Abstract
In this study, utilizing a Corynebacterium glutamicum ΔpimB′ ΔmgtA double deletion mutant, we unequivocally assign the in vivo functions of Rv2188c as an Ac1PIM1:mannosyltransferase (originally termed PimB′Mt [Mycobacterium tuberculosis PimB′]) and Rv0557 as a GlcAGroAc2:mannosyltransferase (originally termed PimBMt), which we have reassigned as PimBMt and MgtAMt, respectively, in Mycobacterium tuberculosis.
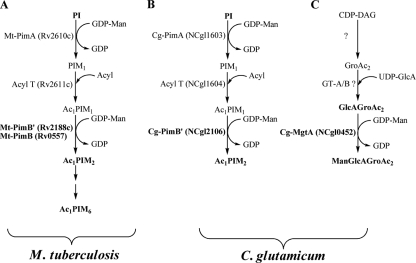
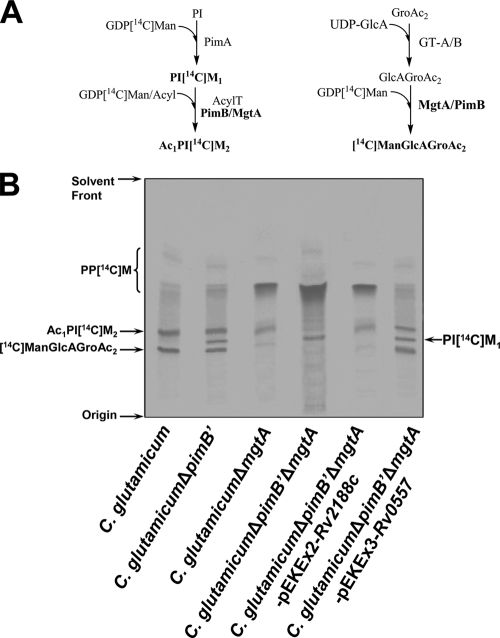
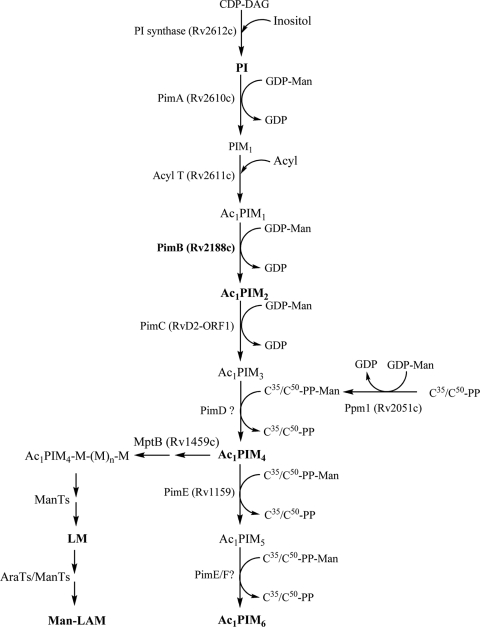
The current model of mycobacterial phosphatidyl-myo-inositol mannoside (PIM) biosynthesis, supported by biochemical and genetic studies, follows a linear pathway from phosphatidylinositol (PI) → Ac1PIM2 → Ac1PIM4 → Ac1PIM6 (4, 17, 19) as shown in Fig. 1. In this pathway, mycobacterial PI is glycosylated by an α-mannopyranosyl residue at the 2-OH position of inositol, followed by the acylation and mannosylation at the 6-OH position of PI to form Ac1PIM2 (3), which is further mannosylated to form Ac1PIM4 and Ac1PIM6, extending the 6-OH position of Ac1PIM2 (19).
FIG. 1.
Glycolipid biosynthetic pathways in Corynebacterineae. (A) PIM synthesis in M. tuberculosis; (B) PIMs; (C) ManGlcAGroAc2 synthesis in C. glutamicum.
