Abstract
In Escherichia coli the genome must be compacted ∼1,000-fold to be contained in a cellular structure termed the nucleoid. It is proposed that the structure of the nucleoid is determined by a balance of multiple compaction forces and one major expansion force. The latter is mediated by transertion, a coupling of transcription, translation, and translocation of nascent membrane proteins and/or exported proteins. In supporting this notion, it has been shown consistently that inhibition of transertion by the translation inhibitor chloramphenicol results in nucleoid condensation due to the compaction forces that remain active in the cell. Our previous study showed that during optimal growth, RNA polymerase is concentrated into transcription foci or “factories,” analogous to the eukaryotic nucleolus, indicating that transcription and RNA polymerase distribution affect the nucleoid structure. However, the interpretation of the role of transcription in the structure of the nucleoid is complicated by the fact that transcription is implicated in both compacting forces and the expansion force. In this work, we used a new approach to further examine the effect of transcription, specifically from rRNA operons, on the structure of the nucleoid, when the major expansion force was eliminated. Our results showed that transcription is necessary for the chloramphenicol-induced nucleoid compaction. Further, an active transcription from multiple rRNA operons in chromosome is critical for the compaction of nucleoid induced by inhibition of translation. All together, our data demonstrated that transcription of rRNA operons is a key mechanism affecting genome compaction and nucleoid structure.
An Escherichia coli cell is small, measuring approximately 2 to 4 μm in length and 1 μm in diameter. The bacterial genome is 4.6 million bp, which would be approximately 1.5 mm in length if stretched fully. In a rapidly growing cell, there are multiple genome equivalents. Thus, the genome must be compressed at least 1,000-fold to fit into the cell. The bacterial chromosome forms a cellular structure named the nucleoid (25, 42). Normally the E. coli nucleoid shows a characteristic “flexible doublet” shape (49) and is membrane associated (2, 46). Despite great advances being made in understanding the biochemistry and molecular biology of E. coli, the structure of the bacterial nucleoid remains poorly defined.
Woldringh et al. proposed that the structure of the nucleoid is determined by a balance of expansion and compaction forces (44). Suggested compaction forces include (i) DNA binding proteins (9, 17), (ii) DNA supercoiling (29, 35, 38), (iii) macromolecular crowding (20, 23, 51), and (iv) entropy-driven depletion attraction (18). One of the proposed forces that significantly contributes to expansion of the nucleoid is called transertion. During this process, coupled transcription and translation of membrane proteins and/or periplasmic exported proteins pull and anchor the transcribed bacterial nucleoid onto the cytoplasmic membrane (3, 43). In addition, RNA polymerase (RNAP) is thought to be a driving force in nucleoid segregation (12). This notion is further supported by the report of an interaction between RNAP and the actin-like MreB protein during chromosome segregation (15). According to the proposition (44), inhibition of both translation and transcription would lead to the disruption of transertion. It is expected that when transertion is blocked, the nucleoid will be condensed due to the remaining compaction forces in the cell. In support of this concept, it is consistently reported that chloramphenicol, a translation inhibitor, induces nucleoid compaction in the cell (41, 50). However, there are conflicting results regarding the effect of rifampin on nucleoid structure: both rifampin-induced nucleoid expansion (6, 13, 26, 36) and nucleoid compaction (3, 40, 52, 53) have been reported. Thus, the exact role of transcription in the structure of the nucleoid remains poorly understood and understudied (28).
