Abstract
The aims of this study were to functionally characterize and analyze the transcriptional regulation and transcriptome of the Rhizobium etli rpoE4 gene. An R. etli rpoE4 mutant was sensitive to oxidative, saline, and osmotic stresses. Using transcriptional fusions, we determined that RpoE4 controls its own transcription and that it is negatively regulated by rseF (regulator of sigma rpoE4; CH03274), which is cotranscribed with rpoE4. rpoE4 expression was induced not only after oxidative, saline, and osmotic shocks, but also under microaerobic and stationary-phase growth conditions. The transcriptome analyses of an rpoE4 mutant and an rpoE4-overexpressing strain revealed that the RpoE4 extracytoplasmic function sigma factor regulates about 98 genes; 50 of them have the rpoE4 promoter motifs in the upstream regulatory regions. Interestingly, 16 of 38 genes upregulated in the rpoE4-overexpressing strain encode unknown putative cell envelope proteins. Other genes controlled by RpoE4 include rpoH2, CH00462, CH02434, CH03474, and xthA1, which encode proteins involved in the stress response (a heat shock sigma factor, a putative Mn-catalase, an alkylation DNA repair protein, pyridoxine phosphate oxidase, and exonuclease III, respectively), as well as several genes, such as CH01253, CH03555, and PF00247, encoding putative proteins involved in cell envelope biogenesis (a putative peptidoglycan binding protein, a cell wall degradation protein, and phospholipase D, respectively). These results suggest that rpoE4 has a relevant function in cell envelope biogenesis and that it plays a role as a general regulator in the responses to several kinds of stress.
In eubacteria, gene expression is controlled at the transcriptional level by the combined actions of sigma factors, activators, and repressors. Sigma factors bind to core RNA polymerase (α2ββ′ω) and recognize specific promoters. The replacement of one sigma factor with another allows the controlled transcription of different genes. Gene expression in exponentially growing bacterial cells depends on a single sigma factor (the σ70 factor) aimed at transcribing housekeeping genes (8, 23). A variable number of alternative sigma factors coordinate the expression of genes required for defined growth conditions and/or responses to specific stimuli (23). Therefore, alternative sigma factors play relevant roles in responding and adapting to different kinds of stresses and environments.
Based on sequence similarities and conserved regions, sigma factors are grouped into two families: σ54 and σ70. In general, bacterial cells have several members from the σ70 family and usually only one or two members from the σ54 family. Members of the diverse σ70 family have four conserved regions; the 2.4 and 4.2 subregions are significantly conserved and recognize the −10 and −35 promoter elements, respectively (8, 23, 32). Moreover, the σ70 family is divided into four phylogenetic groups (23, 26, 32): group 1, the primary sigma factors (σ70-related factors); group 2, nonessential proteins highly similar to primary sigma factors (σS-related factors); group 3, secondary sigma factors (σ28-related factors); and group 4, the extracytoplasmic function (ECF) subfamily (ECF σ, or σE-related, factors). Among these groups, the ECF σ subfamily is the largest and most diverse, and it is involved in a wide range of stress responses and environmental adaptation processes, such as alginate production, carotenoid biosynthesis, starvation responses, and resistance to high temperatures, reactive oxygen species, and antibiotics, etc. (3, 4, 5, 6, 7, 17, 26, 35, 44, 50).
The ECF σ subfamily comprises small proteins (24 to 50 kDa) that in most cases are cotranscribed with an inner membrane anti-σ protein. Anti-σ binds to the sigma factor, reducing the potential interaction with the RNA polymerase (2, 24, 26, 43, 44). After receiving a stimulus, the sigma factor is released and can bind to the promoter regions of specific genes, enabling transcription. Most ECF σ factors positively regulate their own transcription, while coexpression with an anti-σ factor results in the switching off of the ECF σ response (2, 24, 26). The best-characterized ECF σ factors are the homologs of σE, encoded by a gene essential in Escherichia coli (12, 43) but not in Azotobacter vinelandii, Pseudomonas aeruginosa, or Salmonella enterica serovar Typhimurium (6, 17, 35, 50). This protein regulates a large number of genes involved in both the biogenesis and stress responses of the cell envelope (2, 24, 43, 44). Under different conditions, it also controls the transcription of other sigma factors, such as those encoded by rpoD, rpoH, and rpoN (24, 43).
Free-living rhizobia are able to establish symbiotic relationships with roots of leguminous plants and to form nodules in which differentiated bacterial cells reduce atmospheric nitrogen to ammonia (19, 33). In the rhizosphere, but also inside the root nodule, rhizobia may suffer from different kinds of stress, e.g., oxidative stress during the infection process and oxygen limitation after nodule formation (11, 31, 46, 47, 52). In the free-living state, rhizobia face several kinds of stress, such as starvation and changes in osmolarity, pH, and temperature; also, oxidant species are generated as by-products of aerobic respiration or as products from other microorganisms (53).
Rhizobium etli is a gram-negative, free-living soil bacterium able to form nodules on roots of Phaseolus vulgaris and harbors one chromosome and six large plasmids (ranging from 184 to 642 kb) (21). Analysis of the R. etli genome revealed 23 sigma factor-encoding genes: 1 σ70 (rpoD) gene, a housekeeping gene; 2 σ54 (rpoN) genes, involved in symbiosis and nitrogen assimilation; 2 σ32 (rpoH) genes, involved in responses to oxidative and heat stresses; and 18 ECF σ factor genes. Interestingly, none of the encoded sigma factors seem to be homologous to the general stress sigma factor σS, while four of them show high levels of similarity to the E. coli σE factor (21).
In this study, we describe the characterization of a gene encoding an ECF σ factor in R. etli, rpoE4 (CH03273), and evaluate its role under several biological and abiotic stress conditions. The results suggest that RpoE4 is an important general regulator involved in the responses to several stresses, as well as in cell envelope biogenesis.
MATERIALS AND METHODS
Bacterial strains and microbiological methods.
The bacterial strains and plasmids used in this study are listed in Table 1. E. coli strains were grown at 37°C in Luria-Bertani medium (45). R. etli strains were grown at 25°C in either PY medium (40) or minimal medium (MM) (20). Microaerobic (1% O2) conditions were generated as described previously (20). Antibiotics were added at the following final concentrations (in micrograms per milliliter): gentamicin, 5; carbenicillin, 100; nalidixic acid, 20; streptomycin, 100; spectinomycin, 100; kanamycin, 30; and tetracycline, 5. For positive selection of the sacB gene, sucrose was used at concentrations ranging from 7.5 to 10% (wt/vol). To determine the survival rates in the presence of H2O2, R. etli strains were grown in PY medium at 25°C. Aliquots of cultures with optical densities at 600 nm (OD600) of ∼0.3 were taken and incubated with different concentrations of H2O2 (Sigma) for 45 min at 25°C. After treatment, samples were diluted in 10 mM MgSO4-20 mM Tween 40 and plated onto PY medium. To evaluate sensitivity to sucrose, NaCl, and methyl viologen (Paraquat; Sigma), overnight cultures (OD600, ∼1.0) were diluted and plated onto PY medium containing sucrose at 15%, NaCl at 80 mM, or methyl viologen at 40 μM.
TABLE 1.
Strains and plasmids used in this work
| Strain or plasmid | Description and/or genotypea | Reference or source |
|---|---|---|
| R. etli strains | ||
| CE3 | CFN42 derivative; Strr Nalr | 40 |
| CFNXE1Sp | CE3 derivative; rpoE1::loxPSp | This work |
| CFNXE2Sp | CE3 derivative; rpoE2::loxPSp | This work |
| CFNXE3Sp | CE3 derivative; rpoE3::loxPSp | This work |
| CFNXE4Sp | CE3 derivative; rpoE4::loxPSp | This work |
| CFNXTXSp | CE3 derivative; tcrX::loxPSp | This work |
| CFNXΔ3274Sp | CE3 derivative with deletion/substitution ΔCH03274::loxPSp allele | This work |
| CFNXΔ3274lox | CFNXΔ3274Sp derivative with loxPSp interposon deletion; ΔCH03274::loxP | This work |
| E. coliDH5α | Nalr; host for recombinant plasmids | Stratagene |
| Plasmids | ||
| pBBMCS53 | pBBR1MCS-5 derivative carrying a promoterless β-glucuronidase gene | 20 |
| pRK2013 | Conjugation helper plasmid; Kmr | 51 |
| pFAJ1708 | RK2 derivative carrying nptII promoter; Tcr | 14 |
| pK18mobsacB | Kmr Sacs; used for gene replacement | 49 |
| pGUSprpoH2 | pBBMCS53 derivative carrying the promoter of the rpoH2 gene | 37 |
| pJMS2 | Plasmid harboring loxPSp interposon | 36 |
| pJMS8 | pRK7813 derivative harboring cre gene | 38 |
| pJMS24 | pBBMCS53 derivative harboring 570 bp upstream of R. etli CH03274; rpoE4-uidA fusion vector | This work |
| pJMS25 | pBBMCS53 derivative harboring 570 bp upstream of R. etli CH03274; tcrX-uidA fusion vector | This work |
| pJMS26 | pK18mobsacB derivative harboring 1,701 bp of R. etli rpoE1 region | This work |
| pJMS27 | pK18mobsacB derivative harboring 1,878 bp of R. etli rpoE2 region | This work |
| pJMS28 | pK18mobsacB derivative harboring 1,584 bp of R. etli rpoE3 region | This work |
| pJMS29 | pK18mobsacB derivative harboring 1,804 bp of R. etli tcrX-rpoE4 region | This work |
| pJMS30 | pJMS26 derivative harboring loxP Sp interposon in EcoRV site located at codon 47 of rpoE1 | This work |
| pJMS31 | pJMS27 derivative harboring loxP Sp interposon in XhoI site located at codon 147 of rpoE2 | This work |
| pJMS32 | pJMS28 derivative harboring loxP Sp interposon in EcoRV site located at codon 89 of rpoE3 | This work |
| pJMS33 | pJMS29 derivative harboring loxP Sp interposon in ClaI site located at codon 90 of rpoE4 | This work |
| pJMS34 | pJMS29 derivative harboring loxP Sp interposon in NarI site located at bp 282 of trcX | This work |
| pJMS35 | pK18mobsacB derivative harboring 2,517 bp of R. etli rpoE4 region with CH03274 deletion | This work |
| pJMS36 | pJMS35 derivative harboring the deletion/substitution ΔCH03274::loxPSp allele | This work |
| pJMS37 | pFAJ1708 derivative harboring 1,267 bp of rpoE4; PntpII-rpoE4 expression vector | This work |
Nalr, nalidixic acid resistant.
