Abstract
An intracellular multiplication F (IcmF) family protein is a conserved component of a newly identified type VI secretion system (T6SS) encoded in many animal and plant-associated Proteobacteria. We have previously identified ImpLM, an IcmF family protein that is required for the secretion of the T6SS substrate hemolysin-coregulated protein (Hcp) from the plant-pathogenic bacterium Agrobacterium tumefaciens. In this study, we characterized the topology of ImpLM and the importance of its nucleotide-binding Walker A motif involved in Hcp secretion from A. tumefaciens. A combination of β-lactamase-green fluorescent protein fusion and biochemical fractionation analyses revealed that ImpLM is an integral polytopic inner membrane protein comprising three transmembrane domains bordered by an N-terminal domain facing the cytoplasm and a C-terminal domain exposed to the periplasm. impLM mutants with substitutions or deletions in the Walker A motif failed to complement the impLM deletion mutant for Hcp secretion, which provided evidence that ImpLM may bind and/or hydrolyze nucleoside triphosphates to mediate T6SS machine assembly and/or substrate secretion. Protein-protein interaction and protein stability analyses indicated that there is a physical interaction between ImpLM and another essential T6SS component, ImpKL. Topology and biochemical fractionation analyses suggested that ImpKL is an integral bitopic inner membrane protein with an N-terminal domain facing the cytoplasm and a C-terminal OmpA-like domain exposed to the periplasm. Further comprehensive yeast two-hybrid assays dissecting ImpLM-ImpKL interaction domains suggested that ImpLM interacts with ImpKL via the N-terminal cytoplasmic domains of the proteins. In conclusion, ImpLM interacts with ImpKL, and its Walker A motif is required for its function in mediation of Hcp secretion from A. tumefaciens.
Many pathogenic gram-negative bacteria employ protein secretion systems formed by macromolecular complexes to deliver proteins or protein-DNA complexes across the bacterial membrane. In addition to the general secretory (Sec) pathway (18, 52) and twin-arginine translocation (Tat) pathway (7, 34), which transport proteins across the inner membrane into the periplasm, at least six distinct protein secretion systems occur in gram-negative bacteria (28, 46, 66). These systems are able to secrete proteins from the cytoplasm or periplasm to the external environment or the host cell and include the well-documented type I to type V secretion systems (T1SS to T5SS) (10, 15, 23, 26, 30) and a recently discovered type VI secretion system (T6SS) (4, 8, 22, 41, 48, 49). These systems use ATPase or a proton motive force to energize assembly of the protein secretion machinery and/or substrate translocation (2, 6, 41, 44, 60).
Agrobacterium tumefaciens is a soilborne pathogenic gram-negative bacterium that causes crown gall disease in a wide range of plants. Using an archetypal T4SS (9), A. tumefaciens translocates oncogenic transferred DNA and effector proteins to the host and ultimately integrates transferred DNA into the host genome. Because of its unique interkingdom DNA transfer, this bacterium has been extensively studied and used to transform foreign DNA into plants and fungi (11, 24, 40, 67). In addition to the T4SS, A. tumefaciens encodes several other secretion systems, including the Sec pathway, the Tat pathway, T1SS, T5SS, and the recently identified T6SS (72). T6SS is highly conserved and widely distributed in animal- and plant-associated Proteobacteria and plays an important role in the virulence of several human and animal pathogens (14, 19, 41, 48, 56, 63, 74). However, T6SS seems to play only a minor role or even a negative role in infection or virulence of the plant-associated pathogens or symbionts studied to date (5, 37-39, 72).
T6SS was initially designated IAHP (IcmF-associated homologous protein) clusters (13). Before T6SS was documented by Pukatzki et al. in Vibrio cholerae (48), mutations in this gene cluster in the plant symbiont Rhizobium leguminosarum (5) and the fish pathogen Edwardsiella tarda (51) caused defects in protein secretion. In V. cholerae, T6SS was responsible for the loss of cytotoxicity for amoebae and for secretion of two proteins lacking a signal peptide, hemolysin-coregulated protein (Hcp) and valine-glycine repeat protein (VgrG). Secretion of Hcp is the hallmark of T6SS. Interestingly, mutation of hcp blocks the secretion of VgrG proteins (VgrG-1, VgrG-2, and VgrG-3), and, conversely, vgrG-1 and vgrG-2 are both required for secretion of the Hcp and VgrG proteins from V. cholerae (47, 48). Similarly, a requirement of Hcp for VgrG secretion and a requirement of VgrG for Hcp secretion have also been shown for E. tarda (74). Because Hcp forms a hexameric ring (41) stacked in a tube-like structure in vitro (3, 35) and VgrG has a predicted trimeric phage tail spike-like structure similar to that of the T4 phage gp5-gp27 complex (47), Hcp and VgrG have been postulated to form an extracellular translocon. This model is further supported by two recent crystallography studies showing that Hcp, VgrG, and a T4 phage gp25-like protein resembled membrane penetration tails of bacteriophages (35, 45).
Little is known about the topology and structure of T6SS machinery subunits and the distinction between genes encoding machinery subunits and genes encoding regulatory proteins. Posttranslational regulation via the phosphorylation of Fha1 by a serine-threonine kinase (PpkA) is required for Hcp secretion from Pseudomonas aeruginosa (42). Genetic evidence for P. aeruginosa suggested that the T6SS may utilize a ClpV-like AAA+ ATPase to provide the energy for machinery assembly or substrate translocation (41). A recent study of V. cholerae suggested that ClpV ATPase activity is responsible for remodeling the VipA/VipB tubules which are crucial for type VI substrate secretion (6). An outer membrane lipoprotein, SciN, is an essential T6SS component for mediating Hcp secretion from enteroaggregative Escherichia coli (1). A systematic study of the T6SS machinery in E. tarda revealed that 13 of 16 genes in the evp gene cluster are essential for secretion of T6S substrates (74), which suggests the core components of the T6SS. Interestingly, most of the core components conserved in T6SS are predicted soluble proteins without recognizable signal peptide and transmembrane (TM) domains.
The intracellular multiplication F (IcmF) and H (IcmH) proteins are among the few core components with obvious TM domains (8). In Legionella pneumophila Dot/Icm T4SSb, IcmF and IcmH are both membrane localized and partially required for L. pneumophila replication in macrophages (58, 70, 75). IcmF and IcmH are thought to interact with each other in stabilizing the T4SS complex in L. pneumophila (58). In T6SS, IcmF is one of the essential components required for secretion of Hcp from several animal pathogens, including V. cholerae (48), Aeromonas hydrophila (63), E. tarda (74), and P. aeruginosa (41), as well as the plant pathogens A. tumefaciens (72) and Pectobacterium atrosepticum (39). In E. tarda, IcmF (EvpO) interacted with IcmH (EvpN), EvpL, and EvpA in a yeast two-hybrid assay, and its putative nucleotide-binding site (Walker A motif) was not essential for secretion of T6SS substrates (74).
In this study, we characterized the topology and interactions of the IcmF and IcmH family proteins ImpLM and ImpKL, which are two essential components of the T6SS of A. tumefaciens. We adapted the nomenclature proposed by Cascales (8), using the annotated gene designation followed by the letter indicated by Shalom et al. (59). Our data indicate that ImpLM and ImpKL are both integral inner membrane proteins and interact with each other via their N-terminal domains residing in the cytoplasm. We also provide genetic evidence showing that ImpLM may function as a nucleoside triphosphate (NTP)-binding protein or nucleoside triphosphatase to mediate T6S machinery assembly and/or substrate secretion.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Strains, plasmids, and primer sequences used in this study are listed in Table 1 and Table 2. For Hcp secretion assays, A. tumefaciens cells grown overnight in 523 broth (31) with appropriate antibiotics were harvested by centrifugation (8,000 × g, 10 min) and resuspended in AB-MES medium (pH 5.5) (33) without any antibiotics at an optical density at 600 nm (OD600) of ∼0.1. After growth for 6 h at 25°C, the cells were harvested, and proteins secreted into the culture medium were precipitated with trichloroacetic acid as described previously (72). The plasmids were maintained by addition of 250 μg/ml carbenicillin, 50 μg/ml gentamicin, and 200 μg/ml spectinomycin for A. tumefaciens and by addition of 100 μg/ml ampicillin, 100 μg/ml spectinomycin, 20 μg/ml kanamycin, and 50 μg/ml gentamicin for E. coli.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| A. tumefaciens strains | ||
| C58 | Wild-type virulent strain containing nopaline-type Ti plasmid pTiC58 | E. Nester |
| EML1068 | impLM in-frame deletion mutant, C58ΔimpLM | This study |
| EML1073 | impKL in-frame deletion mutant, C58ΔimpKL | This study |
| E. coli strains | ||
| DH10B | Host for DNA cloning | Invitrogen |
| BL21(DE3) | Host for overexpressing proteins driven by T7 promoter | 62 |
| S. cerevisiae AH109 | Host for yeast two-hybrid analysis | Clontech |
| Plasmids | ||
| pRL662 | Gmr, broad-host-range vector derived from pBBR1MCS-2 | 69 |
| pJQ200KS | Gmr, suicide plasmid encoding Gmr and containing sacB gene for selection of double crossover | 50 |
| pET22b(+) | Apr, E. coli overexpression vector to generate C-terminal His-tagged protein | Novagen |
| pTrc200 | Smr Spr, pVS1 origin lacIq, trc promoter expression vector | 57 |
| pGADT7 | Apr, AD vector used in yeast two-hybrid assay | Clontech |
| pGBKT7 | Kanr, DNA-BD vector used in yeast two-hybrid assay | Clontech |
| pImpLM | Gmr, pRL662 expressing ImpLM driven by lacZp | 72 |
| pRU1156 | Apr Tcr, stable broad-host-range promoter probe vector containing gfpmut3.1 and gusA | 32 |
| pImpKL | Gmr, pRL662 expressing ImpKL driven by lacZp | This study |
| pImpLMK145A | Gmr, pRL662 expressing ImpLM containing a K145A substitution | This study |
| pImpLMG144A K145A | Gmr, pRL662 expressing ImpLM with both G144A and K145A substitutions | This study |
| pImpLMΔWalker A | Gmr, pRL662 expressing ImpLM with Walker A deletion | This study |
| pGFP-ImpLM | Gmr, pRL662 expressing GFP-ImpLM fusion protein | This study |
| pImpLM-GFP | Gmr, pRL662 expressing ImpLM-GFP fusion protein | This study |
| pTM1-GFP-TM2+3 | Gmr, pRL662 expressing fusion protein with GFP inserted between first and second TM domains of ImpLM | This study |
| pTM1+2-GFP-TM3 | Gmr, pRL662 expressing fusion protein with GFP inserted between second and third TM domains of ImpLM | This study |
| pTM1+2-GFP | Gmr, pRL662 expressing truncated ImpLM protein with GFP fused after the second TM domain | This study |
| pGFP-ImpKL | Gmr, pRL662 expressing GFP-ImpKL fusion protein | This study |
| pImpKL-GFP | Gmr, pRL662 expressing ImpKL-GFP fusion protein | This study |
| pTM1+2_BlaM | Gmr, pRL662 expressing fusion protein with BlaM fused after second TM domain of pImpLM | This study |
| pBlaM | Gmr, pRL662 expressing BlaM protein without signal peptide | This study |
| pTM1-BlaM-TM2+3 | Gmr, pRL662 expressing fusion protein with BlaM inserted between first and second TM domains of ImpLM | This study |
| pTM1+2-BlaM-TM3 | Gmr, pRL662 expressing fusion protein with BlaM inserted between second and third TM domains of ImpLM | This study |
| pImpLM-BlaM-end | Gmr, pRL662 expressing fusion protein with BlaM fused at the C terminus of ImpLM | This study |
| pBlaM-ImpKL | Gmr, pRL662 expressing BlaM-ImpKL fusion protein | This study |
| pImpKL-BlaM | Gmr, pRL662 expressing ImpKL-BlaM fusion protein | This study |
| pET-ImpKL-His | Apr, pET22b overexpressing His-tagged ImpKL in E. coli | This study |
| pET-C-ImpLM-His | Apr, pET22b overexpressing His-tagged C terminus of ImpLM in E. coli | This study |
| pTrc-ImpLM | Smr Spr, pTrc200 expressing ImpLM driven by trc promoter | This study |
| pJQ200KS-ΔimpLM | Gmr, used in generating impLM in-frame deletion mutant of A. tumefaciens | This study |
| pJQ200KS-ΔimpKL | Gmr, used in generating impKL in-frame deletion mutant of A. tumefaciens | This study |
| pAD-ImpLM | Apr, AD vector expressing ImpLM | This study |
| pBD-ImpLM | Kanr, DNA-BD vector expressing ImpLM | This study |
| pAD-ImpKL | Apr, AD vector expressing ImpKL | This study |
| pBD-ImpKL | Kanr, DNA-BD vector expressing ImpKL | This study |
| pAD-N-ImpLM | Apr, AD vector expressing N terminus of ImpLM (residues 1 to 495) | This study |
| pBD-N-ImpLM | Kanr, DNA-BD vector expressing N terminus of ImpLM (residues 1 to 495) | This study |
| pAD-N-ImpKL | Apr, AD vector expressing N terminus of ImpKL (residues 1 to 255) | This study |
| pBD-N-ImpKL | Kanr, DNA-BD vector expressing N terminus of ImpKL (residues 1 to 255) | This study |
| pAD-C-ImpLM | Apr, AD vector expressing C terminus of ImpLM (residues 466 to 1159) | This study |
| pBD-C-ImpLM | Kanr, DNA-BD vector expressing C terminus of ImpLM (residues 466 to 1159) | This study |
| pAD-C-ImpKL | Apr, AD vector expressing C terminus of ImpKL (residues 279 to 501) | This study |
| pBD-C-ImpKL | Kanr, DNA-BD vector expressing C terminus of ImpKL (residues 279 to 501) | This study |
TABLE 2.