Imaging of RNAP inside the cell containing the chromosomal rpoC-gfp fusion has provided a new tool to study the effects of transcription and RNAP (re)distribution on the structure of the nucleoid (6). Our recent studies suggest that transcription, in particular that from rRNA (rrn) operons, is linked to the dynamic structure of the nucleoid (14). In E. coli, there are seven rrn operons, four of which are located near the origin of replication in the chromosome. Despite the fact that collectively the rrn operons represent only about 1% of the genome, transcription of the genes in the rrn operons accounts for approximately 85% of the total transcription in an E. coli cell during optimal growth conditions (4). We previously showed that the distribution of RNAP is sensitive to environmental cues and that active rRNA synthesis is a driving force for the distribution of RNAP inside the cell (5, 6). For example, in rapidly growing cells cultured in rich medium, RNAP molecules are concentrated on synthesizing rRNA and form a transcription focus or “factory,” a structure analogous to the eukaryotic nucleolus. However, the role of transcription in determining the structure of the nucleoid is complicated by the fact that transcription could influence both expansion and compaction forces as detailed above. In the work described here, we used a new approach to further analyze the role of transcription, particularly of active rRNA synthesis, on the structure of the nucleoid. This was done under conditions where transertion, the major expansion force, was eliminated by inhibiting translation in the cell. Our results demonstrated that active rRNA synthesis is a key mechanism affecting nucleoid structure.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All of the strains used in the study are derivatives of MG1655. DJ2599 contains a chromosomal rpoC-gfp gene fusion linked closely to an Ampr marker (6). The DJ2650 and rpoB3443 strains are DJ2599 derivatives containing relAΔ251::kan and rpoB3443 mutations linked to btuB::Tn10, respectively (6). The DJ2759 (Δ6 rrn) strain was made by transduction of the rpoC-gfp allele from DJ2599 into the SQZ5 strain. The SQZ5 strain contains a single rRNA operon, rrnC, in the chromosome, because the other six rrn operons were deleted by a sequential allelic-exchange technique (10). SQZ5 also harbors plasmid ptRNA67 (p15A replicon) expressing tRNA genes that were present in the deleted rrn operons (a detailed description of the strain construction will be published elsewhere).
The bacterial techniques used are described elsewhere (19). All cultures were grown in Luria-Bertani (LB) medium with vigorous agitation in a water bath at 32°C. Fresh overnight cultures were diluted 1/250 into fresh medium. Samples used for microscopic observation were removed from cultures at an optical density at 600 nm (OD600) of approximately 0.35, and when indicated, rifampin (100 μg/ml), chloramphenicol (100 μg/ml), and freshly made serine hydroxamate (SHX; 1 mM) were added to the cultures at that time as time zero. These inhibitors stopped cell growth almost immediately. The antibiotics and chemicals were from Sigma.
Microscopy and relative nucleoid size (RNS) measurements.
The procedure of culture sampling for imaging was as described previously (6). Microscopy was performed with a Zeiss Axio Observer microscope equipped with a Plan-Apo 100× objective, epifluorescence filters, and a 1.6 optovar. Images were captured with an electron microscopy-charge-coupled device camera (Hamamatsu) working at 1 × 1 binning. The images were processed with Adobe Photoshop.
RNS within a cell was determined operationally as the ratio between the length of the nucleoid(s) and the length of the cell, assuming that the width of the nucleoid and that of the cell are similar with no significant differences as indicated by images of cells. Cell size and shape may be different among different strains (wild type and mutants); however, they were similar within the same strain examined. Because the images are two-dimensional and the cells are small, the values of RNS are merely approximations indicating the trend of compaction or expansion of the nucleoid under the conditions studied. For each condition, at least 30 cells were analyzed using NIH ImageJ software, and the data were compiled and analyzed using Microsoft Office Excel to obtain a mean RNS value with error bars under the condition used.
RESULTS AND DISCUSSION
Transcription is necessary for the compaction of the nucleoid induced by chloramphenicol.
To further study the role of transcription in nucleoid structure, we reexamined the effects of the two antibiotics, chloramphenicol and rifampin, on the nucleoid structure in the cells. Our results showed that while chloramphenicol condensed the nucleoid, rifampin expanded the nucleoid as reported elsewhere (6, 13, 26, 36, 41, 50). Further, the effect of rifampin is specific to RNAP and transcription because the antibiotic did not cause nucleoid expansion in the isogenic Rifr mutant cells (6; also data not shown). Thus, the conflicting reports that rifampin condenses the nucleoid are likely due to differences in experimental conditions, analyses, and/or strains used. The two antibiotics disrupt transertion by different modes of action. Chloramphenicol inhibits translation but does not prevent transcription (37). In contrast, rifampin inhibits transcription and thereby also prevents translation. We interpret the counteracting effects of the two antibiotics on the structure of nucleoids as an indication that transcription is important for nucleoid compaction in E. coli.