In view of the identification of genes involved in PIM, lipomannan (LM), and lipoarabinomannan (LAM) biosynthesis, Schaeffer et al. (22) proposed Rv0557 as an α-d-mannose-α-(1→6)-phosphatidyl-myo-inositol-mannosyltransferase that transfers mannose from GDP-Man to Ac1PIM1 to form Ac1PIM2, a precursor of the immunomodulatory lipoglycans LM and LAM (4, 17). The study was based on a cell-free assay using GDP[14C]Man, Ac1PIM1, Mycobacterium smegmatis membranes, and/or partially purified recombinant Rv0557. On the basis of these in vitro studies, Rv0557 was assigned as PimBMt (Mycobacterium tuberculosis PimB) in the synthesis of Ac1PIM2. However, on the disruption of Rv0557 in Mycobacterium tuberculosis, PIM biosynthesis remains unaffected (G. S. Besra and L. S. Schlesinger, unpublished data), suggesting that either gene duplication or Rv0557 performed another function in M. tuberculosis. Interestingly, in a recent study, Rv0557 was also shown to be involved in the biosynthesis of 1,2-di-O-C16/C18:1-(α-d-mannopyranosyl)-(1→4)-(α-d-glucopyranosylu- ronic acid)-(1→3)-glycerol (ManGlcAGroAc2) and an LM-like molecule in Corynebacterium glutamicum and was termed MgtAMt (M. tuberculosis MgtA) (25). More recently, Rv2188c was also proposed to be involved in the synthesis of Ac1PIM2 as the second α-d-mannose-α-(1→6)-phosphatidyl-myo-inositol-mannosyl transferase (termed PimB′Mt) (13, 16), which has augmented ongoing confusion in the field. Due to the essentiality of M. tuberculosis PIM biosynthesis (3) in this study, we have generated C. glutamicum ΔpimB′ ΔmgtA, deficient in pimB′Cg and mgtACg (C. glutamicum pimB′ and mgtA) and subsequently overexpressed Rv2188c and Rv0557 individually to identify their true in vivo and in vitro biochemical activities.
Construction, growth, and complementation of C. glutamicum ΔpimB′ ΔmgtA.
Rv2188c and Rv0557 both belong to the glycosyltransferase B family (14) and are part of subgroup GT4 according to the Carbohydrate-Active EnZymes (CAZy) classification system (6). Using Rv0557 as a query sequence in a BLAST comparison, the next paralog among the six members of M. tuberculosis within the GT4 family was Rv2188c (identity score, 35%), revealing a structural similarity between the two proteins. Both proteins possess orthologs in C. glutamicum, and previous genetic and biochemical studies confirmed that the orthologous proteins have identical functions (13, 16, 25). When either pimB′Cg or mgtACg was deleted, no reliable growth defect was observed (data not shown). We therefore transformed C. glutamicum ΔpimB′ (16) with the allele replacement vector pK19mobsacBΔmgtA (21, 25) (Table 1) to kanamycin resistance, and after two rounds of positive selection, small colonies on brain heart infusion (BHI) medium plates were obtained. The mgtACg locus was analyzed by PCR, and finally, 1 of 18 positive clones was identified as C. glutamicum ΔpimB′ ΔmgtA, exhibiting the double deletion phenotype. We analyzed the growth of this strain on complex medium (BHI) and found a significantly reduced growth rate in the exponential phase from 0.43 h−1 to 0.32 h−1, whereas the final optical density reached was not influenced (data not shown).
TABLE 1.
Strains, plasmids, and oligonucleotides used in this study
| Strain, plasmid, or oligonucleotidea | Relevant characteristics or sequencea | Source, reference, or purpose |
|---|---|---|
| C. glutamicum strains | ||
| ATCC 13032 | Wild type | Culture collection |
| C. glutamicum ΔmgtA | C. glutamicum devoid of mgtACg | 25 |
| C. glutamicum ΔpimB′ | C. glutamicum devoid of pimB′Cg | 16 |
| C. glutamicum ΔpimB′ΔmgtA | C. glutamicum devoid of pimB′Cg and mgtACg | This work |
| Plasmids | ||
| pK19mobsacBΔmgtA | Vector enabling deletion of 1,094 bp of mgtACg | 25 |
| pEKEx3-Rv0557 | Expression of Rv0557 | 25 |
| pEKEx2-Rv2188c | Expression of Rv2188c | 16 |
| pET16b | Expression vector, His-tag fusion | Novagen |
| pET16b-Cg-pimB′ | Expression of His-PimB′Cg | This work |
| Primers | ||
| Cg-pimB′forprot | CTCCATATGTCTGCATCCCGAAAAACTCTCGTTG | Expression of His6-PimB′Cg (NdeI)b |
| Cg-pimB′revprot | GAGCATATGTTATCGTGGTTCACTCTGCAAAA TATTG | Expression of His6-PimB′Cg (NdeI)c |
Primers are given in their 5′ to 3′ direction.
The linker endonuclease restriction site in the previous column is italicized.
The restriction endonuclease restriction site in the previous column is italicized.
To enable the expression of Rv2188c for functional studies, the open reading frame was amplified and cloned in pEKEx2 (8), producing pEKEx2-Rv2188c (Table 1). This vector, as well as pEKEx3-Rv0557 (25), was introduced into C. glutamicum ΔpimB′ ΔmgtA via electroporation. Complementation with the Rv0557 gene restored growth, whereas the Rv2188c gene was unable to reverse the growth defect but is apparently expressed based on glycolipid and lipoglycan analysis (see below). The complementation of C. glutamicum ΔpimB′ ΔmgtA by pimB′Cg and mgtACg gave similar phenotypes to those of the Rv2188c and Rv0557 genes. The above growth rates for all strains were similar to those for all strains in salt medium CGXII (data not shown).
In vivo glycolipid and lipoglycan analysis.
