Abstract
The slr1192 (adhA) gene from Synechocystis sp. strain PCC 6803 encodes a member of the medium-chain alcohol dehydrogenase/reductase family. The gene product AdhA exhibits NADP-dependent alcohol dehydrogenase activity, acting on a broad variety of aromatic and aliphatic primary alcohols and aldehydes but not on secondary alcohols or ketones. It exhibits superior catalytic efficiency for aldehyde reduction compared to that for alcohol oxidation. The enzyme is a cytosolic protein present in photoautotrophically grown Synechocystis cells. The expression of AdhA is enhanced upon the exposure of cells to different environmental stresses, although it is not essential for survival even under such stress conditions. The induction of the expression of the adhA gene is dependent on the Hik34-Rre1 two-component system, as it is severely impaired in mutant strains lacking either the histidine kinase Hik34 or the response regulator Rre1. In vitro DNA-protein interaction analysis reveals that the response regulator Rre1 binds specifically to the promoter region of the adhA gene.
Medium-chain dehydrogenases/reductases (MDR) constitute a superfamily of alcohol dehydrogenases that catalyze the reversible NAD(P)-dependent oxidation of alcohols to aldehydes or ketones. It includes a large number of structurally related proteins, which catalyze several types of enzymatic activity (23, 41, 44). Screening of complete genome sequences has revealed that this family is widespread, complex, and of ancient origin (22, 44). MDR alcohol dehydrogenases are found in mammals, plants, fungi, and bacteria (52). The alcohol dehydrogenases fulfill an astonishing variety of functions in cell metabolism (21), also being a key enzyme in ethanol generation by Saccharomyces cerevisiae (6) and bacteria (10). Furthermore, the generation of biofuels by photoautotrophic microorganisms is of great biotechnological interest (43). Complementation of a cyanobacterium's enzyme machinery with a specific exogenous gene(s) can result in the ability to generate bioethanol from photosynthetically fixed CO2 (11). Notwithstanding, current knowledge of cyanobacterial alcohol dehydrogenases is rather limited. In the cyanobacterium Synechocystis sp. strain PCC 6803 (referred here as Synechocystis), the slr1192 gene encodes a putative MDR alcohol dehydrogenase. According to in silico analyses (38, 44), the slr1192 protein has similarity with two subfamilies of MDRs: the yeast ADH family (Y-ADH) and the cinnamyl ADH family (CADH). Y-ADH-related enzymes have catabolic functions and are involved mainly in the metabolism of ethanol or short-chain alcohols for which they exhibit broad substrate specificity. CADH and related enzymes, on the other hand, perform anabolic functions and participate in biosynthetic pathways in plants and bacteria (5, 25, 44).
In Synechocystis, the expression of slr1192 is induced by osmotic (35) or salt (48) stress. In higher plants, alcohol dehydrogenase activity appears to be involved in aerobic metabolism under certain stress conditions (26, 56) such as low temperature, water deficit, or ozone exposure, but its function remains unknown. A temperature decrease seems to induce the accumulation of alcohol dehydrogenase mRNA in Arabidopsis thaliana (20), corn, and rice (9).
In general, cyanobacteria perceive and respond to environmental changes by means of two-component regulatory systems, a ubiquitous signal transduction pathway that represents a prevalent signaling mechanism in bacteria (8, 61). Two-component systems consist of a histidine kinase (Hik) and a response regulator (Rre) and generally induce or repress the expression of specific genes in response to environmental stimuli. The histidine kinase autophosphorylates a conserved histidine residue in response to the environmental signal and then transfers the phosphate group to a conserved aspartate residue of the response regulator, which mediates the transfer of the signal. In Synechocystis, different Hik-Rre systems have been identified as being regulators of the response to different environmental stresses (37). A membrane-bound histidine kinase, Hik33, is involved in the perception of cold, salt, and osmotic stress (33, 35, 39, 48, 54). A cytosolic histidine kinase, Hik34, has been shown to be involved in the perception of salt and hyperosmotic stress (33, 39, 48) as well as heat shock (53). Specifically, the couples Hik33-Rre31, Hik10-Rre13, Hik16 Hik41-Rre17, Hik34-Rre1, and a putative Hik2-Rre1 have been identified as being elements involved in the perception and transduction of signals promoted by hyperosmotic and salt stress (39, 48).
In the present work, a biochemical characterization of the slr1192 protein (designated AdhA here) from Synechocystis has been performed, revealing that the tetrameric 140-kDa enzyme is active toward linear and aromatic primary alcohols and that it preferentially reduces aldehydes rather than oxidizing alcohols. In addition, an extensive analysis of the expression of the adhA gene has verified its induction in response to heat shock, hyperosmotic stress, salt stress, and the addition of benzyl alcohol (BA). The Hik34-Rre1 two-component system has been shown to play a relevant role in the regulation of the expression of the adhA gene under these stress conditions. A specific interaction of Rre1 with the promoter region of the adhA gene has also been demonstrated. In light of this finding and additional information presented here, the physiological role of AdhA in Synechocystis is discussed.
MATERIALS AND METHODS
Strains and culture conditions.
Synechocystis sp. strain PCC 6803 cells were grown photoautotrophically on BG-11c medium (42) at 30°C under conditions of continuous illumination (50 μE m−2 s−1) and bubbled with 1% CO2 in air. Heterotrophic growth was performed as described previously (4). The Δhik34 and Δrre1 mutant strains were kindly provided by Iwane Suzuki (Graduate School of Life and Environmental Sciences, University of Tsukuba, Japan) (55). For precultures of the ΔadhA or Δrre1 strain, kanamycin was added to a final concentration of 100 μg ml−1, and for precultures of the Δhik34 strain, spectinomycin was added to a final concentration of 5 μg ml−1. Experiments were performed using cultures from the mid-logarithmic phase (optical density at 730 nm of 0.6 to 0.7 and 3 to 5 μg chlorophyll ml−1). For high-light treatment, cell suspensions were diluted with fresh medium to an optical density at 730 nm of 0.35, and the cultures were placed in a temperature-controlled chamber at 30°C and exposed to an irradiance of 500 μE m−2 s−1. Under heat shock conditions, cultures were incubated in a water bath at 45°C. For salt or hyperosmotic shock, NaCl or sorbitol, respectively, was added to the cell suspension to a final concentration of 0.5 M. BA was added to a final concentration of 30 mM. For oxidative stress, H2O2 was added to a final concentration of 1 mM. Glucose and 3-(3P,4P-dichlorophenyl)-1,1P-dimethylurea were added to final concentrations of 5 mM and 10 μM, respectively.
Escherichia coli DH5α and E. coli BL21(DE3) cells were grown in LB broth medium as described previously (46) and supplemented with 100 μg ml−1 of ampicillin or 50 μg ml−1 of kanamycin when required. M9 minimal medium (46) was used for the metal dependence analysis of His-AdhA expression.
Construction of the adhA disruption mutant.
To inactivate the adhA gene, the entire coding region was amplified by PCR to yield a fragment of 1,011 bp using primer pairs ADH1 and ADH2. All the primers used in this work are listed in Table 1. The PCR product was cloned into the pGEM-T vector to generate plasmid pADH1. A DNA fragment containing the npt gene (neomycin phosphotransferase) under the control of the Synechocystis sp. strain PCC 6803 psbA promoter (13) was introduced into the HindIII site of the coding region of adhA (712 bp downstream of the start codon), generating a construct that was used to transform Synechocystis sp. strain PCC 6803 (14). Transformants were selected by screening for resistance to 50 μg ml−1 of kanamycin in BG-11 medium plates. Segregation of the inactivated adhA gene was monitored by Southern blotting using standard procedures (46). Total DNA from cyanobacteria was isolated as previously described (7).
