Abstract
Bifidobacteria are important members of the human gut flora, especially in infants. Comparative genomic analysis of two Bifidobacterium animalis subsp. lactis strains revealed evolution by internal deletion of consecutive spacer-repeat units within a novel clustered regularly interspaced short palindromic repeat locus, which represented the largest differential content between the two genomes. Additionally, 47 single nucleotide polymorphisms were identified, consisting primarily of nonsynonymous mutations, indicating positive selection and/or recent divergence. A particular nonsynonymous mutation in a putative glucose transporter was linked to a negative phenotypic effect on the ability of the variant to catabolize glucose, consistent with a modification in the predicted protein transmembrane topology. Comparative genome sequence analysis of three Bifidobacterium species provided a core genome set of 1,117 orthologs complemented by a pan-genome of 2,445 genes. The genome sequences of the intestinal bacterium B. animalis subsp. lactis provide insights into rapid genome evolution and the genetic basis for adaptation to the human gut environment, notably with regard to catabolism of dietary carbohydrates, resistance to bile and acid, and interaction with the intestinal epithelium. The high degree of genome conservation observed between the two strains in terms of size, organization, and sequence is indicative of a genomically monomorphic subspecies and explains the inability to differentiate the strains by standard techniques such as pulsed-field gel electrophoresis.
Actinobacteria, Firmicutes, Proteobacteria, and Bacteroidetes are dominant microbial phyla widely distributed in diverse ecosystems on the planet (10, 13, 20, 23, 33, 40, 51). Metagenomic analyses of the microbial landscape inhabiting various mammalian environments, notably the human gastrointestinal tract (GIT) and skin, have specifically identified Actinobacteria as an important and occasionally dominant phylum (18, 21, 33). Among the members of the large, diverse, and dynamic microbial community residing in the human GIT, Bifidobacterium is a dominant genus considered beneficial to humans and includes probiotic strains (live microorganisms which, when administered in adequate amounts, confer a health benefit on the host) (11). The population of bifidobacteria in the human intestine varies over time. Following vaginal delivery, the GIT of healthy newborns is typically colonized by bifidobacteria, especially in breast-fed infants, during the first few days of life (12). Interindividual variation, however, is remarkable in the human infant intestinal flora (41), and dominant genera are not always consistent across metagenomic analyses of the human gut flora (18, 30, 33, 41). Over time, the infant intestinal ecosystem becomes more complex as the diet becomes more diverse, with bifidobacteria typically remaining dominant until weaning (30).
Bifidobacterium animalis subsp. lactis is a gram-positive lactic acid bacterium commonly found in the guts of healthy humans and has been identified in the infant gut biota, particularly in ileal, fecal, and mucosal samples (52, 56). Some strains of B. animalis subsp. lactis are able to survive in the GIT, to adhere to human epithelial cells in vitro, to modify fecal flora, to modulate the host immune response, or to prevent microbial gastroenteritis and colitis (4, 15, 20, 40, 52, 56). Additionally, B. animalis subsp. lactis has been reported to utilize nondigestible oligosaccharides, which may contribute to the organism's ability to compete in the human gut. Carbohydrates resistant to enzymatic degradation and not absorbed in the upper intestinal tract are a primary source of energy for microbes residing in the large intestine. The benefits associated with probiotic strains of B. animalis subsp. lactis have resulted in their inclusion in the human diet via formulation into a large array of dietary supplements and foods, including dairy products such as yogurt. Deciphering the complete genome sequences of such microbes will provide additional insight into the genetic basis for survival and residence in the human gut, notably with regard to the ability to survive gastric passage and utilize available nutrients. Also, these genomes provide reference sequences for ongoing metagenomic analyses of the human environment, including the gut metagenome.
Bifidobacterium animalis subsp. lactis is the most common bifidobacterium utilized as a probiotic in commercial dairy products in North America and Europe (22, 38). However, despite this commercial and probiotic significance, strain-level differentiation of B. animalis subsp. lactis strains has been hindered by the high genetic similarity of these organisms, as determined by pulsed-field gel electrophoresis and other nucleic acid-based techniques (6, 55, 56), and the lack of available genomic sequence information. The genome sequence of strain BB-12 (17) is not currently publicly available, and only a draft genome sequence in 28 contigs is available for strain HN019 (GenBank project 28807). The complete B. animalis subsp. lactis genome for strain AD011 (28) was only recently (2009) published. While this was an important first step, a single genome does not allow identification of unique targets for strain differentiation or comparative analyses within the subspecies.
The objectives of this study were to determine the complete genome sequences of two B. animalis subsp. lactis strains, the type strain and a widely used commercial strain, to provide insights into the functionality of this species and into species identification and strain specialization.
MATERIALS AND METHODS
Bacterial strains.
Bifidobacterium animalis subsp. lactis Bl-04 (also known as DGCC2908 and RB 4825) was originally isolated from a fecal sample from a healthy adult, is a widely used commercial strain, and has been deposited at the American Type Culture Collection safe deposit as strain SD5219. DSM 10140, which is the B. animalis subsp. lactis type strain, was originally isolated from a commercial product (39) and was obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (Braunschweig, Germany). Additionally, B. animalis subsp. lactis strains Bi-07 (ATCC SD5220), HN019, and B420, from the Danisco Global Culture Collection, were used. DSM 10140 was propagated at 37°C anaerobically in LL (31). All other cultures were propagated at 37°C anaerobically in MRS supplemented with 0.05% cysteine.
Genome sequencing.
The B. animalis subsp. lactis Bl-04 genome draft sequence was obtained by whole-genome shotgun sequencing carried out at Macrogen (Rockville, MD). A DNA library was prepared in pC31 and shotgun sequenced using an ABI 3730XL sequencer (Roche, Nutley, NJ), targeting 6× coverage. To complement the Bl-04 draft genome, a 454 pyrosequencing run was performed, targeting 15× coverage, using a GS-20 sequencer.
Independently, the B. animalis subsp. lactis DSM 10140 genome draft was sequenced at The Pennsylvania State University, using 454 pyrosequencing targeting 30× coverage, followed by de novo assembly in Newbler (Roche).