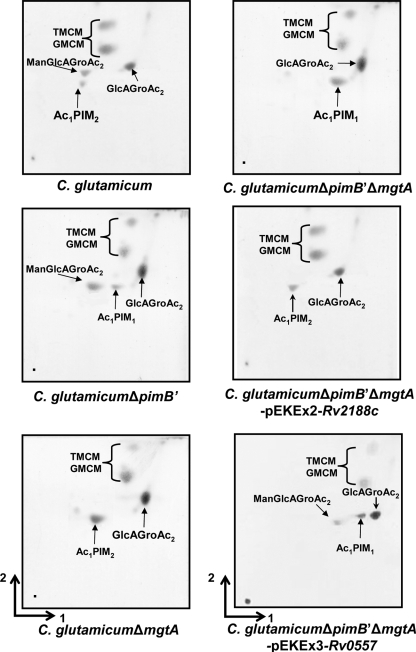
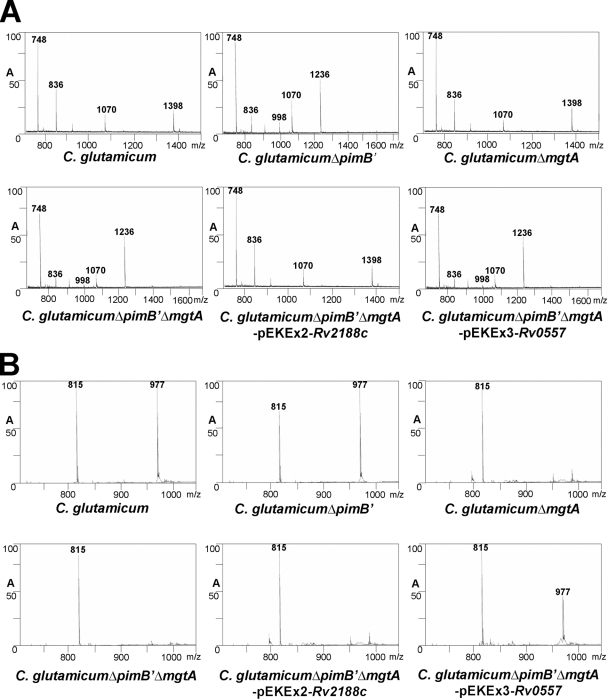
Polar lipids containing PIMs and other glycolipids were extracted from C. glutamicum, C. glutamicum ΔpimB′ (16), C. glutamicum ΔmgtA (25), C. glutamicum ΔpimB′ ΔmgtA, C. glutamicum ΔpimB′ ΔmgtA-pEKEx2-Rv2188c, and C. glutamicum ΔpimB′ ΔmgtA-pEKEx3-Rv0557, using an established chloroform-methanolic saline procedure (7). The extracted lipids were examined by two-dimensional thin-layer chromatography (2D-TLC) and matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS). The lipid extracts from C. glutamicum possessed a typical profile of ManGlcAGroAc2, GlcAGroAc2, Ac1PIM2, trehalose monocorynomycolate (TMCM), and glucose monocorynomycolate (GMCM) by α-naphthol/sulfuric acid staining (Fig. 2). As shown previously the corresponding Ac1PIM2 (negative-ion-mode MALDI-TOF MS, m/z 1398 [M-H]−, fatty acyl groups C16 and C18:1; Fig. 3A) and ManGlcAGroAc2 (positive-ion-mode MALDI-TOF MS, m/z 977 [M-H + 2Na]+, fatty acyl groups C16 and C18:1; Fig. 3B) (25) were confirmed by MS. In addition, as reported earlier, Ac1PIM2 and ManGlcAGroAc2 were completely absent in C. glutamicum ΔpimB′ (16) and C. glutamicum ΔmgtA (25), respectively. Therefore, the absence of both types of lipids in C. glutamicum ΔpimB′ ΔmgtA would be anticipated. Indeed, lipid extracts from the C. glutamicum ΔpimB′ ΔmgtA double knockout were found to be devoid of both Ac1PIM2 and ManGlcAGroAc2 by 2D-TLC (Fig. 2) and MALDI-TOF MS (Fig. 3A and B) and accumulated Ac1PIM1 (negative-ion-mode MALDI-TOF MS, m/z 1236 [M-H]−; Fig. 3A). Therefore, C. glutamicum ΔpimB′ ΔmgtA was utilized to study the role of the orthologous Rv2188c and Rv0557 proteins in this background strain. In the current study, we again establish the inherent usefulness of C. glutamicum in the identification of genes involved in indispensable biochemical pathways in mycobacteria (1-2, 5, 9, 16-18, 23-24).
FIG. 2.