Primers used in this study
| Primer | Sequence (5′-3′)a |
|---|---|
| 1 | GAATCATATGGCCGAAGCGGATCGCAA |
| 2 | AATAAACTCGAGGAACTGCGCAGGGCA |
| 3 | TACGCATATGAGCACGGACAACCCCT |
| 4 | TAGCTCAATGCTCGAGTGGCTGGGCCTCT |
| 5 | GGTGGTACCGTAGAGAACAGTTCG |
| 6 | CCCAAGCTTAAACACCACCGCCTCGTT |
| 7 | CCGCCGCATATGAATCCATTGAGCTATT |
| 8 | TCCCCCGGGCTTTTGAAACACCA |
| 9 | TTCGCGGGCAAAGCCGGAATTGA |
| 10 | AAACTGCAGGAACTGCGCAGGGCA |
| 11 | GAATCCATATGAGCACGGACAACCCCT |
| 12 | CGCGGATCCAATAGCTCAATGGATTC |
| 13 | GGAGGATCCGGTCATCGTCCGCAGT |
| 14 | GGATTCCATATGAACAACGCATCCGATG |
| 15 | GCGACGACGGCGCTGACCAATT |
| 16 | AATTGGTCAGCGCCGTCGTCGCGCCGGA |
| 17 | GTCGTCTAGACCTCGAGTAGAGAACAGTTC |
| 18 | CAGCGACGACGGCGCTGACCAATT |
| 19 | AATTGGTCAGCGCCGTCGTCGCTGCGGAGC |
| 20 | TACGTGATCTTCACGACGGCGCTGAC |
| 21 | CGCCGTCGTGAAGATCACGTACCA |
| 22 | CCGCTCGAGTGCCATTGCCCTGCATCTGG |
| 23 | GCTCTAGATCTCGACATAGGAACGGATCG |
| 24 | GGTGGTACCTTTGAAATCGCCGAG |
| 25 | GGAGGTACCATCGATCACCCAGAAA |
| 26 | CGCCCAAGCTTCCAATGCTTAATCAGTG |
| 27 | CACAAAGCTTCCGCTTGGAACGGT |
| 28 | CTAGTCTAGATTTGAAACACCACCGCCT |
| 29 | GGTGGTACCCAATGCGCCATTGAT |
| 30 | CACAAAGCTTGTCACGCTGTCCATTC |
| 31 | GGAGGTACCCGTAAAGGAGAAGAACTT |
| 32 | CGCCCAAGCTTTTTGTATAGTTCATCCA |
| 33 | TCCGCTCGAGAATCCATTGAGCTATT |
| 34 | TACTCGAGCGTAAAGGAGAAGAACTTTTC |
| 35 | GATCTAGAACGGACCATGATTACCTCAGT |
| 36 | AATTAAAGCTTGACAGCAGCCGGGAGAGG |
| 37 | CACAAAGCTTATGATCATCGATCACCCA |
| 38 | TTCCGCTCGAGCCAATGCTTAATCAGTG |
| 39 | ATCCGCTCGAGATGAGCACGGACAA |
| 40 | GTCGTCTAGACCTCGAGTCAGGTGCCATTG |
| 41 | GGAGGTACCTGGCTGGGCCTCT |
| 42 | CGCCCAAGCTTTTACCAATGCTTAA |
| 43 | GCTCTAGAAGCTGATGGCCTTCCTGCAG |
| 44 | CGGGATCCATTCATGGCTGGGCCTCTCC |
| 45 | CGGGATCCCAGTTCTAGTGACGGAACGAG |
| 46 | TCCCCCCGGGCGAAATCGCCGGTCATCGATG |
| 47 | GCTCTAGACGAAGACTGCATCCAGCTTGC |
| 48 | CGGGATCCGCTCATTCGCGTAACGCCCAC |
| 49 | CGGGATCCCAGCCATGAATCCATTGAGCTA |
| 50 | TCCCCCCGGGAGTGTCGATAAGGATCGCCTC |
Restriction enzyme sites are underlined, and the mutated sequences are indicated by bold type.
Construction of impLM and impKL in-frame deletion mutants and complementing plasmids.
Plasmids pJQ200KS-ΔimpLM and pJQ200KS-ΔimpKL were used to generate ΔimpLM (EML1068) and ΔimpKL (EML1073) in-frame deletion mutants of A. tumefaciens C58, which was also used as the template for PCR unless otherwise indicated. Plasmid pJQ200KS-ΔimpLM was created by ligating XbaI/BamHI-digested impLM PCR product 1 (∼500-bp DNA fragment upstream of the impLM open reading frame [ORF]; primers 43 and 44) and BamHI/XmaI-digested impLM PCR product 2 (∼500-bp DNA fragment downstream of the impLM ORF; primers 45 and 46) into XbaI/XmaI sites of pJQ200KS. Plasmid pJQ200KS-ΔimpKL was created by ligating XbaI/BamHI-digested impKL PCR product 1 (∼500-bp DNA fragment upstream of the impKL ORF; primers 47 and 48) and BamHI/XmaI-digested impKL PCR product 2 (∼500-bp DNA fragment downstream of the impKL ORF; primers 49 and 50) into the XbaI/XmaI sites of suicide plasmid pJQ200KS.
Plasmid pJQ200KS-ΔimpKL or pJQ200KS-ΔimpLM was transformed into A. tumefaciens C58 via electroporation and selected on sucrose-free 523 agar containing gentamicin at 28°C for 2 days for the first recombination events. Positive colonies were confirmed by PCR and were grown further in sucrose-free 523 broth without antibiotics at 28°C overnight. Serial dilutions (up to 10−4) were plated onto 523 agar containing 5% sucrose without antibiotics and grown at 28°C for 2 days to select for colonies with a second recombination event. Sucrose-resistant, gentamicin-sensitive colonies were confirmed by PCR to obtain the ΔimpLM and ΔimpKL deletion mutants. For complementation, plasmid pImpKL was constructed in the same way as pImpLM (or pIcmF), in which impLM (or icmF) was driven by a lac promoter in the broad-host-range vector pRL662. The impKL ORF and the corresponding ribosomal binding sequences were amplified with primers 22 and 23, and the PCR product was digested with XhoI/XbaI and cloned into the same sites of pRL662, resulting in plasmid pImpKL.
Topology analysis using β-lactamase (BlaM) and green fluorescent protein (GFP) fusions.
To generate β-lactamase fusions in pImpLM by transposon mutagenesis, an EZ-Tn5 transposon assay was carried out according to the manufacturer's instructions using an EZ-Tn5 β-lactamase fusion kit (Epicentre Biotechnologies, Madison, WI). Fusions were also created by cloning. Plasmid pBlaM was generated to express BlaM without a signal peptide by ligating a PCR fragment amplified from a plasmid containing the EZ-Tn5 transposon into the HindIII site of pRL662. Plasmid pTM1-BlaM-TM2+3 was generated by ligating XhoI/KpnI-digested impLM PCR product 1 (primers 17 and 24), KpnI/HindIII-digested blaM (primers 25 and 26), and HindIII/XbaI-digested impLM PCR product 2 (primers 27 and 28) into XhoI/XbaI sites of pRL662. For plasmid pTM1+2-BlaM-TM3, XhoI/KpnI-digested impLM PCR product 1 (primers 17 and 29), KpnI/HindIII-digested blaM (primers 25 and 26), and HindIII/XbaI-digested impLM PCR product 2 (primers 28 and 30) were ligated into XhoI/XbaI sites of pRL662. For plasmid pImpLM-BlaM-end, the XhoI/PstI-digested impLM PCR product (primers 17 and 10) was ligated with PstI/HindIII-digested blaM and cloned into the XhoI/HindIII sites of pRL662. For plasmids pTM1+2-BlaM and pTM1+2-GFP, XhoI/KpnI-digested impLM PCR product 1 (primers 17 and 29) was ligated with KpnI/HindIII-digested blaM (primers 25 and 42) and with the KpnI/XbaI-digested gfp ORF PCR product (primers 31 and 35) and cloned into the XhoI/HindIII and XhoI/XbaI sites of pRL662, respectively. For plasmid pGFP-ImpLM, the BamHI/XhoI-digested gfp ORF PCR product amplified from the promoter probe vector pRU1156 containing the gfp reporter and the XhoI/XbaI-digested impLM ORF PCR product (primers 33 and 28) were ligated into the BamHI/XbaI sites of pRL662. For plasmid pImpLM-GFP, the XhoI/XbaI-digested gfp ORF PCR product (primers 34 and 35) and the HindIII/XhoI-digested impLM ORF PCR product (primers 36 and 2) were ligated into the HindIII/XbaI sites of pRL662. For plasmids pTM1-GFP-TM2+3 and pTM1+2-GFP-TM3, a gfp ORF was amplified with primers 31 and 32 to replace blaM in pTM1-BlaM-TM2+3 and pTM1+2-BlaM-TM3, respectively.