TABLE 1.
Primers used in this work
| Oligonucleotide | Sequence (5′→3′) |
|---|---|
| ADH1 | ACTCTATTACATATGATTAAAGCC |
| ADH2 | TGACCATTCCACGTCGACAGAAGC |
| ADP1 | TATTCATCGGATCCGATAACA |
| ADP2 | GGCAGCGTAGGCTTGGATCCTGG |
| PEX1 | TTCCGTTGGCTTCCAGGGCAGCG |
| PEX2 | CCTCTGCCTTGTCTTCATCATCGT |
| RR1 | CAAGGATCCGTGGGGTTGAGTTTGCTG |
| RR2 | TAATTGCTGCAGACTTGTCATAGTTAT |
| S061 | CATGGAGCCTAGTAATCACC |
| S062 | GCGCTGGCATAAACCAGACC |
| S071 | GATCGTTAGGATCATTGCG |
| S072 | CGTCTAGAACTGTCTGCGGGC |
Expression and purification of the AdhA protein.
To generate a plasmid for the expression of recombinant N-terminal His-tagged AdhA, the PCR fragment obtained using primers ADH1 and ADH2 was digested with NdeI and SalI and cloned into the same restriction sites of the pET-28a(+) vector (Novagen) to generate plasmid pETAdhA. The DNA fragments cloned were totally sequenced to ensure that no modifications in the nucleotide sequence occurred during cloning. For the expression of the AdhA protein, E. coli BL21(DE3) was transformed with plasmid pETAdhA. For large-scale protein production, the transformant cells were grown at 37°C in LB broth to an optical density at 580 nm of 0.5, 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) was added, and the culture was incubated for 2.5 h. Thereafter, cells were harvested by centrifugation, resuspended in 20 mM Tris-HCl (pH 7.9) supplemented with 50 mM NaCl and 1 mM phenylmethylsulfonyl fluoride, and broken by sonication. The insoluble debris was pelleted by centrifugation at 18,000 × g for 30 min, and the supernatant was used immediately for protein purification. AdhA was purified by anion-exchange chromatography using DEAE-cellulose resin equilibrated with 20 mM Tris-HCl (pH 7.5) and applying a 0 to 0.5 M NaCl gradient. The eluted fractions containing the protein were loaded onto Ni-nitrilotriacetic acid (NTA) resin (Novagen) and eluted with 0.4 M imidazole. Protein samples were examined by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (46).
Gel filtration.
A 200-μl aliquot of native AdhA (30 μM) was applied to a Superose 6 10/300 GL (GE Healthcare) column connected to a fast-performance liquid chromatograph (Äkta; Amersham Biosciences) at 0.8 ml min−1. The column was previously equilibrated with 30 mM Tris-HCl (pH 8.0) supplemented with 0.5 M NaCl. The molecular mass standards used were bovine thyroglobulin (670,000 Da), bovine γ-globulin (158,000 Da), chicken ovalbumin (44,000 Da), horse myoglobin (17,000 Da), and vitamin B12 (1,350 Da).
Expression and purification of the Rre1 protein.
A DNA fragment covering from +1 to +846 bp of slr1783 (Cyanobase) was amplified by PCR using primers RR1 and RR2, digested with BamHI and PstI, and cloned into the same restriction sites of vector pQE-80L to generate plasmid pQR1, which was used to produce a recombinant N-terminal His-tagged Rre1 protein. The DNA fragment cloned was totally sequenced to ensure that no modifications in the nucleotide sequence occurred during cloning. For the expression of the Rre1 protein, IPTG-inducible E. coli strain DH5α transformed with plasmid pQR1 was grown at 37°C in LB broth to an optical density at 580 nm of 0.5, 1 mM IPTG was then added, and the culture was incubated for 2.5 h. Thereafter, cells were harvested by centrifugation, resuspended in 20 mM Tris-HCl (pH 7.9) supplemented with 50 mM NaCl and 1 mM phenylmethylsulfonyl fluoride, and broken by sonication. The insoluble debris was pelleted by centrifugation at 18,000 × g for 30 min, and the supernatant was used immediately for protein purification. Rre1 was purified by affinity chromatography using Ni-NTA resin (Novagen).
Alcohol dehydrogenase assay.
Enzyme activity was assayed at 25°C by monitoring changes in the absorbance of NAD(P)H at 340 nm in a Pharmacia LKB Ultrospec Plus spectrophotometer with a 1-cm-light-path cuvette. Reactions were performed using 30 mM Tris-HCl buffer (pH 8.0) containing 0.2 mM NAD(P)H for the reduction of aldehydes or 0.5 mM NAD(P)+ for the oxidation of alcohols. The kinetic constants were determined by fitting the initial rates, calculated in the linear range of protein concentration and reaction time, to the Michaelis-Menten equation, and the corresponding values are expressed as the means ± standard deviations for three independent determinations.
RNA isolation and Northern blot analysis.
Total RNA was isolated from 30-ml samples of Synechocystis sp. strain PCC 6803 cultures in the mid-exponential growth phase (3 to 5 μg chlorophyll ml−1) as previously described (17). For Northern blotting, 15 μg of total RNA was loaded per lane in 1% (wt/vol) agarose denaturing formaldehyde gels and transferred onto nylon membranes (Hybond N+; Amersham Biosciences). Hybridization was performed at 65°C. The adhA, sll1106, and sll1107 probes were synthesized by PCR using primer pairs ADH1/ADH2, S061/S062, and S071/S072, respectively, and 32P labeled with a random-primer kit (Amersham Biosciences) with [α-32P]dCTP (3,000 Ci mmol−1). All filters were stripped and rehybridized with the constitutively expressed rnpB gene from Synechocystis sp. strain PCC 6803 (59).
Preparation of AdhA polyclonal antibodies.
A homogeneous sample of the protein was used for the production of antibodies at the Animal Production and Experimentation Service of Seville University (Seville, Spain).
Preparation of cytosolic extracts from Synechocystis and Western blot analysis.
Cytosolic protein extracts from Synechocystis were obtained as described previously (40). Cytosolic proteins (5 μg) from Synechocystis cultures exposed to different conditions were separated by SDS-PAGE and transferred onto nitrocellulose membranes. Membranes were incubated with diluted anti-AdhA (1:1,000) overnight at 4°C. Detection was performed using ECL Plus Western blotting detection reagents (Amersham Biosciences) according to instructions provided by the manufacturer.
Gel mobility shift assay.
A probe was synthesized by PCR using oligonucleotide pair ADP1 and ADP2, both containing a BamHI restriction site. The PCR product was digested with BamHI, generating a fragment from positions −215 to +22 with respect to the transcription start point of adhA, which was end labeled with [α-32P]dCTP (3,000 Ci mmol−1) by using the Klenow fragment of DNA polymerase. The binding reaction was carried out using a final volume of 30 μl containing 0.1 ng of labeled DNA (2 pM) and 3 μg of poly(dI-dC) in 50 mM Tris-HCl (pH 8.0), 100 mM KCl, 15 mM dithiothreitol, 10% glycerol, and different amounts of protein. Reaction mixtures were incubated for 30 min at room temperature, and samples were loaded onto a nondenaturing 6% polyacrylamide gel. Electrophoresis was carried out at 4°C and 180 V in 22 mM Tris-borate-0.5 mM EDTA. The gel was transferred onto a Whatman 3-mm filter and dried prior to autoradiography. For competition assays, a 222-bp DNA fragment from pGEM-T digested with SspI was used as nonspecific competitor DNA.
Primer extension analysis.