Glycolipid profiles of C. glutamicum, C. glutamicum ΔpimB′, C. glutamicum ΔmgtA, C. glutamicum ΔpimB′ ΔmgtA, C. glutamicum ΔpimB′ ΔmgtA-pEKEx2-Rv2188c, and C. glutamicum ΔpimB′ ΔmgtA-pEKEx3-Rv0557. The polar lipid extracts were examined by 2D-TLC on aluminum-backed plates of silica gel 60 F254 (Merck 5554), using CHCl3/CH3OH/H2O (60:30:6, vol/vol/vol) in the first direction and CHCl3/CH3COOH/CH3OH/H2O (40:25:3:6, vol/vol/vol/vol) in the second direction. Glycolipids were visualized by spraying plates with α-naphthol/sulfuric acid, followed by gentle charring of the plates.
FIG. 3.
MALDI-TOF MS analyses of glycolipids from C. glutamicum, C. glutamicum ΔpimB′, C. glutamicum ΔmgtA, C. glutamicum ΔpimB′ ΔmgtA, C. glutamicum ΔpimB′ ΔmgtA-pEKEx2-Rv2188c, and C. glutamicum ΔpimB′ ΔmgtA-pEKEx3-Rv0557. (A) Negative-ion-mode MALDI-TOF MS analysis of total glycolipid extract from strains. The peaks observed are m/z 836 (M-H)− [PI with C16/C18:1 fatty acyl groups], m/z 998 (M-H)− [PIM1 with C16/C18:1 fatty acyl groups], m/z 1236 (M-H)− [Ac1PIM1with 2C16/C18:1 fatty acyl groups], and m/z 1,398 (M-H)− [Ac1PIM2 with 2C16/C18:1 fatty acyl groups]. The peak m/z 748 was not attributable to any PIM species and, as such, may represent unidentified lipid species and/or plasticizer. (B) Positive-ion MALDI-TOF MS spectrum of the cationized, sodiated precursor ion (M-H + 2Na)+ of GlcAGroAc2 and ManGlcAGroAc2 at m/z 815 and m/z 977, respectively.
The plasmid-borne overexpression of Rv2188c in C. glutamicum ΔpimB′ ΔmgtA restored the synthesis of Ac1PIM2 by 2D-TLC (Fig. 2) and MALDI-TOF MS analysis (Fig. 3A), while ManGlcAGroAc2 was still absent (Fig. 2 and 3B), which suggests that Rv2188c is solely involved in the synthesis of Ac1PIM2. In contrast, the plasmid-borne overexpression of Rv0557 in C. glutamicum ΔpimB′ ΔmgtA restored the synthesis of only ManGlcAGroAc2 as observed by 2D-TLC (Fig. 2) and MALDI-TOF MS analysis (Fig. 3B), which suggests a specific role in ManGlcAGroAc2 synthesis. Surprisingly, it did not complement the synthesis of Ac1PIM2 (Fig. 2 and 3A), a function previously assigned by Schaeffer et al. using in vitro studies (22).
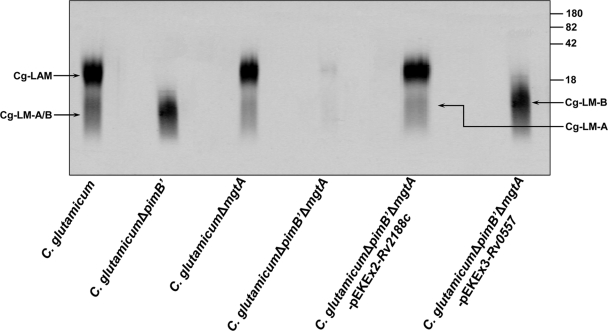
Ac1PIM2 and ManGlcAGroAc2 have been shown to be precursors of the cell wall components LM and LAM in Corynebacterineae. Therefore, lipoglycans were extracted by refluxing delipidated cells in 50% ethanol, followed by hot-phenol treatment, protease digestion, and dialysis. The extracted lipoglycans were examined on 15% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels (Fig. 4) using a Pro-Q emerald glycoprotein stain according to an established protocol (15, 25). Extracts from C. glutamicum showed the presence of LAMCg as well as LM-ACg and LM-BCg (which comigrates with LM-ACg), as shown previously (16, 25). The lipoglycan extract from C. glutamicum ΔpimB′ showed the absence of LAMCg and LM-ACg and the presence of ManGlcAGroAc2-based LM-BCg (16), while C. glutamicum ΔmgtA showed the presence of the PI-based lipoglycans LAMCg and LM-ACg and the absence of LM-BCg (25). Interestingly, C. glutamicum ΔpimB′ ΔmgtA was shown to be devoid of all three species of lipoglycans (Fig. 4). The lipoglycans from C. glutamicum ΔpimB′ ΔmgtA-pEKEx2-Rv2188c were analyzed, and as expected, the synthesis of PI-based LAMCg was restored by Rv2188c, supporting the in vivo lipid studies and the specific role of Rv2188c (Fig. 4). Similarly, C. glutamicum ΔpimB′ ΔmgtA-pEKEx3-Rv0557 restored the synthesis of ManGlcAGroAc2-based LM-BCg akin to the phenotype of C. glutamicum ΔpimB′, again illustrating the specific role of Rv0557 with respect to LM-BCg synthesis.