For plasmids used in ImpKL topology studies, plasmid pBlaM-ImpKL was created by ligating the HindIII/XhoI-digested blaM ORF (primers 37 and 38) and the XhoI/BamHI-digested impKL ORF (primers 39 and 12) into the HindIII/BamHI sites of pRL662. Plasmid pImpKL-BlaM was created by ligating the XhoI/KpnI-digested impKL ORF (primers 40 and 41) and the KpnI/HindIII-digested blaM ORF (primers 25 and 42) into the XhoI/HindIII sites of pRL662. Plasmid pGFP-ImpKL was generated by ligating the HindIII/XhoI-digested gfp ORF and the XhoI/BamHI-digested impKL ORF (primers 39 and 12) into the HindIII/BamHI sites of pRL662. Plasmid pImpKL-GFP was created by ligating the XhoI/KpnI-digested impKL ORF (primers 40 and 41) and the KpnI/XbaI-digested gfp ORF (primers 31 and 35) into the XhoI/XbaI sites of pRL662.
β-Lactamase activity was examined by determining the ability of E. coli strains to grow on LB medium with 100 μg/ml ampicillin at 37°C overnight or the ability of A. tumefaciens strains to grow on 523 medium with 250 μg/ml carbenicillin at 28°C for 24 h.
Generating mutations in the Walker A motif of ImpLM by site-directed mutagenesis.
Mutations in the Walker A motif of ImpLM were created by site-directed mutagenesis as described previously (27). For plasmid pImpLMK145A, two complementary PCR products encoding replacement of lysine 145 by alanine flanked by 10 to 15 bases of overlapping sequence were amplified separately using two pairs of primers (primers 15 and 9 and primers 16 and 17). For plasmid pImpLMG144A K145A, two complementary PCR products encoding replacement of double mutations at glycine 144 and lysine 145 with two alanines were amplified using primers 18 and 9 and primers 19 and 17. For plasmid pImpLMΔWalker A, two complementary PCR products with deletion of the Walker A motif were amplified using primers 20 and 9 and primers 21 and 17. The two PCR products were combined and amplified using primers 9 and 17, resulting in a 1.5-kb DNA fragment. The DNA fragment was further digested with XhoI/BamHI and ligated at the same sites in pImpLM to create the K145A, G144A K145A, and ΔWalker A mutations in pImpLM.
Yeast two-hybrid assay.
The Matchmaker yeast two-hybrid system was used according to the instructions described in the user's manual (Clontech, Mountain View, CA). The impLM ORF PCR amplified with primers 7 and 2 or primers 7 and 8 was digested with NdeI/XhoI or NdeI/SmaI and cloned into the NdeI/XhoI sites of pGADT7 for N-terminal fusion to the activation domain (AD) or into the NdeI/SmaI sites of pGBKT7 for N-terminal fusion to the DNA-binding domain (BD), resulting in pAD-ImpLM and pBD-ImpLM, respectively. For plasmids pAD-N-ImpLM and pBD-N-ImpLM, a PCR fragment encoding the N terminus of ImpLM (amino acids 1 to 495) was amplified using primers 7 and 9, and the NdeI/BamHI-digested PCR product was cloned into the NdeI/BamHI sites of pGADT7 and pGBKT7. For plasmids pAD-C-ImpLM and pBD-C-ImpLM, a PCR fragment encoding the C terminus of ImpLM (amino acids 466 to 1159) was amplified using primers 1 and 2 or primers 1 and 10 and digested with NdeI/XhoI or NdeI/PstI before ligation into the NdeI/XhoI and NdeI/PstI sites of pGADT7 and pBGKT7, respectively. For plasmids pAD-ImpKL and pBD-ImpKL, the impKL ORF was PCR amplified using primers 11 and 12, digested with NdeI/BamHI, and cloned into the same sites of pGADT7 and pGBKT7. For plasmids pAD-N-ImpKL and pBD-N-ImpKL, a PCR fragment encoding the N terminus of ImpKL (amino acids 1 to 255) was amplified using primers 11 and 13, digested with NdeI/BamHI, and cloned into the same sites of both pGADT7 and pGBKT7. For pAD-C-ImpKL and pBD-C-ImpKL, a PCR fragment encoding the C terminus of ImpKL (amino acids 279 to 501) was amplified using primers 14 and 12, digested with NdeI/BamHI, and cloned into the same sites of pGADT7 and pGBKT7.
The resulting prey and bait plasmids were used to cotransform Saccharomyces cerevisiae strain AH109. The transformants were selected on the basis of their growth on synthetic dextrose (SD) minimal medium lacking tryptophan (Trp) and leucine (Leu). Positive interactions of expressed fusion proteins were then determined by growth on SD medium lacking Trp, Leu, adenine, and histidine (His) at 30°C for 3 days.
Biochemical fractionation.
A. tumefaciens cellular fractions were isolated as described previously (16), with minor modifications. Cells were harvested and resuspended in osmotic shock buffer (50 mM Tris-Cl, 20% sucrose, 2 mM EDTA, 0.5 mg/ml lysozyme, 1 mM phenylmethylsulfonyl fluoride [PMSF]; pH 7.5) to an OD600 of 5. After incubation at room temperature for 1 h, the cells whose lysates are referred to below as total protein were centrifuged twice at 10,000 × g at 4°C to obtain a supernatant solution containing the periplasmic proteins. The resulting pellet, resuspended in the same volume of sonication buffer (50 mM Tris-HCl [pH 7.5], 0.2 M KCl, 1 mM PMSF), was subjected to cell lysis via sonication and then centrifugation twice at 10,000 × g at 4°C. The supernatant solution containing both the cytoplasmic and membrane proteins was then centrifuged at 150,000 × g for 1 h at 4°C to obtain the cytosolic proteins (supernatant) and membrane proteins (pellet). For protein solubilization studies, the membrane fractions were resuspended (final protein concentration, 1 mg/ml) by sonication in 50 mM Tris-HCl (pH 7.5) containing 1 mM PMSF and incubated on ice for 30 min with or without one of the following compounds: 1 M NaCl, 0.1% Na2CO3, 6 M urea, 3% Triton X-100, 1.5% N-lauroylsarcosine, or 1% sodium dodecyl sulfate (SDS). Each reaction mixture was centrifuged at 150,000 × g for 1 h at 4°C. The resulting supernatant solution and the pellet, resuspended in an equivalent volume of 50 mM Tris-HCl (pH 7.5), were analyzed by immunoblotting. To separate the inner and outer membranes by sucrose density gradient centrifugation, cells were harvested and resuspended in lysis buffer (50 mM Tris-HCl [pH 7.5], 20% sucrose, 0.2 M KCl, 0.2 mM dithiothreitol, 0.2 mg/ml RNase A, 0.2 mg/ml DNase, 1 mM PMSF) to an OD600 of 10. The cells were lysed by two passages through a French pressure cell (Aminco, Silver Spring, MD) at 16,000 lb/in2, followed by lysozyme (0.5 mg/ml) treatment on ice for 30 min. The cell lysate was centrifuged at 20,000 × g at 4°C for 15 min, and the supernatant solution was then centrifuged at 150,000 × g for 1 h at 4°C. The pellet, enriched with inner and outer membrane proteins, was resuspended in 2.5 ml of buffer containing 20% sucrose, 0.2 mM dithiothreitol, and 5 mM EDTA by sonication on ice. Two milliliters of membrane proteins was layered onto an EDTA (5 mM)-sucrose gradient containing 6 ml of 53% sucrose on top of 2 ml of 70% sucrose, and this was followed by centrifugation at 150,000 × g for 16 h at 4°C. One-milliliter fractions were collected from the top of the gradient and analyzed by immunoblotting. The quality of fractionation was assessed by measuring the absorption at 280 nm of fractions, the enzymatic activity of the inner membrane marker NADH oxidase (16, 43), and the density of the fractions.
ImpLM antibody generation and immunoblot analysis.
Plasmid pET-C-ImpLM-His was constructed by ligating a PCR fragment encoding the C terminus of ImpLM (amino acids 466 to 1159), amplified using primers 1 and 2 to overexpress the C terminus of ImpLM, in E. coli BL21(DE3). The procedure used for protein purification was described previously (72). The major 66-kDa protein band, corresponding to the putative C terminus of ImpLM-His, was cut out from an SDS gel and used to generate polyclonal antibodies in rabbits. Immunoblot analysis was performed as described previously (33) using primary polyclonal antibodies against C-ImpLM, Hcp (72), ActC (36), or His6 (LTK BioLaboratories, Taipei, Taiwan) followed by a secondary antibody using horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G (chemichem), and the results were detected using the Western Lightning system (Perkin Elmer, Boston, MA). Chemiluminescent bands were visualized using X-ray film (Kodak, Rochester, NY).
Copurification of ImpKL-His and ImpLM.
Plasmid pET-ImpKL-His used for copurification was created by PCR amplifying the impKL ORF using primers 3 and 4, and the NdeI/XhoI-digested PCR product was cloned into the NdeI/XhoI sites of pET22b(+). Plasmid pTrc-ImpLM used for copurification was created by PCR amplifying the impLM ORF using primers 5 and 6, and the KpnI/HindIII-digested PCR product was cloned into the KpnI/HindIII sites of pTrc200.
An overnight culture of E. coli BL21(DE3) cells harboring pET-ImpKL-His and pTrc-ImpLM was subcultured at a 1:100 dilution into fresh LB medium and grown to an OD600 of 0.4 to 0.6. Isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 0.4 mM, and the cells were incubated at 28°C for 2 h. The harvested cells were resuspended in lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, 1 mM PMSF; pH 8.0) and subjected to sonication on ice. The cell lysate was centrifuged at 20,000 × g for 15 min at 4°C. The soluble fraction was loaded onto an Ni2+-nitrilotriacetic acid (NTA) column (Novagen) and washed with washing buffer (50 mM NaH2PO4, 0.3 M NaCl, 50 mM imidazole; pH 8.0), and the bound proteins were eluted with elution buffer (50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole; pH 8.0). The fractions were analyzed by immunoblotting.
RESULTS
ImpLM is tightly associated with the inner membrane.
