24), the halotolerant green alga Dunaliella sp. (3), and some haloarchaea (12) exclude cytoplasmic salts and rely on organic solutes such as ectoine, glycine betaine, and glycerol to provide osmotic balance. Glycerol, in particular, is accumulated in molar quantities by Dunaliella as an organic osmotic solute. Due to leakage from healthy Dunaliella cells (1, 3, 26) and/or cellular lysis, glycerol is released into the surrounding environment, where it serves as a primary energy source for haloarchaea. Upon uptake, halophilic microorganisms assimilate glycerol into dihydroxyacetone phosphate (DHAP) by one of two catabolic routes (Fig. 1). In one route, glycerol is first phosphorylated by glycerol kinase to form sn-glycerol-3-phosphate, which is subsequently oxidized by sn-glycerol-3-phosphate dehydrogenase to produce DHAP. Alternatively, glycerol can be first oxidized by glycerol dehydrogenase to form dihydroxyacetone (DHA), which is subsequently phosphorylated by an ATP- or phosphoenolpyruvate:phosphotransferase system-dependent DHA kinase to yield DHAP. Once generated from glycerol, DHAP can be channeled into pyruvate and other metabolic intermediates, including sn-glycerol-1-phosphate, used as a phospholipid backbone in archaea (16).
FIG. 1.
Halophilic microorganisms assimilate glycerol into DHAP by one of two catabolic routes. In the route depicted on the right, glycerol is phosphorylated by glycerol kinase and subsequently converted into DHAP via a dehydrogenation reaction. In the route depicted on the left, glycerol is oxidized via glycerol dehydrogenase to form DHA, which is subsequently phosphorylated by an ATP- or phosphoenolpyruvate:phosphotransferase system (PEP:PTS)-dependent DHA kinase to yield DHAP. DHAP is channeled into pyruvate and other metabolic intermediates.
Although glycerol is an important carbon and energy source for members of halophilic, heterotrophic communities, little is known regarding glycerol metabolism, especially in haloarchaea. Halophilic archaea such as Haloferax volcanii have been demonstrated previously to metabolize glycerol (22), and specific activities of glycerol-metabolizing enzymes in various haloarchaea have been determined (18, 22, 25); however, the metabolic pathways surrounding glycerol utilization at the molecular level have not been described. Here, we provide genetic and biochemical evidence that H. volcanii metabolizes glycerol via a glycerol kinase (encoded by glpK) and, most likely, a glpK-linked sn-glycerol-3-phosphate dehydrogenase (encoded by a gpdABC operon). We also present data suggesting that glycerol may serve as a catabolite repressor of glucose metabolism in H. volcanii. Our results provide insight into the central metabolic pathways of heterotrophic haloarchaea such as H. volcanii.
MATERIALS AND METHODS
Materials.
Biochemicals and α-glycerophosphate dehydrogenase used for glycerol kinase activity analyses were purchased from Sigma-Aldrich (St. Louis, MO). Other organic and inorganic analytical-grade chemicals were from Fisher Scientific (Atlanta, GA) and Bio-Rad (Hercules, CA). Desalted oligonucleotides were from Integrated DNA Technologies (Coralville, IN). dUTP coupled to digoxigenin (DIG) by an 11-atom spacer, alkaline phosphatase-conjugated antibody raised against DIG, disodium 3-{4-methoxyspiro[1,2-dioxetane-3,2′-(5′-chloro)tricyclo(3.3.1.13,7)decan]-4-yl} phenyl phosphate (CSPD), and other DIG-related biochemicals were from Roche Molecular Biochemicals (Indianapolis, IN). Positively charged membranes for Southern hybridization were from Ambion (Austin, TX). Phusion and Taq DNA polymerases, restriction enzymes, T4 polynucleotide kinase, and T4 DNA ligase were from New England Biolabs (Ipswich, MA). Standard agarose used for the separation of DNA for Southern blotting and routine analysis was from Bio-Rad. Hi-Lo DNA standards were from Minnesota Molecular, Inc. (Minneapolis).
Strains, media, and plasmids.
Strains, oligonucleotide primers used for cloning, and plasmids are summarized in Table 1 and Table S1 in the supplemental material. Escherichia coli DH5α was used for routine recombinant DNA experiments. H. volcanii strains were transformed (8) using plasmid DNA isolated from E. coli GM2163. Liquid cultures were aerated at 200 rpm. E. coli strains were grown at 37°C in Luria-Bertani medium supplemented with 100 mg per liter ampicillin as needed. H. volcanii strains were grown at 42°C in various media, including yeast extract-peptone-Casamino Acids, Casamino Acids, glycerol minimal medium (Gly MM), Gly MM supplemented with glucose (Gly Glu MM), and glucose minimal medium (Glu MM). Medium formulae were according to The Halohandbook (9), with the following exception: 20 mM glycerol and/or glucose served as the sole carbon source(s) for the minimal medium. Media were supplemented as needed with novobiocin (0.1 μg·ml−1), 5-fluoroorotic acid (50 μg·ml−1), and uracil (10 and 50 μg·ml−1 for growth in the presence and absence of 5-fluoroorotic acid, respectively). Uracil and 5-fluoroorotic acid were solubilized in 100% dimethyl sulfoxide at 50 mg·ml−1 prior to addition to the growth medium.
TABLE 1.