Concurrently, an optical map was generated for the Bl-04 strain at OpGen (Madison, WI), using NotI. Assembled contigs were aligned with the optical map, and remaining gaps were closed by walking across PCR products. PCR amplicons were sequenced at the Huck Institute Nucleic Acid Facility at The Pennsylvania State University, using 3′ BigDye-labeled dideoxynucleotide triphosphates (v 3.1 dye terminators; Applied Biosystems, Foster City, CA) and running on an ABI 3730XL DNA analyzer using the ABI data collection program (v 2.0), or at Davis Sequencing (Davis, CA). Data were analyzed with ABI sequence analysis software (version 5.1.1). Sequencing reactions were performed using protocol 4303237 from Applied Biosystems. Complete genome sequences were annotated at Integrated Genomics, using the ERGO package (Integrated Genomics, Chicago, IL) without manual curation, and at NCBI.
In silico analyses.
The two B. animalis subsp. lactis genomes were aligned using BioEdit (Isis Biosciences) and SeqMan (DNASTAR, Madison, WI). Single nucleotide polymorphism (SNP) reports were generated to identify insertions, deletions, and mutations. Comparative genomic analyses for clusters of orthologous groups (COGs) (51) were carried out by similarity clustering in the ERGO package and were visualized using MeV (J. Craig Venter Institute, MD) (45).
SNP resequencing.
After identification of putative SNPs between the two genome drafts, primers were designed to amplify regions containing SNPs from both genomes. PCR amplicons were purified using a Qiaquick kit (Qiagen, Valencia, CA) and were sequenced at The Pennsylvania State University or the Davis Sequencing facilities.
CRISPR analyses.
Clustered regularly interspaced short palindromic repeat (CRISPR) loci were identified in the Bl-04 and DSM 10140 genome sequences by use of Dotter (49). An internal segment of the putative CRISPR locus was amplified by PCR, using primers 5′-TTGGATGCAAGCCCTCAATGAAGC-3′ and 5′-TGAGGGAAGCCGAACTCAATCACA-3′. The PCR amplicons were subsequently sequenced (Davis Sequencing) from both ends with each primer. The CRISPR spacers were visualized as two-tone color combinations as previously described (3, 26).
Nucleotide sequence accession numbers.
The genome sequences for Bl-04 and DSM 10140 were deposited at NCBI under accession numbers CP001515 and CP001606, respectively.
RESULTS
B. animalis subsp. lactis genome.
Bl-04 shotgun sequencing generated 1,824,258 bp assembled in 121 contigs. Complementation by pyrosequencing generated 1,948,770 bp assembled in 16 contigs. Concurrently and independently, DSM 10140 pyrosequencing generated 33× coverage of the 1.9-Mbp genome, assembled in 30 contigs. Following a few rounds of gap closing (by contig end pairing), the remaining contigs (3 for Bl-04 and 10 for DSM 10140) from both genome drafts were aligned, using the Bl-04 optical map as a template. Remaining gaps were closed subsequently and independently in both genomes.
The distinct sequencing strategies used for the two genomes generated comparable assemblies, with the exception of one misassembly in the pyrosequencing-only genome draft. Upon comparison of the two genomes, a total of 211 putative SNPs were identified. Fourteen of the putative SNPs were located in rRNA operons or insertion sequence (IS) elements and were not resequenced. After resequencing of the remaining 197 SNPs, 150 were found to be the results of sequencing errors, while 47 SNP sites were confirmed.
Overall, the Bl-04 and DSM 10140 genomes are 1,938,709 bp and 1,938,483 bp, respectively. Using the largest genome as a reference (Bl-04), 1,655 open reading frames (ORFs) were identified, and putative functions were assigned to 1,072 of them, which is typical across sequenced Bifidobacterium genomes (Table 1). The features of the B. animalis subsp. lactis genomes are detailed in Table 1 and in Fig. SA in the supplemental material.
TABLE 1.
General characteristics of selected bifidobacterial genomesa
| Strain | Genome size (bp) | Coding % | % G+C | Avg ORF length (bp) | No. of predicted ORFs | No. of ORFs with assigned function | No. of COG matches | No. of Pfam domains | No. of rRNA operons | No. of tRNAs | No. of transposases |
|---|---|---|---|---|---|---|---|---|---|---|---|
| B. animalis subsp. lactis Bl-04 | 1,938,709 | 90.45 | 60.48 | 1,053 | 1,655 | 1,072 | 773 | 890 | 4 | 52 | 9 |
| B. animalis subsp. lactis DSM 10140 | 1,938,483 | 90.34 | 60.48 | 1,056 | 1,658 | 1,071 | 773 | 890 | 4 | 51 | 9 |
| B. adolescentisATCC 15703 | 2,089,645 | 87.57 | 59.18 | 1,122 | 1,631 | 1,165 | 791 | 936 | 5 | 55 | 15 |
| B. longumNCC2705 | 2,260,266 | 86.07 | 60.13 | 1,503 | 1,729 | 1,305 | 839 | 969 | 4 | 54 | 1 |
Numbers were extracted from the ERGO Database at the time of analysis.
The origin and terminus of replication in the B. animalis subsp. lactis genome were predicted (see Fig. SA and SB in the supplemental material) based on dnaA (Balac_0001), DnaA boxes, and base composition asymmetry between the leading and lagging strands (16, 24, 34). The putative oriC origin of replication was identified in an AT-rich intergenic region upstream of dnaA, in the vicinity of a cluster of hypothetical DnaA boxes (see Fig. SB in the supplemental material). Similar putative oriC's with comparable DnaA boxes were also identified in other bifidobacterial genomes (see Fig. SB in the supplemental material), were consistent with previously observed degeneracy at certain sites, and reflected the high G+C content of the genome (24, 34). These conserved elements might be used to locate the origin of replication in Bifidobacterium species and other genomes of Actinobacteria, since GC skew analysis alone does not necessarily identify an unequivocal oriC (46).
Comparative analysis of two B. animalis subsp. lactis genomes.