Oligonucleotides PEX1 and PEX2 were end labeled with T4 polynucleotide kinase and [γ-32P]ATP (3,000 Ci mmol) and used for primer extension analysis of the adhA and sll1106 genes, respectively. Primer extension reactions were carried out as previously described (31). One-half of the reaction mixture was subjected to electrophoresis on a 6% polyacrylamide sequencing gel, and the adhA and sll1106 promoter regions were subjected to a sequencing reaction using the oligonucleotides described above and plasmid pEXAdh as a template.
RESULTS
AdhA is a tetrameric medium-chain Zn-containing alcohol dehydrogenase.
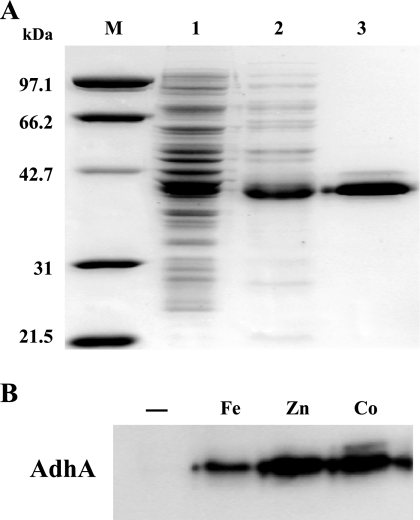
In order to gain knowledge on the activity and properties of the AdhA protein, the adhA gene was cloned, and a His6-tagged version of the protein was expressed in E. coli cells. Homogeneous preparations of AdhA were obtained by DEAE-cellulose ion-exchange chromatography followed by affinity chromatography on Ni-NTA (Fig. 1A). Purification attempts using extracts of E. coli cells grown on M9 chemically defined medium revealed that the presence of divalent metals in the medium was a requisite for the availability of the enzyme in a soluble form (Fig. 1B). Thus, when metals were absent from the growth medium, expressed AdhA did not appear in the soluble fraction but appeared there, however, if the medium was supplemented with zinc, cobalt, or ferrous ions (Fig. 1B). Differences in the specific activities of the enzyme purified from cultures grown on media with different metal contents showed that AdhA can use either zinc or cobalt with analogous effectivenesses (2 ± 0.2 or 1.9 ± 0.12 mU·mg total protein−1, respectively), while ferrous ion was less efficient (0.3 ± 0.05 mU·mg total protein−1). Analysis of the amino acid sequences (see Fig. S1 in the supplemental material) revealed the presence of conserved residues involved in the binding of both catalytic (Cys55, His78, Cys176, Asp58, and Glu79) and structural (Cys111, Cys114, Cys117, and Cys125) zinc ions (23, 30, 52).
FIG. 1.
Purification and metal dependence of AdhA. (A) Coomassie-stained SDS-PAGE gel of different fractions corresponding to steps of AdhA purification (0.5 μg total protein per lane). M, molecular mass markers. Lane 1, crude extract from E. coli cells that overexpress His-tagged AdhA; lane 2, pooled DEAE-cellulose chromatography fractions containing His-tagged AdhA; lane 3, pooled affinity chromatography (Ni-NTA) fractions containing His-tagged AdhA. (B) Western blotting of soluble extracts of AdhA-overexpressing E. coli cells grown on M9 medium. −, no metal added; Fe, 5 μM (NH4)2Fe(SO4)2; Zn, 5 μM ZnSO4; Co, 5 μM CoCl2.
To establish the quaternary structure of the protein, gel filtration on a Superose 6 10/300 GL column were performed using purified preparations of recombinant AdhA, and an apparent molecular mass of 140 kDa was determined. Taking into account the polypeptide mass (36.7 kDa) of the recombinant protein calculated from migration on SDS-PAGE gels (Fig. 1A), a tetrameric structure is suggested for the AdhA active enzyme.
A thorough substrate specificity analysis as well as a kinetic characterization of AdhA have been performed. As shown in Table 2, the enzyme was active toward a wide variety of primary alcohols and their corresponding aldehydes, but neither ketones nor secondary alcohols were effective substrates. Aliphatic compounds were efficiently processed by the enzyme, with the activity increasing with the length of the chain up to five carbon atoms and decreasing for longer chains. Aromatic alcohols and aldehydes were also substrates for the enzyme, as was the case for the BA/benzaldehyde or cinnamyl alcohol/cinnamaldehyde. Branched-chain primary alcohols, such as 2-methyl-butanol, were processed less efficiently than the linear substrate (Table 2). The Km and kcat values were calculated for several substrate pairs of alcohols and their corresponding aldehydes, and they are shown in Table 3. The values of kcat were significantly higher for aldehydes than for alcohols. Cinnamaldehyde and pentanal behaved as the substrates most efficiently processed by the AdhA protein, with the highest kcat/Km ratios. The enzyme could use both NADPH and NADH as electron donors, although NADPH (Km = 0.025 mM) was preferred to NADH (Km = 1.3 mM) as the reductant for aldehydes. Equally, that affinity for NADP+ was higher than that for NAD+ as an alcohol oxidant. Thus, it is likely that AdhA works to reduce aldehydes to alcohols.
TABLE 2.
Substrate specificity of AdhA from Synechocystisc
| Reductiona | Relative activity (%)
|
Oxidationb | Relative activity (%)
|
||
|---|---|---|---|---|---|
| 0.1 mM | 1 mM | 1 mM | 10 mM | ||
| Acetaldehyde | 1 | 23 | Ethanol | 19 | 91 |
| Propanal | 26 | 87 | Propanol | 21 | 84 |
| Butanal | 73 | 120 | Butanol | 23 | 70 |
| Pentanal | 100 | 107 | Pentanol | 31 | 52 |
| Hexanal | 53 | 100 | Hexanol | 27 | 40 |
| Octanal | 42 | 80 | Octanol | 8 | 17 |
| Decanal | 21 | 22 | Decanol | 3 | 6 |
| Benzaldehyde | 30 | 14 | Benzyl alcohol | 42 | 79 |
| Cinnamaldehyde | 61 | 7 | Cinnamyl alcohol | 100 | 116 |
| Acetone | ND | ND | Phenylethanol | 15 | 24 |
| 3-Pentanone | ND | ND | 2-Methylbutanol | 24 | 22 |
| d-Glucose | ND | ND | 3-Pentanol | ND | ND |
The reduction activities were measured using 30 mM Tris-HCl (pH 8.0) containing 0.1 mM or 1 mM substrate in the presence of 0.2 mM NADPH. The activity toward 0.1 mM pentanal (67.7 U mg−1 protein) was considered to be 100%.
The oxidation activities were measured using 30 mM Tris-HCl (pH 8.0) containing 1 mM or 10 mM substrate in the presence of 0.5 mM NADP+. The activity toward 1 mM cinnamyl alcohol (26.1 U mg−1 protein) was considered to be 100%.
ND, not detected.
TABLE 3.