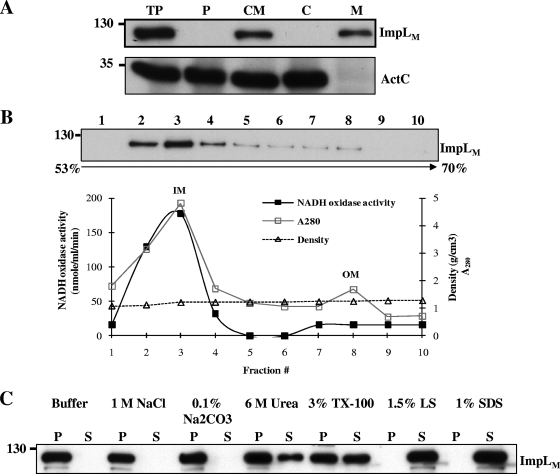
IcmF is a conserved component that is essential for T6SS-mediated Hcp secretion. However, there is little biochemical evidence that can be used to understand its function, except the prediction that it is an inner membrane protein containing a Walker A nucleotide-binding motif (8). A. tumefaciens produces an IcmF family protein, ImpLM, that is essential for Hcp secretion from A. tumefaciens (72). We first examined the subcellular localization and biochemical properties of ImpLM by performing fractionation and protein solubilization assays, followed by immunoblot analysis. ImpLM was detected exclusively in the membrane fraction of A. tumefaciens wild-type virulent strain C58. A citrate transporter, ActC (36), served as a soluble protein marker in the periplasmic and cytosolic fractions (Fig. 1A). Ultracentrifugation through sucrose density gradients was used to separate inner and outer membrane fractions and to determine the membrane localization of ImpLM. Most ImpLM was present in the inner membrane fractions (Fig. 1B), suggesting that ImpLM is localized to the inner membrane.
FIG. 1.
ImpLM is an integral inner membrane protein. (A) Equal volumes of total proteins (TP), periplasmic proteins (P), cytoplasmic and membrane proteins (CM), cytoplasmic proteins (C), and membrane proteins (M) of wild-type strain A. tumefaciens C58 grown in AB-MES (pH 5.5) for 6 h at 25°C were resolved by 12% glycine-SDS-polyacrylamide gel electrophoresis (PAGE). The proteins were analyzed by immunoblotting with antibodies against C-ImpLM and ActC, which served as a soluble protein marker. (B) Membrane fractions separated by sucrose density gradient centrifugation were collected from the top of the gradient and analyzed by immunoblotting with C-ImpLM antibody. The fractions containing the outer membranes (OM) and inner membranes (IM) were identified on the basis of the activity of the inner membrane marker NADH oxidase. The values for sucrose density and A280 for each of the fractions of the 53% to 70% sucrose gradient are indicated. (C) Total membranes were incubated with various chemical reagents and centrifuged to separate soluble (S) and pellet (P) (insoluble) fractions. The fractions were analyzed by immunoblotting with C-ImpLM antibody. The positions of the molecular mass markers used (in kDa) are indicated on the left in each panel. TX-100, Triton X-100; LS, N-lauroylsarcosine.
We attempted to solubilize ImpLM by incubating the membrane fractions of A. tumefaciens C58 with various chemicals. ImpLM was not solubilized by high salt (1 M NaCl) or by basic buffer (0.1% Na2CO3), but it was slightly solubilized by 6 M urea (Fig. 1C), suggesting that it is tightly associated with membranes. The partial solubilization of ImpLM by the nonionic detergent Triton X-100 at a concentration of 3% and the complete solubilization of ImpLM by the strong ionic detergents SDS and N-lauroylsarcosine (Fig. 1C) indicated that this protein is associated with membranes mainly via hydrophobic interactions. The biochemical fractionation and solubilization data coincided with the predicted features of ImpLM as an integral inner membrane protein; this prediction was further confirmed as follows.
ImpLM is a polytopic integral inner membrane protein with the N terminus facing the cytoplasm and the C terminus exposed to the periplasmic space.
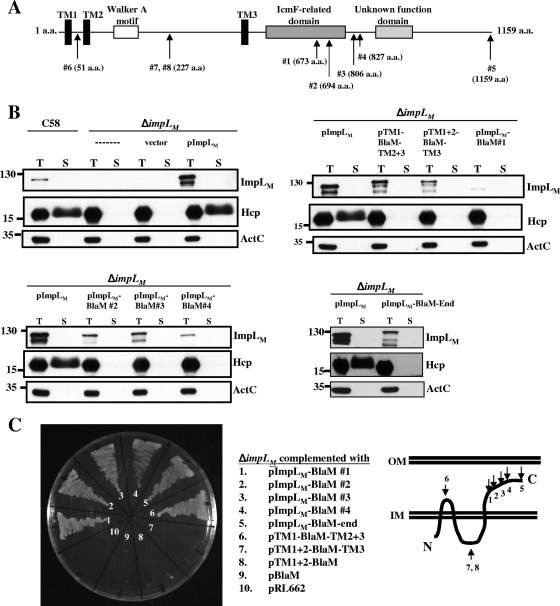
ImpLM is a 128-kDa protein which is predicted to be localized to the inner membrane with three TM domains and a Walker A motif located between the second and third TM domains (Fig. 2A). To investigate further the biochemical functions of ImpLM, it is important to validate its membrane topology. We previously demonstrated that trans expression of impLM from plasmid pImpLM (previously designated pIcmF) in a ΔimpLM mutant could restore Hcp secretion from the virulent strain A. tumefaciens NT1RE(pJK270) (72). In this study, the loss of Hcp secretion from the ΔimpLM in-frame deletion mutant created with A. tumefaciens C58 was restored in the presence of pImpLM (Fig. 2B). To determine the topology of ImpLM, we used transposon EZ-Tn5 randomly to insert a gene encoding β-lactamase (BlaM) into pImpLM to generate various EZ-Tn5 transposon-mutagenized plasmids which may produce ImpLM-BlaM fusion proteins. BlaM serves as a periplasmic reporter by conferring resistance to ampicillin or carbenicillin because it is active only after translocation to the periplasm (68). EZ-Tn5-mutagenized plasmids were first transformed into E. coli DH10B to select Apr colonies, following which resistance to carbenicillin was confirmed for the A. tumefaciens ΔimpLM mutant. We obtained seven independent Cbr clones, which resulted in four different insertion sites. Sequence analysis revealed that BlaM was inserted into the C-terminal domain of ImpLM in all four plasmids at positions 1 to 4 (Fig. 2A), which suggested that the C terminus of ImpLM is localized to the periplasm. (Fig. 2C).
FIG. 2.
ImpLM topology analysis using β-lactamase fusions and immunoblotting of fusion proteins. (A) ImpLM domain organization predicted with the SMART software (http://smart.embl-heidelberg.de/). ImpLM is predicted to be an inner membrane protein with three TM domains (TM1 to TM3) (black bars) containing a Walker A motif (open bar) located between TM2 and TM3. A conserved domain with an unknown function (light gray bar) and an IcmF-related domain (dark gray bar) are located at the C terminus of ImpLM. The positions of BlaM insertions or fusions are indicated by arrows labeled with amino acid (a.a.) positions. (B) Total (T) and secreted (S) proteins isolated from wild-type, ΔimpLM, and various ImpLM-complemented strains grown in AB-MES (pH 5.5) for 6 h at 25°C were resolved by 12% glycine-SDS-PAGE. BlaM fusion proteins were analyzed to determine their expression and function in Hcp secretion by immunoblot analysis with C-ImpLM, Hcp, and ActC antibodies. The secreted proteins were collected from 1 ml of culture medium after removal of bacterial cells and were concentrated by trichloroacetic acid precipitation and analyzed as described previously (72). The nonsecreted soluble protein ActC served as an internal control. (C) A. tumefaciens ΔimpLM strains containing either the vector (pRL662) or one of the complementing plasmids containing BlaM fusions were grown in 523 medium with 250 μg/ml carbenicillin at 28°C for 24 h to examine their resistance to carbenicillin. The membrane topology of ImpLM is based on the BlaM and GFP fusion results. The positions of molecular mass markers (in kDa) are indicated to the left of the blots. IM, inner membrane; OM, outer membrane.
If the ImpLM C-terminal domain is localized to the periplasm, one can propose that the N terminus is localized in the cytosol with three TM domains integrated in the inner membrane (Fig. 2C). To validate this topology, we first created the pImpLM-BlaM-end construct, in which BlaM lacking a signal peptide is fused to the C terminus of ImpLM. The ability of pImpLM-BlaM-end to confer Apr or Cbr when it was expressed in E. coli or A. tumefaciens indicated that the entire C-terminal domain was located in the periplasm. Next, we created constructs in which BlaM was fused internally in the region between the first and second TM domains (position 6 for pTM1-BlaM-TM2+3 [Fig. 2A and 2C]) and in the region between the second and third TM domains (position 7 for pTM1+2-BlaM-TM3 and position 8 for pTM1+2-BlaM [Fig. 2A and 2C]). pTM1-BlaM-TM2+3, but not pTM1+2-BlaM-TM3 or pTM1+2-BlaM, was able to confer Apr or Cbr when it was expressed in E. coli or A. tumefaciens, suggesting that the region between TM1 and TM2 is located in the periplasm (Fig. 2C). The inability of pTM1+2-BlaM-TM3 to confer Cbr did not result from instability of the fusion protein because we were able to detect expression of this protein at levels comparable to those of other proteins by immunoblotting (Fig. 2B). None of the BlaM fusion proteins were able to complement the loss of Hcp secretion in the ΔimpLM strain (Fig. 2B), suggesting that the sequences between TM1/TM2, TM2/TM3, and the C-terminal domain of ImpLM, as well as the size and distance between these domains, are important for retaining the function of ImpLM in mediation of Hcp secretion.
The ImpLM topology suggested by BlaM fusion protein analysis was further confirmed using GFP as a cytoplasmic reporter. GFP folds correctly and is fluorescent in the cytoplasm, but it does not fold into an active form when it is targeted to the periplasm by the Sec general secretory pathway (18, 52). GFP signals were detected in E. coli or A. tumefaciens when GFP was fused to the N terminus of ImpLM (pGFP-ImpLM) or fused after the second TM domain (pTM1+2-GFP; the same position as pTM1+2-BlaM, position 8 [Fig. 2A and 2C]) (data not shown). However, no GFP signal was detected when GFP was fused to the C terminus of ImpLM (pImpLM-GFP) or inserted into the region between the first and second TM domains (pTM1-GFP-TM2+3; the same position as pTM1-BlaM-TM2+3, position 6). Thus, ImpLM is a polytopic integral inner membrane protein with the N terminus facing the cytoplasmic side and the C terminus exposed to the periplasmic space.
The Walker A motif of ImpLM is required for Hcp secretion.
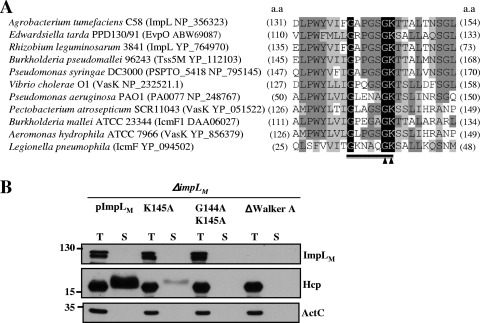
Amino acid sequence alignment of ImpLM and its orthologs in several T6SS as well as T4SSb revealed a highly conserved Walker A motif (Fig. 3A), suggesting that ImpLM and its orthologs may function as nucleotide-binding proteins and nucleoside triphosphatases. The Walker A motif of ImpLM is located between amino acid residues 139 and 146. Two conserved glycines are separated by 4 amino acids and followed by a conserved lysine and threonine/serine (GXXXXGKT/S). The conserved lysine typically binds the β and γ phosphoryl groups of ATP or GTP (71) and is required for full ATPase activity in many traffic ATPases (64).
FIG. 3.