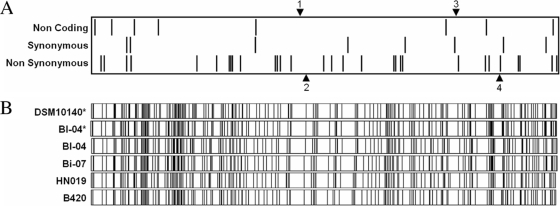
Comparison of the two B. animalis subsp. lactis genomes revealed nearly perfect alignment. Of the 47 SNPs validated for the two genome sequences, 39 were in predicted coding sequences and 8 were in intergenic regions (Fig. 1; see Table S2 in the supplemental material). Of the 39 coding SNPs, 31 represent nonsynonymous mutations and 8 are synonymous (Fig. 1), indicating positive selection and/or recent divergence. Four distinct insertion/deletion sites (INDELs) were also identified, totaling 443 bp (see Table S3 in the supplemental material). INDEL1 is a 121-bp sequence encoding tRNA-Ala-GGC which is present in the Bl-04 genome (bp 881,420 to 881,540) but absent in the DSM 10140 genome. INDEL2 is a 54-bp sequence within the long-chain-fatty-acid-coenzyme A ligase gene (Balac_0771; EC 6.2.1.3; COG1022) which is present in the DSM 10140 genome (bp 902,893 to 902,946) but absent in the Bl-04 genome. INDEL2 yields an in-frame deletion of 18 amino acids in the Bl-04 protein. Interestingly, the importance of long-chain-fatty-acid-coenzyme A synthetases in bifidobacteria has been noted previously (46). INDEL3 is a 214-bp sequence within the CRISPR locus which is present in Bl-04 (bp 1,512,373 to 1,512,586) but absent in the DSM 10140 genome, and it corresponds to three repeat-spacer units. INDEL4 is a 54-bp sequence which is present in an intergenic region in the DSM 10140 genome (bp 1,715,507 to 1,715,560) but absent in the Bl-04 genome.
FIG. 1.
Alignment of B. animalis subsp. lactis genomes. Differences observed between the two B. animalis subsp. lactis genomes are shown in the figure. (A) SNPs are indicated using bars, with noncoding SNPs at the top, synonymous SNPs in the middle, and nonsynonymous SNPs at the bottom. INDELs are shown by arrows, with insertions in the Bl-04 genome shown on top and insertions in the DSM 10140 genome shown on the bottom. (B) Optical maps of various B. animalis subsp. lactis strains, visualized using OpGen MapViewer, based on a NotI digest. The first two optical maps were generated in silico (indicated by asterisks) based on the genome sequences. The other optical maps represent experimental data.
Overall, notwithstanding the 47 SNPs and the 443 bp of INDELs, the two genomes were 99.975% identical. In addition to the two complete genome sequences, optical mapping was used to analyze the genome layouts of four B. animalis subsp. lactis strains. The highly similar optical maps of this strain set (comprised of the type strain, commercial strains, and isolates used in functional and clinical studies) indicate a high degree of genome conservation in terms of size, organization, and sequence (Fig. 1). The lack of polymorphism observed is indicative of a genomically monomorphic subspecies.
Comparative genomic analysis of bifidobacteria.
Analysis of the distribution of annotated ORFs over COG categories revealed a high overall conservation of COG representation across B. animalis subsp. lactis, B. longum, and B. adolescentis (Table 1), which belong to three different Bifidobacterium clusters (14, 52, 53). Specifically, the top four functional categories, namely, general prediction, translation, carbohydrate metabolism, and amino acid metabolism, were identical, which is typical of lactic acid bacteria (35, 37). However, fewer ORFs were associated with carbohydrate utilization in B. animalis subsp. lactis than in B. longum and B. adolescentis. Overall, most genes (1,117 [67%]) were conserved in all three species, representing the “core” bifidobacterial genome (see Fig. SC in the supplemental material), whereas 416 (25%), 368, and 298 genes were unique to B. animalis subsp. lactis, B. longum, and B. adolescentis, respectively. Interestingly, a relatively small proportion of genes were shared between two genomes only, while a larger proportion of genes were unique to each of the three genomes examined, suggesting distinct speciations (see Fig. SC in the supplemental material). Alignment of the three chromosomes indicated that B. animalis subsp. lactis shares a relatively high level of synteny with B. adolescentis but exhibits little colinearity with B. longum (see Fig. SD in the supplemental material). This further confirms the observed differences between the genomes of B. longum and B. adolescentis (47).
Comparative analysis of gene conservation across the three Bifidobacterium species through BLASTp (see Table S1 in the supplemental material) revealed eight areas containing consecutive sets of ORFs present in B. animalis subsp. lactis and absent in both B. adolescentis and B. longum (Fig. 2). Notably, a prophage remnant is located in area 5, including a phage-related integrase gene (Balac_1179; COG0582) and a phage-related prohead protein gene (Balac_1191; COG3740). Although there is only anecdotal evidence of bacteriophages in bifidobacteria (48), a previous analysis of bifidobacterial genome sequences revealed the presence of prophages integrated in a tRNAMet gene (53, 54). The observed remnants of a prophage-like element in the B. animalis subsp. lactis genome do not seem to be adjacent to a tRNAMet sequence. An eps cluster with a low GC content was identified in area 7 (Balac_1383 to Balac_1391) and may be involved in the synthesis of membrane-associated exopolysaccharides. Two genes potentially involved in oxalic acid catabolism (13), namely, oxc (Balac_1453) and frc (Balac_1449), which encode an oxalyl-coenzyme A decarboxylase (EC 4.1.1.8) and a formyl-coenzyme A transferase (EC 2.8.3.16), respectively, were identified in area 8.
FIG. 2.

Comparative genomic analysis of bifidobacteria. Sequences homologous to B. animalis subsp. lactis (Bla) proteins are shown in black, whereas the absence of a homolog in the B. adolescentis ATCC 15703 (Bad) or B. longum NCC2705 (Blo) genome appears in white, represented using MeV. (Left) Data are presented in chromosomal ORF order, with the origin of replication at the top and with numbers indicating locations of areas of unique content in B. animalis subsp. lactis. (Right) Hierarchical representation of homologs across the three genomes.
Analysis of B. animalis subsp. lactis CRISPR locus.