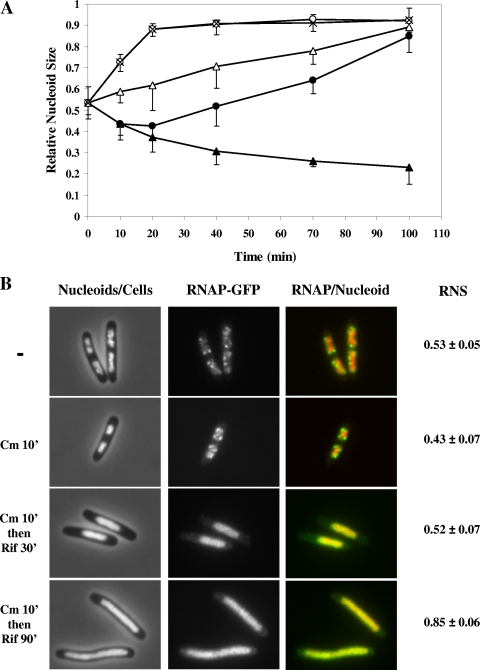
The nucleoid-compacting effect induced by chloramphenicol suggested to us a convenient way to determine the role of transcription in nucleoid compaction by sequential treatments of cells with chloramphenicol first and then with rifampin (Fig. 1). Because transertion will be disrupted by chloramphenicol first in the cell, the role of transcription in nucleoid compaction can be dissected with rifampin in the absence of the major expansion force for the nucleoid. To this end, we first analyzed the kinetics of nucleoid compaction and nucleoid expansion in wild-type DJ2599 cells, induced by chloramphenicol and rifampin, respectively, as no such analysis was previously performed. The changes in nucleoid structure were monitored by measuring the mean RNS values at several time points after the addition of an antibiotic. Prior to the antibiotic treatment, the mean RNS value was 0.53 in cells grown in LB medium. After the addition of chloramphenicol, the nucleoids in cells compacted gradually over time until they reached a mean RNS value of 0.23 at 100 min (Fig. 1A). In contrast, the nucleoids of cells treated with rifampin expanded relatively quickly and approached a plateau with a mean RNS value of about 0.90 at 20 min (Fig. 1A).
FIG. 1.
(A) Measurements of the RNS over time of the wild-type DJ2599 cells treated with chloramphenicol first, followed by rifampin. At time zero, the antibiotic(s) was added into cultures with an OD600 of ∼0.35. Open circles, rifampin; closed triangles, chloramphenicol; open triangles, chloramphenicol and rifampin simultaneously; crosses, rifampin for 10 min and then chloramphenicol; closed circles, chloramphenicol for 10 min and then rifampin. Note that the open circles overlap with crosses. The error bars in the figure indicate the deviations from the means. (B) Cell morphology, nucleoid size, and distribution of RNAP of the wild-type DJ2599 cells before and after sequential treatment with chloramphenicol (Cm) for 10 min and then rifampin (Rif) for 30 min and 90 min, respectively. In false-colored overlay images, RNAP is green and nucleoid is red.
To determine if the gradual compaction of nucleoids induced by chloramphenicol required transcription, the cells were first treated with chloramphenicol for 10 min, and then rifampin was added into the culture at that time. At different times thereafter, the mean RNS value in the cells was measured (Fig. 1A) and transcription foci were also examined (Fig. 1B). A modest compaction of nucleoids was observed, with a mean RNS value of 0.43, when chloramphenicol was added for 10 min compared to the cells prior to the addition of the antibiotics, and at this time the transcription foci were visible. However, shortly after the sequential addition of rifampin to the cultures, the chloramphenicol-induced compaction of the nucleoid ceased and the transcription foci disappeared. Subsequently, the nucleoid expanded and the mean RNS value gradually increased to 0.85 at 100 min, similar to the value obtained in the cells treated with rifampin only (Fig. 1B). Evidently, nucleoid compaction required transcription in the absence of the expansion force for the nucleoid.
Control experiments showed that a sequential treatment of cells with rifampin for 10 min followed by chloramphenicol led to the same expansion rate of the nucleoid as that in the cells treated with rifampin only (Fig. 1A). Further, after simultaneous treatment with rifampin and chloramphenicol, the nucleoids of the cells expanded incrementally over time to reach a final mean RNS value of 0.89 after 100 min (Fig. 1A). Together, these results suggested that transcription was required for chloramphenicol-induced nucleoid compaction.