Kinetic parameters of AdhA from Synechocystisa
| Substrate | Mean Km (mM) ± SDa | Mean kcat (min−1) ± SD | kcat/Km ratio (min−1 mM−1) |
|---|---|---|---|
| Acetaldehyde | 0.510 ± 0.050 | 9,458 ± 138 | 18,545 |
| Butanal | 0.110 ± 0.008 | 9,398 ± 181 | 84,664 |
| Pentanal | 0.075 ± 0.002 | 9,547 ± 185 | 123,295 |
| Cinnamaldehyde | 0.017 ± 0.001 | 6,475 ± 131 | 380,882 |
| Ethanol | 18.800 ± 3.300 | 1,641 ± 33 | 87 |
| Butanol | 3.280 ± 0.100 | 1,023 ± 21 | 312 |
| Pentanol | 1.540 ± 0.060 | 789 ± 15 | 512 |
| Cinnamyl alcohol | 0.270 ± 0.010 | 3,647 ± 73 | 13,507 |
| NADPH | 0.025 ± 0.007 | ||
| NADP | 0.050 ± 0.007 | ||
| NADH | 1.310 ± 0.060 | ||
| NAD | 9.700 ± 0.800 |
Reactions were performed using 30 mM Tris-HCl (pH 8.0). For the reduction of aldehydes and oxidation of alcohols, 0.15 mM NAD(P)H and 0.4 mM NAD(P)+ were used, respectively. Kinetic constants for NAD(P)+ and NAD(P)H were determined with 100 mM ethanol and 25 mM acetaldehyde, respectively.
Expression of adhA is induced under various stress conditions.
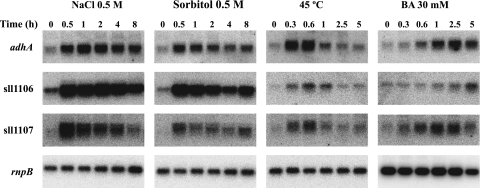
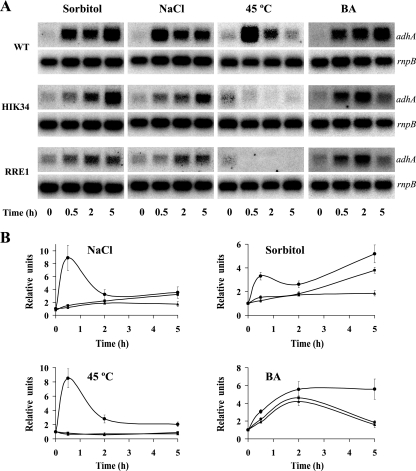
The adhA gene was previously identified by DNA microarray analysis to be a hyperosmotic-stress- and salt stress-inducible gene (35, 48). Using Northern blot hybridization (Fig. 2), we have confirmed that the level of transcription of adhA increases in response to either hyperosmotic stress (0.5 M sorbitol) or salt stress (0.5 M NaCl). In order to determine whether adhA expression is specifically enhanced in response to the osmotic stress induced by the addition of sorbitol or NaCl or as a part of a general stress response, Synechocystis cells were exposed to different stress conditions such as heat shock, high-light conditions, and oxidative stress, and changes in levels of adhA transcripts were monitored. The exposure of cells to a sudden increase in the ambient temperature (45°C) also resulted in enhanced adhA transcript levels (Fig. 2). However, neither high-light treatment nor oxidative stress (1 mM H2O2) had an influence on the transcription of the adhA gene (data not shown). It was previously reported (19) that many of the genes that respond to heat shock are also induced when BA, a membrane-fluidizing agent (18), is added to the culture medium. As shown in Fig. 2, we have confirmed that BA also induces adhA gene expression, although the kinetics of adhA transcript induction were different. Thus, the induction of adhA expression was transient under conditions of heat shock exposure, with the maximum transcript level being reached in less than 1 h and returning to the steady-state level in 2 h (Fig. 2). However, for the response to BA, the maximum transcript level was reached at 5 h. On the other hand, the maximum level of adhA transcript was reached in less than 1 h and then decreased at 2 h under conditions of salt stress. In response to hyperosmotic shock, the kinetics were very similar to those of salt stress, but the decrease in the transcript level at 2 h was followed by another increase to reach the maximum level at 5 h. In both cases, the transcription of the adhA gene remained induced at least 8 h after treatment and returned to basal levels after 24 h (data not shown). In addition to the information on the induction of the expression of adhA, Fig. 2 includes also data for the expression of the nearby sll1106 and sll1107 genes, which exhibited a pattern analogous to that of adhA. This circumstance is considered below.
FIG. 2.
Northern blotting analysis of the stress-inducible expression of the adhA, sll1106, and sll1107 genes. Synechocystis sp. strain PCC 6803 cells at the exponential phase of growth were subjected to various conditions, and total RNA was extracted at different time points. Fifteen micrograms of total RNA was denatured, separated by electrophoresis in a 1.2% agarose gel, blotted, and hybridized with probes for the adhA, sll1106, and sll1107 genes. The filters were stripped and hybridized with an rnpB gene probe as a loading control. All probes were synthesized as described in Materials and Methods. Three independent experiments were performed, and data plotted are the averages ± standard errors.
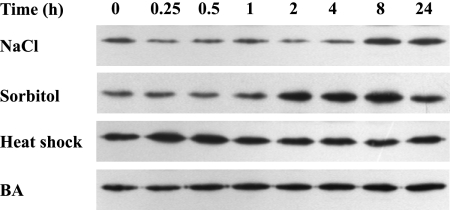
In order to analyze the relationship between adhA transcript and AdhA protein levels, Western blot experiments were performed. A basal amount of AdhA was detected in Synechocystis cells grown under photoautotrophic conditions. As shown in Fig. 3, the AdhA protein level increased under conditions of salt and hyperosmotic stresses in accordance with the observed induction of transcript levels. The kinetics of AdhA accumulation differed depending on the stress condition imposed (Fig. 3). While under hyperosmotic conditions, an increase in AdhA levels was readily observed after 2 h of treatment, under salt stress conditions, the protein started to accumulate after 8 h (Fig. 3). This difference could be explained by the fact that protein synthesis was previously reported to be nearly blocked within 4 h after the onset of salt stress (15). In contrast, the amount of the AdhA protein remained virtually unaltered under conditions of heat shock and upon the addition of BA. Induction experiments performed in the presence of chloramphenicol confirmed that no degradation of AdhA took place under any condition tested (data not shown).
FIG. 3.
Western blotting analysis of the stress-inducible expression of the AdhA protein. Synechocystis sp. strain PCC 6803 cells at the exponential phase of growth were subjected to various conditions as indicated. Cells were harvested at the times indicated, and 5 μg of total protein from soluble extracts was separated by 12% SDS-PAGE and subjected to Western blotting.
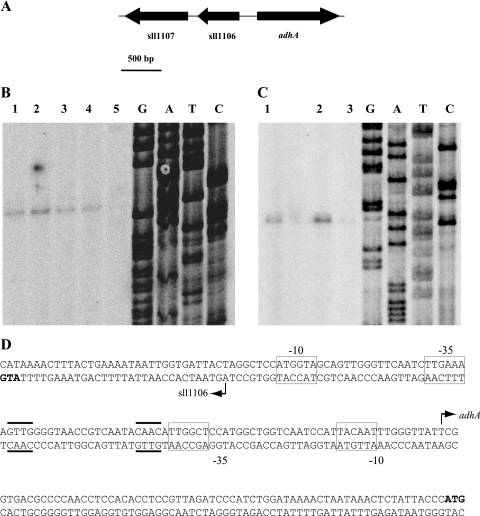
The sll1106 and sll1107 genes share the same expression pattern as adhA.