List of strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Source or reference |
|---|---|---|
| E. coli strains | ||
| DH5α | F−recA1 endA1 hsdR17(rK− mK+) supE44 thi-1 gyrA relA1 | Life Technologies |
| GM2163 | F−ara-14 leuB6 fhuA31 lacY1 tsx78 glnV44 galK2 galT22 mcrA dcm-6 hisG4 rfbD1 rpsL136 dam13::Tn9 xylA5 mtl-1 thi-1 mcrB1 hsdR2 | New England Biolabs |
| H. volcanii strains | ||
| DS70 | Wild-type isolate DS2 cured of plasmid pHV2 | 21 |
| H26 | DS70 pyrE2 | 2 |
| KS4 | H26 glpK (devoid of GlpK) | This study |
| Plasmids | ||
| pTA131 | Apr; pBluescript II containing Pfdx-pyrE2 | 2 |
| pJAM202c | Apr Nvr; control plasmid derived from pBAP5010 | 22 |
| pJAM2055 | Apr Nvr; pJAM202c-derived expression plasmid including HpaI site for C-terminal fusion to StrepII tag | G. Y. Zhou, unpublished data |
| pJAM2658 | Apr; pTA131-derived presuicide plasmid containing glpK with ∼500-bp genomic DNA sequences flanking 5′ and 3′ ends of glpK | This study |
| pJAM2675 | Apr; pJAM2658-derived glpK suicide plasmid | This study |
| pJAM2666 | Apr Nvr; pJAM2055-derived expression plasmid containing P2rrn-glpK-StrepII tag | This study |
The StrepII tag is a peptide that binds to the biotin binding site of streptavidin. Apr, ampicillin resistance; Nvr, novobiocin resistance.
For growth assays, cells were grown in yeast extract-peptone-Casamino Acids, Gly MM, Gly Glu MM, and Glu MM as indicated below. Cells from −80°C glycerol stocks were freshly inoculated onto the appropriate agar-based media on plates. Cells were thrice subcultured and used as inocula for the final analyses of growth under various conditions as described below. Each subculture was inoculated to a final optical density at 600 nm (OD600) of 0.03 to 0.04. For the analyses of growth rates and cell yields, cells were grown in 20 ml of medium in 250-ml baffled Erlenmeyer flasks. For enzyme activity assays, cells were grown in 100 ml of Gly MM, Glu MM, or Gly Glu MM in 1,000-ml flasks. For RNA preparation, cells were grown in 3 ml of medium in 13- by 100-cm2 culture tubes. Cell growth was monitored by an increase in OD600 (where 1 OD600 unit equals approximately 109 CFU·ml−1 for all strains used in this study). All experiments were performed at least in triplicate.
DNA isolation and analysis.
DNA was separated by electrophoresis using 0.8% (wt/vol) agarose gels in 1× TAE electrophoresis buffer (40 mM Tris acetate, 2 mM EDTA, pH 8.5). Plasmid DNA was isolated from E. coli strains by using the QIAprep spin miniprep kit (Qiagen, Valencia, CA). PCR products were purified by using the MinElute kit (Qiagen) prior to modification by a restriction enzyme (BamHI, HindIII, KpnI, XbaI, HpaI, or NdeI) or T4 DNA polynucleotide kinase. For rapid PCR screening, template DNA was extracted from H. volcanii mutant and parent strains and recombinant E. coli DH5α as described previously (28). For Southern blotting, H. volcanii genomic DNA was isolated from 5-ml cultures by DNA spooling (9).
PCRs.
High-fidelity double-stranded DNA used for the construction of the plasmids listed in Table 1 was amplified by PCR using Phusion DNA polymerase. Taq DNA polymerase was used for screening mutant strains and for generating the DIG-labeled double-stranded DNA probes for Southern blotting. All PCRs were performed according to the instructions of the material suppliers, with the following modifications: 3% (vol/vol) dimethyl sulfoxide and a DIG-labeling mix of 1.1 mM dATP, 1.1 mM dCTP, 1.1 mM d6TP, 0.75 mM dTTP, and 0.35 mM DIG-11 dUTP (catalog no. 1277065; Roche) were included as needed. The primer pairs and DNA templates used for the PCRs are outlined in Table S1 in the supplemental material. PCR was performed using an iCycler or GeneCycler (Bio-Rad Laboratories), and products were analyzed on 0.8% (wt/vol) agarose gels in TAE buffer. Gels were stained with ethidium bromide at 0.5 μg·ml−1 and photographed with a MiniVisionary imaging system (Fotodyne, Hartland, WI). The sizes of the fragments were estimated using the Hi-Lo DNA molecular weight markers.
DNA sequencing.
The fidelity of all PCR-amplified products was confirmed by sequencing both DNA strands of the plasmid inserts listed in Table 1, as well as those of the mutant strains, by Sanger automated DNA sequencing using an Applied Biosystems model 3130 genetic analyzer (ICBR Genomics Division, University of Florida).
Southern blotting.
Genomic DNA isolated from H. volcanii parent and mutant KS4 strains was subjected to digestion with SphI and BspHI and analyzed by CSPD-mediated chemiluminescence using DIG-labeled probes specific for the 500-bp region flanking the 5′ or 3′ end of the target coding region as described previously (28).
Chromosomal gene knockout.