The Walker A motif of ImpLM is required for Hcp secretion. (A) Alignment of the GXXXXGKT/S Walker A motifs conserved in selected T6SS and T4SSb IcmF family proteins, indicated by protein name and accession number, in several gram-negative bacteria, with the conserved amino acid residues highlighted. The arrowheads indicate the amino acids targeted for mutagenesis, and the underlining indicates the Walker A motif targeted for deletion. a.a, amino acid. (B) Total (T) and secreted (S) proteins isolated from ΔimpLM strains harboring plasmids expressing either the wild-type (pImpLM) or mutated ImpLM (pImpLMK145A, pImpLMG144A K145A, or pImpLMΔWalker A) protein. Cells grown in AB-MES (pH 5.5) for 6 h at 25°C were resolved by 12% glycine-SDS-PAGE and subjected to immunoblot analysis with C-ImpLM, Hcp, and ActC antibodies for expression and secretion analysis. The positions of molecular mass markers (in kDa) are indicated on the left.
To investigate the importance of the Walker A motif of ImpLM in Hcp secretion from A. tumefaciens, we generated mutants by site-directed mutagenesis using a single point mutation in the Walker A motif that replaced Lys with Ala (K145A), using double mutations (G144A K145A), or using deletion of the Walker A motif (ΔWalker A) to replace the wild-type impLM gene on pImpLM. The resulting mutant plasmids (pImpLMK145A, pImpLMG144A K145A, and pImpLMΔWalker A) were tested to determine their abilities to complement Hcp secretion from the Agrobacterium ΔimpLM mutant strain. Despite the fact that there was no significant difference in the level of intracellular Hcp among the mutants and the wild-type bacteria, the level of Hcp secreted into the culture medium was significantly lower for the K145A mutant than for wild-type cells (Fig. 3B). The double mutation or deletion of the Walker A motif eliminated secretion of Hcp (Fig. 3B). The reduction and lack of Hcp secretion from the K145A and G144A K145A mutants, respectively, did not result from instability of the mutant proteins because the ImpLM protein levels in the ΔimpLM strains complemented with pImpLMK145A and pImpLMG144A K145A were similar to that in the ΔimpLM strain complemented with pImpLM (Fig. 3B). In contrast, we were unable to detect any ImpLM upon complementation with pImpLMΔWalker A, suggesting that an intact Walker A motif is important for the stability of ImpLM proteins. Taken together, these data provide genetic evidence that the Walker A motif of an IcmF family protein is important in T6SS-mediated Hcp secretion and that ImpLM may bind and/or hydrolyze NTP necessary for T6SS machinery assembly and/or substrate secretion.
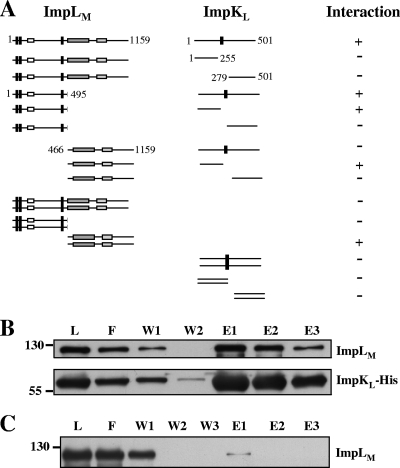
ImpLM interacts with another essential T6SS component, ImpKL.
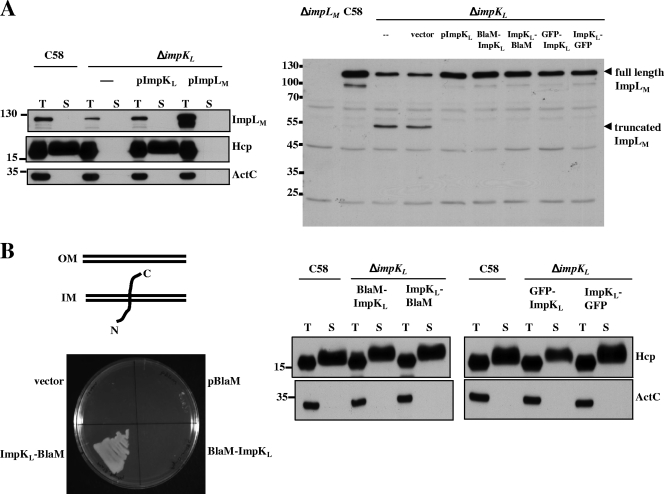
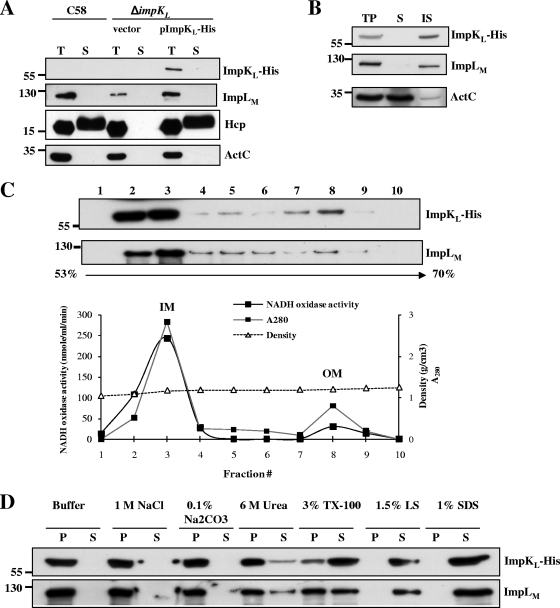
It is conceivable that ImpLM could interact with other T6SS components to assemble a functional secretory apparatus. We therefore investigated whether ImpLM interacts with another putative T6SS component, ImpKL, which is a predicted inner membrane protein containing a C-terminal OmpA-like domain. An interaction between these two proteins was suggested by a yeast two-hybrid analysis of E. tarda T6SS (74) and genetic evidence for L. pneumophila T4SSb (58). We first determined whether ImpKL is required for Hcp secretion from A. tumefaciens. Indeed, Hcp was expressed by but not secreted into culture medium from the ΔimpKL mutant (Fig. 4A). trans expression of impKL in the ΔimpKL mutant could restore the level of Hcp secretion to wild-type levels (Fig. 4A), indicating that ImpKL is a T6SS component essential for mediation of Hcp secretion. Interestingly, in the ΔimpKL mutant the level of full-length ImpLM was significantly reduced, along with accumulation of a ∼55-kDa truncated form. The truncated ImpLM contains at least part of the periplasmic C-terminal domain since this is the domain used to generate antibody for immunoblot analysis of ImpLM. The instability of ImpLM was corrected by trans expression of impKL in the ΔimpKL mutant (Fig. 4A), suggesting that that there is an interaction between ImpLM and ImpKL because the loss of one protein by a mutation affecting the stability of another protein is a common trait of proteins that interact (21, 25).
FIG. 4.
Immunoblotting and ImpKL topology analyses with β-lactamase fusions. (A) Total (T) and secreted (S) proteins isolated from wild-type C58, ΔimpKL, and ΔimpKL strains harboring either the vector or a plasmid expressing wild-type ImpKL, ImpLM, or various fusions were resolved by 12% glycine-SDS-PAGE and analyzed to determine Hcp secretion. Ten percent glycine-SDS-PAGE was employed to examine both full-length and truncated C-ImpLM (indicated by arrowheads) by immunoblotting with C-ImpLM antibody. (B) ΔimpKL mutant strains harboring the vector or plasmids expressing only BlaM, BlaM-ImpKL, or ImpKL-BlaM were streaked on 523 medium containing 200 μg/ml carbenicillin and incubated at 28°C for 24 h to examine their resistance to carbenicillin. The membrane topology of ImpKL is based on the BlaM and GFP fusion results. Total (T) and secreted (S) proteins isolated from wild-type strain C58 or ΔimpKL strains containing various ImpKL proteins fused with BlaM or GFP grown in AB-MES (pH 5.5) for 6 h at 25°C were resolved by 12% glycine-SDS-PAGE and analyzed to determine Hcp secretion by immunoblotting. The positions of molecular mass markers (in kDa) are indicated on the left. IM, inner membrane; OM, outer membrane.
ImpKL may function as an essential T6SS component or may play only an accessory role to stabilize ImpLM in mediating Hcp secretion. We therefore tested whether the deficiency in Hcp secretion from the ΔimpKL mutant could be restored by overexpressing ImpLM in the ΔimpKL mutant. As shown in Fig. 4A, no Hcp was detected in the culture medium when ImpLM was overexpressed in the ΔimpKL mutant, suggesting that ImpKL plays a role in T6SS-mediated Hcp secretion in addition to its function in stabilizing ImpLM.
To determine whether ImpLM interacts directly with ImpKL, we first performed a yeast two-hybrid assay using full-length ImpLM and ImpKL fused with the GAL4 AD and BD, respectively. Whereas no cell growth could be detected when each of the fusion proteins was expressed alone with the paired empty vector (see Table S1 in the supplemental material), we are able to detect an interaction between ImpLM and ImpKL (Fig. 5A). To investigate further the interaction between ImpLM and ImpKL, we engineered E. coli to coexpress ImpKL-His and ImpLM and used an Ni-NTA column to purify ImpKL-His and the interaction proteins. ImpLM coeluted with ImpKL-His from the column, and its level was comparable to that of purified ImpKL-His (Fig. 5B). In contrast, only a trace amount of ImpLM was eluted from the Ni column by imidazole when ImpLM was expressed alone (Fig. 5C), suggesting that ImpLM copurified with ImpKL-His due to their physical interaction. Together, these data strongly suggest that ImpLM interacts directly with ImpKL.
FIG. 5.
Interactions of ImpLM and ImpKL. (A) Yeast two-hybrid protein-protein interaction results obtained using combinations of various full-length and truncated ImpLM and ImpKL. The fragments of ImpLM and ImpKL used for each interaction in the assay are shown schematically. Yeasts expressing interacting proteins (+) were selected on the basis of their growth on SD medium in the absence of adenine, His, Leu, and Trp at 30°C for 3 days. Full-length ImpLM contained amino acid residues 1 to 1159, N-ImpLM contained amino acid residues 1 to 495, C-ImpLM contained amino acid residues 466 to 1159, full-length ImpKL contained amino acid residues 1 to 501, N-ImpKL contained amino acid residues 1 to 255, and C-ImpKL contained amino acid residues 279 to 501. E. coli BL21(DE3) containing both pImpLM and pImpKL-His (B) or only pImpLM (C) was induced by IPTG, and the soluble protein extracts were passed through Ni-NTA His-binding resins to purify ImpKL-His and its interacting proteins. The load (L), flowthrough (F), wash (W1 and W2), and elution (E1 to E3) fractions were analyzed by immunoblotting with antisera against His6 and C-ImpLM. The positions of molecular mass markers (in kDa) are indicated on the left.
ImpKL is a bitopic integral inner membrane protein with its N terminus localized in the cytoplasm and its C terminus exposed to the periplasmic space.