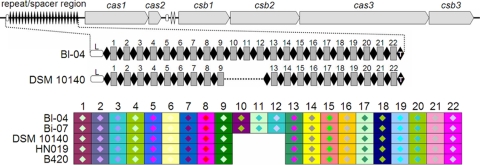
A novel CRISPR-Cas system, Bala1, was identified in the B. animalis subsp. lactis genome in area 6 (Fig. 2), and this system is different from other CRISPR loci previously identified in bifidobacteria. It is the ninth CRISPR family identified in lactic acid bacterial genomes and the fourth CRISPR family identified in bifidobacteria, in addition to Blon1 (in B. longum), Lhel1 (in B. adolescentis), and Ldbu1 (present in Bifidobacterium catenulatum) (25). Novelty was observed in terms of both the CRISPR repeat sequence and cas (CRISPR-associated) gene content. In Bl-04, the typical 36-bp CRISPR repeat, 5′-ATCTCCGAAGTCTCGGCTTCGGAGCTTCATTGAGGG-3′, which is partially palindromic, is present 23 times and separated by 22 unique spacer sequences (Fig. 3; see Fig. SF in the supplemental material). The typical repeat sequence is conserved in the first 21 repeats, while the last 2 have SNPs (see Fig. SF in the supplemental material), notably at the 3′ end, as previously shown in other CRISPR systems (26). Two remnant CRISPR repeats were also identified between cas2 and csb1 (Fig. 3). The co-occurrence of remnant CRISPR repeats and a typical CRISPR repeat-spacer array has been observed previously, notably in Streptococcus thermophilus CRISPR3 (26) and B. adolescentis (25). Six cas genes were identified downstream of the repeat-spacer region, including the cas1 universal nuclease gene (Balac_1308; COG1518; TIGR00287), the cas2 endonuclease gene (Balac_1307; COG1343; TIGR01573), and the cas3 helicase gene (Balac_1304; COG1203; TIGR02621). Additionally, three novel putative cas genes, csb1 to -3 (Cas subtype Bifidobacterium Bala1, with nomenclature derived from reference 23), were also identified in this CRISPR locus. Two of them, csb1 (Balac_1306) and csb2 (Balac_1305), contain cas-type conserved elements, i.e., cas_GSU0053 and cas_GSU0054, respectively. The spacer size varied from 34 to 39 bp. Some Bl-04 CRISPR spacers showed similarity to viral sequences (S3 homology with Streptomyces phage phi-BT1 AJ550940, S17 homology with frog virus sequence AY548484, and S20 homology with a phage capsid protein in Chromohalobacter salexigens CP000285) and metagenome-derived sequences (S19 homology with human gut metagenome BABA01032251 and S20 homology with marine metagenome AACY021620797). However, the analysis of CRISPR spacers was limited by the absence of bifidobacterial phage sequences in public databases.
FIG. 3.
Overview of B. animalis subsp. lactis CRISPR locus. The Bala1 CRISPR locus, as it appears in the genome, is shown at the top, with the repeat-spacer region on the left (repeats are represented as black diamonds, and spacers are represented as gray boxes) and cas genes on the right. The repeat-spacer region is shown below for Bl-04 and DSMZ 10140, with repeats shown as black diamonds, spacers shown as numbered gray boxes, the leader shown as a white box, and the terminal repeat shown as a black diamond annotated with a “T.” The bottom panel shows a CRISPR spacer overview where repeats are not included and only spacers are represented, using two-color combinations.
While the Bala1 CRISPR locus was identified in both Bl-04 and DSM 10140 (Fig. 3), polymorphism was observed in terms of spacer content. Although the spacer contents were identical at both the leader and the trailer end of the locus, three consecutive internal repeat-spacer units were unique to Bl-04 (Fig. 3). The Bala1 CRISPR locus was sequenced for several B. animalis subsp. lactis strains, and spacer content analysis revealed only two versions of the CRISPR locus, as seen in the two genomes, with the presence or absence of three consecutive internal repeat-spacer units (Fig. 3). The Bala1 CRISPR locus was subjected to PCR amplification from several bifidobacterial species, including the B. animalis subsp. animalis type strain. This locus was present exclusively in B. animalis subsp. lactis strains and was absent in B. animalis subsp. animalis, B. longum, B. adolescentis, B. dentium, B. catenulatum, Bifidobacterium breve, and Bifidobacterium bifidum, suggesting that it is subspecies specific. Interestingly, the GC content of the Bala1 CRISPR locus was approximately 49.74%, while that of the genome is 60.19%, suggesting that it may have been acquired laterally from a low-GC microbe, as previously discussed for CRISPR loci (19, 25).
DISCUSSION
The relatively small size of the B. animalis subsp. lactis genome compared to those of other bifidobacteria is consistent with a genome simplification process that reduces biosynthetic capabilities and favors the retention and acquisition of genes involved in the utilization of a broad repertoire of carbon and nitrogen sources (35, 37). This is typical of the genomic evolution of microbes that live in nutritionally rich environments, such as the human gut, and that rely on the host and other members of the intestinal flora for energy sources and metabolic intermediates (35, 37). This likely explains the smaller number of hydrolases in B. animalis subsp. lactis than in B. longum and B. adolescentis (see Table S4 in the supplemental material). Notably, the genes involved in the catabolism of human milk oligosaccharides in B. longum subsp. infantis (47) and in the degradation and utilization of mucin (endo-α-N-acetylgalactosaminidase) were not identified. Also, unlike the case for most bacteria, ABC transporters do not appear in the typical operon configuration, and only two copies of a carbohydrate-specific ATP-binding protein, Balac_0062 (COG1129) and Balac_1610 (COG3839), were identified in Bifidobacterium animalis subsp. lactis. No phosphotransferase system (PTS) component was identified, contradicting a previous report of fructose-PTS activity in the DSM 10140 strain (42) and in contrast to the presence of PTS in B. longum and B. adolescentis.
While the number of carbohydrate hydrolases is smaller for B. animalis subsp. lactis, the variety of hydrolases present suggests the ability to utilize a wide range of complex carbohydrates, including milk galactosides and undigestible plant-derived oligosaccharides (see Table S4 in the supplemental material). This is consistent with a previous report that genes in the carbohydrate transport and metabolism COG are overrepresented in the human gut microbiome (30). This may also reflect the strong influence of the host diet on bacterial gut communities in mammals (33).