Immediately upstream of the adhA gene in the opposite DNA strand are two open reading frames, sll1106 and sll1107 (Fig. 4A). One of them, sll1107, was also identified, together with the adhA gene, as being a salt-inducible gene in a genome-wide DNA microarray assay (48); both genes were shown to be under the control of the Hik34-Rre1 two-component system. The proximity between sll1106 and adhA and the coregulation of sll1107 prompted us to test for a possible coordinated regulation of the sll1106, sll1107, and adhA genes. In order to test this hypothesis, filters used for Northern hybridizations with adhA were rehybridized with probes corresponding to sll1106 and sll1107. The results (Fig. 2) showing the sll1106 and sll1107 genes to be induced under the same conditions that induced the expression of adhA suggest a possible coregulation in which the sll1106-adhA intergenic region could contain the promoter and the regulatory sequences for the expression of the three genes. However, transcripts of different sizes (516 nucleotides for sll1106 and 777 nucleotides for sll1107) were detected by Northern blotting, indicating that both genes are probably transcribed under the control of independent promoters. On the other hand, the sll1106 and adhA genes are separated by a 200-bp region. In order to establish the location of the adhA and sll1106 promoter(s), starting points of adhA and sll1106 transcripts were mapped by primer extension experiments. One single extension product, which appeared as a double band, was obtained for both genes when RNA extracted from cells subjected to stress conditions was used (Fig. 4B and C). The adhA transcription start site was located 69 bp upstream of the ATG start codon, while the sll1106 start site was located 31 bp upstream of the corresponding ATG start codon (Fig. 4D). Both sites were separated by a region of 100 bp, which might contain the promoters for both genes.
FIG. 4.
Primer extension analysis of adhA and sll1106 transcripts. (A) Schematic representation of the genomic region corresponding to the adhA, sll1106, and sll1107 genes. (B) Primer extension analysis of adhA transcripts from Synechocystis cells exposed for 1 h to 30 mM BA (lane 1), heat shock (45°C) (lane 2), 0.5 M NaCl (lane 3), and 0.5 M sorbitol (lane 4) and from nonexposed cells (lane 5). (C) Primer extension analysis of sll1106 transcripts from Synechocystis cells exposed to heat shock (45°C) (lane 1) and 0.5 M sorbitol (lane 2) and from nonexposed cells (lane 3). (D) Sequence of the adhA-sll1106 intergenic region, with translation start codons in boldface type. The transcripts start points are marked with arrows. Putative −10 and −35 boxes are indicated with rectangles.
The two-component system Hik34-Rre1 regulates the induction of adhA expression in response to stress conditions.
The adhA gene was previously identified as being a salt-stress-inducible gene under the control of the Hik34-Rre1 two-component system (48). In order to clarify if the histidine kinase Hik34 and the response regulator Rre1 regulate the expression of the adhA gene in response to the other conditions tested (Fig. 2), the pattern of expression of the adhA gene in wild-type cells was compared to that of Δhik34 and Δrre1 mutant strains. As shown in Fig. 5, the expression levels of the adhA gene in response to heat shock or BA were similar for both the Δhik34 and Δrre1 mutants and differed from that of the wild-type strain. In the case of heat shock, the induction of the expression of the adhA gene was completely abolished in both mutants. In the case of BA, the effect of either mutation was noticeable only at 5 h after treatment (Fig. 5). When mutant cells were exposed to either salt or hyperosmotic shock, the early induction of the adhA gene (30 min after exposure to stress) was abolished. In the case of the Δrre1 mutant, this situation was maintained with time, although this was not the case for the Δhik34 mutant.
FIG. 5.
Expression of the adhA gene under stress conditions in wild-type (WT), Δhik34, and Δrre1 strains. (A) Northern blot analysis. Cultures at the exponential phase of growth were subjected to various stress conditions, and total RNA was extracted at different time points. Fifteen micrograms of total RNA was denatured, separated by electrophoresis in a 1.2% agarose gel, blotted, and hybridized with adhA. As a loading control, all the filters were rehybridized with rnpB. (B) Kinetics of expression of the adhA transcripts. The values in the graphs indicate the ratio of adhA transcripts in stressed cells to those in control cells. Three independent experiments were performed, and data plotted are the averages ± standard errors. •, wild-type strain; ▪, Δhik34 mutant strain; ▴, Δrre1 mutant strain.
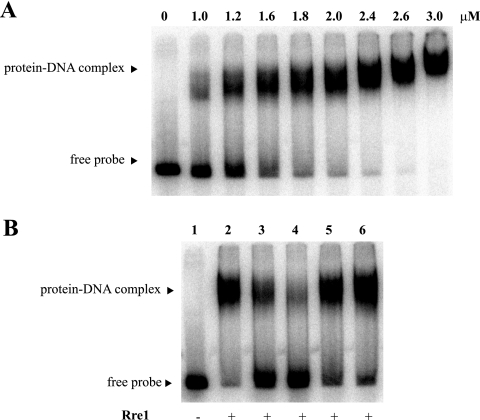
Rre1 binds to the promoter region of the adhA gene.
In order to test if the response regulator Rre1 affects the expression of the adhA gene directly by interactions with its promoter region, a DNA binding shift assay was performed. A radiolabeled DNA fragment corresponding to the sll1106-adhA intergenic region was incubated with increasing amounts of the purified Rre1 protein (see Materials and Methods). A single shifted band representing a DNA-protein complex was observed upon the addition of Rre1 (Fig. 6A). Sequence-specific binding was evident from competition assays in the presence of different concentrations of either specific or nonspecific competitor DNA. The Rre1-dependent band shift was diminished in the presence of a 200-fold excess of the same unlabeled fragment but was unaffected by the presence of an excess of the unrelated DNA fragment (Fig. 6B).
FIG. 6.
Binding of Rre1 to the adhA-sll1106 promoter region. (A) Mobility shift assay of the adhA-sll1106 promoter-operator region with increasing amounts of Rre1. (B) Competitive assays were performed with Rre1 (1.6 μM) in the presence of either a 50-fold (lanes 3 and 5) or a 200-fold (lanes 4 and 6) excess of unlabeled self-competitor fragment (lanes 3 and 4) or unlabeled nonspecific competitor fragment (lanes 5 and 6).
DISCUSSION
To the best of our knowledge, this is the first report of the molecular characterization of an alcohol dehydrogenase in cyanobacteria. We show that the protein encoded by the Synechocystis adhA gene is a zinc-dependent homotetrameric enzyme with NADP-dependent alcohol dehydrogenase activity acting on a broad variety of substrates. The higher catalytic efficiency of Synechocystis AdhA for aldehyde reduction, with pentanal and cinnamaldehyde as the preferred substrates, points to the reduction of aromatic and medium-chain aliphatic aldehydes as a most plausible in vivo activity of the enzyme. Synechocystis AdhA exhibits a preferential specificity for NADP(H) but is also able to use NAD(H). This may be explained by the presence of a threonine at position 226 (see Fig. S1 in the supplemental material), which is more similar to the serine residue of the known NADPH-dependent ADH (52) than to the aspartate or glutamate residue of NADH-dependent ADH (29, 34). Thus, the Synechocystis AdhA enzyme shares biochemical features with both Y-ADH enzymes, which are tetrameric NAD-dependent enzymes, and CADH enzymes, which are dimeric and NADP dependent.
A search performed with the AdhA sequence (slr1192 protein) as a query using a nonredundant protein sequence database of the NBCI revealed more than 60% identity with putative MDRs from gammaproteobacteria. Among cyanobacteria, homologous sequences showing about 50 to 60% identity were found in some strains of Microcystis and Synechococcus. Sequence alignment of AdhA with related members of the MDR family from bacteria, plants, and yeasts yielded different clusters, with bacterial sequences forming a closed group independent from CADH and Y-ADH groups (see Fig. S2 in the supplemental material).