FIG. 4.
Lipoglycan profiles of C. glutamicum, C. glutamicum ΔpimB′, C. glutamicum ΔmgtA, C. glutamicum ΔpimB′ ΔmgtA, C. glutamicum ΔpimB′ ΔmgtA-pEKEx2-Rv2188c, and C. glutamicum ΔpimB′ ΔmgtA-pEKEx3-Rv0557. Lipoglycans were analyzed using SDS-PAGE and visualized using a Pro-Q emerald glycoprotein stain (Invitrogen) specific for carbohydrates. The three major bands represented by LAMCg, LM-ACg, and LM-BCg (which comigrates with LM-ACg) are indicated. The CandyCane glycoprotein molecular weight standards (Invitrogen) are provided on the right for comparison. The four major bands represent glycoproteins of 180, 82, 42, and 18 kDa, respectively.
In vitro mannolipid biosynthesis.
Reaction mixtures containing GDP[14C]Man, ATP, and membrane protein from wild-type, mutant, and/or complemented C. glutamicum strains were incubated at 37°C for 30 min as described previously (10). Membrane preparations from wild-type C. glutamicum synthesized C50-polyprenylmonophospho[14C]mannose (PP[14C]M), Ac1PI[14C]M2, and [14C]ManGlcAGroAc2 utilizing endogenous acceptors and GDP[14C]Man as a sugar donor, consistent with previous studies (Fig. 5A) (10, 17, 25). In assays performed with C. glutamicum ΔpimB′ membranes, an additional minor species migrating between Ac1PI[14C]M2 and 14[C]ManGlcAGroAc2 was observed and was confirmed as PI[14C]M1 based on previous studies and in comparison with authentic standards (11). Surprisingly, a radiolabeled band corresponding to Ac1PI[14C]M2 was also detected, which suggests a relaxed acceptor specificity for MgtACg in a C. glutamicum ΔpimB′ background. Assays utilizing membrane preparations from C. glutamicum ΔmgtA synthesized Ac1PI[14C]M2 but surprisingly also possessed a faint radiolabeled band corresponding to [14C]ManGlcAGroAc2, again due to the relaxed substrate specificity of PimB′Cg present in membrane preparations of C. glutamicum ΔmgtA (Fig. 5B). The synthesis of Ac1PI[14C]M2 and [14C]ManGlcAGroAc2 was totally abrogated in assays with membranes prepared from C. glutamicum ΔpimB′ ΔmgtA (Fig. 5B), while the accumulation of PI[14C]M1 and PP[14C]M was observed. Interestingly, Rv2188c from membrane preparations from C. glutamicum ΔpimB′ ΔmgtA-pEKEx2-Rv2188c showed substrate specificity toward Ac1PIM1 and also a weak recognition for the substrate GlcAGroAc2, resulting in the synthesis of Ac1PI[14C]M2 and [14C]ManGlcAGroAc2 (Fig. 5B), respectively. In contrast with the above studies of Rv2188c, assays performed using membranes prepared from C. glutamicum ΔpimB′ ΔmgtA-pEKEx3-Rv0557 illustrated that Rv0557 possessed a broader relaxed substrate specificity, as both Ac1PIM1 and GlcAGroAc2 were equally efficient substrates for the enzyme affording Ac1[14C]PIM2 and [14C]ManGlcAGroAc2 synthesis (Fig. 5B). These results explain the previous misinterpretation of the function of Rv0557 on the basis of in vitro data that Rv0557 was involved in the synthesis of Ac1PIM2 and annotated as PimBMt (22).
FIG. 5.
In vitro mannolipid biosynthesis. (A) Biosynthetic reaction scheme of products formed in in vitro assays utilizing GDP[14C]Man and corynebacterial membranes. (B) TLC-autoradiography of synthesized mannolipids, using GDP[14C]Man and membrane extracts from C. glutamicum, C. glutamicum ΔpimB′, C. glutamicum ΔmgtA, C. glutamicum ΔpimB′ ΔmgtA, C. glutamicum ΔpimB′ ΔmgtA-pEKEx2-Rv2188c, and C. glutamicum ΔpimB′ ΔmgtA-pEKEx3-Rv0557. Enzymatically synthesized products PP[14C]M, [14C]ManGlcAGroAc2, Ac1PI[14C]M2, and PI[14C]M1 were isolated and subjected to TLC/autoradiography using CHCl3/CH3OH/NH4OH/H2O (65:25:0.4:3.6, vol/vol/vol/vol).
Mannolipid synthesis using recombinant PimB′Cg.