Active synthesis of rRNA is required for the compaction of nucleoids induced by chloramphenicol.
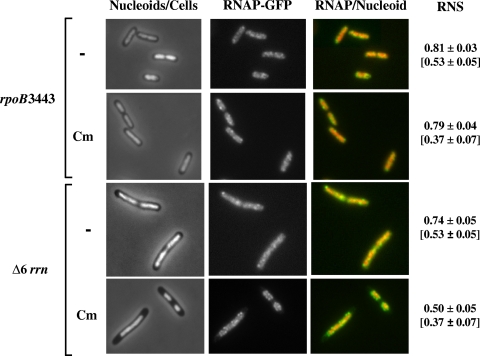
Rifampin inhibits total RNA synthesis in E. coli cells. To further study the effect of partial transcription inhibition, in particular the effect of reduced synthesis of rRNA operons on the compaction of nucleoids induced by chloramphenicol, we took advantage of two classes of E. coli mutants, both of which have reduced rRNA synthesis compared to the wild type. The first is a “stringent” RNAP mutant, the rpoB3443 strain, which harbors the L533P mutation in the β subunit and exhibits a “constitutive” stringent response phenotype even when cells are grown in a rich LB medium (47, 48). The other was the strain DJ2759 (Δ6 rrn), in which six out of the seven rrn operons were deleted from the chromosome. These two mutants had a lower growth rate than did wild-type DJ2599 in LB medium, likely due to their defects in rRNA synthesis. Because initiation in DNA replication occurs more frequently in a fast-growing cell than in a slow-growing cell (4), the DNA contents in these two mutant cells are expected to be lower than that in the wild type. While the rpoB3443 mutant cells were smaller than wild-type cells, the Δ6 rrn mutant cells were close in size to the wild type. The rpoC-gfp allele was introduced into the two mutants so that nucleoid compaction and RNAP distribution could be examined simultaneously. We studied the effects of these two mutations on nucleoid structure when cells were grown either in LB medium, where transertion is functional, or in LB medium plus chloramphenicol, where transertion is disrupted (Fig. 2).
FIG. 2.
Cell morphology, nucleoid size, and distribution of RNAP in the two E. coli mutants defective in rRNA synthesis: the constitutive “stringent” RNAP rpoB3443 mutant and the Δ6 rrn mutant (DJ2759). Cells were grown in LB medium to an OD600 of ∼0.35, and chloramphenicol (Cm) was added for 20 min when indicated. Mean values of the RNS of the two mutant cells in LB medium without (−) or with chloramphenicol (Cm) are indicated. For comparison, the respective RNS values of the wild type (DJ2599) are shown in brackets. In false-colored overlay images, RNAP is green and nucleoid is red.
The distribution of RNAP in the rpoB3443 cells grown in LB medium, in contrast to that in wild-type cells (Fig. 1B), was relatively homogeneous, with no distinctive transcription foci as previously reported (6). The nucleoids in these small “stringent” RNAP mutant cells, notwithstanding the smaller cells, approached full expansion, occupying a much larger mean RNS value of 0.81 than in the wild type (0.53). It is important to note that after transertion was disrupted by adding chloramphenicol into the culture for 20 min, the nucleoids still maintained a high RNS value in the rpoB3443 mutant cells (0.79) with minimal compaction in contrast to the wild type (0.37). Similarly, for the Δ6 rrn mutant that contains only one rRNA operon in the chromosome, the transcription foci were absent as the distribution of RNAP-GFP (green fluorescent protein) was relatively homogeneous within the nucleoids when cells were grown in LB medium. Also, the nucleoids in the Δ6 rrn mutant cells occupied a higher mean RNS value of 0.74 than in the wild type (0.53). Likewise, after treatment with chloramphenicol for 20 min, the nucleoids of the Δ6 rrn mutant cells were only partially compacted but maintained a high mean RNS value (0.50) compared to the wild type (0.37). The lack of complete nucleoid compaction upon treatment of chloramphenicol in both of the mutants is particularly significant considering that there would be less DNA content in the cells of the two slow-growing mutants than in the wild type as detailed above. Because both mutants defective in rRNA synthesis had more expanded nucleoids than did the wild type when transertion was either functional or inhibited by chloramphenicol, these results are consistent with the explanation that a wild-type level of rRNA synthesis from multiple rrn operons in the genome was required for the nucleoid compaction in the cell.