Recombinant-DNA procedures.
Genomic DNA was isolated using the GenomicPrep cell and tissue DNA isolation kit according to the instructions of the manufacturer (Amersham Biosciences). Plasmid DNA was isolated by an alkaline-sodium dodecyl sulfate lysis method, and CaCl2-treated E. coli cells were transformed with the DNA; other general DNA methods were carried out according to standard protocols (45). Restriction enzymes and T4 DNA ligase were used as specified by the manufacturer (Invitrogen). Pfu DNA polymerase (Altaenzymes) was used for PCRs. The conjugative mobilization of plasmids from E. coli to R. etli was done by triparental mating, using the pRK2013 plasmid as a helper (51).
Plasmid and R. etli mutant construction.
The sequences of the oligonucleotides (Unidad de Biosíntesis, Instituto de Biotecnología/UNAM, Mexico) used for R. etli gene amplifications are available in the supplemental material (see Table S1). Primers tcrX-lw and ch3274wb were both used to obtain a PCR product of 570 bp from the region upstream of ATG in rseF (CH03274; 246 bp of a noncoding region and 324 bp of the trcX gene) from R. etli CE3, and the product was cloned in both orientations into the EcoRV site of pBBMCS53 (20) to create pJMS24 (carrying an rpoE4-uidA fusion) and pJMS25 (carrying a tcrX-uidA fusion).
The 1.70-kb R. etli rpoE1, 1.87-kb R. etli rpoE2, 1.58-kb R. etli rpoE3, and 1.80-kb R. etli tcrX-rpoE4 regions were amplified from strain CE3 by PCR and cloned into the appropriate sites in pK18mobsac (49) to give pJMS26, pJMS27, pJMS28, and pJMS29, respectively. pJMS30 is a pJMS26 derivative with a 2.3-kb fill-out ClaI-digested loxP spectinomycin resistance (Sp) interposon (36) inserted into the EcoRV site located at codon 47 of rpoE1. pJMS31 is a pJMS27 derivative with a 2.3-kb SalI-digested loxP Sp interposon inserted into the XhoI site located at codon 147 of rpoE2. pJMS32 is a pJMS28 derivative with a 2.3-kb fill-out ClaI-digested loxP Sp interposon inserted into the EcoRV site located at codon 89 of rpoE3. pJMS33 is a pJMS29 derivative with a 2.3-kb ClaI-digested loxP Sp interposon inserted into the ClaI site located at codon 90 of rpoE4. pJMS34 is a pJMS29 derivative with a 2.3-kb ClaI-digested loxP Sp interposon inserted into the NarI site located at codon 94 of tcrX.
The primer pairs RechE4uh-ch3274wb and ch3274E4ub-RerpoE4we were used to amplify fragments containing the tcrX (1,257-bp) and rpoE4 (1,253-bp) genes, respectively, from strain CE3 by PCR. Both fragments were digested with BamHI, ligated with T4 DNA ligase, and used as templates to amplify a fragment with the RechE4uh and RerpoE4we primers. The 2,510-bp amplification product, which contained a deletion of the rseF (CH03274) gene, was cloned into the HindIII/EcoRI site of pK18mobsacB to give pJMS35. The pJMS36 plasmid is a pJMS35 derivative with a 2.3-kb BamHI-digested loxP Sp interposon inserted into the BamHI site located 33 bp upstream of rpoE4. This construct has rseF deleted by replacement with the loxP Sp interposon (ΔCH03274::loxP Sp).
The 1,267-bp BamHI-EcoRI PCR product generated by using ch3274ub and RerpoE4we primers was cloned into the BamHI/EcoRI sites of pFAJ1708 to produce pJMS37; this construct has the rpoE4 gene under the control of the nptII promoter.
The R. etli rpoE1, rpoE2, rpoE3, rpoE4, trcX, and rseF mutants were obtained by the replacement of the wild-type allele with the rpoE1::loxP Sp (pJMS30), rpoE2::loxP Sp (pJMS31), rpoE3::loxP Sp (pJMS32), rpoE4::loxP Sp (pJMS33), tcrX::loxP Sp (pJMS34), and ΔCH03274::loxP Sp (pJMS35) alleles, respectively. For this purpose, the corresponding plasmid was mobilized from E. coli to R. etli by triparental mating, the single recombinants (exhibiting a streptomycin-resistant [Strr], Spr, kanamycin-resistant [Kmr] phenotype) obtained by plasmid cointegration were plated onto PY medium with sucrose at 7.5 or 10% (wt/vol), and double recombinants were screened by detection of the Spr Kms sucrose-resistant (Sacr) phenotype.
To generate a nonpolar rseF mutation, the loxP Sp interposon was excised from the CFNX3274Sp strain by using the loxP-specific Cre recombinase expressed by the pJMS8 plasmid (38). Losses of the Spr marker and pJMS8 were screened by detection of the Sps, tetracycline-sensitive (Tcs) phenotype.
Microarray hybridizations and analysis.
For microarrays, 70-mer oligonucleotides representing all (6,034) predicted R. etli open reading frames (ORFs) were designed by E. Salazar et al. (submitted for publication), synthesized by MWG-Biotech (Ebersberg, Germany), and spotted in duplicate onto superamide-coated slides (25 by 75 mm; TeleChem International, Inc.) by means of a high-speed robot at the microarray facility at the Instituto de Fisiología Celular/UNAM. Total RNAs from exponential-phase cultures grown in MM were obtained using the RNeasy minikit (Qiagen). The RNA was reverse transcribed and labeled using the CyScribe first-strand cDNA labeling kit according to the instructions of the manufacturer. Briefly, the RNA concentration was determined by measuring the absorbance at 260 nm, and RNA integrity was evaluated by electrophoresis on 1.3% agarose. Samples of 10 μg of RNA were reverse transcribed and differentially labeled with Cy3-dCTP and Cy5-dCTP by using the CyScribe first-strand cDNA labeling kit (Amersham Biosciences). Pairs of Cy3- and Cy5-labeled cDNA samples were mixed and hybridized to the array as described by Hegde et al. (25). After being washed, the array was scanned with a Scan Array Lite microarray scanner (Perkin-Elmer, Boston, MA) using a 10-μm pixel size. Microarray spot detection was carried out, mean signals and mean local background intensities were determined, and image segmentation and signal quantification were performed by using the Array-Pro Analyzer 4.0 software (Media Cybernetics, L.P). Microarray data analysis was performed with GenArise software, developed in the computing unit at the Instituto de Fisología Celular/UNAM (http://www.ifc.unam.mx/genarise/). This software identifies the genes expressed differentially by calculating an intensity-dependent Z-score. It uses a sliding-window algorithm to calculate the mean and standard deviation within the window surrounding each data point and defines a Z-score, where Z represents the number of standard deviations of a data point from the mean, as follows: Zi = [Ri·mean(R)]/sd(R), where Zi is the Z-score for an individual element, mean(R) is the mean log2 ratio of the duplicate of the same element, Ri is the log2 ratio for the individual element, and sd(R) is the standard deviation of the log2 ratio. With these criteria, the elements with Z-scores of >2 or <−2 were considered to be significantly differentially expressed. The DNA microarray experiments were performed three times with the RNAs isolated from independent cultures.
Consensus promoter sequences were obtained from consensus matrices generated and selected by using WCONSENSUS v5c (28). Briefly, sequences upstream (bp −350 to +10 relative to the translation start site) from the genes identified in the microarray analysis were used as input for CONSENSUS to identify the motifs. Sequences containing promoters were aligned using SeqView version 1.0.1 (ftp://gimr.garvan.unsw.edu.au/pub/) and displayed using WebLogo version 2.8.2 (10). The best matrix was selected to deduce the consensus sequence. The prediction of other promoters was done using the genome-scale gene pattern program from the Regulatory Sequence Analysis Tools website (http://embnet.ccg.unam.mx/rsa-tools/) (54).