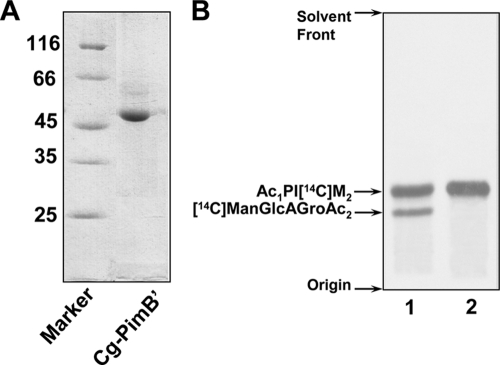
Initial attempts to develop an in vitro assay using either purified recombinant Rv2188c or Rv0557 have thus far proved unsuccessful. Therefore, their C. glutamicum orthologs were cloned into pET16b and transformed into Escherichia coli BL21(DE3); cultures were grown at 30°C in Luria-Bertani medium (Difco) supplemented with ampicillin (100 μg/ml). The expression of PimB′Cg was induced by the addition of 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at an A600 of 0.4 to 0.6 for 4 h and purified to near homogeneity (>95%) as observed on a 12% SDS-PAGE gel using Ni2+-affinity chromatography with a negligible effect on activity (see below). While PimB′Cg (Rv2188c ortholog) was expressed as a soluble protein (Fig. 6A) and shown to be active in an in vitro assay (see below), MgtACg resulted in an inactive protein (data not shown). The activity of purified PimB′Cg was initially determined in a well-established in vitro assay utilizing GDP[14C]Man and purified polar lipid extracts from C. glutamicum ΔpimB′ ΔmgtA, which possess Ac1PIM1 and GlcAGroAc2. The resulting products from the assay involving PimB′Cg showed a high substrate specificity of the enzyme toward Ac1PIM1 and a relaxed specificity toward GlcAGroAc2 (Fig. 6B, lane 1). In addition, the assay performed with highly purified Ac1PIM1 (Fig. 6B, lane 2) resulted in the formation of Ac1PI[14C]M2. Altogether, the data support the findings from the previous section and the redundant features of these enzymes in vitro (22).
FIG. 6.
Mannolipid synthesis using recombinant PimB′Cg. (A) Recombinant PimB′Cg was purified using Ni2+-affinity chromatography and purity determined on a 12% SDS-PAGE gel. (B) TLC-autoradiography of synthesized mannolipids, using GDP[14C]Man and lipid extracts from C. glutamicum ΔpimB′ ΔmgtA (lane 1) and purified Ac1PIM1 (lane 2) with purified PimB′Cg. Enzymatically synthesized products were isolated and subjected to TLC/autoradiography using CHCl3/CH3OH/NH4OH/H2O (65:25:0.4:3.6, vol/vol/vol/vol).
It has been previously shown by us and others that a high degree of functional redundancy exists in a number of biosynthetic pathways in mycobacteria, e.g., MptB (17), PimC (12), and EmbA and EmbB (3) in PIM/LM/LAM and arabinogalactan biosynthesis and the antigen 85 complex in mycolic acid biosynthesis (20). The generation of the C. glutamicum ΔpimB′ ΔmgtA mutant has clearly enabled the assignment of the precise function of the mycobacterial glycosyltransferases Rv0557 and Rv2188c. It is surprising that a glucuronosyl diacylglycerol-based lipid or lipoglycan has not been identified in mycobacteria or a potential role for Rv0557 in M. tuberculosis. However, it is possible that Rv0557 might supplement for the “loss of function” of Rv2188c, as suggested by our in vitro mannolipid studies, due to the essentiality of PIM biosynthesis (3). The identification of a precise role for Rv0557 in M. tuberculosis will require the generation of a conditional mutant in M. tuberculosis devoid of Rv0557/Rv2188c or novel methods to fractionate polar lipids and lipoglycans from M. tuberculosis in search of such glycolipids. A revised biosynthetic pathway for PIM synthesis that takes into account the findings of the current study is presented in Fig. 7. On the basis of the biochemical studies, we have assigned the functions of Rv2188c as an Ac1PIM1:α-d-mannose-α-(1→6)-phosphatidyl-myo-inositol-mannosyltransferase (originally termed PimB′Mt) and Rv0557 as a GlcAGroAc2: α-d-mannose-α-(1→4)-α-d-glucpyranosyl-uronicacid-mannosyltransferase (originally termed PimBMt), which we have reassigned as PimBMt and MgtAMt, respectively, in M. tuberculosis.
FIG. 7.
Revised PIM and LAM biosynthetic pathway in M. tuberculosis.
Acknowledgments
A.K.M. is a Darwin Trust-sponsored Ph.D. student. G.S.B. acknowledges support in the form of a personal research chair from James Bardrick and a Royal Society Wolfson Research Merit award, as a former Lister Institute-Jenner research fellow, and from the Medical Research Council and The Wellcome Trust (081569/Z/06/Z).