Active rRNA synthesis in a “relaxed” relA mutant is critical for the compaction of nucleoid induced by amino acid starvation that mimics the effect of chloramphenicol.
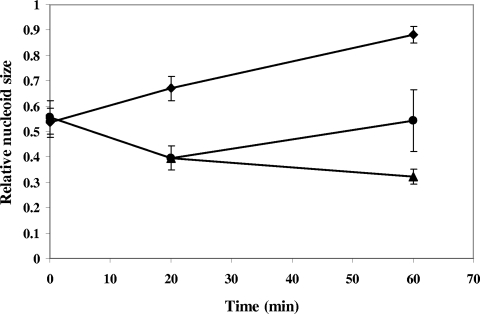
We previously showed that the nucleoid structures of wild-type cells and the relA mutant cells were different when cells were starved for an amino acid (6). SHX, an amino acid analogue, causes starvation for the amino acid serine, leading to inhibition of translation, similar to the effect of chloramphenicol (37). This compound thereby should interrupt transertion, thus eliminating the major nucleoid expansion force. Upon amino acid starvation, transcription of rRNA operons is blocked in wild-type cells due to the stringent response (7, 24). However, transcription of rRNA operons is fully active in the “relaxed” relA mutant cells (7, 11, 33). While the nucleoid of wild-type cells treated with SHX is expanded, the nucleoid of the relA mutant cells is condensed (6). Taking advantage of the phenotype of the relA mutant, we decided to determine the effect of transcription on the nucleoid compaction induced by amino acid starvation in the relA mutant by sequential treatments of cells with SHX first and then with rifampin (Fig. 3). We first analyzed the kinetics of nucleoid compaction in the relA mutant after the addition of SHX up to 60 min, as we previously examined only earlier points and did not measure RNS (6). The nucleoid was compacted gradually over time and reached a mean RNS value of 0.32 at 60 min (Fig. 3). In contrast, the nucleoid expanded to a mean RNS value of 0.88 in the wild-type cells (Fig. 3). Sequential addition of rifampin for 40 min to the relA mutant cells, which were pretreated with SHX for 20 min, resulted in expansion of the nucleoid to a mean RNS value of 0.54 (Fig. 3). Clearly, active rRNA synthesis is required for the nucleoid compaction induced by amino acid starvation.
FIG. 3.
Measurements of the RNS over time of the “relaxed” relA mutant after addition of SHX and then treatment with rifampin. Results are for the “relaxed” relAΔ251::kan mutant (DJ2650) cells after addition of SHX only (triangles) or after treatment with SHX for 20 min followed by the addition of rifampin (circles). The wild-type DJ2599 cells after addition of SHX only (diamonds) were used as a control. The error bars in the figure indicate the deviations from the means.
The synthesis of rRNA from multiple rrn operons plays a central role in E. coli biology for quick response to the influx of nutrients and fast growth in rich medium (8, 22, 24, 34). It is conceivable that E. coli has evolved in such a way that the dynamic structure of the nucleoid is coupled to rRNA synthesis and RNAP (re)distribution as proposed previously (6, 14). Their roles in nucleoid compaction could be direct, indirect, or a combination of the two. Transcription could affect the multiple compaction forces described in the introduction. For example, transcription and supercoiling of the DNA are linked (16). Transcription and/or transcription-induced supercoiling could also interfere with the binding of nucleoid-associated proteins to DNA (1, 17), which in turn could affect DNA supercoiling. The supercoiling introduced by transcription, however, is unlikely to account for the effect of transcription on the structure of the nucleoid described in this study, because the nucleoid of the cell remains compact when treated with either coumermycin or nalidixate, an inhibitor of gyrase (35, 36, 52), in contrast to fully expanded nucleoids induced by rifampin or the stringent response. The E. coli chromosome consists of ∼50 large distinctive domains of supercoiling, each of which is topologically independent of the others (31, 45). These loop domains are further organized into larger macrodomains (21, 39). It is conceivable that active synthesis of multiple rRNA operons in the chromosome would play a direct role in nucleoid compaction by (i) promoting interactions among macrodomains by RNAP-RNAP aggregation (30); (ii) promoting formation of antitermination elongation complexes (27, 32); (iii) coupling the synthesis, processing, and maturation of rRNA to ribosomal assembly within these transcription factories; and (iv) affecting RNAP distribution in the cell (6, 14). Future studies are warranted to address these issues.