A gene (glpK; HVO_1541) encoding a glycerol kinase homolog was targeted for markerless deletion from the chromosome of H. volcanii H26 by using the pyrE2-based “pop-in/pop-out” method (2, 5). Colonies were screened for the absence of a readily generated 500-bp PCR product by using internal primers (Negative-Forward and Negative-Reverse primer pairs) specific for the coding region of the gene. Strains which did not generate this PCR product were confirmed to be mutant strains by Southern blotting (as described above) and PCR with Confirm-Forward and Confirm-Reverse primer pairs, which anneal to a site outside the H. volcanii genomic region cloned into the suicide plasmid (pJAM2675). The latter PCR products were separated by agarose gel electrophoresis and sequenced to confirm DNA fidelity (as described above). The sequences of primers used for the cloning and screening of mutant colonies are provided in Table S1 in the supplemental material.
Enzyme activity assays.
All experiments were performed in biological triplicate, and the means ± standard deviations (SD) of the results were calculated. Exponential-growth-phase cells were harvested by centrifugation (15 min at 6,000 × g and 4°C), resuspended in 100 mM potassium phosphate buffer (pH 7.4) with 3 M KCl, and lysed by sonication (four pulses of 20 s at 140 W). Debris was removed by centrifugation (10 min at 12,000 × g and 4°C), and the protein concentration in the cell extract was estimated using the Bradford assay with bovine serum albumin as a standard.
Glycerol kinase (ATP:glycerol 3-phosphotransferase; EC 2.7.1.30) specific activity was assayed by a coupled photometric reaction as described previously (23), with the following modifications. The reaction mixture (5 ml) contained 0.5 ml of cell extract (0.5 to 1 mg of protein·ml−1), 30 μmol ATP, and 100 μmol l-cysteine in 100 mM potassium phosphate buffer (pH 7.4) supplemented with 3 M KCl. The reaction (at 42°C to parallel cell growth) was started by the addition of 125 μmol glycerol. Negative control mixtures lacked ATP or glycerol, or the reactions were performed using boiled cell lysate from the parent strain H26. Samples (0.45 ml) were withdrawn at 10, 20, 30 35, 40, 45, 50, 60, and 90 min after the start of the reaction, which was terminated by the addition of 0.45 ml of 0.2 N H3PO4, and the reaction mixture was used to measure the formation of glycerol-3-phosphate with rabbit muscle glycerol-3-phosphate dehydrogenase as described previously (23).
RNA purification.
Total RNA was isolated from H. volcanii parent H26 cells (exponential phase; OD600, 0.4 to 0.6) by using RNeasy RNA purification columns (Qiagen). RNA was treated with amplification-grade DNase I according to the recommendations of the supplier (Sigma-Aldrich), with the following modifications: 3 U of enzyme per μg of RNA was added, and the mixture was incubated for 15 min at 22°C. The integrity of RNA was determined by agarose gel electrophoresis. The RNA concentration was determined by assessing A260 with a Bio-Rad SmartSpec 3000 instrument.
qRT-PCR and RT-PCR.
Quantitative reverse transcriptase PCR (qRT-PCR) and RT-PCR were performed using H. volcanii total RNA (0.1 μg) as the template, appropriate primers (see Table S1 in the supplemental material), iQSYBR green supermix (Bio-Rad), and an iCycler (Bio-Rad). RNA was reverse transcribed to generate cDNA by using an iSCRIPT kit according to the instructions of the manufacturer (Bio-Rad). After cDNA synthesis (at 25°C for 5 min, 42°C for 30 min, and 85°C for 5 min), qRT-PCR mixtures were preheated to 95°C (4 min) and subjected to 40 amplification cycles consisting of denaturation (95°C for 30 s), annealing (1 min at temperatures listed in Table S1 in the supplemental material), and elongation (72°C for 17 s). Final extension was performed at 72°C (10 min). For RT-PCR, reaction mixtures were preheated to 95°C (4 min) and subjected to 40 amplification cycles consisting of denaturation (95°C for 30 s), annealing (58°C for 1 min), and elongation (72°C for 41 s). For each primer pair, negative and positive controls were included to exclude genomic DNA contamination and confirm primer pair function, respectively. For the controls, reactions were identical, with the following exceptions: the negative control sample was maintained on ice during the reverse transcription step, and for the positive control, H. volcanii genomic DNA was used as a template.
HPLC.
At various time points during growth on Glu MM and Gly Glu MM, 1-ml samples of both parent (H26) and glycerol kinase glpK mutant (KS4) cultures were withdrawn and centrifuged (10 min at 10,000 × g and 4°C). Supernatant fractions were filtered and subsequently analyzed by high-performance liquid chromatography (HPLC) using a Bio-Rad HPX-87H column with a 4 mM H2SO4 eluent. All experiments were performed at least in triplicate.
RESULTS AND DISCUSSION
Glycerol is metabolized through glycerol kinase.
To analyze glycerol catabolism in H. volcanii, a gene encoding a glycerol kinase homolog (HVO_1541; glpK) was targeted for knockout in a pyrE2 mutant strain (H26). The deduced product of this gene, GlpK, was most closely related (with 74 to 78% identity) to and clustered in dendrograms with other putative glycerol kinases of haloarchaea, including those of Halorubrum lacusprofundi, Haloarcula marismortui, Haloquadratum walsbyi, Natrialba magadii, and Halobacterium salinarum (sp. NRC-1), with the notable absence of GlpK homologs in the haloalkaliphilic archaeon Natronomonas pharaonis and other archaea (see Fig. S1 in the supplemental material). The H. volcanii and other haloarchaeal GlpK proteins also clustered with the bacterial glycerol kinases with the greatest degrees of identity (up to 58%) to those of the “Thermoanaerobacterales” and Thermotogales (see Fig. S1 in the supplemental material).