We are thankful for the technical assistance provided by Peter Ashton for the MALDI-TOF MS analysis.
Footnotes
Published ahead of print on 24 April 2009.
REFERENCES
- 1.Alderwick, L. J., E. Radmacher, M. Seidel, R. Gande, P. G. Hitchen, H. R. Morris, A. Dell, H. Sahm, L. Eggeling, and G. S. Besra. 2005. Deletion of Cg-emb in Corynebacterineae leads to a novel truncated cell wall arabinogalactan, whereas inactivation of Cg-ubiA results in an arabinan-deficient mutant with a cell wall galactan core. J. Biol. Chem. 28032362-32371. [DOI] [PubMed] [Google Scholar]
- 2.Alderwick, L. J., M. Seidel, H. Sahm, G. S. Besra, and L. Eggeling. 2006. Identification of a novel arabinofuranosyltransferase (AftA) involved in cell wall arabinan biosynthesis in Mycobacterium tuberculosis. J. Biol. Chem. 28115653-15661. [DOI] [PubMed] [Google Scholar]
- 3.Berg, S., D. Kaur, M. Jackson, and P. J. Brennan. 2007. The glycosyltransferases of Mycobacterium tuberculosis; roles in the synthesis of arabinogalactan, lipoarabinomannan, and other glycoconjugates. Glycobiology 1735R-56R. [DOI] [PubMed] [Google Scholar]
- 4.Besra, G. S., and P. J. Brennan. 1997. The mycobacterial cell wall: biosynthesis of arabinogalactan and lipoarabinomannan. Biochem. Soc. Trans. 25845-850. [DOI] [PubMed] [Google Scholar]
- 5.Birch, H. L., L. J. Alderwick, A. Bhatt, D. Rittmann, K. Krumbach, A. Singh, Y. Bai, T. L. Lowary, L. Eggeling, and G. S. Besra. 2008. Biosynthesis of mycobacterial arabinogalactan: identification of a novel α(1→3) arabinofuranosyltransferase. Mol. Microbiol. 691191-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cantarel, B. L., P. M. Coutinho, C. Rancurel, T. Bernard, V. Lombard, and B. Henrissat. 2008. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37D233-D238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dobson, G., D. E. Minnikin, S. M. Minnikin, J. H. Parlett, M. Goodfellow, M. Ridell, and M. Magnusson. 1985. Systematic analysis of complex mycobacterial lipids, p. 237-265. In M. Goodfellow and D. E. Minnikin (ed.), Chemical methods in bacterial systematics. Academic Press, London, United Kingdom.
- 8.Eikmanns, B. J., E. Kleinertz, W. Liebl, and H. Sahm. 1991. A family of Corynebacterium glutamicum/Escherichia coli shuttle vectors for cloning, controlled gene expression and promoter probing. Gene 10293-98. [DOI] [PubMed] [Google Scholar]
- 9.Gande, R., K. J. Gibson, A. K. Brown, K. Krumbach, L. G. Dover, H. Sahm, S. Shioyama, T. Oikawa, G. S. Besra, and L. Eggeling. 2004. Acyl-CoA carboxylases (accD2 and accD3), together with a unique polyketide synthase (Cg-Pks), are key to mycolic acid biosynthesis in Corynebacterineae such as Corynebacterium glutamicum and Mycobacterium tuberculosis. J. Biol. Chem. 27944847-44857. [DOI] [PubMed] [Google Scholar]
- 10.Gibson, K. J., L. Eggeling, W. N. Maughan, K. Krumbach, S. S. Gurcha, J. Nigou, G. Puzo, H. Sahm, and G. S. Besra. 2003. Disruption of Cg-ppm1, a polyprenyl monophosphomannose synthase, and the generation of lipoglycan-less mutants in Corynebacterium glutamicum. J. Biol. Chem. 27840842-40850. [DOI] [PubMed] [Google Scholar]
- 11.Kordulakova, J., M. Gilleron, G. Puzo, P. J. Brennan, B. Gicquel, K. Mikusova, and M. Jackson. 2003. Identification of the required acyltransferase step in the biosynthesis of the phosphatidylinositol mannosides of Mycobacterium species. J. Biol. Chem. 27836285-36295. [DOI] [PubMed] [Google Scholar]
- 12.Kremer, L., S. S. Gurcha, P. Bifani, P. G. Hitchen, A. Baulard, H. R. Morris, A. Dell, P. J. Brennan, and G. S. Besra. 2002. Characterization of a putative α-mannosyltransferase involved in phosphatidylinositol trimannoside biosynthesis in Mycobacterium tuberculosis. Biochem. J. 363437-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lea-Smith, D. J., K. L. Martin, J. S. Pyke, D. Tull, M. J. McConville, R. L. Coppel, and P. K. Crellin. 2008. Analysis of a new mannosyltransferase required for the synthesis of phosphatidylinositol mannosides and lipoarabinomannan reveals two lipomannan pools in Corynebacterineae. J. Biol. Chem. 2836773-6782. [DOI] [PubMed] [Google Scholar]