To carry out semiquantitative reverse transcriptase PCR (RT-PCR) experiments, the RNA was incubated with 1 U μg−1 RNase-free DNase I according to the instructions of the manufacturer (Fermentas Life Sciences) and the absence of DNA contamination was confirmed by PCR. Amplifications were performed with the Thermoscript RT-PCR system (Invitrogen) and with a reduced number of cycles, in order to avoid the plateau of the DNA amplification reaction. The relative quantification of gene expression was done using the CH02950 gene as an endogenous control. The PCR products were separated by agarose gel electrophoresis, and the intensities of the products were quantified by using the Gel Logic 100 imaging system and molecular imaging software (Kodak).
Nodulation and nitrogen fixation determination.
P. vulgaris cv. Negro Jamapa seed surfaces were sterilized, and the seeds were germinated on sterile trays containing sterile vermiculite. Three-day-old seedlings were transferred into 1-liter plastic pots with sterile vermiculite and inoculated with 1 ml of overnight culture (in PY medium). Plants were kept in a culture room at 25°C under a 12-h light/dark cycle and watered with a nitrogen-free nutrient solution (16). Acetylene reduction assays for nitrogenase activity were carried out as described by Girard et al. (20).
β-Glucuronidase activity measurements.
Exponential cultures of strains harboring the uidA transcriptional fusions were grown on MM (to an OD600 of ∼0.3) and incubated with H2O2 at 1 mM, NaCl at 100 mM, or sucrose at 10% (wt/vol) for 45 min. Bacterial cultures containing the pJMS24, pJMS25, or pGUSprpoH2 plasmid were grown in MM under aerobic or microaerobic conditions, and samples at exponential growth phase (OD600, ∼0.3) and stationary growth phase (OD600, ∼0.8) were collected. Quantitative β-glucuronidase assays were performed with 4-nitrophenyl-β-d-glucuronide as the substrate as described previously (20). Nodules were isolated and stained for β-glucuronidase activity as described previously (42).
RESULTS
Phenotypes associated with rpoE homologs in R. etli.
The four R. etli RpoE factors are phylogenetically related to E. coli RpoE and have the 2.1, 2.4, 4.1, and 4.2 subregions, typical domains of the ECF σ factors, conserved (see Fig. S1 in the supplemental material). In addition, rpoE1, rpoE2, and rpoE3 are present downstream of, and oriented in the same transcriptional direction as, an ORF encoding a product with anti-σ domain-transmembrane, or COG5662, domains, common in proteins that perform as anti-σ factors (9). The fourth homolog, rpoE4, is present upstream of, and oriented in same transcriptional direction as, the ORF CH03274 (rseF). Upstream of rseF, transcribed in the opposite direction, is ORF CH03275 (tcrX) (see Fig. S1 in the supplemental material). Genes similar to rseF were identified in Sinorhizobium meliloti as ORF SMc01505, a negative regulator of S. melitoli rpoE2 (homologous to R. etli rpoE4) (48), and in Caulobacter crescentus as ORF CC3476, which has activity similar to that of the S. meliloti ORF and is present upstream of the σT gene (homologous to R. etli rpoE4) (4). On the other hand, TcrX is highly similar to Methylobacterium extorquens PhyR and includes regions homologous to RpoE-like (COG1595) and CheY-like receiver sensor (COG0784) domains. In M. extorquens, PhyR regulates a large number of genes involved in responses to different stresses (22).
In order to evaluate the roles of the four putative R. etli rpoE homologs, we constructed individual interruptions with the loxP Sp interposon in each one of them (see Materials and Methods). The mutant strains had growth rates, colony morphologies, and levels of viability similar to those of the parental strain at 25°C. They were also able to establish symbiotic interactions with P. vulgaris roots, and 28 days after inoculation, the nodules could perform nitrogen fixation at levels similar to the wild-type strain (data not shown). Additionally, none of the mutations affected the sensitivities of the carrier strains to heat shock (20 min at 42 or 55°C in exponential or stationary phase) compared with that of the parental strain (data not shown).
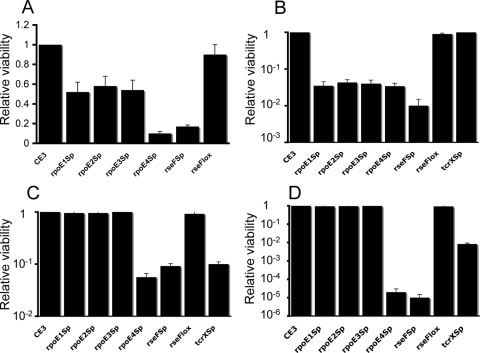
Since ECF σ factors are involved in the responses to environmental stresses, we evaluated whether any of the R. etli rpoE homologs were implicated in the responses to different challenges: oxidative, saline, and osmotic. The oxidative challenge was exerted by exposure of the cells to the strong oxidant H2O2 or to the superoxide generator methyl viologen. The rpoE1::Sp, rpoE2::Sp, and rpoE3::Sp mutants displayed the same degrees of tolerance toward sodium chloride (80 mM) and sucrose (15%) as the wild-type strain, but they were sensitive to oxidative agents, with approximately 50% decreases in viability after H2O2 shock (exposure to 5 mM H2O2 for 45 min during exponential phase) and 23-fold decreases in viability in the presence of methyl viologen (40 μM) compared to the viability of the parental strain (Fig. 1). In contrast, the rpoE4::Sp mutant was significantly more vulnerable than the wild type to all challenges, exhibiting 10-fold-higher sensitivity to H2O2, 24-fold-higher sensitivity to methyl viologen, 17-fold-higher sensitivity to NaCl, and 50,000-fold-higher sensitivity to sucrose (Fig. 1). These results indicate that rpoE1, rpoE2, and rpoE3 participate in the oxidative stress responses elicited by methyl viologen and H2O2 and that rpoE4 may be involved in more general responses to different kinds of environmental challenges, implying an important and general role. With these results and with the aim of pinpointing other elements involved in the regulatory network comprising the rpoE4 ECF σ factor, we decided to explore the regulation of rpoE4, as well as identify the genes regulated by its product.
FIG. 1.
Roles of the tcrX and rpoE genes of R. etli in responses to oxidative and osmotic stresses. The viabilities of the CFNXE1Sp (rpoE1 Sp), CFNXE2Sp (rpoE2 Sp), CFNXE3Sp (rpoE3 Sp), CFNXE4Sp (rpoE4 Sp), CFNXΔ3274Sp (rseF Sp), CFNXΔ3274lox (rseF lox), and CFNXTXSp (tcrX Sp) strains relative to that of wild-type strain CE3 in the presence of H2O2 (A), methyl viologen (B), NaCl (C), and sucrose (D) are expressed as the surviving fractions of the mutant strain populations divided by the surviving fraction of the CE3 population. The surviving fraction was calculated as the number of viable cells after treatment with or in the presence of the compound, divided by the number of viable cells in the absence of stress. The surviving fractions of the CE3 populations in the presence of H2O2, methyl viologen, NaCl, and sucrose were 0.02, 0.92, 0.93, and 0.93, respectively. The data presented are the averages of results from at least three independent experiments. For H2O2 treatment, exponential-phase cultures (OD600, ∼0.3) were incubated with 5 mM H2O2 for 45 min. For the other treatments, the bacteria were grown in PY medium containing methyl viologen (40 μM), NaCl (80 mM), or sucrose (15%, wt/vol).
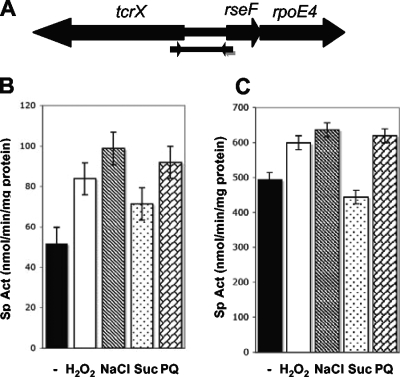
Different mutants were constructed with the aim of identifying the physiological roles of rseF (regulator of sigma rpoE4; CH03274) and trcX, genes located in the rpoE4 region (see Fig. 2A). The mutations included two deletions/substitutions, expressed in the mutant strains CFNXΔ3274Sp and CFNXΔ3274lox (carrying polar and nonpolar mutations in relation to rpoE4, respectively). A third mutant (the CFNXTXSp strain) harbored a tcrX (CH03275) gene inactivated by the insertion of a loxP Sp interposon. The rseF::loxP Sp polar insertion mutant exhibited degrees of sensitivity to all environmental challenges similar to those observed in the rpoE4 mutant. In contrast, the sensitivities of the rseF::loxP nonpolar insertion mutant were similar to those of the wild-type strain (Fig. 1), indicating that rseF and rpoE4 form an operon. The methyl viologen sensitivity caused by the inactivation of trcX was similar to that of the parental strain. Interestingly, the trcX inactivation mutant was significantly more sensitive than the wild-type strain to exposure to sucrose and NaCl, suggesting a role for trcX in the response to osmotic stress (Fig. 1).
FIG. 2.