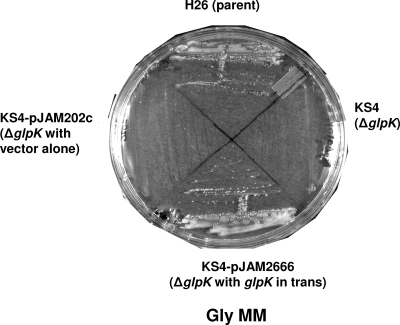
The glpK gene was deleted from the chromosome of H. volcanii by a markerless knockout strategy as described previously (2, 5). Gene deletion was confirmed by PCR, Southern blotting, and sequencing analysis (Table 1; see also Table S1 and Fig. S2 in the supplemental material). The resultant glycerol kinase mutant, H26 ΔglpK (KS4), was incapable of growth either on Gly MM plates (Fig. 2) or in liquid culture (data not shown). A pHV2-based self-replicating plasmid containing the glpK gene under the control of a strong rRNA P2 promoter (pJAM2666) complemented KS4 for growth on Gly MM, while the plasmid vector alone (pJAM202c) did not complement this glpK mutant (Fig. 2).
FIG. 2.
H. volcanii metabolizes glycerol through glycerol kinase. Both the H26 parent strain and the H26 ΔglpK mutant KS4 transformed with a complementary plasmid carrying the glycerol kinase gene homolog glpK (pJAM2666) grew on Gly MM. In contrast, the glpK mutant strain (KS4) and KS4 transformed with vector alone (pJAM202c) were unable to grow on Gly MM. Cells were transferred with a loop from liquid Glu MM cultures onto plates of solid Gly MM, and the plates were incubated at 42°C for 3.5 days.
Based on these results, we conclude that the glycerol kinase homolog encoded by glpK is required for the growth of H. volcanii on glycerol. Previous studies reported glycerol kinase activity in the lysate of H. volcanii cells grown in the presence of glycerol, while glycerol dehydrogenase activity remained undetectable (e.g., reference 22). These early findings support our claim that in H. volcanii, glycerol metabolism proceeds through the glpK-encoded glycerol kinase rather than the conversion of glycerol to DHA by a glycerol dehydrogenase. Thus, our results also suggest that genes HVO_1546 to HVO_1544, which are predicted to encode putative DHA kinase subunits K, L, and M based on the H. volcanii genome sequence (communicated by J. Eisen, TIGR; http://archaea.ucsc.edu/, April 2007 version), function in DHA metabolism and not glycerol metabolism. Consistent with this possibility, recent evidence suggests that Salinibacter ruber mediates the incomplete oxidation of glycerol to yield DHA as an overflow product, which may then be taken up by different types of heterotrophs present in hypersaline environments (4).
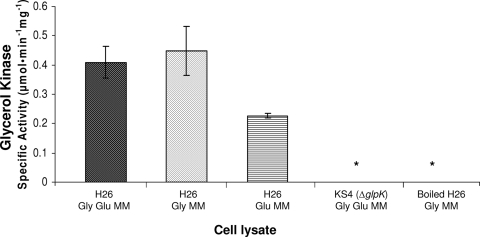
To biochemically confirm that the glpK homolog HVO_1541 codes for glycerol kinase, the specific activities of glycerol kinase in cell lysates from H26 and KS4 (H26 ΔglpK) were measured as described previously (6, 20). In brief, the generation of glycerol-3-phosphate (the product of the phosphorylation reaction catalyzed by glycerol kinase) was evaluated by a photometric, coupled assay that included glycerol as a substrate. Glycerol-3-phosphate dehydrogenase, which catalyzes the NAD+-dependent oxidation of glycerol-3-phosphate to form DHAP, was also included in the assay. The reduction of NAD+ to NADH was monitored at A340, and product formation was quantified using standards of glycerol-3-phosphate. By using this assay, glycerol kinase activity was readily detected in cells of the parent strain H26 grown in medium containing glycerol (Gly MM or Gly Glu MM), regardless of the presence of glucose (Fig. 3). Significant levels of glycerol kinase activity were also detected in H26 cells grown on medium with glucose alone (Glu MM), although the levels were reduced about twofold compared to those in H26 cells grown on media with glycerol (i.e., Gly MM and Gly Glu MM). Glycerol kinase activity was not detected in the glycerol kinase mutant (KS4) or boiled cell lysate (the negative control) (Fig. 3). The specific activity of glycerol kinase in parent strain H26 grown in the presence of glycerol was measured as 430 ± 30 nmol·min−1 mg−1; although fivefold or more higher than previously reported specific activities of glycerol kinase proteins in Salinibacter ruber (90 nmol·min−1 mg−1), Halobacterium cutirubrum (14 nmol·min−1 mg−1), and H. volcanii (31 nmol·min−1 mg−1 for cells grown in complex medium with peptides), determined in similar assays (20, 23, 25), this value was within a reasonable range of measurement for glycerol kinase enzymes.
FIG. 3.
H. volcanii glycerol kinase activity is dependent on glpK (HVO_1541), stimulated by growth on glycerol, and uninhibited by glucose supplementation. H. volcanii H26 (parent) and KS4 (glpK mutant) cells were grown in Gly Glu MM, Gly MM, or Glu MM. Measurements of glycerol kinase specific activities were performed using lysates prepared from log-phase cells as specified in Materials and Methods. * indicates that enzyme activity was not detected. Experiments were performed in biological triplicate, and the means ± SD were calculated.