In plants, the function of the CADH family is related to lignin biosynthesis (5, 25). In bacteria, from which lignin is absent, CADH-like genes are related to the biosynthesis of lipids of the cell envelope (16, 62), but it is possible that they fulfill other specific functions. An alcohol dehydrogenase from Acinetobacter that shares 46% identity with the adhA gene from Synechocystis has been proposed to be involved in the biosynthesis of wax esters as cell reserves when cells are grown on n-alkanes (57). On the other hand, some Y-ADH-related enzymes, which show broad substrate specificity, fulfill a function related to primordial metabolic pathways such as alcohol fermentative activity (49). A variety of different fermentation pathways yielding products such as CO2, H2, formate, acetate, lactate, and ethanol have been reported to exist in cyanobacteria (50). A number of cyanobacteria are capable of heterotrophic growth under dark conditions at the expense of either endogenous glycogen or exogenous substrates. In most cases, heterotrophic growth takes place under aerobic conditions, but some of the facultative heterotrophic cyanobacteria are able to survive under dark anoxic conditions via fermentation of the glucose derived from endogenous substrates such as glycogen or some osmoprotectant compounds (50). Thus, AdhA might participate in any of such fermentative pathways in Synechocystis.
Here we show that a soluble AdhA protein is present in Synechocystis cells and that its expression is enhanced upon the exposure of cells to stress conditions. Although the level of the adhA transcript increases, the AdhA protein level remains unchanged under conditions of heat shock, making it unlikely that AdhA plays a role in the response to this condition. On the other hand, the induction of adhA transcription under conditions of salt stress or hyperosmotic stress is accompanied by an increase in the amount of AdhA. Both salt and hyperosmotic stress cause alterations in the physical integrity of the plasma membrane (24) and affect the activity of photosystem I (PSI) and photosystem II in cyanobacteria (1-3, 32), thus altering ATP production and the NADP/NADPH balance. In Synechocystis, salt stress has been reported to enhance PSI-dependent cyclic electron flow (58) that generates ATP but not NADPH (36), thus participating in determining the ATP/NADPH ratio when photosynthesis operates under changing environmental conditions (45). The fact that AdhA from Synechocystis oxidizes NADPH much more efficiently than it reduces NADP+ supports a possible role for this enzyme in oxidizing NADPH under such conditions and thus contributing to the maintenance of the ATP/NADPH balance, as was also previously proposed for two members of the CADH family in Saccharomyces (27, 28).
We have compared the growth of an ΔadhA mutant with that of the wild-type strain under a variety of conditions. No differences in responses to heat shock, salt, or hyperosmotic stress have been observed. Both strains exhibit analogous growth rates when tested in the light under autotrophic, mixotrophic (added glucose), or photoheterotrophic [added glucose and 3-(3P,4P-dichlorophenyl)-1,1P-dimethylurea] conditions. Nevertheless, the adhA mutant strain was unable to grow heterotrophically (added glucose) in darkness, contrary to the wild-type strain (our unpublished data). These results strongly suggest an involvement of AdhA in heterotrophic metabolism in Synechocystis, which calls for further investigation.
The histidine kinase Hik34 is involved in the regulation of the expression of heat shock genes in Synechocystis (53). Four two-component systems are involved in the perception and transduction of salt and hyperosmotic stress signals in Synechocystis, with the Hik34-Rre1 system being the one responsible for controlling the expression of the adhA and sll1107 genes under conditions of salt stress (39, 48). The results obtained in the present work concerning the effect of a mutation of either hik34 or rre1 on the expression of the adhA gene show that the Hik34-Rre1 system is involved in the early induction of adhA expression under conditions of heat shock and hyperosmotic stress, a fact not yet reported. Although this system is essential for the response of the adhA gene to heat shock, the enhancement of transcription of adhA in response to salt stress, hyperosmotic stress, or the presence of BA is only in part dependent on the operation of the Hik34-Rre1 system. Moreover, the differences observed between hik34 and rre1 mutants with regard to adhA gene transcription in response to hyperosmotic and salt stress suggest the involvement of an additional factor in the activation of Rre1.
We have demonstrated that the Rre1 protein isolated from E. coli binds specifically to the promoter region of the adhA gene in vitro. Although in general, it is accepted that response regulators exist in two conformations, an inactive state and an active state, with phosphorylation serving to shift the equilibrium toward the active state (60), recent findings (12) support the contention of an intermediate conformation between inactive and active states (51). This is the case for the CheY response regulator, which binds to its target in the unphosphorylated form and displays a basal level of kinase-independent activity in vivo (47). The phosphoacceptor domain of Rre1 is a homolog of that of CheY. The affinity for the specific DNA sequence in the adhA promoter of unphosphorylated Rre1 would be sufficient for the observed binding. Nevertheless, the specific sequence for binding remains to be elucidated. The upstream divergently transcribed sll1106 gene, which shows the same expression pattern, could share the regulatory sequences with the adhA gene, as suggested by the proximity of their corresponding −35 boxes. Two repeated palindromic GTTG sequences located between the putative −35 boxes of both the adhA and sll1106 promoters (Fig. 4D) might be the actual binding sites for Rre1, allowing the activation of both promoters. In silico analysis predicts that the sll1106 gene encodes a membrane protein that highly conserved among cyanobacteria. Unfortunately, the lack of information about its function hampers the establishment of any functional relationship between the sll1106 and AdhA proteins. On the other hand, the assigned function of the sll1107 gene product, a type IV pilus biogenesis protein, excludes a direct functional relationship with the AdhA protein.
In conclusion, Synechocystis posses a zinc-dependent alcohol dehydrogenase (AdhA), which has a preference for NADPH as a reductant and aromatic and medium-chain aliphatic aldehydes as oxidants. The corresponding gene is upregulated in response to certain stress conditions via the Hik34-Rre1 two-component system. Its specific role in the heterotrophic growth of Synechocystis remains to be established.
Supplementary Material
Acknowledgments
This work was supported by a grant from the Ramón Areces Foundation (to M.G.G.), grants BFU2004-0635 and BFU2007-60300 from the Ministry of Science and Education of Spain (to F.J.F.), and funds of Junta de Andalucia (BIO131 and BIO284).
We acknowledge Iwane Suzuky for kindly providing the Δhik34 and Δrre1 mutant strains. We acknowledge Anna Lindahl for the critical reading of the manuscript.
Footnotes
Published ahead of print on 1 May 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Allakhverdiev, S. I., Y. Nishiyama, I. Suzuki, Y. Tasaka, and N. Murata. 1999. Genetic engineering of the unsaturation of fatty acids in membrane lipids alters the tolerance of Synechocystis to salt stress. Proc. Natl. Acad. Sci. USA 965862-5867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allakhverdiev, S. I., A. Sakamoto, Y. Nishiyama, M. Inaba, and N. Murata. 2000. Ionic and osmotic effects of NaCl-induced inactivation of photosystems I and II in Synechococcus sp. Plant Physiol. 1231047-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allakhverdiev, S. I., A. Sakamoto, Y. Nishiyama, and N. Murata. 2000. Inactivation of photosystems I and II in response to osmotic stress in Synechococcus. Contribution of water channels. Plant Physiol. 1221201-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson, S. L., and L. McIntosh. 1991. Light-activated heterotrophic growth of the cyanobacterium Synechocystis sp. strain PCC 6803: a blue-light-requiring process. J. Bacteriol. 1732761-2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boudet, A. M. 2007. Evolution and current status of research in phenolic compounds. Phytochemistry 682722-2735. [DOI] [PubMed] [Google Scholar]
- 6.Boulton, B., V. L. Singleton, L. F. Bisson, and R. E. Kunkee. 1996. Principles and practices of winemaking, p.139-172. Chapman and Hall, New York, NY.