Physical organization of R. etli rseF-rpoE4 and tcrX genes and expression under different stress conditions. (A) Genomic organization of the rseF-rpoE4 and tcrX region. The fragment cloned upstream of uidA into pBBMCS53 is indicated by the lower diagram. (B and C) Levels of expression of the rpoE4-uidA (B) and trcX-uidA (C) transcriptional fusions under oxidative, saline, or osmotic stress. Exponential-phase cultures (OD600, ∼0.3) were grown on MM, and CE3/pJMS24 and CE3/pJMS25 strains were incubated with 1 mM H2O2, 100 mM NaCl, 10% (wt/vol) sucrose (Suc), or 40 μM methyl viologen (PQ) for 45 min. Specific activity (Sp Act) was determined by using β-glucuronidase activity. The data shown are the averages of results from at least three independent experiments, and the vertical bars represent the standard deviations. −, no stress treatment.
Expression profile of R. etli rpoE4.
To evaluate rseF-rpoE4 and tcrX expression under different free-living conditions, we cloned the 570-bp region upstream of rseF into pBBMCS53 in both orientations relative to the uidA gene (see Materials and Methods) to create rpoE4-uidA (pJMS24) and tcrX-uidA (pJMS25) transcriptional fusions (Fig. 2A). In the wild-type strain CE3, the rpoE4 promoter was induced 1.6-fold by H2O2, 1.9-fold by NaCl, 1.4-fold by sucrose, and 1.8-fold by methyl viologen (Fig. 2B) while the tcrX promoter was induced 1.2-fold by H2O2, 1.3-fold by NaCl, and 1.2-fold by methyl viologen but was not induced by sucrose (Fig. 2C). In addition, induction of the rpoE4 promoter (2.2- and 2.9-fold) in the stationary phase under aerobic and microaerobic conditions, respectively, was observed (Fig. 3A). In contrast, the levels of tcrX-uidA expression under the different conditions were similar and independent of the oxygen concentration, with a slight induction (1.4-fold) only in stationary phase under microaerobic conditions (Fig. 3B).
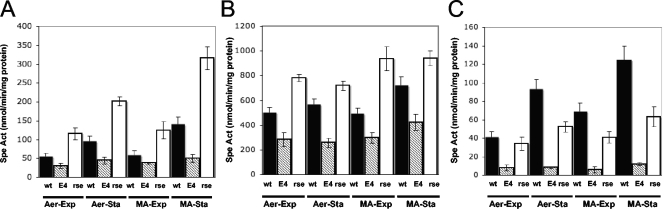
FIG. 3.
The R. etli rpoE4 gene is autoregulated and controls the expression of the rpoE4, trcX, and rpoH2 genes under different conditions. The rpoE4-uidA (A), trcX-uidA (B), and rpoH2-uidA (C) transcriptional fusions were expressed in the wild-type (wt), rpoE4::Sp (E4), and ΔrseF::lox (rse) genetic backgrounds. The strains containing pJMS24, pJMS25, or pGUSprpoH2 were grown in MM under aerobic (Aer) or microaerobic (MA) conditions, and samples were collected at exponential (Exp; OD600, ∼0.3) and stationary (Sta; OD600, ∼0.8) growth phases. Specific activity (Sp Act) was determined by using β-glucuronidase activity. The data shown are the averages of results from at least three independent experiments, and the vertical bars represent the standard deviations.
In other alphaproteobacteria, such as Brucella melitensis, C. crescentus, and S. meliloti, rpoE4 homologs regulate the expression of rpoH (4, 13, 48). In order to elucidate whether the same cascade is present in R. etli, the expression of an rpoH2-uidA fusion was evaluated. With the aim of characterizing the genetic regulation of rpoE4, tcrX, and rpoH2, transcriptional fusions comprising these genes were conjugated into two different mutant backgrounds: those of the CFNXE4Sp strain (lacking rpoE4) and the CFNXΔ3274lox strain (carrying a nonpolar rseF interruption). Under all tested conditions, the three constructs presented reduced expression (35 to 65%) in the CFNXE4Sp mutant background relative to wild-type levels, with the decline of rpoH2 expression being the most dramatic (Fig. 3). These results indicate that RpoE4 regulates the expression of the transcriptional regulators tcrX and rpoH2, which are involved in the responses to heat, oxidative, saline, and osmotic stresses. Moreover, rpoE4 expression, which was induced by microaerobic and stationary growth conditions, was autoregulated. The basal activity in the rpoE4 mutant suggests that RpoE4, along with another sigma factor(s), transcribes the tcrX gene under aerobic and microaerobic conditions; also, tcrX induction under microaerobic conditions seems to be independent of RpoE4. In clear contrast, the expression of the three fusions in the CFNXΔ3274lox mutant background was enhanced under all testing conditions. Moreover, the expression of rpoE4, tcrX, and rpoH2 in rpoE4 and rseF mutant backgrounds was not affected by oxidative or osmotic stress under the same growth conditions (data not shown). These results suggest that rseF, either directly or indirectly, is a negative regulator of the rseF-rpoE4 operon and also of rpoH2.
Identification of genes regulated by the rpoE4 sigma factor.
With the aim of identifying the R. etli genes regulated by RpoE4, transcriptome analyses of the rpoE4 mutant and the rpoE4-overexpressing strain (CE3/pJMS37) were performed. Total RNAs from exponential-phase cultures, grown on MM without treatment, were obtained and used in microarray experiments. A total of 64 genes in the rpoE4 mutant were downregulated with respect to those in the wild type (Z-score < −2); 48 of them were located on the chromosome, and the other 16 were carried in large plasmids (mainly pSym and pCFN42e) (Table 2). Thirteen of the genes code for unknown proteins, although some of these proteins present domains, such as the PCR barrel domain (CH00479 and CH00851) and the MutT/nudix domain (CH00806 and CH01407), similar to those of proteins involved in the stress response. Moreover, putative zinc and iron transport proteins (CH02683, CH02712, and CH04056) may also be implicated in the stress response. Fifteen genes, including agaL1, atpH, ggt2, and aglE, encoding proteins potentially involved in metabolism and 13 genes encoding products, including PheA, RpmG, and RpsC, etc., involved in protein biosynthesis were downregulated in the rpoE4 mutant, suggesting differences in metabolism between the rpoE4 mutant and the wild type. Five rpoE4-regulated genes, three (pE00108, pE00148, and pE00373) located in the pCFN42e plasmid and two (CH01253 and CH03555) on the chromosome, encode proteins with a putative role in cell wall biogenesis, while four genes encode unknown proteins with domains associated with the cell envelope. Interestingly, eight genes encode proteins involved in gene regulation, as components of signal transduction systems (PF00269, PE00176, CH02204, and tcrX) and as transcriptional regulators (CH00371, CH3529, CH04025, and rpoH2).
TABLE 2.