Additionally, genes involved in the organism's adaptation to and survival in the human GIT were identified. Specifically, two paralogs encoding putative N-acetylmuramidases (COG3757; Balac_1516 and Balac_1517), which may be involved in the degradation of bacterial cell wall components available in the intestinal environment, were identified. Four genes encoding putative cell surface proteins that could be involved in interactions with human epithelial cells were identified in the B. animalis subsp. lactis genomes, including two putative collagen adhesion proteins (encoded by capA [Balac_1456 and COG4932] and capB [Balac_1484]), an elastin-binding protein (encoded by ebpS), and a fibronectin-binding protein (encoded by fbp [Balac_0271]). A typical LPXTG anchor motif was identified only in CapB (LPLTG). An alternative putative anchor motif, VAATG, was identified in silico in the vicinity of a poly(R) sequence at the C termini of CapA and Fbp. The presence of two distinct sortase-encoding genes, namely, srtB (Balac_1349) and srtA (Balac_1485), in the genome of B. animalis subsp. lactis is consistent with the presence of two distinct anchor motifs.
Numerous genes commonly associated with the stress response were identified in the B. animalis subsp. lactis genome (see Table S5 in the supplemental material). Notably, a bile salt hydrolase gene, bsh (Balac_0863; EC 3.5.1.24), encoding a member of the Ntn-PVA family of enzymes (COG3049), was identified. This gene family is enriched in the human gut microbiome (27) and is likely involved in the ability of B. animalis subsp. lactis to tolerate bile and to survive in the human gut environment.
Genomic content was compared across members of the following three distinct Bifidobacterium phylogenetic clusters: B. adolescentis, B. longum, and B. animalis subsp. lactis (a member of the Bifidobacterium pseudolongum group) (14, 53). Comparative analysis revealed a high degree of conservation and synteny overall (Fig. 2; see Fig. SD in the supplemental material), as previously observed in Bifidobacterium genomes (47). However, there are significant, functionally relevant differences, notably in CRISPR content, catabolism of dietary compounds, cell surface proteins and polysaccharides, and prophages. Notably, cell surface proteins encoded in the B. animalis subsp. lactis genome may be involved in its ability to adhere to human intestinal epithelial cells (20) and may possess immunomodulation properties.
Overall, it appears that the genomes of the B. animalis subsp. lactis strains included in this study are highly monomorphic, consistent with previous reports indicating a high degree of similarity among B. animalis subsp. lactis strains isolated from infant samples and various commercial products, as determined by pulsed-field gel electrophoresis (6, 55, 56), which may suggest clonal ancestry.
Comparison of the Bl-04 genome to the recently published genome sequence of B. animalis subsp. lactis AD011 (28) revealed the AD011 genome to be 5,014 bp shorter than the Bl-04 genome and indicated a marked difference in assembly between the two genomes. An advantage of the approach taken in the current work—independently sequencing two related strains—is the increased confidence and ability to identify strain-level differences, which has been a challenge for this particular subspecies. In this particular case, the number of SNPs initially identified between Bl-04 and DSM 10140 was decreased approximately fivefold by resequencing. Thus, by sequencing two genomes independently, using both traditional and pyrosequencing methods, and resequencing SNPs regardless of quality score, a low error rate (approximately 0.00035%) was obtained, leading to increased confidence in the genome sequences. Comparison of the Bl-04 genome to the HN109 draft genome (currently in 28 contigs) did not reveal any novel content.
Despite the genetic homogeneity among B. animalis subsp. lactis strains, interstrain functional differences have been observed, such as variability in immunogenic properties (15) and the ability to catabolize glucose (5). Interestingly, a SNP was identified in glcU (Balac_1097; COG4975), encoding a putative glucose uptake protein. Bl-04 has a reduced ability to grow using glucose as the primary carbohydrate source, which is linked to a significant reduction in its ability to transport glucose into the cell (5). In silico analysis of the putative transmembrane domains of the two glcU variants indicated that the nonsynonymous CGT (Arg in Bl-04)-to-GGT (Gly in DSM 10140) mutation impacts the predicted structural arrangement of the resulting proteins in the cell membrane (see Fig. SE in the supplemental material). Perhaps this SNP resulting in the loss of the ability to transport glucose is indicative of rapid evolution driven either by the lack of selective pressure to maintain a functional glucose transporter in the human gut environment, where glucose is likely absent due to its absorption in the upper GIT, or by selective pressure against maintaining an arguably useless gene.
While it is difficult to study the evolutionary changes in such monomorphic bacterial genomes, SNPs and INDELs provide insights into the evolution and diversity of clonal bacteria (10). Based on the size and number of SNPs identified in these two B. animalis subsp. lactis genomes (32, 43, 57), these strains are separated by 1,268 to 304,414 generations, or 6 to 1,522 years (based on 200 generations per year). The primary observed genomic difference between the two strains consists of three CRISPR repeat-spacer units present in the Bl-04 genome and absent in the DSM 10140 genome. CRISPR represents a family of repeated DNA elements that provide acquired immunity against foreign genetic elements (3, 7, 36, 50). Since it was previously shown that CRISPR loci evolve primarily by polarized addition of novel spacers at the leader end of the locus and by internal deletion of contiguous vestigial spacers (3, 9, 26), the observed difference between the two B. animalis subsp. lactis genomes is likely the result of an internal deletion within the Bala1 CRISPR locus. This suggests that Bl-04 is the ancestral strain from which a variety of B. animalis subsp. lactis clonal offspring may have recently derived.
The determination of two B. animalis subsp. lactis genome sequences provides insights into the genetic basis for survival in the human GIT. The ability of microbes to survive in the human environment has been associated with resistance to acid and bile, as well as their opportunistic capacity to utilize undigested dietary compounds. This can be achieved by encoding the enzymatic machinery necessary to catabolize either a wide range of carbohydrates or niche-specific nutrients, as documented for the cariogenic organism Streptococcus mutans in the oral cavity (1), the adaptation of Lactobacillus plantarum to a variety of environmental niches (29), the presence of Lactobacillus johnsonii and Lactobacillus acidophilus in the GIT (2, 44), and the residence of B. longum in the colon (46). The comparison of two nearly identical genome sequences revealed rapid evolution via internal deletion of three consecutive repeat-spacer units within a CRISPR locus, three small INDELs, and 47 SNPs, including a nonsynonymous mutation resulting in the loss of the ability to utilize glucose efficiently, perhaps resulting from directed evolutionary pressure. In addition, the availability and analyses of these genome sequences may allow for the development of molecular methods for strain differentiation and will aid in metagenomic analyses of the human microbiome.