Our yeast two-hybrid and copurification data suggested that there is an interaction between ImpLM and ImpKL, which led us to characterize further the subcellular localization, topology, and biochemical properties of ImpKL. ImpKL, a 55-kDa protein (501 amino acids), contains one putative TM domain and a carboxyl-terminal domain homologous to the peptidoglycan-binding motif of OmpA from E. coli. We determined ImpKL topology by creating constructs to express a BlaM protein fused with the N or C terminus of ImpKL and examined the resistance to carbenicillin in A. tumefaciens. The ΔimpKL strain complemented with pImpKL-BlaM but not with pBlaM-ImpKL could grow on 523 medium containing carbenicillin (Fig. 4B), suggesting that the C terminus of ImpKL is exposed to the periplasm and its N terminus is localized in the cytoplasm. This suggested topology was further confirmed using GFP as a cytoplasmic reporter as described above. A GFP signal was detected in the ΔimpKL strain expressing GFP-ImpKL but not in the ΔimpKL strain containing ImpKL-GFP (data not shown). These data suggest that ImpKL is localized to the inner membrane, with its N terminus facing the cytoplasm and its C terminus exposed to the periplasmic space.
We next examined whether the ImpLM protein level in the ΔimpKL strain is increased by complementation with BlaM or GFP fusion proteins. Figure 4A shows that the ImpLM protein level was restored by complementation with a BlaM or GFP fusion. Furthermore, Hcp secretion was also restored for all complemented strains (Fig. 4B), suggesting that ImpKL fused with BlaM or GFP at either the N or C terminus retains its functionality for stabilizing ImpLM and mediating Hcp secretion.
To investigate further the subcellular localization and biochemical properties of ImpKL, we constructed pImpKL-His to express His-tagged ImpKL in the ΔimpKL strain. ImpKL-His was fully functional for complementing Hcp secretion from ΔimpKL. (Fig. 6A). Similar to ImpLM, ImpKL was localized in the membrane fraction (Fig. 6B). Further sucrose density gradient analysis revealed that most ImpKL cofractionated with ImpLM in the inner membrane fractions (Fig. 6C). Protein solubilization analysis indicated that both ImpKL-His and ImpLM are tightly associated with membranes, mainly via hydrophobic interactions, because they are significantly solubilized only by detergents (Fig. 6D). Thus, our data suggest that ImpKL is a bitopic integral inner membrane protein with the N terminus exposed to the cytoplasm and the C terminus localized in the periplasm.
FIG. 6.
ImpKL is an integral inner membrane protein. (A) Total (T) and secreted (S) proteins isolated from wild-type strain C58, the ΔimpKL strain, and the ΔimpKL strain complemented with pImpKL-His were resolved by 12% glycine-SDS-PAGE and analyzed to determine Hcp secretion by immunoblotting. (B) Equal volumes of total proteins (T), the soluble fraction (S), and the insoluble membrane fraction (IS) of the ΔimpKL strain containing pImpKL-His were analyzed by immunoblotting. (C) Membrane fractions isolated from the ΔimpKL strain containing pImpKL-His were separated by sucrose density gradient centrifugation and analyzed by immunoblotting. The fractions containing outer membranes (OM) and inner membranes (IM) were identified on the basis of the activity of the inner membrane marker NADH oxidase and the protein density in each of the fractions from a 53% to 70% sucrose gradient. (D) Total membranes of the ΔimpKL strain containing pImpKL-His were incubated with various chemical reagents and centrifuged to separate soluble (S) and pellet (P) (insoluble) fractions. The fractions were subsequently analyzed by immunoblotting using antibodies against His6, C-ImpLM, Hcp, and ActC. The positions of molecular mass markers (in kDa) are indicated on the left. TX-100, Triton X-100; LS, N-lauroylsarcosine.
DISCUSSION
In this study, we show that ImpLM and ImpKL interact with each other and are both integral inner membrane proteins essential for Hcp secretion from A. tumefaciens. We also provide genetic evidence that the Walker A motif of the IcmF family protein ImpLM is essential for its function in Hcp secretion, suggesting the importance of its NTP-binding and hydrolysis activities in T6SS machinery assembly and/or substrate secretion.
Our topology and biochemical fractionation analyses indicated that ImpLM and ImpKL are both tightly embedded in the inner membrane. Topology analysis with BlaM and GFP fusions suggested that ImpLM consists of three TM domains bordered by an N-terminal domain facing the cytoplasm and a C-terminal domain exposed to the periplasm and that ImpKL contains one TM domain with the N terminus facing the cytoplasm and the C terminus exposed to the periplasmic space (Fig. 2 and Fig. 4). The same topological results for the fusion proteins expressed in E. coli or A. tumefaciens suggested that the translocation of these proteins into the inner membrane does not require the assistance of other T6SS components but instead depends on a conserved secretion pathway, such as the Sec pathway (18, 52). Despite the fact that ImpLM and ImpKL are essentially integral inner membrane proteins, small amounts of them were detected in the outer membrane fraction, where low levels of the inner membrane protein marker NADH oxidase was detected (Fig. 1 and Fig. 6). It seems inevitable that trace amounts of inner membrane proteins are retained in outer membrane fractions because the use of similar techniques with many gram-negative bacteria, including A. tumefaciens, resulted in the same observations (17, 20, 61, 65). These results may be caused by the presence of several multiprotein complexes spanning both the inner and outer membranes to form protein channels for substrate translocation. However, we do not exclude the possibility that a small portion of ImpKL and/or ImpLM may associate with the outer membrane in a stable or dynamic manner. Because ImpKL possesses a C-terminal periplasmic OmpA-like domain, which contains a peptidoglycan-binding motif, it is possible that the ImpLM-ImpKL interaction complex or ImpKL alone may be linked to the outer membrane via its interactions with the cell wall.
We also obtained strong evidence, by using the yeast two-hybrid assay and copurification from E. coli, of the physical interaction between ImpLM and ImpKL (Fig. 5; see Table S1 in the supplemental material). This interaction was also supported by the fact that ImpKL is required for ImpLM protein stability. Full-length ImpLM protein levels were greatly reduced along with an increase in the amount of a C-terminal truncated form in the ΔimpKL mutant (Fig. 4A). Similar results were obtained for L. pneumophila T4SSb, in which IcmF is required to stabilize IcmH (or DotU), and the interaction of the proteins may be important in stabilization of the T4SS complex in L. pneumophila (58). The expression of ImpLM from a heterologous lacZ promoter in the absence of ImpKL (in either A. tumefaciens or E. coli) can overcome the requirement of ImpKL for ImpLM stability but cannot restore Hcp secretion from the ΔimpKL mutant of A. tumefaciens (Fig. 4A), suggesting that ImpKL plays an essential role in the T6SS in addition to stabilizing ImpLM. In addition, the data also suggested that ImpKL is not absolutely required for the stability of ImpLM but that interaction of ImpLM and ImpKL may be required to stabilize each protein in order to assemble a functional T6SS.
We also conducted a comprehensive yeast two-hybrid analysis to determine in which cellular compartment the interaction occurs and which protein domains of ImpLM and ImpKL are important for these two proteins to interact with each other. As shown in Fig. 5A, full-length ImpKL could interact with either full-length ImpLM or N-ImpLM (residues 1 to 495 without the C-terminal periplasmic domain). N-ImpLM also interacted with both full-length ImpKL and N-ImpKL (N-terminal cytoplasmic domain, residues 1 to 255) but not with C-ImpKL (C-terminal periplasmic domain, residues 279 to 501). These data suggest that ImpLM interacts with ImpKL via the cytoplasmic N-terminal domains. Interestingly, C-ImpLM (C-terminal periplasmic domain, residues 466 to 1159) also interacted with N-ImpKL but not with C-ImpKL. Interaction of these two domains was also observed in the yeast two-hybrid system by Zheng and Leung (74), who found that the N terminus of EvpN (an ortholog of ImpKL) interacted with the C terminus of EvpO (an ImpLM ortholog) from E. tarda (74). However, we did not find any interaction between C-ImpLM and full-length ImpKL using yeast two-hybrid assays (Fig. 5A) and E. coli coexpression experiments in which hemagglutinin-tagged ImpKL did not copurify with C-ImpLM-His (data not shown). Therefore, the interaction between N-ImpKL and C-ImpLM observed in yeast may not occur in A. tumefaciens where the intact proteins are expressed. Indeed, these two domains likely do not interact in A. tumefaciens because they are not localized in the same compartment (Fig. 2 and Fig. 4). The self-interaction of C-ImpLM detected by yeast two-hybrid assays (Fig. 5) also suggested that there is oligomerization of ImpLM, a biochemical property similar to the self-oligomerization or hexamer formation of several ATPases, such as ClpV (6), VirB11 (55, 73), and VirB4 (12), all of which are energizers of the T6SS and T4SS.
During the course of the yeast two-hybrid assays, we found that all interactions of pairs occur only in one direction (see Table S1 in the supplemental material). Because many factors can determine the outcomes of these assays (e.g., protein stability and structural constraints), we are not surprised that certain fusion proteins may be unstable or expressed in a form that does not permit interaction. Nevertheless, our protein-protein interaction studies, along with the topology data, strongly suggest that ImpLM may self-oligomerize via its C-terminal periplasmic domain and interact with ImpKL via the N-terminal domains of the proteins facing the cytoplasm. Further resolution of the crystal structure of the ImpLM-ImpKL interaction complex in combination with mutational analysis should unravel the interaction sites and the importance of the ImpLM-ImpKL interaction in the function of the T6SS.
We also obtained genetic evidence that the Walker A motif of the IcmF family protein is important in T6SS-mediated Hcp secretion (Fig. 3). The conserved Lys residue (K145) in the Walker A motif of ImpLM is important for T6SS-mediated Hcp secretion from A. tumefaciens because the ImpLMK145A mutant protein resulted in a significant reduction in Hcp secretion (Fig. 3B). The T2SS ATPase XpsE of Xanthomonas campestris with substitution of Met for Lys (K331M) (60) and SecA2 of Mycobacterium tuberculosis with substitution of Ala for Lys (K115A) (29) resulted in reductions in but not loss of ATP-binding and ATPase activities. Mutation of the R64 PilQ ATPase by substitution of Gln for Lys (K238Q) resulted in a significant decrease in the ATPase activity and function in R64 thin pilus biogenesis (54). In the case of R388 TrwD, a T4SS VirB11 family ATPase, substitution of Gln for Lys (K203Q) resulted in undetectable ATPase activity and loss of function in conjugation (53). Thus, the defects of ImpLMK145A for mediating Hcp secretion may be caused by a reduction in its NTP-binding and hydrolysis activity. The complete loss of Hcp secretion from the ImpLM G144A K145A double mutant suggests that G144 and K145 are essential for the function of ImpLM. In the future, it would be interesting to determine the correlation between the NTP-binding and hydrolysis activities of ImpLMK145A and ImpLMG144A K145A and their effects on mediation of Hcp secretion. Interestingly, an intact Walker A motif is important for ImpLM stability because of the absence of ImpLMΔWalker A when the protein is expressed in a ΔimpLM mutant.