Genes downregulated in the rpoE4::Sp mutant with respect to those in the wild type
| Genea | Z-scoreb | Gene name and/or description of gene productc | Function(s) |
|---|---|---|---|
| RHE_CH00131 | −2.79 | pheA; prephenate dehydratase | Phe biosynthesis |
| RHE_CH00180* | −2.05 | Putative inner membrane protein | Cell envelope protein with unknown function |
| RHE_CH00265* | −2.57 | Hypothetical conserved protein | Unknown |
| RHE_CH00371 | −2.94 | HTH_XRE transcriptional regulator protein | Gene regulation |
| RHE_CH00479* | −2.79 | Protein with conserved PCR barrel domain | Unknown |
| RHE_CH00494 | −2.64 | Aldo/keto dehydrogenase protein | Metabolism |
| RHE_CH00696 | −2.39 | aglE; alpha-glucoside ABC transporter | Metabolism |
| RHE_CH00725* | −2.59 | Putative nucleotidyltransferase protein | Unknown |
| RHE_CH00806* | −2.04 | NTP pyrophosphohydrolase, MutT/nudix family | Stress response |
| RHE_CH00821* | −2.07 | Histidyl-tRNA synthetase protein | Protein biosynthesis |
| RHE_CH00851* | −2.12 | PCR barrel domain membrane protein | Cell envelope protein with unknown function |
| RHE_CH00900 | −2.37 | Hypothetical protein | Unknown |
| RHE_CH00926 | −2.11 | Hypothetical protein | Unknown |
| RHE_CH00996 | −2.86 | Putative dehalogenase-like hydrolase protein | Unknown |
| RHE_CH01253 | −2.08 | Putative peptidoglycan binding protein | Cell wall degradation |
| RHE_CH01407 | −2.12 | NTP pyrophosphohydrolase protein, MutT/nudix family | Stress response |
| RHE_CH01620 | −2.11 | birA; biotin-acetyl-CoA carboxylase ligase | Coenzyme biosynthesis |
| RHE_CH01634 | −2.29 | rpmG; 50S ribosomal protein L33 | Protein biosynthesis |
| RHE_CH01663 | −2.86 | rplK; 50S ribosomal protein L11 | Protein biosynthesis |
| RHE_CH01670 | −2.15 | rpsL; 30S ribosomal protein S12 | Protein biosynthesis |
| RHE_CH01677 | −2.24 | rplW; 50S ribosomal protein L23 | Protein biosynthesis |
| RHE_CH01681 | −2.71 | rpsC; 30S ribosomal protein S3 | Protein biosynthesis |
| RHE_CH01682 | −3.26 | rplP; 50S ribosomal protein L16 | Protein biosynthesis |
| RHE_CH01683 | −2.6 | rpmC; 50S ribosomal protein L29 | Protein biosynthesis |
| RHE_CH01684 | −2.19 | rpsQ; 30S ribosomal protein S17 | Protein biosynthesis |
| RHE_CH01685 | −2.21 | rplN; 50S ribosomal protein L14 | Protein biosynthesis |
| RHE_CH01688 | −2.07 | rpsN; 30S ribosomal protein S14 | Protein biosynthesis |
| RHE_CH01690 | −2.24 | rplF; 50S ribosomal protein L6 | Protein biosynthesis |
| RHE_CH01715* | −2.12 | Carboxymuconolactone decarboxylase family protein | Metabolism |
| RHE_CH01976* | −2.34 | Nucleotide binding protein with TIR-like domain | Unknown |
| RHE_CH02172* | −2.86 | Conserved inner membrane protein | Cell envelope protein with unknown function |
| RHE_CH02204* | −2.33 | Putative signal transduction protein with CBS domain | Gene regulation |
| RHE_CH02683 | −2.44 | Putative ferrichrome ABC transporter | Stress response |
| RHE_CH02712* | −2.86 | Zinc uptake ABC transporter | Stress response |
| RHE_CH02862 | −2.19 | 6-Phosphofructokinase protein | Metabolism |
| RHE_CH03273 | −3.55 | rpoE4; ECF σ factor protein | Gene regulation |
| RHE_CH03274* | −3.74 | rseF; negative regulator of rpoE4 | Gene regulation |
| RHE_CH03275* | −4.89 | tcrX; two-component response regulator protein | Gene regulation |
| RHE_CH03453* | −2.71 | Hypothetical conserved protein | Unknown |
| RHE_CH03529 | −2.5 | HTH homodimeric repressor | Gene regulation |
| RHE_CH03536 | −2.04 | ggt2; gamma-glutamyltranspeptidase protein | Metabolism |
| RHE_CH03555* | −3.33 | LysM domain-containing 5′-nucleotidase/2′,3′-cyclic phosphodiesterase | Cell wall degradation |
| RHE_CH03873* | −2.04 | atpH; ATP synthase delta subunit | Energy production |
| RHE_CH03925 | −2.4 | Short-chain dehydrogenase/reductase | Metabolism, stress response |
| RHE_CH03966 | −2.19 | Probable aldo/keto oxidoreductase protein | Metabolism |
| RHE_CH04025 | −2.67 | Transcriptional regulator, CarD-TRCF family | Gene regulation |
| RHE_CH04026* | −2.8 | rpoH2; heat shock sigma factor | Gene regulation |
| RHE_CH04056 | −2.17 | sfuA; Fe(III) ABC transporter | Stress response |
| RHE_PC00212 | −2.59 | Probable C4-dicarboxylate permease | Metabolism |
| RHE_PD00100 | −2.63 | Insertion sequence transposase | |
| RHE_PD00238 | −2.46 | Insertion sequence transposase | |
| RHE_PD00265 | −2.15 | Hypothetical protein | Unknown |
| RHE_PD00295 | −2.45 | fixO; cytochrome c oxidase | Energy production |
| RHE_PE00016 | −2.11 | Sugar transporter family protein | Metabolism |
| RHE_PE00052 | −2.8 | Putative alpha/beta-hydrolase family protein | Metabolism |
| RHE_PE00089 | −2.56 | agaL1; alpha-galactosidase | Metabolism |
| RHE_PE00099 | −2.67 | Sugar ABC transporter protein | Metabolism |
| RHE_PE00108 | −2.83 | PDZ_CTP periplasm protease | Cell envelope biogenesis |
| RHE_PE00148 | −2.31 | Pilus assembly Flp-like protein | Cell envelope biogenesis |
| RHE_PE00163 | −4.79 | Conserved outer membrane protein | Cell envelope protein with unknown function |
| RHE_PE00176 | −2.75 | Two-component response sensor regulator | Gene regulation |
| RHE_PE00373 | −3.3 | Fasciclin domain protein | Cell envelope biogenesis |
| RHE_PF00264 | −2.62 | Two-component sensor histidine | Gene regulation |
| RHE_PF00478* | −2.15 | Hypothetical protein | Unknown |
RHE_CH, RHE_PD, RHE_PE, and RHE_PF indicate genes located on the chromosome and in pCFN42d (pSym), pCFN42e, and pCFN42f, respectively. * denotes genes with RpoE4 promoter motifs.
Values are based on the log2 ratios of hybridization signals. Only genes with Z-scores of ≤−2 are listed. The results shown are the averages of results from independent experiments.
NTP, nucleoside triphosphate; acetyl-CoA, acetyl coenzyme A; TIR, Toll-interleukin-1 receptor; CBS, cystathionine-beta-synthase; HTH, helix-turn-helix; TRCF, transcription-repair coupling factor.
To overexpress the rpoE4 sigma factor, the gene from strain CE3 was amplified by PCR and cloned under the control of the ntpII promoter in pFAJ1708 (14), generating plasmid pJMS37 (see Materials and Methods). Total RNAs from CE3/pJMS37 and CE3/pFAJ1708 in exponential-phase cultures grown in MM were obtained and used in the microarray experiments. Thirty-eight genes in the rpoE4-overexpressing strain were upregulated (Z-score > 2) with respect to those in the wild type; 28 of them are located on the chromosome, while 10 are in large plasmids (Table 3). Six rpoE4-regulated genes may be involved in stress responses, CH02434 and xthA1 in DNA repair and CH00462 and CH03474 in the response to oxidative stress, while CH00965 and CH01802 encode CsbD-like proteins. Three genes encode proteins potentially implicated in metabolism, and two encode proteins possibly involved in gene regulation. Curiously, a large number (25 of 38) of the rpoE4-upregulated genes encode proteins with unknown functions, and some of the genes (15 of 38) encode proteins without domains or motifs. Interestingly, 16 of the unknown proteins were predicted (using the PSORT program) to be associated with the cell envelope. These data suggest that rpoE4 regulates the transcription of the genes related to stress responses, as well as those related to cell envelope biogenesis.
TABLE 3.
Genes upregulated in the rpoE4-overexpressing strain with respect to those in the wild type
| Genea | Z-scoreb | Gene name and/or description of gene product | Function(s) |
|---|---|---|---|
| RHE_CH00180* | 3.48 | Putative inner membrane protein | Cell envelope protein with unknown function |
| RHE_CH00268* | 3.85 | Conserved hypothetical protein | Unknown |
| RHE_CH00462* | 3.43 | Putative Mn-catalase | Oxidative stress response |
| RHE_CH00479* | 3.93 | PCR barrel domain protein | Unknown |
| RHE_CH00965* | 3.48 | CsbD-like protein, stress inducible | Stress response |
| RHE_CH01004* | 2.90 | Putative inner membrane protein | Cell envelope protein with unknown function |
| RHE_CH01231* | 3.25 | Conserved inner membrane protein | Cell envelope protein with unknown function |
| RHE_CH01335* | 3.07 | Attachment or secretion protein | Cell envelope protein with unknown function |
| RHE_CH01537* | 4.2 | Hypothetical periplasm protein | Cell envelope protein with unknown function |
| RHE_CH01731* | 3.00 | xthA1; exonuclease III | DNA repair |
| RHE_CH01732* | 3.57 | Conserved inner membrane protein | Cell envelope protein with unknown function |
| RHE_CH01778* | 3.94 | Conserved outer membrane protein | Cell envelope protein with unknown function |
| RHE_CH01802* | 3.15 | CsbD-like protein, stress inducible | Stress response |
| RHE_CH02152 | 3.23 | Hypothetical inner membrane protein | Cell envelope protein with unknown function |
| RHE_CH02153 | 3.36 | Conserved cytoplasm protein | Unknown |
| RHE_CH02434* | 2.19 | Putative DNA alkylation repair protein | DNA repair |
| RHE_CH02435* | 2.85 | Putative signal transduction protein with CBSc domain | Gene regulation |
| RHE_CH02503* | 3.65 | Hypothetical conserved protein | Unknown |
| RHE_CH02573* | 3.03 | Conserved inner membrane protein | Cell envelope protein with unknown function |
| RHE_CH02632* | 4.01 | Hypothetical protein | Unknown |
| RHE_CH02635* | 3.47 | Putative inner membrane protein | Cell envelope protein with unknown function |
| RHE_CH02742 | 2.4 | Beta subunit of coenzyme F420 dehydrogenase | Energy production |
| RHE_CH02743* | 3.75 | Hypothetical conserved protein | Unknown |
| RHE_CH03254* | 3.77 | Conserved inner membrane protein | Cell envelope protein with unknown function |
| RHE_CH03273* | 2.95 | rpoE4; ECF σ factor | Gene regulation |
| RHE_CH03274* | 1.9 | rseF; negative regulator of rpoE4 | Gene regulation |
| RHE_CH03404* | 2.11 | Conserved inner membrane protein | Cell envelope protein with unknown function |
| RHE_CH03474* | 3.20 | Putative pyridoxine-phosphate oxidase | B6 biosynthesis, stress response |
| RHE_PB00004* | 2.40 | Hypothetical protein | Unknown |
| RHE_PB00039* | 2.83 | Putative inner membrane protein | Cell envelope protein with unknown function |
| RHE_PB00040* | 3.87 | Conserved periplasm protein | Cell envelope protein with unknown function |
| RHE_PE00299* | 3.48 | Short-chain dehydrogenase/reductase family protein | Metabolism, stress response |
| RHE_PF00044* | 4.34 | Hypothetical outer membrane protein | Cell envelope protein with unknown function |
| RHE_PF00051* | 2.32 | Hypothetical conserved protein | Unknown |
| RHE_PF00085* | 2.04 | Hypothetical inner membrane protein | Cell envelope protein with unknown function |
| RHE_PF00247* | 3.33 | Phospholipase D, PLDc family | Lipid biosynthesis, cell envelope biogenesis |
| RHE_PF00261* | 4.15 | Conserved cytoplasm protein | Unknown |
| RHE_PF00277* | 2.60 | Putative alpha-amilase | Metabolism |
RHE_CH, RHE_PB, RHE_PE, and RHE_PF indicate genes located on the chromosome and in pCFN42b, pCFN42e, and pCFN42f, respectively. * denotes genes with RpoE4 promoter motifs.