In summary, by eliminating the major nucleoid expansion force mediated by transertion in the cell, the effect of transcription on nucleoid compaction is easily interpreted. Results from this new approach, coupled with an advanced imaging system for RNAP-GFP, clarified the role of transcription in the process and demonstrated that transcription, in particular active synthesis of multiple rRNA operons in the chromosome, plays a pivotal role in the compaction of the nucleoid in the cell.
Acknowledgments
We are grateful to Tim Durfee and Yan Ning Zhou for enthusiastic discussions during the course of study. We thank Don Court, Mikhail Kashlev, and Julie Torruellas Garcia for helpful comments on the manuscript.
This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research and National Institutes of Health grant GM24751 to C.L.S.
Footnotes
Published ahead of print on 24 April 2009.
REFERENCES
- 1.Ali Azam, T., A. Iwata, A. Nishimura, S. Ueda, and A. Ishihama. 1999. Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J. Bacteriol. 1816361-6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Binenbaum, Z., E. Klyman, and I. Fishov. 1999. Division-associated changes in membrane viscosity of Escherichia coli. Biochimie 81921-929. [DOI] [PubMed] [Google Scholar]
- 3.Binenbaum, Z., A. H. Parola, A. Zaritsky, and I. Fishov. 1999. Transcription- and translation-dependent changes in membrane dynamics in bacteria: testing the transertion model for domain formation. Mol. Microbiol. 321173-1182. [DOI] [PubMed] [Google Scholar]
- 4.Bremer, H., and P. P. Dennis. 1996. Modulation of chemical composition and other parameters of the cell by growth rate, p. 1553-1569. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 2. ASM Press, Washington, DC. [Google Scholar]
- 5.Cabrera, J. E., and D. J. Jin. 2006. Active transcription of rRNA operons is a driving force for the distribution of RNA polymerase in bacteria: effect of extrachromosomal copies of rrnB on the in vivo localization of RNA polymerase. J. Bacteriol. 1884007-4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cabrera, J. E., and D. J. Jin. 2003. The distribution of RNA polymerase in Escherichia coli is dynamic and sensitive to environmental cues. Mol. Microbiol. 501493-1505. [DOI] [PubMed] [Google Scholar]
- 7.Cashel, M., D. R. Gentry, V. J. Hernandez, and D. Vinella. 1996. The stringent response, p. 1458-1496. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, DC. [Google Scholar]
- 8.Condon, C., C. Squires, and C. L. Squires. 1995. Control of rRNA transcription in Escherichia coli. Microbiol. Rev. 59623-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dame, R. T. 2005. The role of nucleoid-associated proteins in the organization and compaction of bacterial chromatin. Mol. Microbiol. 56858-870. [DOI] [PubMed] [Google Scholar]
- 10.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 976640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Durfee, T., A. M. Hansen, H. Zhi, F. R. Blattner, and D. J. Jin. 2008. Transcription profiling of the stringent response in Escherichia coli. J. Bacteriol. 1901084-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dworkin, J., and R. Losick. 2002. Does RNA polymerase help drive chromosome segregation in bacteria? Proc. Natl. Acad. Sci. USA 9914089-14094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dworsky, P., and M. Schaechter. 1973. Effect of rifampin on the structure and membrane attachment of the nucleoid of Escherichia coli. J. Bacteriol. 1161364-1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jin, D. J., and J. E. Cabrera. 2006. Coupling the distribution of RNA polymerase to global gene regulation and the dynamic structure of the bacterial nucleoid in Escherichia coli. J. Struct. Biol. 156284-291. [DOI] [PubMed] [Google Scholar]