- 7.Cai, Y. P., and C. P. Wolk. 1990. Use of a conditionally lethal gene in Anabaena sp. strain PCC 7120 to select for double recombinants and to entrap insertion sequences. J. Bacteriol. 1723138-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang, C., and R. C. Stewart. 1998. The two-component system. Regulation of diverse signaling pathways in prokaryotes and eukaryotes. Plant Physiol. 117723-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christie, P. J., M. Hahn, and V. Walbot. 1991. Low-temperature accumulation of alcohol dehydrogenase-1 mRNA and protein activity in maize and rice seedlings. Plant Physiol. 95699-706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dawes, E. A., D. W. Ribbons, and P. J. Large. 1966. The route of ethanol formation in Zymomonas mobilis. Biochem. J. 98795-803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deng, M. D., and J. R. Coleman. 1999. Ethanol synthesis by genetic engineering in cyanobacteria. Appl. Environ. Microbiol. 65523-528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dyer, C. M., and F. W. Dahlquist. 2006. Switched or not?: the structure of unphosphorylated CheY bound to the N terminus of FliM. J. Bacteriol. 1887354-7363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elhai, J., and C. P. Wolk. 1988. A versatile class of positive-selection vectors based on the nonviability of palindrome-containing plasmids that allows cloning into long polylinkers. Gene 68119-138. [DOI] [PubMed] [Google Scholar]
- 14.Ferino, F., and F. Chauvat. 1989. A promoter-probe vector-host system for the cyanobacterium, Synechocystis PCC6803. Gene 84257-266. [DOI] [PubMed] [Google Scholar]
- 15.Fulda, S., S. Mikkat, F. Huang, J. Huckauf, K. Marin, B. Norling, and M. Hagemann. 2006. Proteome analysis of salt stress response in the cyanobacterium Synechocystis sp. strain PCC 6803. Proteomics 62733-2745. [DOI] [PubMed] [Google Scholar]
- 16.Galamba, A., K. Soetaert, P. Buyssens, D. Monnaie, P. Jacobs, and J. Content. 2001. Molecular and biochemical characterisation of Mycobacterium smegmatis alcohol dehydrogenase C. FEMS Microbiol. Lett. 19651-56. [DOI] [PubMed] [Google Scholar]
- 17.Garcia-Dominguez, M., and F. J. Florencio. 1997. Nitrogen availability and electron transport control the expression of glnB gene (encoding PII protein) in the cyanobacterium Synechocystis sp. PCC 6803. Plant Mol. Biol. 35723-734. [DOI] [PubMed] [Google Scholar]
- 18.Horvath, I., A. Glatz, V. Varvasovszki, Z. Torok, T. Pali, G. Balogh, E. Kovacs, L. Nadasdi, S. Benko, F. Joo, and L. Vigh. 1998. Membrane physical state controls the signaling mechanism of the heat shock response in Synechocystis PCC 6803: identification of hsp17 as a “fluidity gene.” Proc. Natl. Acad. Sci. USA 953513-3518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inaba, M., I. Suzuki, B. Szalontai, Y. Kanesaki, D. A. Los, H. Hayashi, and N. Murata. 2003. Gene-engineered rigidification of membrane lipids enhances the cold inducibility of gene expression in Synechocystis. J. Biol. Chem. 27812191-12198. [DOI] [PubMed] [Google Scholar]
- 20.Jarillo, J. A., A. Leyva, J. Salinas, and J. M. Martinez-Zapater. 1993. Low temperature induces the accumulation of alcohol dehydrogenase mRNA in Arabidopsis thaliana, a chilling-tolerant plant. Plant Physiol. 101833-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jornvall, H., and J. O. Hoog. 1995. Nomenclature of alcohol dehydrogenases. Alcohol Alcohol. 30153-161. [PubMed] [Google Scholar]
- 22.Jornvall, H., J. O. Hoog, and B. Persson. 1999. SDR and MDR: completed genome sequences show these protein families to be large, of old origin, and of complex nature. FEBS Lett. 445261-264. [DOI] [PubMed] [Google Scholar]
- 23.Jornvall, H., B. Persson, and J. Jeffery. 1987. Characteristics of alcohol/polyol dehydrogenases. The zinc-containing long-chain alcohol dehydrogenases. Eur. J. Biochem. 167195-201. [DOI] [PubMed] [Google Scholar]
- 24.Kanesaki, Y., I. Suzuki, S. I. Allakhverdiev, K. Mikami, and N. Murata. 2002. Salt stress and hyperosmotic stress regulate the expression of different sets of genes in Synechocystis sp. PCC 6803. Biochem. Biophys. Res. Commun. 290339-348. [DOI] [PubMed] [Google Scholar]
- 25.Kim, S. J., K. W. Kim, M. H. Cho, V. R. Franceschi, L. B. Davin, and N. G. Lewis. 2007. Expression of cinnamyl alcohol dehydrogenases and their putative homologues during Arabidopsis thaliana growth and development: lessons for database annotations? Phytochemistry 681957-1974. [DOI] [PubMed] [Google Scholar]
- 26.Kimmerer, T. W., and T. T. Kozlowski. 1982. Ethylene, ethane, acetaldehyde, and ethanol production by plants under stress. Plant Physiol. 69840-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Larroy, C., M. R. Fernandez, E. Gonzalez, X. Pares, and J. A. Biosca. 2002. Characterization of the Saccharomyces cerevisiae YMR318C (ADH6) gene product as a broad specificity NADPH-dependent alcohol dehydrogenase: relevance in aldehyde reduction. Biochem. J. 361163-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Larroy, C., X. Pares, and J. A. Biosca. 2002. Characterization of a Saccharomyces cerevisiae NADP(H)-dependent alcohol dehydrogenase (ADHVII), a member of the cinnamyl alcohol dehydrogenase family. Eur. J. Biochem. 2695738-5745. [DOI] [PubMed] [Google Scholar]
- 29.Lauvergeat, V., K. Kennedy, C. Feuillet, J. H. McKie, L. Gorrichon, M. Baltas, A. M. Boudet, J. Grima-Pettenati, and K. T. Douglas. 1995. Site-directed mutagenesis of a serine residue in cinnamyl alcohol dehydrogenase, a plant NADPH-dependent dehydrogenase, affects the specificity for the coenzyme. Biochemistry 3412426-12434. [DOI] [PubMed] [Google Scholar]
- 30.Leskovac, V., S. Trivic, and D. Pericin. 2002. The three zinc-containing alcohol dehydrogenases from baker's yeast, Saccharomyces cerevisiae. FEMS Yeast Res. 2481-494. [DOI] [PubMed] [Google Scholar]
- 31.Lopez-Maury, L., M. Garcia-Dominguez, F. J. Florencio, and J. C. Reyes. 2002. A two-component signal transduction system involved in nickel sensing in the cyanobacterium Synechocystis sp. PCC 6803. Mol. Microbiol. 43247-256. [DOI] [PubMed] [Google Scholar]
- 32.Marin, K., Y. Kanesaki, D. A. Los, N. Murata, I. Suzuki, and M. Hagemann. 2004. Gene expression profiling reflects physiological processes in salt acclimation of Synechocystis sp. strain PCC 6803. Plant Physiol. 1363290-3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marin, K., I. Suzuki, K. Yamaguchi, K. Ribbeck, H. Yamamoto, Y. Kanesaki, M. Hagemann, and N. Murata. 2003. Identification of histidine kinases that act as sensors in the perception of salt stress in Synechocystis sp. PCC 6803. Proc. Natl. Acad. Sci. USA 1009061-9066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McKie, J. H., R. Jaouhari, K. T. Douglas, D. Goffner, C. Feuillet, J. Grima-Pettenati, A. M. Boudet, M. Baltas, and L. Gorrichon. 1993. A molecular model for cinnamyl alcohol dehydrogenase, a plant aromatic alcohol dehydrogenase involved in lignification. Biochim. Biophys. Acta 120261-69. [DOI] [PubMed] [Google Scholar]