Values are based on the log2 ratios of hybridization signals. Only genes with Z-scores of ≥2 are listed. The results shown are the averages of results from independent experiments.
CBS, cystathionine-beta-synthase.
In order to validate the results of the microarray experiments, semiquantitative RT-PCR was performed with six selected genes identified as putative members of the rpoE4 sigmulon. For this analysis, the gene expression levels in the CE3/pFAJ1708 and CE36/pJMS37 strains in the absence of treatment were compared. Increases of 1.99-, 2.15-, 1.85-, 2.05-, 2.43-, and 2.59-fold in the expression of the genes CH00268, CH00479, CH01335, CH01732, CH02453, and tcrX, respectively, in the rpoE4-overexpressing strain with respect to the expression in the parental strain were observed (see Fig. S2 in the supplemental material). In the rpoE4 mutant, a 4.28-fold reduction in the expression of the tcrX gene with respect to that in the wild type was observed, while the genes CH00479, CH01732, CH00268, CH01335, and CH02453 were not expressed in the rpoE4 mutant (after 35 cycles, RT-PCR products were not observed; data not shown). These results validate the findings of our microarray experiments, indicating that the genes are directly or indirectly regulated by rpoE4.
A search for an rpoE4 consensus promoter in the regions from −350 to +10 relative to the translation start sites of the rpoE4-regulated genes identified by transcriptome analysis was performed with both the rpoE4 Sp mutant and the rpoE4-overexpressing strain. WCONSENSUS v5s (28) was used to search for a conserved motif. The sequence GGAAC-N16/17-CGTT was found in 50 of 98 genes; 19 of these 50 genes were identified in the rpoE4 mutant, and 35 were identified in the rpoE4-overexpressing strain (Fig. 4).
FIG. 4.
R. elti rpoE4 motifs located in the regulatory regions of rpoE4-regulated genes. (A) Alignment of nucleotide sequences of upstream regions of rpoE4-regulated genes (motifs are shown in boldface, uppercase letters and are underlined); (B) WebLogo of the rpoE4 consensus motifs; (C) alignment of R. etli rpoE4 consensus motifs with promoter sequences of S. meliloti rpoE2, C. crescentus sigT, M. ex torquens AM1 phyR, M. tuberculosis sigH, E. coli rpoE, S. coelicolor sigR, R. sphaeroides rpoE, B. subtilis sigW, and P. aeruginosa algU (4, 5, 17, 22, 34, 39, 41, 48).
DISCUSSION
Several sigma factor genes are present in the alphaproteobacterial genomes; most of them belong to the ECF σ subfamily. Twenty-three sigma factors are found in R. etli (20): 1 σ70 factor (encoded by rpoD), 2 σ54 factors (encoded by rpoN), 2 σ32 factors (encoded by rpoH), and 18 ECF σ factors. Intriguingly, four ECF σ factors are homologous to the E. coli σE factor, while no homolog of the general stress sigma factor σS is found (21). In this study, we describe the role, regulation, and sigmulon of the RpoE4 sigma factor in R. etli.
Our initial approach was to generate mutations in the R. etli rpoE genes. The inactivation of rpoE1, rpoE2, and rpoE3 resulted in only slight sensitivity to oxidative stress caused by methyl viologen and H2O2, while the disruption of rpoE4 resulted in sensitivities to agents that cause saline, osmotic, and oxidative stresses. To date, few studies of the roles of ECF σ factors in alphaproteobacteria have been described. The most closely related homolog of R. etli rpoE4, rpoE2 in S. meliloti, is not associated with any phenotype (48), while in C. crescentus, the inactivation of σT provokes sensitivities to osmotic and oxidative stresses (4). Also in C. crescentus, σF is involved in the oxidative response of stationary-phase cultures (3).
In several free-living alphaproteobacteria, the rpoE4 region is highly syntenic (4, 48; http://mbgd.genome.ad.jp/), suggesting important roles for the tcrX, rseF, and rpoE4 genes. We studied the roles of these genes, as well as their transcriptional regulation. Using the ΔCH03274::loxP Sp allele, we generated a mutant with a phenotype similar to the one shown by the rpoE4 Sp strain; this finding implies that the mutation has a polar effect relative to rpoE4 and that CH03274 and rpoE4 form an operon. In addition, the deletion of the loxP Sp interposon by using the loxP-specific Cre recombinase generates a nonpolar mutation, and the resulting strain displays a phenotype similar to that of the parent strain. In the closely related species S. meliloti, the homologous genes SMc01505 and SMc01506 also form an operon, but SMc01505 cannot be disrupted (48). With respect to trcX, the associated phenotype indicates that this gene is involved in the responses to saline and osmotic stresses but not in the response to oxidative stress. In M. extorquens AM1, the phyR gene (highly homologous to trcX) is involved in the responses to heat shock, desiccation, and oxidative, UV, ethanol, and osmotic stresses, as well as in phyllosphere colonization (22). In addition, the phosphorylated PhyR form interacts with the anti-sigma factor NepR and therefore acts as anti-anti-sigma factor (18). Due to the high level of similarity between PhyR and TcrX, more studies are needed to clarify whether TcrX acts as a transcriptional regulator, as a sigma factor, or as an anti-anti-sigma factor for rseF.
Our results demonstrate that R. etli rpoE4 regulates its own transcription and that it is negatively regulated by rseF. They also demonstrate that the gene is inducible by saline and osmotic and oxidative stresses, as well as microaerobic and stationary-phase growth conditions, but not heat shock. Moreover, at 21 days after inoculation, nodules formed by CE3/pJMS24 (carrying the rpoE4-uidA fusion) exhibited significant β-glucuronidase activity (data not shown), indicating that rpoE4 is expressed during the symbiotic process and may have some role during symbiosis (see above). Similar results for rpoE2 in S. meliloti (48) and the σT gene in C. crescentus (4) (homologous to R. etli rpoE4) have been reported previously. Furthermore, the products of rseF homologs in S. meliloti (48) and C. crescentus (4), as well as in other alphaproteobacteria, do not have transmembrane or anti-σ domains, suggesting a new regulation mechanism for the ECF σ factor.
In several bacteria, RpoE-like factors control the expression of other sigma factors under different conditions. RpoE regulates the expression of rpoN, rpoH, and rpoD in E. coli (2, 43, 44), σT controls σU and σR expression in C. crescentus (4), rpoHII in Rhodobacter sphaeroides is RpoE dependent (5), and the rpoE5 and rpoH2 genes in S. meliloti are RpoE2 dependent (48). We found that the expression of rpoH2 in R. etli was RpoE4 dependent under aerobic and microaerobic conditions. This pattern was clearly observed in the rpoE4 mutant, but not in the rseF::lox mutant and in the rpoE4-overexpressing strain (transcriptome data), suggesting that a second regulator is needed for transcription or for mRNA stability. Similar results for E. coli rpoS transcription, which is cyclic AMP-catabolite activator protein dependent, were reported previously (27); moreover, the promoter sequence recognized by RpoE4 is present in the upstream regions of rpoH1 and ORF PF00052 (encoding an ECF σ factor) (see Table S2 in the supplemental material). In addition, in R. etli, rpoH1 is involved mainly in oxidative and heat shock responses while rpoH2 is involved in osmotic tolerance and oxidative stress. Both genes are also involved in the senescence of nodules in the symbiotic processes (37). These data suggest that rpoE4 plays, directly or indirectly, a relevant role in survival under free-living conditions and possibly in the senescence of nodules as a master regulator of rpoH1 and rpoH2 (37).