- 15.Kruse, T., B. Blagoev, A. Lobner-Olesen, M. Wachi, K. Sasaki, N. Iwai, M. Mann, and K. Gerdes. 2006. Actin homolog MreB and RNA polymerase interact and are both required for chromosome segregation in Escherichia coli. Genes Dev. 20113-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu, L. F., and J. C. Wang. 1987. Supercoiling of the DNA template during transcription. Proc. Natl. Acad. Sci. USA 847024-7027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luijsterburg, M. S., M. C. Noom, G. J. Wuite, and R. T. Dame. 2006. The architectural role of nucleoid-associated proteins in the organization of bacterial chromatin: a molecular perspective. J. Struct. Biol. 156262-272. [DOI] [PubMed] [Google Scholar]
- 18.Marenduzzo, D., C. Micheletti, and P. R. Cook. 2006. Entropy-driven genome organization. Biophys. J. 903712-3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 20.Murphy, L. D., and S. B. Zimmerman. 2001. A limited loss of DNA compaction accompanying the release of cytoplasm from cells of Escherichia coli. J. Struct. Biol. 13375-86. [DOI] [PubMed] [Google Scholar]
- 21.Niki, H., Y. Yamaichi, and S. Hiraga. 2000. Dynamic organization of chromosomal DNA in Escherichia coli. Genes Dev. 14212-223. [PMC free article] [PubMed] [Google Scholar]
- 22.Nomura, M., R. Gourse, and G. Baughman. 1984. Regulation of the synthesis of ribosomes and ribosomal components. Annu. Rev. Biochem. 5375-117. [DOI] [PubMed] [Google Scholar]
- 23.Odijk, T. 1998. Osmotic compaction of supercoiled DNA into a bacterial nucleoid. Biophys. Chem. 7323-29. [DOI] [PubMed] [Google Scholar]
- 24.Paul, B. J., W. Ross, T. Gaal, and R. L. Gourse. 2004. rRNA transcription in Escherichia coli. Annu. Rev. Genet. 38749-770. [DOI] [PubMed] [Google Scholar]
- 25.Pettijohn, D. E. 1996. The nucleoid, p. 158-166. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, DC.
- 26.Pettijohn, D. E., and R. Hecht. 1974. RNA molecules bound to the folded bacterial genome stabilize DNA folds and segregate domains of supercoiling. Cold Spring Harbor Symp. Quant. Biol. 3831-41. [DOI] [PubMed] [Google Scholar]
- 27.Quan, S., N. Zhang, S. French, and C. L. Squires. 2005. Transcriptional polarity in rRNA operons of Escherichia coli nusA and nusB mutant strains. J. Bacteriol. 1871632-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saier, M. H., Jr. 2008. The bacterial chromosome. Crit. Rev. Biochem. Mol. Biol. 4389-134. [DOI] [PubMed] [Google Scholar]
- 29.Sawitzke, J. A., and S. Austin. 2000. Suppression of chromosome segregation defects of Escherichia coli muk mutants by mutations in topoisomerase I. Proc. Natl. Acad. Sci. USA 971671-1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shaner, S. L., D. M. Piatt, C. G. Wensley, H. Yu, R. R. Burgess, and M. T. Record, Jr. 1982. Aggregation equilibria of Escherichia coli RNA polymerase: evidence for anion-linked conformational transitions in the protomers of core and holoenzyme. Biochemistry 215539-5551. [DOI] [PubMed] [Google Scholar]
- 31.Sinden, R. R., and D. E. Pettijohn. 1981. Chromosomes in living Escherichia coli cells are segregated into domains of supercoiling. Proc. Natl. Acad. Sci. USA 78224-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Squires, C. L., and D. Zaporojets. 2000. Proteins shared by the transcription and translation machines. Annu. Rev. Microbiol. 54775-798. [DOI] [PubMed] [Google Scholar]
- 33.Stent, G. S., and S. Brenner. 1961. A genetic locus for the regulation of ribonucleic acid synthesis. Proc. Natl. Acad. Sci. USA 472005-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stevenson, B. S., and T. M. Schmidt. 2004. Life history implications of rRNA gene copy number in Escherichia coli. Appl. Environ. Microbiol. 706670-6677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stuger, R., C. L. Woldringh, C. C. van der Weijden, N. O. Vischer, B. M. Bakker, R. J. van Spanning, J. L. Snoep, and H. V. Westerhoff. 2002. DNA supercoiling by gyrase is linked to nucleoid compaction. Mol. Biol. Rep. 2979-82. [DOI] [PubMed] [Google Scholar]