Our results are consistent with the findings for L. pneumophila T4SSb, in which substitution of Ser for Gly38 (G38S) or substitution of Ala for Lys39 (K39A) in the Walker A motif of IcmF resulted in attenuated replication in macrophages (75). In contrast, in E. tarda, replacing one or two conserved amino acids (G118A, G123A, K124A, G118A G123A, or K124A S125A) in the Walker A motif of EvpO (IcmF) had no effect on T6SS substrate secretion (74), suggesting that this NTP-binding motif does not play an important role in the E. tarda T6SS. The difference between the importance of the IcmF Walker A motif in the T6SS of A. tumefaciens and the importance of the IcmF Walker A motif in the T6SS E. tarda is intriguing and may be explainable. First, the ClpV-like AAA+ ATPase, another conserved T6SS ATPase found in P. aeruginosa (41) and V. cholerae (6), may be sufficient to provide energy to mediate Hcp secretion from E. tarda but not Hcp secretion from A. tumefaciens. Alternatively, there may be another so-far-unidentified IcmF ortholog(s) encoded by the E. tarda genome which could not fully replace the function of the entire EvpO protein but may compensate for its NTP-binding activity. Nevertheless, our mutant studies provide strong genetic evidence that ImpLM may bind and/or hydrolyze NTP in mediating Hcp secretion. Exactly how ImpLM and another putative ATPase, ImpOH (ClpV-like AAA+ ATPase), provide the energy or conformation change that facilitates assembly of the T6SS machinery and/or substrate translocation across membranes remains to be elucidated. ClpV was recently demonstrated to have an unexpected role in protein secretion involving remodeling of VipA/VipB tubules in T6SS of V. cholerae (6). Because ClpV interacts with VipA and VipB, the T6SS components essential for Hcp and VgrG secretion, but not with the secreted proteins Hcp and VgrG (6), one could argue that ClpV may regulate the assembly of the T6SS. It would be of great interest to determine whether IcmF provides energy to direct the secretion of Hcp, VgrG, or another substrate(s) yet to be determined.
Despite the conservation of T6SS in pathogenic Proteobacteria and its involvement in the virulence of several human and animal pathogens (14, 19, 41, 48, 56, 63, 74), the importance of T6SS in infection by or the virulence of plant-associated bacteria is still obscure (5, 37, 39, 72). In A. tumefaciens, deletion of hcp resulted in reduced efficiency of tumorigenesis. However, no effects on virulence were observed in Hcp secretion-deficient mutants with icmF or the entire t6ss/imp operon deleted (72). It would be interesting to determine the role of T6SS in Agrobacterium infection by testing the mutants in different host plants and employing infection assays mimicking the natural environment. With such assays, various impLM mutants with defects in Hcp secretion and NTP-binding and hydrolysis activity may be further tested to determine their roles during the course of Agrobacterium-plant interactions.
In conclusion, ImpLM and ImpKL are two integral inner membrane proteins which interact with each other, likely via their N-terminal cytoplasmic domains. The nucleotide-binding Walker A motif located in the cytoplasmic domain of ImpLM is important for mediating Hcp secretion from A. tumefaciens. The inner membrane localization of the ImpLM-ImpKL complex may provide a scaffold for linking the T6SS cytoplasmic and outer membrane components through the peptidoglycan layer to build a TM protein channel for substrate translocation.
Supplementary Material
Acknowledgments
We thank Stan Gelvin, Yun-Long Tsai, Yin-Ru Chiang, and Hung-Yi Wu for critically reading the manuscript and Lai lab members for discussions and technical assistance. We also appreciate the technical support provided by the IPMB DNA Sequencing Laboratory and IPMB/ABRC Proteomics Core Laboratory of Academia Sinica for confirming the DNA sequences of PCR products and the identity of purified C-ImpLM, respectively.
This work was supported by a grant from Academia Sinica to E. M. Lai, by a Ph.D. fellowship from the Academia Sinica Taiwan International Graduate Program to L. S. Ma, and by an Academia Sinica postdoctoral fellowship to J. S. Lin.
Footnotes
Published ahead of print on 24 April 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Aschtgen, M. S., C. S. Bernard, S. De Bentzmann, R. Lloubes, and E. Cascales. 2008. SciN is an outer membrane lipoprotein required for type VI secretion in enteroaggregative Escherichia coli. J. Bacteriol. 1907523-7531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atmakuri, K., E. Cascales, and P. J. Christie. 2004. Energetic components VirD4, VirB11 and VirB4 mediate early DNA transfer reactions required for bacterial type IV secretion. Mol. Microbiol. 541199-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ballister, E. R., A. H. Lai, R. N. Zuckermann, Y. Cheng, and J. D. Mougous. 2008. In vitro self-assembly of tailorable nanotubes from a simple protein building block. Proc. Natl. Acad. Sci. USA 1053733-3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bingle, L., C. Bailey, and M. Pallen. 2008. Type VI secretion: a beginner's guide. Curr. Opin. Microbiol. 113-8. [DOI] [PubMed] [Google Scholar]
- 5.Bladergroen, M. R., K. Badelt, and H. P. Spaink. 2003. Infection-blocking genes of a symbiotic Rhizobium leguminosarum strain that are involved in temperature-dependent protein secretion. Mol. Plant-Microbe Interact. 1653-64. [DOI] [PubMed] [Google Scholar]
- 6.Bonemann, G., A. Pietrosiuk, A. Diemand, H. Zentgraf, and A. Mogk. 2009. Remodelling of VipA/VipB tubules by ClpV-mediated threading is crucial for type VI protein secretion. EMBO J. 28315-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bruser, T. 2007. The twin-arginine translocation system and its capability for protein secretion in biotechnological protein production. Appl. Microbiol. Biotechnol. 7635-45. [DOI] [PubMed] [Google Scholar]
- 8.Cascales, E. 2008. The type VI secretion toolkit. EMBO Rep. 9735-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christie, P. J., K. Atmakuri, V. Krishnamoorthy, S. Jakubowski, and E. Cascales. 2005. Biogenesis, architecture, and function of bacterial type IV secretion systems. Annu. Rev. Microbiol. 59451-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cianciotto, N. P. 2005. Type II secretion: a protein secretion system for all seasons. Trends Microbiol. 13581-588. [DOI] [PubMed] [Google Scholar]
- 11.Citovsky, V., S. V. Kozlovsky, B. Lacroix, A. Zaltsman, M. Dafny-Yelin, S. Vyas, A. Tovkach, and T. Tzfira. 2007. Biological systems of the host cell involved in Agrobacterium infection. Cell. Microbiol. 99-20. [DOI] [PubMed] [Google Scholar]
- 12.Dang, T. A., X. R. Zhou, B. Graf, and P. J. Christie. 1999. Dimerization of the Agrobacterium tumefaciens VirB4 ATPase and the effect of ATP-binding cassette mutations on the assembly and function of the T-DNA transporter. Mol. Microbiol. 321239-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Das, S., and K. Chaudhuri. 2003. Identification of a unique IAHP (IcmF associated homologous proteins) cluster in Vibrio cholerae and other proteobacteria through in silico analysis. In Silico Biol. 3287-300. [PubMed] [Google Scholar]
- 14.de Bruin, O., J. Ludu, and F. Nano. 2007. The Francisella pathogenicity island protein IgIA localizes to the bacterial cytoplasm and is needed for intracellular growth. BMC Microobiol. 71-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delepelaire, P. 2004. Type I secretion in gram-negative bacteria. Biochim. Biophys. Acta 1694149-161. [DOI] [PubMed] [Google Scholar]
- 16.de Maagd, R. A., and B. Lugtenberg. 1986. Fractionation of Rhizobium-leguminosarum cells into outer-membrane, cytoplasmic membrane, periplasmic, and cytoplasmic components. J. Bacteriol. 1671083-1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doerrler, W. T., H. S. Gibbons, and C. R. Raetz. 2004. MsbA-dependent translocation of lipids across the inner membrane of Escherichia coli. J. Biol. Chem. 27945102-45109. [DOI] [PubMed] [Google Scholar]
- 18.Driessen, A. J., and N. Nouwen. 2008. Protein translocation across the bacterial cytoplasmic membrane. Annu. Rev. Biochem. 77643-667. [DOI] [PubMed] [Google Scholar]
- 19.Dudley, E. G., N. R. Thomson, J. Parkhill, N. P. Morin, and J. P. Nataro. 2006. Proteomic and microarray characterization of the AggR regulon identifies a pheU pathogenicity island in enteroaggregative Escherichia coli. Mol. Microbiol. 611267-1282. [DOI] [PubMed] [Google Scholar]
- 20.Fernandez, D., T. A. Dang, G. M. Spudich, X. R. Zhou, B. R. Berger, and P. J. Christie. 1996. The Agrobacterium tumefaciens virB7 gene product, a proposed component of the T-complex transport apparatus, is a membrane-associated lipoprotein exposed at the periplasmic surface. J. Bacteriol. 1783156-3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fernandez, D., G. M. Spudich, X. R. Zhou, and P. J. Christie. 1996. The Agrobacterium tumefaciens VirB7 lipoprotein is required for stabilization of VirB proteins during assembly of the T-complex transport apparatus. J. Bacteriol. 1783168-3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Filloux, A., A. Hachani, and S. Bleves. 2008. The bacterial type VI secretion machine: yet another player for protein transport across membranes. Microbiology 1541570-1583. [DOI] [PubMed] [Google Scholar]
- 23.Galan, J. E., and H. Wolf-Watz. 2006. Protein delivery into eukaryotic cells by type III secretion machines. Nature 444567-573. [DOI] [PubMed] [Google Scholar]
- 24.Gelvin, S. B. 2003. Agrobacterium-mediated plant transformation: the biology behind the “gene-jockeying” tool. Microbiol. Mol. Biol. Rev. 6716-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hapfelmeier, S., N. Domke, P. C. Zambryski, and C. Baron. 2000. VirB6 is required for stabilization of VirB5 and VirB3 and formation of VirB7 homodimers in Agrobacterium tumefaciens. J. Bacteriol. 1824505-4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Henderson, I. R., F. Navarro-Garcia, M. Desvaux, R. C. Fernandez, and D. Ala'Aldeen. 2004. Type V protein secretion pathway: the autotransporter story. Microbiol. Mol. Biol. Rev. 68692-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 7751-59. [DOI] [PubMed] [Google Scholar]
- 28.Holland, I. B. 2004. Translocation of bacterial proteins—an overview. Biochim. Biophys. Acta 16945-16. [DOI] [PubMed] [Google Scholar]
- 29.Hou, J. M., N. G. D'Lima, N. W. Rigel, H. S. Gibbons, J. R. McCann, M. Braunstein, and C. M. Teschke. 2008. ATPase activity of Mycobacterium tuberculosis SecA1 and SecA2 proteins and its importance for SecA2 function in macrophages. J. Bacteriol. 1904880-4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Juhas, M., D. W. Crook, and D. W. Hood. 2008. Type IV secretion systems: tools of bacterial horizontal gene transfer and virulence. Cell. Microbiol. 102377-2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kado, C. I., and M. G. Heskett. 1970. Selective media for isolation of Agrobacterium, Carynebacterium, Erwinia, Pseudomonas, and Xanthomonas. Phytopathology 60969-976. [DOI] [PubMed] [Google Scholar]