Our results demonstrate that the glpK homolog HVO_1541 encodes a glycerol kinase and provide strong evidence that this gene is required for the catabolism of glycerol in H. volcanii. Note that glycerol kinase activity is not universal among the archaea, unlike bacteria, and is absent in those organisms that cannot use glycerol as an energy source (e.g., autotrophic methanogens [16]). Thus, glycerol kinase is not thought to be involved in the synthesis of the backbone of archaeal phospholipids and is instead mediated by sn-glycerol-1-phosphate dehydrogenase, which generates sn-glycerol-1-phosphate from DHAP. Therefore, glycerol does not appear to be channeled into DHA by a glycerol dehydrogenase for the growth of H. volcanii.
Our results also reveal that glycerol kinase activity in H. volcanii, although stimulated by growth on glycerol, is not inhibited by the presence of glucose in the growth medium. The detection of comparable levels of glycerol kinase activity in H. volcanii cells grown on Gly MM regardless of glucose supplementation directly contrasts with results for the E. coli model, in which glucose exhibits catabolite repression of the glpK regulon encoding glycerol kinase (10).
Glycerol serves as a catabolite repressor of glucose metabolism.
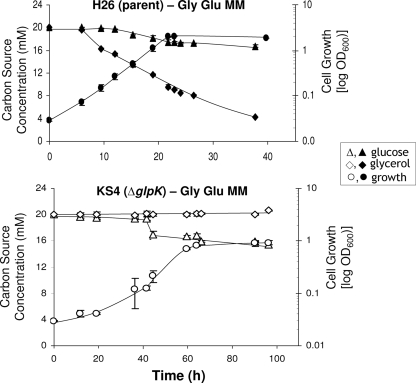
To determine if the growth defect of the glycerol kinase mutant (KS4) was exclusive to glycerol and to further investigate the observed differences in catabolite repression between H. volcanii and E. coli, the growth of and utilization of carbon by the H. volcanii H26 parent and KS4 mutant strains on minimal medium supplemented with either 20 mM glycerol and 20 mM glucose or 20 mM glucose alone (Gly Glu MM or Glu MM, respectively) were measured (Fig. 4 and 5). Although there are other carbon sources (i.e., Tris buffer, uracil, biotin, and thiamine) in the minimal medium, these alone do not support the growth of H. volcanii (data not shown). Thus, any observed growth would be due to glucose and/or glycerol supplementation.
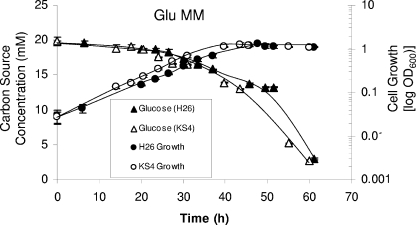
FIG. 4.
The parent strain H26 and glycerol kinase mutant KS4 (H26 ΔglpK) exhibit similar growth rates, cell yields, and carbon utilization patterns when grown in Glu MM. Growth at 42°C (200 rpm) was monitored by an increase in OD600, where 1 U was equivalent to approximately 109 CFU per ml for all strains. At various time points, supernatant fractions were withdrawn from both parent H26 and KS4 cultures and analyzed via HPLC for glucose consumption. Experiments were performed in triplicate, and the means ± SD were calculated. Cell growth and glucose utilization levels are indicated.
FIG. 5.
Parent strain H26 and glycerol kinase mutant KS4 (H26 ΔglpK) exhibit glycerol-induced repression of glucose metabolism. The growth rates of and levels of carbon utilization by parent strain H26 and mutant KS4 cells grown on Gly Glu MM are shown. Growth at 42°C (200 rpm) was monitored by an increase in OD600, where 1 U was equivalent to approximately 109 CFU per ml for all strains. At various time points, supernatant fractions were withdrawn from cultures and analyzed via HPLC for glycerol and glucose consumption. Experiments were performed in triplicate, and the means ± SD were calculated.
The glpK mutant (KS4) and parent strain H26 grew and utilized glucose at nearly identical rates when cultured in minimal medium containing glucose as the sole carbon source (Glu MM) (Fig. 4). In both cases, roughly 85% of the glucose was metabolized within 60 h, contributing to an average overall OD600 of 1.27 and a growth rate of 0.11 doublings per h. The addition of glycerol to the Glu MM (producing Gly Glu MM) enhanced the growth rate and cell yield of the parent H26 approximately twofold, to 0.22 doublings per h and an OD600 of 2.0 (Fig. 5). Consistent with these results, H26 metabolized both of these carbon/energy sources but displayed a preference for glycerol, with 79% of the glycerol and only 16% of the glucose in the Gly Glu MM being metabolized (Fig. 5). Thus, H26 metabolized glucose at a reduced rate when glycerol was included in the growth medium. In contrast to H26, the glpK mutant KS4 was unable to metabolize glycerol and did not induce glucose metabolism when grown in the presence of glycerol until approximately 30 h later than its parent, H26 (Fig. 5), and 20 h later than KS4 grown on medium with glucose alone (Fig. 4). Once KS4 initiated this delayed metabolism of glucose in glycerol-supplemented medium (Gly Glu MM), the growth rate (0.095 doublings per h) and final OD600 (0.93) of these cells were only slightly reduced compared to those of cells grown in medium with glucose alone.