- 36.Sun, Q., and W. Margolin. 2004. Effects of perturbing nucleoid structure on nucleoid occlusion-mediated toporegulation of FtsZ ring assembly. J. Bacteriol. 1863951-3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tosa, T., and L. I. Pizer. 1971. Biochemical bases for the antimetabolite action of l-serine hydroxamate. J. Bacteriol. 106972-982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Travers, A., and G. Muskhelishvili. 2005. Bacterial chromatin. Curr. Opin. Genet. Dev. 15507-514. [DOI] [PubMed] [Google Scholar]
- 39.Valens, M., S. Penaud, M. Rossignol, F. Cornet, and F. Boccard. 2004. Macrodomain organization of the Escherichia coli chromosome. EMBO J. 234330-4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van Helvoort, J. M., P. G. Huls, N. O. Vischer, and C. L. Woldringh. 1998. Fused nucleoids resegregate faster than cell elongation in Escherichia coli pbpB(Ts) filaments after release from chloramphenicol inhibition. Microbiology 1441309-1317. [DOI] [PubMed] [Google Scholar]
- 41.van Helvoort, J. M., J. Kool, and C. L. Woldringh. 1996. Chloramphenicol causes fusion of separated nucleoids in Escherichia coli K-12 cells and filaments. J. Bacteriol. 1784289-4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Woldringh, C., and N. Nanninga. 1985. Structure of nucleoid and cytoplasm in the intact cell, p. 161-197. In N. E. Nanninga (ed.), Molecular cytology of Escherichia coli. Academic Press, London, United Kingdom.
- 43.Woldringh, C. L. 2002. The role of co-transcriptional translation and protein translocation (transertion) in bacterial chromosome segregation. Mol. Microbiol. 4517-29. [DOI] [PubMed] [Google Scholar]
- 44.Woldringh, C. L., P. R. Jensen, and H. V. Westerhoff. 1995. Structure and partitioning of bacterial DNA: determined by a balance of compaction and expansion forces? FEMS Microbiol. Lett. 131235-242. [DOI] [PubMed] [Google Scholar]
- 45.Worcel, A., and E. Burgi. 1972. On the structure of the folded chromosome of Escherichia coli. J. Mol. Biol. 71127-147. [DOI] [PubMed] [Google Scholar]
- 46.Worcel, A., and E. Burgi. 1974. Properties of a membrane-attached form of the folded chromosome of Escherichia coli. J. Mol. Biol. 8291-105. [DOI] [PubMed] [Google Scholar]
- 47.Zhou, Y. N., and D. J. Jin. 1997. RNA polymerase β mutations have reduced σ70 synthesis leading to a hyper-temperature-sensitive phenotype of a σ70 mutant. J. Bacteriol. 1794292-4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou, Y. N., and D. J. Jin. 1998. The rpoB mutants destabilizing initiation complexes at stringently controlled promoters behave like “stringent” RNA polymerases in Escherichia coli. Proc. Natl. Acad. Sci. USA 952908-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zimmerman, S. B. 2006. Shape and compaction of Escherichia coli nucleoids. J. Struct. Biol. 156255-261. [DOI] [PubMed] [Google Scholar]
- 50.Zimmerman, S. B. 2002. Toroidal nucleoids in Escherichia coli exposed to chloramphenicol. J. Struct. Biol. 138199-206. [DOI] [PubMed] [Google Scholar]
- 51.Zimmerman, S. B., and L. D. Murphy. 1996. Macromolecular crowding and the mandatory condensation of DNA in bacteria. FEBS Lett. 390245-248. [DOI] [PubMed] [Google Scholar]
- 52.Zimmerman, S. B., and L. D. Murphy. 2001. Release of compact nucleoids with characteristic shapes from Escherichia coli. J. Bacteriol. 1835041-5049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zusman, D. R., A. Carbonell, and J. Y. Haga. 1973. Nucleoid condensation and cell division in Escherichia coli MX74T2 ts52 after inhibition of protein synthesis. J. Bacteriol. 1151167-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]