Supplementary Material
Acknowledgments
We acknowledge Theresa Walunas at Integrated Genomics for assistance with annotation and Emily Zentz and Buffy Stahl for optical mapping at OpGen.
The work at The Pennsylvania State University was sponsored in part by grants from NutriCorp North East and The Penn State Ice Cream Short Course. Work at Danisco was sponsored by Danisco USA Inc.
This work was performed at The Pennsylvania State University and Danisco USA Inc.
Footnotes
Published ahead of print on 17 April 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Ajdić, D., W. M. McShan, R. E. McLaughlin, G. Savić, J. Chang, M. B. Carson, C. Primeaux, R. Tian, S. Kenton, H. Jia, S. Lin, Y. Qian, S. Li, H. Zhu, F. Najar, H. Lai, J. White, B. A. Roe, and J. J. Ferretti. 2002. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc. Natl. Acad. Sci. USA 9914434-14439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altermann, E., W. M. Russell, M. A. Azcarate-Peril, R. Barrangou, B. L. Buck, O. McAuliffe, N. Souther, A. D. W. Dobson, T. Duong, M. Callanan, S. Lick, A. Hamrick, R. Cano, and T. R. Klaenhammer. 2005. Complete genome sequence of the probiotic lactic acid bacterium Lactobacillus acidophilus NCFM. Proc. Natl. Acad. Sci. USA 1023906-3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barrangou, R., C. Fremaux, H. Deveau, M. Richards, P. Boyaval, S. Moineau, D. A. Romero, and P. Horvath. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 3151709-1712. [DOI] [PubMed] [Google Scholar]
- 4.Bartosch, S., E. J. Woodmansey, J. C. M. Paterson, M. E. T. McMurdo, and G. T. Macfarlane. 2005. Microbiological effects of consuming a synbiotic containing Bifidobacterium bifidum, Bifidobacterium lactis, and oligofructose in elderly persons, determined by real-time polymerase chain reaction and counting of viable bacteria. Clin. Infect. Dis. 4028-37. [DOI] [PubMed] [Google Scholar]
- 5.Briczinski, E. P., A. T. Phillips, and R. F. Roberts. 2008. Transport of glucose by Bifidobacterium animalis subsp. lactis occurs via facilitated diffusion. Appl. Environ. Microbiol. 746941-6948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Briczinski, E. P., and R. F. Roberts. 2006. Technical note: a rapid pulsed-field gel electrophoresis method for analysis of bifidobacteria. J. Dairy Sci. 892424-2427. [DOI] [PubMed] [Google Scholar]
- 7.Brouns, S. J. J., M. M. Jore, M. Lundgren, E. R. Westra, R. J. H. Slijkhuis, A. P. L. Snijders, M. J. Dickman, K. S. Makarova, E. V. Koonin, and J. van der Oost. 2008. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321960-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reference deleted.
- 9.Deveau, H., R. Barrangou, J. E. Garneau, J. Labonté, C. Fremaux, P. Boyaval, D. A. Romero, P. Horvath, and S. Moineau. 2008. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J. Bacteriol. 1901390-1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dos Voltos, T., O. Mestre, J. Rauzier, M. Golec, N. Rastogi, V. Rasolofo, T. Tonjum, C. Sola, I. Matic, and B. Gicquel. 2008. Evolution and diversity of clonal bacteria: the paradigm of Mycobacterium tuberculosis. PLoS ONE 3e1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.FAO/WHO. 2002. Joint FAO/WHO Working Group report on drafting guidelines for the evaluation of probiotics in food, London, Ontario. WHO, Geneva, Switzerland.
- 12.Favier, C. F., E. E. Vaughan, W. M. de Vos, and A. D. L. Akkermans. 2002. Molecular monitoring of succession of bacterial communities in human neonates. Appl. Environ. Microbiol. 68219-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Federici, F., B. Vitali, R. Gotti, M. R. Pasca, S. Gobbi, A. B. Peck, and P. Brigidi. 2004. Characterization and heterologous expression of the oxalyl coenzyme A decarboxylase gene from Bifidobacterium lactis. Appl. Environ. Microbiol. 705066-5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Felis, G. E., and F. Dellaglio. 2007. Taxonomy of lactobacilli and bifidobacteria. Curr. Issues Intest. Microbiol. 844-61. [PubMed] [Google Scholar]
- 15.Foligne, B., S. Nutten, C. Grangette, V. Dennin, D. Goudercourt, S. Poiret, J. Dewulf, D. Brassart, A. Mercenier, and B. Pot. 2007. Correlation between in vitro and in vivo immunomodulatory properties of lactic acid bacteria. World J. Gastroenterol. 13236-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frank, A. C., and J. R. Lobry. 2000. Oriloc: prediction of replication boundaries in unannotated bacterial chromosomes. Bioinformatics 16560-561. [DOI] [PubMed] [Google Scholar]
- 17.Garrigues, C., B. Stuer-Lauridsen, and E. Johansen. 2005. Characterisation of Bifidobacterium animalis subsp. lactis BB-12 and other probiotic bacteria using genomics, transcriptomics and proteomics. Aust. J. Dairy Technol. 6084-92. [Google Scholar]
- 18.Gill, S. R., M. Pop, R. T. DeBoy, P. B. Eckburg, P. J. Turnbaugh, B. S. Samuel, J. I. Gordon, D. A. Relman, C. M. Fraser-Liggett, and K. E. Nelson. 2006. Metagenomic analysis of the human distal gut microbiome. Science 3121355-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Godde, J. S., and A. Bickerton. 2006. The repetitive DNA elements called CRISPRs and their associated genes: evidence of horizontal transfer among prokaryotes. J. Mol. Evol. 62718-729. [DOI] [PubMed] [Google Scholar]