Based on these results, it appears that H. volcanii mediates glycerol-dependent catabolite repression of glucose metabolism. Glucose metabolism by both H. volcanii strains examined was repressed by the presence of glycerol, with the repression in the glpK mutant more pronounced than that in its parent, based on the extreme lag observed in the growth of KS4 on medium with glycerol and glucose compared to that on glucose alone. The observed preference of H. volcanii for glycerol directly contrasts with that of E. coli, which exhibits diauxic growth with glucose as the preferred carbon and energy source (11). Catabolite repression by carbon compounds other than glucose is not novel. For example, members of the genus Pseudomonas exhibit organic acid-induced catabolite repression of glucose metabolism (15). However, glycerol-induced catabolite repression has not been reported previously.
Levels of glycerol-3-phosphate dehydrogenase and glycerol kinase transcripts are upregulated by the addition of glycerol.
Based on the genome sequence (Hartman et al., unpublished), H. volcanii has two putative NAD(P)-linked sn-glycerol-3-phosphate dehydrogenase operons: one (gpdA2B2C2; HVO_A0269 to HVO_A0271) on the minichromosome pHV4 and one (gpdABC; HVO_1538 to HVO_1540) on the chromosome directly upstream of the glycerol kinase gene (Fig. 6A). The protein paralogs encoded by these two operons are closely related in primary amino acid sequence (58 to 74% identical and 67 to 85% similar) and cluster with oxidoreductases such as the A, B, and C subunits of sn-glycerol-3-phosphate dehydrogenase. The products of both operons are distinct from the enantiomeric glycerophosphate synthase (EgsA), an NAD(P)-linked sn-glycerol-1-phosphate dehydrogenase responsible for the formation of the glycerol-1-phosphate backbone of archaeal phospholipids (16) and most likely encoded by HVO_0822 in H. volcanii.
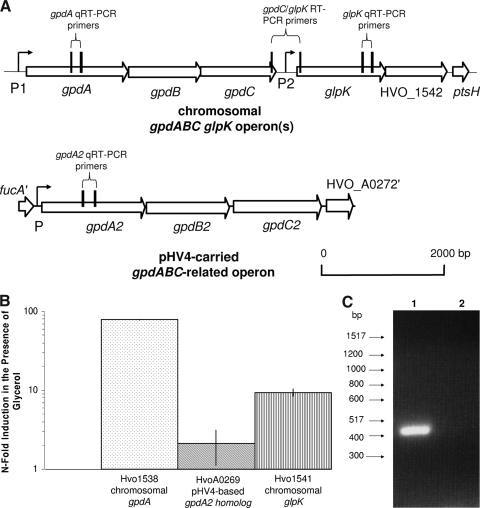
FIG. 6.
Genomic organization and transcript analysis of the glycerol kinase gene and glycerol-3-phosphate dehydrogenase-related operons of H. volcanii. (A) Schematic representations of the glycerol kinase gene (glpK) and the glycerol-3-phosphate dehydrogenase-related operons (gpdABC and gpdA2B2C2) of H. volcanii and location of annealing sites for qRT- and RT-PCR. Representations of the chromosomally located glpK and gpdABC operon(s) and the pHV4-based gpdA2B2C2 operon are presented. Vertical lines indicate annealing sites of primers used for qRT- and RT-PCR analyses. P1, P2, and P signify locations of BRE and TATA box archaeal promoter consensus elements. (B) Relative quantification of transcript levels specific for both the chromosomal gpdA and pHV4-based gpdA2 genes encoding glycerol-3-phosphate dehydrogenase subunit A homologs (Hvo1538 and HvoA0269, respectively) and the glycerol kinase glpK gene (Hvo1541). Transcripts for the chromosomal glpK and gpdA genes are upregulated in the presence of glycerol. Transcript levels were derived by qRT-PCR as described in Materials and Methods. Calculations are based on the n-fold induction of transcription in the presence of Gly Glu MM compared to transcription in the presence of Glu MM. Results were normalized to the n-fold induction of the internal control, ribL. Experiments were performed in triplicate, and the means ± SD were calculated. (C) Chromosomal glycerol kinase (glpK) and glycerol-3-phosphate dehydrogenase (gpdABC) genes are under the control of a common promoter. An RT-PCR primer pair based on the 5′ and 3′ ends of glpK and gpdC, respectively, was designed. Hi-Lo DNA markers and molecular sizes are indicated to the left. Total RNA from parent H26 was extracted and reverse transcribed to generate cDNA, which was used as a template for PCR (lane 1). RNA which had not undergone reverse transcription was used as a negative control template for PCR (lane 2).
In order to determine whether either gpd operon is upregulated in the presence of glycerol, qRT-PCR was performed using gene-specific primers (see Table S1 in the supplemental material for all primer details). Primers were designed to correspond to the first gene (gpdA2 [HVO_A0269] and gpdA [HVO_1538]) in each of the two operons in order to achieve the strongest signal for transcriptional analysis (Fig. 6A). In addition, qRT-PCR primers for glpK were designed to determine if the transcription of this gene was induced by glycerol. A transcript specific to the ribosomal protein L10 gene (ribL) was used as an internal control based on data from previous studies (17) and our confirmation by qRT-PCR that the level of induction of transcripts specific for ribL in cells grown in the presence of glycerol and glucose was close to onefold compared to transcript levels in cells grown in the presence of glucose alone.