The rpoE4 sigmulon was identified using the rpoE4 Sp mutant and the rpoE4-overexpressing strain under no-stress conditions; these analyses revealed that RpoE4 controls at least 98 genes, 50 of them containing a conserved motif in the upstream regulatory regions. Interestingly, the putative promoter consensus was identified in 35 of the 38 genes differentially expressed in the rpoE4-overexpressing strain but in only 19 of the 64 genes differentially expressed in the rpoE4 Sp mutant. This result may be explained by the possibility that RpoE4 controls other regulators, i.e., rpoH2 is repressed in the mutant, and therefore, more than one regulon may be affected. On the other hand, in the overexpressing strain, the large amount of RpoE4 RNA polymerase formed may transcribe principally rpoE4-dependent promoters. The putative consensus sequence identified here resembles the sequences of promoters recognized by the ECF σ factors, such as those of E. coli rpoE, S. meliloti rpoE2, the C. crescentus σT gene, M. extorquens phyR, Mycobacterium tuberculosis sigH, Bacillus subtilis sigW, P. aeruginosa algU, R. sphaeroides rpoE, and Streptomyces coelicolor sigR (4, 5, 7, 17, 22, 26, 34, 39, 41, 43, 48) (Fig. 4). Among the genes regulated by rpoE4 were (i) genes such as CH00371, CH02204, CH03529, CH04025, PE00176, PF00264, tcrX, and rpoH2 that are involved in gene regulation and encode components of putative two-component systems, transcriptional regulators, or sigma factors; (ii) genes such as rpoH2, CH00462, CH02434, CH03474, xthA1, CH02712, and sfuA that are involved in stress responses (encoding a sigma factor, a putative Mn-catalase, an alkylation DNA repair protein, pyridoxine phosphate oxidase, exonuclease III, and Zn and Fe efflux transporters, respectively) and the CH00695 and CH01802 genes, whose products are homologous to CsbD-like proteins and are possibly involved in the general stress response (1); and (iii) genes such as CH01253, CH03555, and PF00247 (encoding peptidoglycan binding and cell wall degradation proteins and phospholipase D, respectively) with putative roles in cell envelope biogenesis. Intriguingly, a large number of genes that encode proteins with unknown functions that bear membrane domains were induced in the rpoE4-overexpressing strain; the gene products are possibly structural proteins from the cell envelope.
The genes regulated by the rpoE2 sigma factor and by σT in two alphaproteobacteria, S. meliloti and C. crescentus, respectively, have been described previously (4, 48). In both cases, the ECF σ factor acts as a general regulator in response to several stress conditions and controls a large number of genes, consistent with our results. However, there are several differences among the sigmulons in S. meliloti, C. crescentus, and R. etli. In S. meliloti, a large number of genes regulated by rpoE2 are located in pSymB (48) and some of them are also involved in the osmoadaptation response (15), while no plasmids are present in C. crescentus (4). In R. etli, a large number of genes controlled by rpoE4 are located on the chromosome and about 25% are located in plasmids; these distributions suggest that the chromosome has an important role in the general stress response, although the large plasmids may also participate. In addition, using the bidirectional best fit, we found that only a few genes are shared among the sigmulons: rpoE4, rseF, tcrX, CH00965 (csbD-like gene), CH01802 (encoding a PCR barrel protein), and CH02435 (gene regulator) in R. etli, corresponding to SmrpoE2, SMc01505, SMc01504, SMa2071, SMb21330, and SMc21441 in S. meliloti and to CC3475, CC3476, CC3477, CC0938, CC0532, and CC2626 in C. crescentus, respectively. In addition, between the R. etli RpoE4 and S. meliloti RpoE2 sigmulons, some genes are shared: rpoH2, PF00247 (encoding phospholipase D), CH00268 (encoding a conserved protein), CH00851 (encoding a PCR barrel protein), CH01335 (encoding a membrane protein), CH02172 (encoding a membrane protein), CH03254 (encoding a membrane protein), and CH03453 (encoding a conserved protein) in R. etli, corresponding to SMc03873, SMb20094, SMc00371, SMc00885, SMc20879, SMc00063, SMb20454, and SMb21454 in S. meliloti, respectively. On the other hand, in S. enterica and E. coli, the csbD-like homolog is stress induced (1) and regulated by rpoS (55, 56). Furthermore, in E. coli the CH00268 homolog, encoding the YciF protein of unknown function, is rpoS dependent (29, 30). These results, along with the gene expression data, suggest that rpoE4 plays an important role in R. etli during the stationary growth phase.
In conclusion, rpoE4 in R. etli is an important general regulator that is involved in the responses to saline and osmotic and oxidative stresses, and it also has a relevant role in cell envelope biogenesis. Moreover, a bioinformatics analysis of the R. etli genome using the Regulatory Sequence Analysis Tools (http://embnet.ccg.unam.mx/rsa-tools/; 54) suggests that RpoE4 may regulate an additional approximately 140 genes (see Table S2 in the supplemental material).
Supplementary Material
Acknowledgments
We thank Laura Cervantes and José Espíritu for their skillful technical support and B. Valderrama for helpful comments on the manuscript.
Partial financial support was from grant no. IN220307 and IN201006 from PAPIIT-DGAPA Universidad Nacional Autónoma de México.
Footnotes
Published ahead of print on 17 April 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Akbar, S., S. Lee, S. Boylan, and C. Price. 1999. Two genes from Bacillus subtilis under the sole control of the general stress transcription factor sigma B. Microbiology 1451069-1078. [DOI] [PubMed] [Google Scholar]
- 2.Alba, B. M., and C. A. Gross. 2004. Regulation of the Escherichia coli σE-dependent envelope stress response. Mol. Microbiol. 52613-619. [DOI] [PubMed] [Google Scholar]
- 3.Alvarez-Martinez, C. E., R. L. Baldini, and S. L. Gomes. 2006. A Caulobacter crescentus extracytoplasmic function sigma factor mediating the response to oxidative stress in stationary phase. J. Bacteriol. 1881835-1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alvarez-Martinez, C. E., R. F. Lourenço, R. L. Baldini, M. T. Laub, and S. L. Gomes. 2007. The ECF sigma factor σT is involved in osmotic and oxidative stress responses in Caulobacter crescentus. Mol. Microbiol. 661240-1255. [DOI] [PubMed] [Google Scholar]
- 5.Anthony, J., K. L. Warczak, and T. J. Donohue. 2005. A transcriptional response to singlet oxygen, a toxic byproduct of photosynthesis. Proc. Natl. Acad. Sci. USA 1026502-6507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bang, I.-S., J. G. Frye, M. McClelland, J. Velayudhan, and F. C. Fang. 2005. Alternative sigma factor interactions in Salmonella: σE and σH promote antioxidant defences by enhancing σS levels. Mol. Microbiol. 56811-823. [DOI] [PubMed] [Google Scholar]
- 7.Braatsch, S., O. V. Moskvin, G. Klug, and M. Gomelsky. 2004. Responses of the Rhodobacter sphaeroides transcriptome to blue light under semiaerobic conditions. J. Bacteriol. 1867726-7735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Browning, F. B., and S. J. Busby. 2004. The regulation of bacterial transcription initiation. Nat. Rev. Microbiol. 21-9. [DOI] [PubMed] [Google Scholar]
- 9.Campbell, E. A., R. Greenwell, J. R. Anthony, S. Wang, L. Lim, K. Das, H. J. Sofia, T. J. Donohue, and S. A. Darst. 2007. A conserved structural module regulates transcriptional responses to diverse stress signals in Bacteria. Mol. Cell 27793-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crooks G.E., G. Hon, J. M. Chandonia, and S. E. Brenner. 2004. WebLogo: a sequence logo generator. Genome Res. 141188-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davis, B. W., and G. C. Walker. 2007. Identification of novel Sinorhizobium meliloti mutants compromised for oxidative stress protection and symbiosis. J. Bacteriol. 1892110-2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Las Peñas, A., L. Connolly, and C. A. Gross. 1997. σE is an essential sigma factor in Escherichia coli. J. Bacteriol. 1796862-6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delory, M., R. Hallez, J. J. Letesson, and X. De Bolle. 2006. An RpoH-like heat shock sigma factor is involved in stress response and virulence in Brucella melitensis 16M. J. Bacteriol. 1887707-7710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dombrecht, B., J. Vanderleyden, and J. Michiels. 2001. Stable RK2-derived cloning vectors for the analysis of gene expression and gene function in gram-negative bacteria. Mol. Plant-Microbe Interact. 14426-430. [DOI] [PubMed] [Google Scholar]
- 15.Domínguez-Ferreras, A., R. Pérez-Arnedo, A. Becker, J. Olivares, M. J. Soto, and J. Sanjuán. 2006. Transcriptome profiling reveals the importance of plasmid pSymB for osmoadaptation of Sinorhizobium meliloti. J. Bacteriol. 1887617-7625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fahraeus, G. 1957. The infection of clover root hairs by nodule bacteria studied by a simple glass slide technique. J. Gen. Microbiol. 16374-381. [DOI] [PubMed] [Google Scholar]