- 14.Liu, J., and A. Mushegian. 2003. Three monophyletic superfamilies account for the majority of the known glycosyltransferases. Protein Sci. 121418-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ludwiczak, P., M. Gilleron, Y. Bordat, C. Martin, B. Gicquel, and G. Puzo. 2002. Mycobacterium tuberculosis phoP mutant: lipoarabinomannan molecular structure. Microbiology 1483029-3037. [DOI] [PubMed] [Google Scholar]
- 16.Mishra, A. K., C. Klein, S. S. Gurcha, L. J. Alderwick, P. Babu, P. G. Hitchen, H. R. Morris, A. Dell, G. S. Besra, and L. Eggeling. 2008. Structural characterization and functional properties of a novel lipomannan variant isolated from a Corynebacterium glutamicum pimB′ mutant. Antonie van Leeuwenhoek 94277-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mishra, A. K., L. J. Alderwick, D. Rittmann, C. Wang, A. Bhatt, W. R. Jacobs, Jr., K. Takayama, L. Eggeling, and G. S. Besra. 2008. Identification of a novel α(1→6) mannopyranosyltransferase MptB, from Corynebacterium glutamicum by deletion of a conserved gene, NCgl1505, affords a lipomannan- and lipoarabinomannan-deficient mutant. Mol. Microbiol. 681595-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mishra, A. K., L. J. Alderwick, D. Rittmann, R. V. Tatituri, J. Nigou, M. Gilleron, L. Eggeling, and G. S. Besra. 2007. Identification of an α(1→6) mannopyranosyltransferase (MptA), involved in Corynebacterium glutamicum lipomannan biosynthesis, and identification of its orthologue in Mycobacterium tuberculosis. Mol. Microbiol. 651503-1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morita, Y. S., C. B. Sena, R. F. Waller, K. Kurokawa, M. F. Sernee, F. Nakatani, R. E. Haites, H. Billman-Jacobe, M. J. McConville, Y. Maeda, and T. Kinoshita. 2006. PimE is a polyprenol-phosphate-mannose-dependent mannosyltransferase that transfers the fifth mannose of phosphatidylinositol mannoside in mycobacteria. J. Biol. Chem. 28125143-25155. [DOI] [PubMed] [Google Scholar]
- 20.Puech, V., C. Guilhot, E. Perez, M. Tropis, L. Y. Armitige, B. Gicquel, and M. Daffe. 2002. Evidence for a partial redundancy of the fibronectin-binding proteins for the transfer of mycoloyl residues onto the cell wall arabinogalactan termini of Mycobacterium tuberculosis. Mol. Microbiol. 441109-1122. [DOI] [PubMed] [Google Scholar]
- 21.Schäfer, A., A. Tauch, W. Jäger, J. Kalinowski, G. Thierbach, and A. Pühler. 1994. Small mobilizable multipurpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 14569-73. [DOI] [PubMed] [Google Scholar]
- 22.Schaeffer, M. L., K. H. Khoo, G. S. Besra, D. Chatterjee, P. J. Brennan, J. T. Belisle, and J. M. Inamine. 1999. The pimB gene of Mycobacterium tuberculosis encodes a mannosyltransferase involved in lipoarabinomannan biosynthesis. J. Biol. Chem. 27431625-31631. [DOI] [PubMed] [Google Scholar]
- 23.Seidel, M., L. J. Alderwick, H. L. Birch, H. Sahm, L. Eggeling, and G. S. Besra. 2007. Identification of a novel arabinofuranosyltransferase AftB, involved in a terminal step of cell wall arabinan biosynthesis in Corynebacterineae, such as Corynebacterium glutamicum and Mycobacterium tuberculosis. J. Biol. Chem. 28214729-147240. [DOI] [PubMed] [Google Scholar]
- 24.Seidel, M., L. J. Alderwick, H. Sahm, G. S. Besra, and L. Eggeling. 2007. Topology and mutational analysis of the single Emb arabinofuranosyltransferase of Corynebacterium glutamicum as a model of Emb proteins of Mycobacterium tuberculosis. Glycobiology 17210-219. [DOI] [PubMed] [Google Scholar]
- 25.Tatituri, R. V., P. A. Illarionov, L. G. Dover, J. Nigou, M. Gilleron, P. Hitchen, K. Krumbach, H. R. Morris, N. Spencer, A. Dell, L. Eggeling, and G. S. Besra. 2007. Inactivation of Corynebacterium glutamicum NCgl0452 and the role of MgtA in the biosynthesis of a novel mannosylated glycolipid involved in lipomannan biosynthesis. J. Biol. Chem. 2824561-4572. [DOI] [PubMed] [Google Scholar]