By qRT-PCR analysis, transcripts specific for glpK, gpdA, and gpdA2 were detected at significant levels under all growth conditions examined (growth on Gly MM and Gly Glu MM) (Fig. 6B). In addition, transcripts specific for gpdA and glpK were upregulated approximately 78- and 9-fold, respectively, in the presence of glycerol and glucose compared to those in the presence of glucose alone (Fig. 6B). In contrast, gpdA2 transcripts were not induced to significant levels (2.0- ± 1.0-fold) by glycerol (Fig. 6 B), indicating that the gpdA2 operon is not likely to be involved in glycerol metabolism. Our results reveal that transcripts specific for glpK and its gene neighbor gpdA are significantly induced by glycerol, supporting the argument that both genes and their encoded enzymes (glycerol kinase and glycerol-3-phosphate dehydrogenase) are involved in glycerol metabolism in H. volcanii.
Glycerol kinase and glycerol-3-phosphate dehydrogenase genes are under the control of a common promoter.
Due to the close proximity of glpK and gpdABC and the likely involvement of the encoded gene products in a common metabolic pathway (14, 21), we investigated whether these genes were cotranscribed in an operon. RT-PCR was performed using primers designed such that the forward primer would anneal to the 3′ coding region of gpdC and the reverse primer would anneal to the 5′ coding region of glpK, amplifying a portion of each gene as well as the 364-bp intergenic region (Fig. 6A). A single PCR product of the expected size was detected using synthesized cDNA (Fig. 6C), and the sequence of the product was later confirmed. No product from the negative control reaction with RNA as a template was detected (Fig. 6C). Thus, glpK and gpdC are linked at the transcriptional level.
Although we have demonstrated that glpK and gpdC of the putative gpdABC operon are transcriptionally linked, the reasons for the significant differences in the levels of induction of glpK- and gpdA-specific transcripts in the presence of glycerol (Fig. 6B) remain to be determined. Multiple promoter elements may be involved in the transcription of this region of the chromosome and account for these differences in induction. Consistent with the possibility that multiple promoters control the expression of the gpdABC-glpK region, BRE and TATA box promoter consensus sequence elements were identified upstream of both gpdA and glpK (Fig. 6A, P1 and P2, respectively). Alternatively, the transcription of glpK (which is distal relative to gpdA) may be reduced by transcriptional polarity, which leads to the premature termination of polycistronic mRNA translation, resulting in the reduced transcription of genes located distally from the operon (27). Another possibility is that the glpK-specific transcripts are more susceptible to degradation than those specific for gpdA.
Conclusion.
We demonstrate in this study that glycerol metabolism in H. volcanii requires glycerol kinase encoded by the glpK gene (HVO_1541) and that this gene is transcriptionally linked to a putative glycerol-3-phosphate dehydrogenase operon (gpdABC; HVO_1538 to HVO_1540) located upstream of glpK on the chromosome. The levels of both glpK- and gpdA-specific transcripts are significantly upregulated in the presence of glycerol, although not to the same extent, with the glycerol-dependent induction of gpdA-specific transcripts being eightfold greater that of glpK-specific transcripts. Promoter consensus elements upstream of both gpdA and glpK suggest that, in addition to sharing a common promoter with gpdA, glpK may be regulated independently of gpdA. Our present model is that glpK and gpdA share a common P1 promoter immediately upstream of gpdA that is tightly regulated in response to glycerol availability and that additional control of glpK transcription is achieved through a gpdA-independent P2 promoter immediately upstream of glpK.
In this study, we also provide evidence that glucose metabolism in H. volcanii is under catabolite control by glycerol and speculate that this control is mediated by a GlpR-type regulator. Future phenotypic and biochemical characterization of a glpR-deficient strain is expected to provide insight into gene function in H. volcanii, as well as other microorganisms with similar gene organization patterns. Overall, the results not only shed light on glycerol metabolism in H. volcanii, but also add to our understanding of central metabolic pathways of haloarchaea.
Supplementary Material
Acknowledgments
We thank T. Allers for providing parent strain H26 and plasmid pTA131 and Jonathan Eisen and the Institute for Genomic Research for completion and annotation of the H. volcanii genome sequence. We also thank Savita Shanker at the UF Genomics Core of UF ICBR for DNA sequencing.
This work was funded in part by NIH R01 GM057498 and DOE DE-FG02-05ER15650 to J.A.M.-F.
Footnotes
Published ahead of print on 1 May 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Al Harabi, N., and D. J. Gilmour. 2006. A comparison of glycerol production and leakage by three strains of the unicellular green alga Dunaliella (Volvocales, Chloropycaea), abstr. P167. 6th Int. Congr. Extremophiles, Brest, France.