- 32.Karunakaran, R., T. H. Mauchline, A. H. Hosie, and P. S. Poole. 2005. A family of promoter probe vectors incorporating autofluorescent and chromogenic reporter proteins for studying gene expression in Gram-negative bacteria. Microbiology 1513249-3256. [DOI] [PubMed] [Google Scholar]
- 33.Lai, E. M., and C. I. Kado. 1998. Processed VirB2 is the major subunit of the promiscuous pilus of Agrobacterium tumefaciens. J. Bacteriol. 1802711-2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee, P. A., D. Tullman-Ercek, and G. Georgiou. 2006. The bacterial twin-arginine translocation pathway. Annu. Rev. Microbiol. 60373-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leiman, P. G., M. Basler, U. A. Ramagopal, J. B. Bonanno, J. M. Sauder, S. Pukatzki, S. K. Burley, S. C. Almo, and J. J. Mekalanos. 2009. Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc. Natl. Acad. Sci. USA 1064154-4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu, A. C., H. W. Shih, T. Hsu, and E. M. Lai. 2008. A citrate-inducible gene, encoding a putative tricarboxylate transporter, is downregulated by the organic solvent DMSO in Agrobacterium tumefaciens. J. Appl. Microbiol. 1051372-1383. [DOI] [PubMed] [Google Scholar]
- 37.Liu, H., S. J. Coulthurst, L. Pritchard, P. E. Hedley, M. Ravensdale, S. Humphris, T. Burr, G. Takle, M. B. Brurberg, P. R. Birch, G. P. Salmond, and I. K. Toth. 2008. Quorum sensing coordinates brute force and stealth modes of infection in the plant pathogen Pectobacterium atrosepticum. PLoS Pathog. 4e1000093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mattinen, L., R. Nissinen, T. Riipi, N. Kalkkinen, and M. Pirhonen. 2007. Host-extract induced changes in the secretome of the plant pathogenic bacterium Pectobacterium atrosepticum. Proteomics 73527-3537. [DOI] [PubMed] [Google Scholar]
- 39.Mattinen, L., P. Somervuo, J. Nykyri, R. Nissinen, P. Kouvonen, G. Corthals, P. Auvinen, M. Aittamaa, J. P. Valkonen, and M. Pirhonen. 2008. Microarray profiling of host-extract-induced genes and characterization of the type VI secretion cluster in the potato pathogen Pectobacterium atrosepticum. Microbiology 1542387-2396. [DOI] [PubMed] [Google Scholar]
- 40.McCullen, C. A., and A. N. Binns. 2006. Agrobacterium tumefaciens and plant cell interactions and activities required for interkingdom macromolecular transfer. Annu. Rev. Cell Dev. Biol. 22101-127. [DOI] [PubMed] [Google Scholar]
- 41.Mougous, J. D., M. E. Cuff, S. Raunser, A. Shen, M. Zhou, C. A. Gifford, A. L. Goodman, G. Joachimiak, C. L. Ordonez, S. Lory, T. Walz, A. Joachimiak, and J. J. Mekalanos. 2006. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 3121526-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mougous, J. D., C. A. Gifford, T. L. Ramsdell, and J. J. Mekalanos. 2007. Threonine phosphorylation post-translationally regulates protein secretion in Pseudomonas aeruginosa. Nat. Cell Biol. 9797-803. [DOI] [PubMed] [Google Scholar]
- 43.Osborn, M. J., J. E. Gander, E. Parisi, and J. Carson. 1972. Mechanisms of assembly of the outer membrane of Salmonella typhimurium. Isolation and characterization of cytoplasmic and outer membrane. J. Biol. Chem. 2473962-3972. [PubMed] [Google Scholar]
- 44.Paul, K., M. Erhardt, T. Hirano, D. F. Blair, and K. T. Hughes. 2008. Energy source of flagellar type III secretion. Nature 451489-492. [DOI] [PubMed] [Google Scholar]
- 45.Pell, L. G., V. Kanelis, L. W. Donaldson, P. L. Howell, and A. R. Davidson. 2009. The phage lambda major tail protein structure reveals a common evolution for long-tailed phages and the type VI bacterial secretion system. Proc. Natl. Acad. Sci. USA 1064160-4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Preston, G. M., D. J. Studholme, and I. Caldelari. 2005. Profiling the secretomes of plant pathogenic Proteobacteria. FEMS Microbiol. Rev. 29331-360. [DOI] [PubMed] [Google Scholar]
- 47.Pukatzki, S., A. T. Ma, A. T. Revel, D. Sturtevant, and J. J. Mekalanos. 2007. Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proc. Natl. Acad. Sci. USA 10415508-15513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pukatzki, S., A. T. Ma, D. Sturtevant, B. Krastins, D. Sarracino, W. C. Nelson, J. F. Heidelberg, and J. J. Mekalanos. 2006. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc. Natl. Acad. Sci. USA 1031528-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pukatzki, S., S. B. McAuley, and S. T. Miyata. 2009. The type VI secretion system: translocation of effectors and effector-domains. Curr. Opin. Microbiol. 1211-17. [DOI] [PubMed] [Google Scholar]
- 50.Quandt, J., and M. F. Hynes. 1993. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene 12715-21. [DOI] [PubMed] [Google Scholar]
- 51.Rao, P. S., Y. Yamada, Y. P. Tan, and K. Y. Leung. 2004. Use of proteomics to identify novel virulence determinants that are required for Edwardsiella tarda pathogenesis. Mol. Microbiol. 53573-586. [DOI] [PubMed] [Google Scholar]
- 52.Rapoport, T. A. 2007. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature 450663-669. [DOI] [PubMed] [Google Scholar]
- 53.Rivas, S., S. Bolland, E. Cabezon, F. M. Goni, and F. de la Cruz. 1997. TrwD, a protein encoded by the IncW plasmid R388, displays an ATP hydrolase activity essential for bacterial conjugation. J. Biol. Chem. 27225583-25590. [DOI] [PubMed] [Google Scholar]
- 54.Sakai, D., T. Horiuchi, and T. Komano. 2001. ATPase activity and multimer formation of PilQ protein are required for thin pilus biogenesis in plasmid R64. J. Biol. Chem. 27617968-17975. [DOI] [PubMed] [Google Scholar]
- 55.Savvides, S. N., H. J. Yeo, M. R. Beck, F. Blaesing, R. Lurz, E. Lanka, R. Buhrdorf, W. Fischer, R. Haas, and G. Waksman. 2003. VirB11 ATPases are dynamic hexameric assemblies: new insights into bacterial type IV secretion. EMBO J. 221969-1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schell, M. A., R. L. Ulrich, W. J. Ribot, E. E. Brueggemann, H. B. Hines, D. Chen, L. Lipscomb, H. S. Kim, J. Mrazek, W. C. Nierman, and D. Deshazer. 2007. Type VI secretion is a major virulence determinant in Burkholderia mallei. Mol. Microbiol. 641466-1485. [DOI] [PubMed] [Google Scholar]
- 57.Schmidt-Eisenlohr, H., N. Domke, and C. Baron. 1999. TraC of IncN plasmid pKM101 associates with membranes and extracellular high-molecular-weight structures in Escherichia coli. J. Bacteriol. 1815563-5571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sexton, J. A., J. L. Miller, A. Yoneda, T. E. Kehl-Fie, and J. P. Vogel. 2004. Legionella pneumophila DotU and IcmF are required for stability of the Dot/Icm complex. Infect. Immun. 725983-5992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shalom, G., J. Shaw, and M. Thomas. 2007. In vivo expression technology identifies a type VI secretion system locus in Burkholderia pseudomallei that is induced upon invasion of macrophages. Microbiology 1532689-2699. [DOI] [PubMed] [Google Scholar]
- 60.Shiue, S. J., K. M. Kao, W. M. Leu, L. Y. Chen, N. L. Chan, and N. T. Hu. 2006. XpsE oligomerization triggered by ATP binding, not hydrolysis, leads to its association with XpsL. EMBO J. 251426-1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sperandeo, P., R. Cescutti, R. Villa, C. Di Benedetto, D. Candia, G. Deho, and A. Polissi. 2007. Characterization of lptA and lptB, two essential genes implicated in lipopolysaccharide transport to the outer membrane of Escherichia coli. J. Bacteriol. 189244-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Studier, F. W., A. H. Rosenberg, J. J. Dunn, and J. W. Dubendorff. 1990. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 18560-89. [DOI] [PubMed] [Google Scholar]
- 63.Suarez, G., J. C. Sierra, J. Sha, S. Wang, T. E. Erova, A. A. Fadl, S. M. Foltz, A. J. Horneman, and A. K. Chopra. 2008. Molecular characterization of a functional type VI secretion system from a clinical isolate of Aeromonas hydrophila. Microb. Pathog. 44344-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thomsen, N. D., and J. M. Berger. 2008. Structural frameworks for considering microbial protein- and nucleic acid-dependent motor ATPases. Mol. Microbiol. 691071-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Thorstenson, Y. R., G. A. Kuldau, and P. C. Zambryski. 1993. Subcellular localization of seven VirB proteins of Agrobacterium tumefaciens: implications for the formation of a T-DNA transport structure. J. Bacteriol. 1755233-5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tseng, T. T., B. M. Tyler, and J. C. Setubal. 2009. Protein secretion systems in bacterial-host associations, and their description in the Gene Ontology. BMC Microbiol. 9(Suppl. 1)S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tzfira, T., and V. Citovsky. 2006. Agrobacterium-mediated genetic transformation of plants: biology and biotechnology. Curr. Opin. Biotechnol. 17147-154. [DOI] [PubMed] [Google Scholar]
- 68.van Geest, M., and J. S. Lolkema. 2000. Membrane topology and insertion of membrane proteins: search for topogenic signals. Microbiol. Mol. Biol. Rev. 6413-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vergunst, A., B. Schrammeijer, A. den Dulk-Ras, C. de Vlaam, T. Regensburg-Tuink, and P. Hooykaas. 2000. VirB/D4-dependent protein translocation from Agrobacterium into plant cells. Science 290979-982. [DOI] [PubMed] [Google Scholar]
- 70.Vincent, C. D., J. R. Friedman, K. C. Jeong, E. C. Buford, J. L. Miller, and J. P. Vogel. 2006. Identification of the core transmembrane complex of the Legionella Dot/Icm type IV secretion system. Mol. Microbiol. 621278-1291. [DOI] [PubMed] [Google Scholar]
- 71.Walker, J. E., M. Saraste, M. J. Runswick, and N. J. Gay. 1982. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1945-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wu, H. Y., P. C. Chung, H. W. Shih, S. R. Wen, and E. M. Lai. 2008. Secretome analysis uncovers an Hcp-family protein secreted via a type VI secretion system in Agrobacterium tumefaciens. J. Bacteriol. 1902841-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yeo, H. J., S. N. Savvides, A. B. Herr, E. Lanka, and G. Waksman. 2000. Crystal structure of the hexameric traffic ATPase of the Helicobacter pylori type IV secretion system. Mol. Cell 61461-1472. [DOI] [PubMed] [Google Scholar]
- 74.Zheng, J., and K. Y. Leung. 2007. Dissection of a type VI secretion system in Edwardsiella tarda. Mol. Microbiol. 661192-1206. [DOI] [PubMed] [Google Scholar]
- 75.Zusman, T., M. Feldman, E. Halperin, and G. Segal. 2004. Characterization of the icmH and icmF genes required for Legionella pneumophila intracellular growth, genes that are present in many bacteria associated with eukaryotic cells. Infect. Immun. 723398-3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.