- 35.Mikami, K., Y. Kanesaki, I. Suzuki, and N. Murata. 2002. The histidine kinase Hik33 perceives osmotic stress and cold stress in Synechocystis sp. PCC 6803. Mol. Microbiol. 46905-915. [DOI] [PubMed] [Google Scholar]
- 36.Munekage, Y., M. Hashimoto, C. Miyake, K. Tomizawa, T. Endo, M. Tasaka, and T. Shikanai. 2004. Cyclic electron flow around photosystem I is essential for photosynthesis. Nature 429579-582. [DOI] [PubMed] [Google Scholar]
- 37.Murata, N., and I. Suzuki. 2006. Exploitation of genomic sequences in a systematic analysis to access how cyanobacteria sense environmental stress. J. Exp. Bot. 57235-247. [DOI] [PubMed] [Google Scholar]
- 38.Nordling, E., H. Jornvall, and B. Persson. 2002. Medium-chain dehydrogenases/reductases (MDR). Family characterizations including genome comparisons and active site modeling. Eur. J. Biochem. 2694267-4276. [DOI] [PubMed] [Google Scholar]
- 39.Paithoonrangsarid, K., M. A. Shoumskaya, Y. Kanesaki, S. Satoh, S. Tabata, D. A. Los, V. V. Zinchenko, H. Hayashi, M. Tanticharoen, I. Suzuki, and N. Murata. 2004. Five histidine kinases perceive osmotic stress and regulate distinct sets of genes in Synechocystis. J. Biol. Chem. 27953078-53086. [DOI] [PubMed] [Google Scholar]
- 40.Perez-Perez, M. E., F. J. Florencio, and M. Lindahl. 2006. Selecting thioredoxins for disulphide proteomics: target proteomes of three thioredoxins from the cyanobacterium Synechocystis sp. PCC 6803. Proteomics 6(Suppl. 1)S186-S195. [DOI] [PubMed] [Google Scholar]
- 41.Persson, B., J. S. Zigler, Jr., and H. Jornvall. 1994. A super-family of medium-chain dehydrogenases/reductases (MDR) sub-lines including ζ-crystallin, alcohol and polyol dehydrogenases, quinone oxidoreductases, enoyl reductases, VAT-1 and other proteins. Eur. J. Biochem. 22615-22. [DOI] [PubMed] [Google Scholar]
- 42.Rippka, R., J. Deruelles, J. B. Waterbury, M. Herman, and R. Y. Stanier. 1979. Genetics assignments, strain histories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 1111-61. [Google Scholar]
- 43.Rittmann, B. E., R. Krajmalnik-Brown, and R. U. Halden. 2008. Pre-genomic, genomic and post-genomic study of microbial communities involved in bioenergy. Nat. Rev. Microbiol. 6604-612. [DOI] [PubMed] [Google Scholar]
- 44.Riveros-Rosas, H., A. Julian-Sanchez, R. Villalobos-Molina, J. P. Pardo, and E. Pina. 2003. Diversity, taxonomy and evolution of medium-chain dehydrogenase/reductase superfamily. Eur. J. Biochem. 2703309-3334. [DOI] [PubMed] [Google Scholar]
- 45.Rumeau, D., G. Peltier, and L. Cournac. 2007. Chlororespiration and cyclic electron flow around PSI during photosynthesis and plant stress response. Plant Cell Environ. 301041-1051. [DOI] [PubMed] [Google Scholar]
- 46.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 47.Schuster, M., R. E. Silversmith, and R. B. Bourret. 2001. Conformational coupling in the chemotaxis response regulator CheY. Proc. Natl. Acad. Sci. USA 986003-6008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shoumskaya, M. A., K. Paithoonrangsarid, Y. Kanesaki, D. A. Los, V. V. Zinchenko, M. Tanticharoen, I. Suzuki, and N. Murata. 2005. Identical Hik-Rre systems are involved in perception and transduction of salt signals and hyperosmotic signals but regulate the expression of individual genes to different extents in Synechocystis. J. Biol. Chem. 28021531-21538. [DOI] [PubMed] [Google Scholar]
- 49.Skulachev, V. P. 1994. Bioenergetics: the evolution of molecular mechanisms and the development of bioenergetic concepts. Antonie van Leeuwenhoek 65271-284. [DOI] [PubMed] [Google Scholar]
- 50.Stal, L. J., and R. Moezelaar. 1997. Fermentation in cyanobacteria. FEMS Microbiol. Rev. 21179-211. [Google Scholar]
- 51.Stock, A. M., and J. Guhaniyogi. 2006. A new perspective on response regulator activation. J. Bacteriol. 1887328-7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sun, H. W., and B. V. Plapp. 1992. Progressive sequence alignment and molecular evolution of the Zn-containing alcohol dehydrogenase family. J. Mol. Evol. 34522-535. [DOI] [PubMed] [Google Scholar]
- 53.Suzuki, I., Y. Kanesaki, H. Hayashi, J. J. Hall, W. J. Simon, A. R. Slabas, and N. Murata. 2005. The histidine kinase Hik34 is involved in thermotolerance by regulating the expression of heat shock genes in Synechocystis. Plant Physiol. 1381409-1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Suzuki, I., Y. Kanesaki, K. Mikami, M. Kanehisa, and N. Murata. 2001. Cold-regulated genes under control of the cold sensor Hik33 in Synechocystis. Mol. Microbiol. 40235-244. [DOI] [PubMed] [Google Scholar]
- 55.Suzuki, I., D. A. Los, Y. Kanesaki, K. Mikami, and N. Murata. 2000. The pathway for perception and transduction of low-temperature signals in Synechocystis. EMBO J. 191327-1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tadege, M., I. I. Dupuis, and C. Kuhlemeier. 1999. Ethanolic fermentation: new functions for an old pathway. Trends Plant Sci. 4320-325. [DOI] [PubMed] [Google Scholar]
- 57.Tani, A., Y. Sakai, T. Ishige, and N. Kato. 2000. Thermostable NADP+-dependent medium-chain alcohol dehydrogenase from Acinetobacter sp. strain M-1: purification and characterization and gene expression in Escherichia coli. Appl. Environ. Microbiol. 665231-5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van Thor, J. J., R. Jeanjean, M. Havaux, K. A. Sjollema, F. Joset, K. J. Hellingwerf, and H. C. Matthijs. 2000. Salt shock-inducible photosystem I cyclic electron transfer in Synechocystis PCC6803 relies on binding of ferredoxin:NADP(+) reductase to the thylakoid membranes via its CpcD phycobilisome-linker homologous N-terminal domain. Biochim. Biophys. Acta 1457129-144. [DOI] [PubMed] [Google Scholar]
- 59.Vioque, A. 1992. Analysis of the gene encoding the RNA subunit of ribonuclease P from cyanobacteria. Nucleic Acids Res. 206331-6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Volkman, B. F., D. Lipson, D. E. Wemmer, and D. Kern. 2001. Two-state allosteric behavior in a single-domain signaling protein. Science 2912429-2433. [DOI] [PubMed] [Google Scholar]
- 61.West, A. H., and A. M. Stock. 2001. Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem. Sci. 26369-376. [DOI] [PubMed] [Google Scholar]
- 62.Wilkin, J. M., K. Soetaert, M. Stelandre, P. Buyssens, G. Castillo, V. Demoulin, G. Bottu, M. A. Laneelle, M. Daffe, and J. De Bruyn. 1999. Overexpression, purification and characterization of Mycobacterium bovis BCG alcohol dehydrogenase. Eur. J. Biochem. 262299-307. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.