- 17.Firoved, A. M., J. C. Boucher, and V. Deretic. 2002. Global genomic analysis of AlgU (σE)-dependent promoters (sigmulon) in Pseudomonas aeruginosa and implications for inflammatory processes in cystic fibrosis. J. Bacteriol. 1841057-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Francez-Charlot, A., J. Frunzke, C. Reichen, J. Z. Ebneter, B. Gourion, and J. A. Vorholt. 2009. Sigma factor mimicry involved in regulation of general stress response. Proc. Natl. Acad. Sci. USA 1063467-3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gage, D. J. 2004. Infection and invasion of roots by symbiotic, nitrogen-fixing rhizobia during nodulation of temperate legumes. Microbiol. Mol. Biol. Rev. 68280-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Girard, L., S. Brown, A. Dávalos, O. López, M. Soberón, and D. Romero. 2000. Differential regulation of fixN-reiterated genes in Rhizobium etli by a novel fixL-fixK cascade. Mol. Plant-Microbe Interact. 131283-1292. [DOI] [PubMed] [Google Scholar]
- 21.González, V., R. I. Santamaría, P. Bustos, I. Hernández-González, A. Medrano-Soto, G. Moreno-Hagelsieb, S. Chandra-Janga, M. A. Ramírez, V. Jiménez-Jacinto, J. Collado-Vides, and G. Dávila. 2005. The partitioned Rhizobium etli genome: genetic and metabolic redundancy in seven interacting replicons. Proc. Natl. Acad. Sci. USA 1033834-3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gourion, B., A. Francez-Charlot, and J. A. Vorholt. 2008. PhyR is involved in the general stress response of Methylobacterium extorquens AM1. J. Bacteriol. 1901027-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gruber, T. M., and C. A. Gross. 2003. Multiple sigma subunits and the partitioning of bacterial transcription space. Annu. Rev. Microbiol. 57441-466. [DOI] [PubMed] [Google Scholar]
- 24.Hayden, J. D., and S. E. Ades. 2008. The extracytoplasmic stress factor, σE, is required to maintain cell envelope integrity in Escherichia coli. PLoS ONE 3e1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hegde, P., R. Qi, K. Abernathy, C. Gay, S. Dharap, R. Gaspard, J. E. Hughes, E. Snesrud, N. Lee, and J. Quackenbush. 2000. A concise guide to cDNA microarray analysis. BioTechniques 29548-562. [DOI] [PubMed] [Google Scholar]
- 26.Helmann, J. D. 2002. The extracytoplasmic function (ECF) sigma factors. Adv. Microb. Physiol. 4647-110. [DOI] [PubMed] [Google Scholar]
- 27.Hengge-Aronis, R. 2002. Signal transduction and regulatory mechanisms involved in control of the σS (RpoS) subunit of RNA polymerase. Microbiol. Mol. Biol. Rev. 66373-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hertz, G. Z., and G. D. Stormo. 1999. Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics 15563-577. [DOI] [PubMed] [Google Scholar]
- 29.Hindupur, A., D. Liu, Y. Zhao, H. D. Bellamy, M. A. White, and R. O. Fox. 2006. The crystal structure of the E. coli stress protein YciF. Protein Sci. 152605-2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ibanez-Ruiz, M., V. Robbe-Saule, D. Hermant, S. Labrude, and F. Nore. 2000. Identification of RpoS (σS)-regulated genes in Salmonella enterica serovar Typhimurium. J. Bacteriol. 1825749-5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jamet, A., S. Sigaud, G. Van de Sype, A. Puppo, and D. Herouart. 2003. Expression of the bacterial catalase genes during Sinorhizobium meliloti-Medicago sativa symbiosis and their crucial role during the infection process. Mol. Plant-Microbe Interact. 16217-225. [DOI] [PubMed] [Google Scholar]
- 32.Lonetto, M., M. Gribskov, and C. A. Gross. 1992. The sigma 70 family: sequence conservation and evolutionary relationships. J. Bacteriol. 1743843-3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Long, S. R. 2001. Genes and signals in the rhizobium-legume symbiosis. Plant Physiol. 12569-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manganelli, R., M. I. Voskuil, G. K. Schoolnik, E. Dubnau, M. Gomez, and I. Smith. 2002. Role of the extracytoplasmic-function σ factor σH in Mycobacterium tuberculosis global gene expression. Mol. Microbiol. 45365-374. [DOI] [PubMed] [Google Scholar]
- 35.Martínez-Salazar, J. M., S. Moreno, R. Najera, J. C. Boucher, G. Espín, G. Soberón-Chávez, and V. Deretic. 1996. Characterization of the genes coding for the putative sigma factor AlgU and its regulators MucA, MucB, MucC, and MucD in Azotobacter vinelandii and evaluation of their role in alginate biosynthesis. J. Bacteriol. 1781800-1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martinez-Salazar, J. M., and D. Romero. 2000. Role of the ruvB gene in homologous and homeologous recombination in Rhizobium etli. Gene 243125-131. [DOI] [PubMed] [Google Scholar]
- 37.Martínez-Salazar, J. M., M. Sandoval-Calderón, X. Guo, S. Castillo-Ramírez, A. Reyes, M. G. Loza, J. Rivera, X. Alvarado-Affantranger, F. Sánchez, V. González, G. Dávila, and M. A. Ramírez-Romero. 2009. The Rhizobium etli RpoH1 and RpoH2 sigma factors are involved in different stress responses. Microbiology 155386-397. [DOI] [PubMed] [Google Scholar]
- 38.Martínez-Salazar, J. M., J. Zuñiga-Castillo, and D. Romero. 2009. Differential roles of proteins involved in migration of Holliday junctions on recombination and tolerance to DNA damaging agents in Rhizobium etli. Gene 43226-32. [DOI] [PubMed] [Google Scholar]
- 39.Mascher, T., A.-B. Hachmann, and J. D. Helmann. 2007. Regulatory overlap and functional redundancy among Bacillus subtilis extracytoplasmic function factors. J. Bacteriol. 1896919-6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Noel, K. D., A. Sánchez, L. Fernández, J. Leemans, and M. A. Cevallos. 1984. Rhizobium phaseoli symbiotic mutants with transposon Tn5 insertions. J. Bacteriol. 158148-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paget, M. S., V. Molle, G. Cohen, Y. Aharonowitz, and M. J. Buttner. 2001. Defining the disulphide stress response in Streptomyces coelicolor A3(2): identification of the sigma R regulon. Mol. Microbiol. 421007-1020. [DOI] [PubMed] [Google Scholar]
- 42.Pichon, M., E. P. Journet, A. Dedieu, F. de Billy, G. Truchet, and D. G. Barker. 1992. Rhizobium meliloti elicits transient expression of the early nodulin gene ENOD12 in the differentiating root epidermis of transgenic alfalfa. Plant Cell 41199-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rhodius, V. A., W. Suh, G. Nonaka, J. West, and C. A. Gross. 2006. Conserved and variable functions of the σE stress response in related genomes. PLoS Biol. 443-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rowley, G., M. Spector, J. Kormanec, and M. Roberts. 2006. Pushing the envelope: extracytoplasmic stress responses in bacterial pathogens. Nat. Rev. Microbiol. 4383-394. [DOI] [PubMed] [Google Scholar]
- 45.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 46.Santos, R., D. Herouart, A. Puppo, and D. Touati. 2000. Critical protective role of bacterial superoxide dismutase in rhizobium-legume symbiosis. Mol. Microbiol. 38750-759. [DOI] [PubMed] [Google Scholar]
- 47.Santos, R., D. Herouart, S. Sigaud, D. Touati, and A. Puppo. 2001. Oxidative burst in alfalfa-Sinorhizobium meliloti symbiotic interaction. Mol. Plant-Microbe Interact. 1486-89. [DOI] [PubMed] [Google Scholar]
- 48.Sauviac, L., H. Philippe, K. Phok, and C. Bruand. 2007. An extracytoplasmic function sigma factor acts as a general stress response regulator in Sinorhizobium meliloti. J. Bacteriol. 1894204-4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schafer, A., A. Tauch, W. Jager, J. Kalinowski, G. Thierbach, and A. Puhler. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 14569-73. [DOI] [PubMed] [Google Scholar]
- 50.Schurr, M. J., H. Yu, J. M. Martinez-Salazar, J. C. Boucher, and V. Deretic. 1996. Control of AlgU, a member of the σE-like family of stress sigma factors, by the negative regulators MucA and MucB and Pseudomonas aeruginosa conversion to mucoidy in cystic fibrosis. J. Bacteriol. 1784997-5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Biotechnology 1784-791. [Google Scholar]
- 52.Soupène, E., M. Foussard, P. Boistard, G. Truchet, and J. Batut. 1995. Oxygen as a key developmental regulator of Rhizobium meliloti N2-fixation gene expression within the alfalfa root nodule. Proc. Natl. Acad. Sci. USA 923759-3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thorne, S. H., and H. D. Williams. 1997. Adaptation to nutrient starvation in Rhizobium leguminosarum bv. phaseoli: analysis of survival, stress resistance, and changes in macromolecular synthesis during entry to and exit from stationary phase. J. Bacteriol. 1796894-6901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van Helden, J. 2003. Regulatory sequence analysis tools. Nucleic Acids Res. 313593-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weber, A., S. A. Kogl, and K. Jung. 2006. Time-dependent proteome alterations under osmotic stress during aerobic and anaerobic growth in Escherichia coli. J. Bacteriol. 1887165-7175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yamashino, T., M. Kakeda, C. Ueguchi, and T. Mizuno. 1994. An analogue of the DnaJ molecular chaperone whose expression is controlled by σS during the stationary phase and phosphate starvation in Escherichia coli. Mol. Microbiol. 13475-483. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.