- 20.Gopal, P. K., J. Prasad, J. Smart, and H. S. Gill. 2001. In vitro adherence properties of Lactobacillus rhamnosus DR20 and Bifidobacterium lactis DR10 strains and their antagonistic activity against an enterotoxigenic Escherichia coli. Int. J. Food Microbiol. 67207-216. [DOI] [PubMed] [Google Scholar]
- 21.Grice, E. A., H. H. Kong, G. Renaud, A. C. Young, NISC Comparative Sequencing Program, G. G. Bouffard, R. W. Blakesley, T. G. Wolfsberg, M. L. Turner, and J. A. Segre. 2008. A diversity profile of the human skin microbiota. Genome Res. 181043-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gueimonde, M., S. Delgado, B. Mayo, P. Ruas-Madiedo, A. Margolles, and C. G. de los Reyes-Gavilán. 2004. Viability and diversity of probiotic Lactobacillus and Bifidobacterium populations included in commercial fermented milks. Food Res. Int. 37839-850. [Google Scholar]
- 23.Haft, D. H., J. Selengut, E. F. Mongodin, and K. E. Nelson. 2005. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput. Biol. 1e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hansen, F. G., B. B. Christensen, and T. Atlung. 2007. Sequence characteristics required for cooperative binding and efficient in vivo titration of the replication initiator protein DnaA in E. coli. J. Mol. Biol. 367942-952. [DOI] [PubMed] [Google Scholar]
- 25.Horvath, P., A.-C. Coûté-Monvoisin, D. A. Romero, P. Boyaval, C. Fremaux, and R. Barrangou. 2008. Comparative analysis of CRISPR loci in lactic acid bacteria genomes. Int. J. Food Microbiol. 13162-70. [DOI] [PubMed] [Google Scholar]
- 26.Horvath, P., D. A. Romero, A.-C. Coûté-Monvoisin, M. Richards, H. Deveau, S. Moineau, P. Boyaval, C. Fremaux, and R. Barrangou. 2008. Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus. J. Bacteriol. 1901401-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones, B. V., M. Begley, C. Hill, C. G. M. Gahan, and J. R. Marchesi. 2008. Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc. Natl. Acad. Sci. USA 10513580-13585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim, J. F., H. Jeong, D. S. Yu, S.-H. Choi, C.-G. Hur, M.-S. Park, S. H. Yoon, D.-W. Kim, G. E. Ji, H.-S. Park, and T. K. Oh. 2009. Genome sequence of the probiotic bacterium Bifidobacterium animalis subsp. lactis AD011. J. Bacteriol. 191678-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kleerebezem, M., J. Boekhorst, R. van Kranenburg, D. Molenaar, O. P. Kuipers, R. Leer, R. Tarchini, S. A. Peters, H. M. Sandbrink, M. W. E. J. Fiers, W. Stiekema, R. M. K. Lankhorst, P. A. Bron, S. M. Hoffer, M. N. N. Groop, R. Kerkhoven, M. de Vries, B. Ursing, W. M. de Vos, and R. J. Siezen. 2003. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc. Natl. Acad. Sci. USA 1001990-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kurokawa, K., T. Itoh, T. Kuwahara, K. Oshima, H. Toh, A. Toyoda, H. Takami, H. Morita, V. K. Sharma, T. P. Srivastava, T. D. Taylor, H. Noguchi, H. Mori, Y. Ogura, D. S. Ehrlich, K. Itoh, T. Takagi, Y. Sakaki, T. Hayashi, and M. Hattori. 2007. Comparative metagenomics revealed commonly enriched gene sets in human gut microbiomes. DNA Res. 14169-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lapierre, L., P. Undeland, and L. J. Cox. 1992. Lithium chloride-sodium propionate agar for the enumeration of bifidobacteria in fermented dairy products. J. Dairy Sci. 751192-1196. [DOI] [PubMed] [Google Scholar]
- 32.Lenski, R. E., C. L. Winkworth, and M. A. Riley. 2003. Rates of DNA sequence evolution in experimental populations of Escherichia coli during 20,000 generations. J. Mol. Evol. 56498-508. [DOI] [PubMed] [Google Scholar]
- 33.Ley, R. E., M. Hamady, C. Lozupone, P. J. Turnbaugh, R. R. Ramey, J. S. Bircher, M. L. Schlegel, T. A. Tucker, M. D. Schrenzel, R. Knight, and J. I. Gordon. 2008. Evolution of mammals and their gut microbes. Science 3201647-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mackiewicz, P., J. Zakrzewska-Czerwińska, A. Zawilak, M. R. Dudek, and S. Cebrat. 2004. Where does bacterial replication start? Rules for predicting the oriC region. Nucleic Acids Res. 323781-3791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Makarova, K., A. Slesarev, Y. Wolf, A. Sorokin, B. Mirkin, E. Koonin, A. Pavlov, N. Pavlova, V. Karamychev, N. Polouchine, V. Shakhova, I. Grigoriev, Y. Lou, D. Rohksar, S. Lucas, K. Huang, D. M. Goodstein, T. Hawkins, V. Plengvidhya, D. Welker, J. Hughes, Y. Goh, A. Benson, K. Baldwin, J.-H. Lee, I. Díaz-Muñiz, B. Dosti, V. Smeianov, W. Wechter, R. Barabote, G. Lorca, E. Altermann, R. Barrangou, B. Ganesan, Y. Xie, H. Rawsthorne, D. Tamir, C. Parker, F. Breidt, J. Broadbent, R. Hutkins, D. O'Sullivan, J. Steele, G. Unlu, M. Saier, T. Klaenhammer, P. Richardson, S. Kozyavkin, B. Weimer, and D. Mills. 2006. Comparative genomics of the lactic acid bacteria. Proc. Natl. Acad. Sci. USA 10315611-15616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Makarova, K. S., N. V. Grishin, S. A. Shabalina, Y. I. Wolf, and E. V. Koonin. 2006. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol. Direct 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Makarova, K. S., and E. V. Koonin. 2007. Evolutionary genomics of lactic acid bacteria. J. Bacteriol. 1891199-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Masco, L., G. Huys, E. De Brandt, R. Temmerman, and J. Swings. 2005. Culture-dependent and culture-independent qualitative analysis of probiotic products claimed to contain bifidobacteria. Int. J. Food Microbiol. 102221-230. [DOI] [PubMed] [Google Scholar]