- 2.Allers, T., H. P. Ngo, M. Mevarech, and R. G. Lloyd. 2004. Development of additional selectable markers for the halophilic archaeon Haloferax volcanii based on the leuB and trpA genes. Appl. Environ. Microbiol. 70943-953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bardavid, R. E., P. Khristo, and A. Oren. 2008. Interrelationships between Dunaliella and halophilic prokaryotes in saltern crystallizer ponds. Extremophiles 125-14. [DOI] [PubMed] [Google Scholar]
- 4.Bardavid, R. E., and A. Oren. 2008. Dihydroxyacetone metabolism in Salinibacter ruber and in Haloquadratum walsbyi. Extremophiles 12125-131. [DOI] [PubMed] [Google Scholar]
- 5.Bitan-Banin, G., R. Ortenberg, and M. Mevarech. 2003. Development of a gene knockout system for the halophilic archaeon Haloferax volcanii by use of the pyrE gene. J. Bacteriol. 185772-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bublitz, C., and E. Kennedy. 1954. Synthesis of phosphatides in isolated mitochondria. III. The enzymatic phosphorylation of glycerol. J. Biol. Chem. 211951-961. [PubMed] [Google Scholar]
- 7.Christian, J., and J. Waltho. 1962. Solute concentrations within cells of halophilic and non-halophilic bacteria. Biochim. Biophys. Acta 65506-508. [DOI] [PubMed] [Google Scholar]
- 8.Cline, S. W., W. L. Lam, R. L. Charlebois, L. C. Schalkwyk, and W. F. Doolittle. 1989. Transformation methods for halophilic archaebacteria. Can. J. Microbiol. 35148-152. [DOI] [PubMed] [Google Scholar]
- 9.Dyall-Smith, M. 2008. The halohandbook: protocols for halobacterial genetics. Mark Dyall-Smith, Martinsried, Germany.
- 10.Freedberg, W. B., and E. C. Lin. 1973. Three kinds of controls affecting the expression of the glp regulon in Escherichia coli. J. Bacteriol. 115816-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holtman, C. K., A. C. Pawlyk, N. D. Meadow, and D. W. Pettigrew. 2001. Reverse genetics of Escherichia coli glycerol kinase allosteric regulation and glucose control of glycerol utilization in vivo. J. Bacteriol. 1833336-3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lai, M. C., K. R. Sowers, D. E. Robertson, M. F. Roberts, and R. P. Gunsalus. 1991. Distribution of compatible solutes in the halophilic methanogenic archaebacteria. J. Bacteriol. 1735352-5358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lanyi, J. K. 1974. Salt-dependent properties of proteins from extremely halophilic bacteria. Bacteriol. Rev. 38272-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lawrence, J. G. 1997. Selfish operons and speciation by gene transfer. Trends Microbiol. 5355-359. [DOI] [PubMed] [Google Scholar]
- 15.Lynch, W. H., and M. Franklin. 1978. Effect of temperature on diauxic growth with glucose and organic acids in Pseudomonas fluorescens. Arch. Microbiol. 118133-140. [DOI] [PubMed] [Google Scholar]
- 16.Nishihara, M., T. Yamazaki, T. Oshima, and Y. Koga. 1999. sn-Glycerol-1-phosphate-forming activities in Archaea: separation of archaeal phospholipid biosynthesis and glycerol catabolism by glycerophosphate enantiomers. J. Bacteriol. 1811330-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Norais, C., M. Hawkins, A. L. Hartman, J. A. Eisen, H. Myllykallio, and T. Allers. 2007. Genetic and physical mapping of DNA replication origins in Haloferax volcanii. PLoS Genet. 3e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oren, A. 1994. Enzyme diversity in halophilic archaea. Microbiologia 10217-228. [PubMed] [Google Scholar]
- 19.Oren, A., M. Heldal, S. Norland, and E. A. Galinski. 2002. Intracellular ion and organic solute concentrations of the extremely halophilic bacterium Salinibacter ruber. Extremophiles 6491-498. [DOI] [PubMed] [Google Scholar]
- 20.Oren, A., and P. Gurevich. 1994. Distribution of glycerol dehydrogenase and glycerol kinase activity in halophilic archaea. FEMS Microbiol. Lett. 118311-316. [Google Scholar]
- 21.Overbeek, R., M. Fonstein, M. D'Souza, G. D. Pusch, and N. Maltsev. 1999. The use of gene clusters to infer functional coupling. Proc. Natl. Acad. Sci. USA 962896-2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rawal, N., S. M. Kelkar, and W. Altekar. 1988. Alternative routes of carbohydrate metabolism in halophilic archaebacteria. Ind. J. Biochem. Biophys. 25674-686. [PubMed] [Google Scholar]
- 23.Sher, J., R. Elevi, L. Mana, and A. Oren. 2004. Glycerol metabolism in the extremely halophilic bacterium Salinibacter ruber. FEMS Microbiol. Lett. 232211-215. [DOI] [PubMed] [Google Scholar]
- 24.Ventosa, A., J. J. Nieto, and A. Oren. 1998. Biology of moderately halophilic aerobic bacteria. Microbiol. Mol. Biol. Rev. 62504-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wassef, M. K., J. Sarner, and M. Kates. 1970. Stereospecificity of the glycerol kinase and the glycerophosphate dehydrogenase in Halobacterium cutirubrum. Can. J. Biochem. 4869-73. [DOI] [PubMed] [Google Scholar]
- 26.Wegmann, K., A. Ben Amotz, and M. Avron. 1980. Effect of temperature on glycerol retention in the halotolerant algae Dunaliella and Asteromonas. Plant Physiol. 661196-1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wek, R. C., J. H. Sameshima, and G. W. Hatfield. 1987. Rho-dependent transcriptional polarity in the ilvGMEDA operon of wild-type Escherichia coli K12. J. Biol. Chem. 26215256-15261. [PubMed] [Google Scholar]
- 28.Zhou, G. Y., D. Kowalczyk, M. A. Humbard, S. Rohatgi, and J. A. Maupin-Furlow. 2008. Proteasomal components required for cell growth and stress responses in the haloarchaeon Haloferax volcanii. J. Bacteriol. 1908096-8105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.