- 39.Meile, L., W. Ludwig, U. Rueger, C. Gut, P. Kaufmann, G. Dasen, S. Wenger, and M. Teuber. 1997. Bifidobacterium lactis sp. nov., a moderately oxygen tolerant species isolated from fermented milk. Syst. Appl. Microbiol. 2057-64. [Google Scholar]
- 40.Paineau, D., D. Carcano, G. Leyer, S. Darquy, M.-A. Alyanakian, G. Simoneau, J.-F. Bergmann, D. Brassart, F. Bornet, and A. C. Ouwehand. 2008. Effects of seven potential probiotic strains on specific immune responses in healthy adults: a double-blind, randomized, controlled trial. FEMS Immunol. Med. Microbiol. 53107-113. [DOI] [PubMed] [Google Scholar]
- 41.Palmer, C., E. M. Bik, D. B. DiGiulio, D. A. Relman, and P. O. Brown. 2007. Development of the human infant intestinal microbiota. PLoS Biol. 5e177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parche, S., J. Amon, I. Jankovic, E. Rezzonico, M. Beleut, H. Barutçu, I. Schendel, M. P. Eddy, A. Burkovski, F. Arigoni, and F. Titgemeyer. 2007. Sugar transport systems of Bifidobacterium longum NCC 2705. J. Mol. Microbiol. Biotechnol. 129-19. [DOI] [PubMed] [Google Scholar]
- 43.Perfeito, L., L. Fernandes, C. Mota, and I. Gordo. 2007. Adaptive mutations in bacteria: high rate and small effects. Science 317813-815. [DOI] [PubMed] [Google Scholar]
- 44.Pridmore, R. D., B. Berger, F. Desiere, D. Vilanova, C. Barretto, A.-C. Pittet, M.-C. Zwahlen, M. Rouvet, E. Altermann, R. Barrangou, B. Mollet, A. Mercenier, T. Klaenhammer, F. Arigoni, and M. A. Schell. 2004. The genome sequence of the probiotic intestinal bacterium Lactobacillus johnsonii NCC 533. Proc. Natl. Acad. Sci. USA 1012512-2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saeed, A. I., V. Sharov, J. White, J. Li, W. Liang, N. Bhagabati, J. Braisted, M. Klapa, T. Currier, M. Thiagarajan, A. Sturn, M. Snuffin, A. Rezantsev, D. Popov, A. Ryltsov, E. Kostukovich, I. Borisovsky, Z. Liu, A. Vinsavich, V. Trush, and J. Quackenbush. 2003. TM4: a free, open-source system for microarray data management and analysis. BioTechniques 34374-378. [DOI] [PubMed] [Google Scholar]
- 46.Schell, M. A., M. Karmirantzou, B. Snel, D. Vilanova, B. Berger, G. Pessi, M.-C. Zwahlen, F. Desiere, P. Bork, M. Delley, R. D. Pridmore, and F. Arigoni. 2002. The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc. Natl. Acad. Sci. USA 9914422-14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sela, D. A., J. Chapman, A. Adeuya, J. H. Kim, F. Chen, T. R. Whitehead, A. Lapidus, D. S. Rokhsar, C. B. Lebrilla, J. B. German, N. P. Price, P. M. Richardson, and D. A. Mills. 2008. The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proc. Natl. Acad. Sci. USA 10518964-18969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sgorbati, B., M. B. Smiley, and T. Sozzi. 1983. Plasmids and phages in Bifidobacterium longum. Microbiologica 6169-173. [PubMed] [Google Scholar]
- 49.Sonnhammer, E. L., and R. Durbin. 1995. A dot-matrix program with dynamic threshold control suited for genomic DNA and protein sequence analysis. Gene 167GC1-GC10. [DOI] [PubMed] [Google Scholar]
- 50.Sorek, R., V. Kunin, and P. Hugenholtz. 2008. CRISPR—a widespread system that provides acquired resistance against phages in bacteria and archaea. Nat. Rev. Microbiol. 6181-186. [DOI] [PubMed] [Google Scholar]
- 51.Tatusov, R. L., M. Y. Galperin, D. A. Natale, and E. V. Koonin. 2000. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2833-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Turroni, F., E. Foroni, P. Pizzetti, V. Giubellini, A. Ribbera, P. Merusi, P. Cagnasso, B. Bizzarri, G. L. de'Angelis, F. Shanahan, D. van Sinderen, and M. Ventura. 2009. Exploring the diversity of the bifidobacterial population in the human intestinal tract. Appl. Environ. Microbiol. 751534-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ventura, M., C. Canchaya, A. Tauch, G. Chandra, G. F. Fitzgerald, K. F. Chater, and D. van Sinderen. 2007. Genomics of Actinobacteria: tracing the evolutionary history of an ancient phylum. Microbiol. Mol. Biol. Rev. 71495-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ventura, M., J.-H. Lee, C. Canchaya, R. Zink, S. Leahy, J. A. Moreno-Munoz, M. O'Connell-Motherway, D. Higgins, G. F. Fitzgerald, D. J. O'Sullivan, and D. van Sinderen. 2005. Prophage-like elements in bifidobacteria: insights from genomics, transcription, integration, distribution, and phylogenetic analysis. Appl. Environ. Microbiol. 718692-8705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ventura, M., and R. Zink. 2002. Rapid identification, differentiation, and proposed new taxonomic classification of Bifidobacterium lactis. Appl. Environ. Microbiol. 686429-6434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wall, R., S. G. Hussey, C. A. Ryan, M. O'Neill, G. Fitzgerald, C. Stanton, and R. P. Ross. 2008. Presence of two Lactobacillus and Bifidobacterium probiotic strains in the neonatal ileum. ISME J. 283-91. [DOI] [PubMed] [Google Scholar]
- 57.Zhang, W., W. Qi, T. J. Albert, A. S. Motiwala, D. Alland, E. K. Hyytia-Trees, E. M. Ribot, P. I. Fields, T. S. Whittam, and B. Swaminathan. 2006. Probing genomic diversity and evolution of Escherichia coli O157 by single nucleotide polymorphisms. Genome Res. 16757-767. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.