Abstract
The replication-associated protein (Rep) of geminiviruses, single-stranded DNA viruses of higher plants, is essential for virus replication. Since these viruses do not encode their own polymerases, Rep induces differentiated plant cells to reenter the cell cycle by interacting with the plant homologues of retinoblastoma proteins in order to activate the host DNA synthesis machinery. We have used fission yeast (Schizosaccharomyces pombe) as a model organism to analyze the impact of ectopically expressed African cassava mosaic virus Rep protein on the cell division cycle in closer detail. Upon expression, Rep showed its characteristic DNA cleavage activity, and about 10% of the cells exhibited morphological changes. They were elongated threefold, on average, and possessed a single but enlarged and less compact nucleus in comparison to noninduced or vector-only control cells. Flow cytometry of Rep-expressing cultures revealed a distinct subpopulation of Rep protein-containing cells with aberrant morphology. The other 90% of the cells were indistinguishable from control cells, and no Rep was detectable. Rep-expressing cells exhibited DNA contents beyond 2C, indicating ongoing replication without intervening mitosis. Because a second open reading frame (ORF), AC4, is present within the Rep gene, the role of AC4 was examined by destroying its start codon within the AC1 ORF. The results confirmed that Rep is necessary and sufficient to induce rereplication in fission yeast. The unique potential of this well-investigated model for dissecting the cell cycle control by geminiviral proteins is discussed.
Viruses of the Geminiviridae family are characterized by their unique geminate capsids formed by two incomplete icosahedra (9, 85). Geminiviruses infect a variety of plants and cause significant losses of economically important crops worldwide (52). The genome of geminiviruses consists of either one or two circular single-stranded DNA molecules. African cassava mosaic virus (ACMV) belongs to the genus Begomovirus (73), one of the four genera of the geminivirus family, and is transmitted by the whitefly Bemisia tabaci (Gennadius). ACMV has two genomic components, which are designated DNA A and DNA B (74). The DNA A component codes for six proteins, and the DNA B component codes for two proteins (19, 20, 76-78). The open reading frames (ORFs) for a replication-associated protein (Rep, or AC1) and a replication enhancer protein (REn, or AC3) are located on the complementary strand of DNA A. Although both proteins are involved in viral replication (20, 78), only Rep is indispensable (16, 32).
Geminivirus replication occurs via double-stranded DNA intermediates in the nucleus of infected cells, and different modes of replication have been described (30, 39-41). Viral replication relies extensively on host cell factors (33) since geminiviruses do not encode a polymerase. During rolling circle replication, Rep cleaves the virion sense strand within the nonanucleotide sequence conserved among geminiviruses (71), binds covalently to the 5′ end, and joins it to the 3′ end after one round of replication. Besides nicking and closing activity (47), Rep possesses ATPase (17) and helicase activities (14, 15) as well as DNA binding properties (23). Additionally, domains involved in interaction with itself and other viral proteins have been mapped (59, 68).
Geminiviruses do not enter meristems (36, 49) and therefore face the task of activating the cell cycle of differentiated plant cells (56) either in the phloem or the mesophyll, depending on the virus species (for a review, see reference 82). Like animal tumor viruses (21), geminiviruses solve this problem by removing a cell cycle arrest and thereby inducing the host replication machinery (31). Wheat dwarf virus RepA protein harbors the canonical LXCXE motif that is essential for its binding to the plant homologue of retinoblastoma protein (pRBR), a key regulator protein for the transition from G1 to S phase in animal cells (28, 83, 84). Although the LXCXE motif is lacking in other geminiviruses, for example, Tomato golden mosaic virus (TGMV), Rep is nevertheless able to bind maize pRBR (2). Kong et al. (43) identified the pRBR binding domain of TGMV Rep between amino acids 101 and 180, a region including domains involved in DNA cleavage/ligation, DNA binding, and Rep oligomerization. Within this region, side chains of the predicted helix 4 have been shown to be involved in efficient pRBR binding by yeast two-hybrid assays (4). By modifying the plant cell cycle via Rep, E2F-family transcription factors are regulated, which in turn activate transcription of S-phase-specific genes, thus providing a favorable environment for viral replication (30, 31, 33).
In differentiated cells, TGMV Rep caused the accumulation of proliferating cell nuclear antigen (PCNA) (56), a versatile processivity factor for DNA polymerases during replication and repair (60, 63, 81). PCNA is able to recruit Rep and REn, as shown for tomato yellow leaf curl Sardinia virus (TYLCSV) by yeast two-hybrid assays (12).
Schizosaccharomyces pombe has been exploited over the last decades as a valuable model organism for cell cycle studies. Many processes and proteins involved in cell cycle regulation in fission yeast resemble those in higher eukaryotes. Often proteins of S. pombe share a higher degree of identity with homologues of higher eukaryotes than those of Saccharomyces cerevisiae. For example, fission yeast PCNA, as one of the geminivirus Rep protein-interacting partners, shares 52% overall amino acid identity with human PCNA and 62% with rice PCNA, whereas the S. cerevisiae PCNA is only 35% identical with the human protein (80). On account of these advantages, we have chosen fission yeast to analyze the impact of ACMV Rep on cell division and DNA content.
MATERIALS AND METHODS
Construction of plasmids.
DNA A of ACMV-[NG], a Nigerian isolate of ACMV, was released from plasmid pUC19-APA9 (kindly provided by Rob Briddon, Faisalabad, Pakistan) with SphI and religated. The AC1 ORF was amplified by PCR (94°C for 3 min, followed by 25 cycles of 94°C for 30 s, 58°C for 30 s, and 72°C for 1 min, with a final step of 72°C for 10 min) using a proofreading DNA polymerase (Qiagen, Hilden, Germany), oligonucleotides 1 and 2 (Table 1) and DNA A as the template. The PCR fragment was introduced into the SmaI-linearized vector pREP2 (ura4+) (kindly provided by Henning Schmidt, TU Braunschweig, Germany) by the BD In-Fusion method (BD In-Fusion Dry-Down PCR Cloning Kit; BD Biosciences) to obtain the plasmid pREP2 expressing AC1 (pREP2:AC1). Correctness of the construct was confirmed by sequencing.
TABLE 1.
Oligonucleotides used for cloning of viral ORFs into fission yeast expression plasmids
| Oligonucleotide no. | Name | Sequence (5′→3′)a |
|---|---|---|
| 1 | AC1-for | GACTCTAGAGGATCCCCGGGATGCGAACTCCGCGTTTTAGA |
| 2 | AC1-rev | GGGAGACATTCCTTTTACCCGGGCTACGCCGGATGGCTCGCTT |
| 3 | AC1mAC4-for | CCAAAGTGTTCTATACCCAAAGAACAC |
| 4 | AC1mAC4-rev | GTATGTGAGAAAGACGTTCTTGGCTTGAACTC |
| 5 | ACMV-12 | TAATATT^ACCGG |
Start and stop codons of the viral ORFs are shown in boldface, and the nucleotide changed for construction pREP2:AC1mAC4 is underlined. In ACMV-12, the caret indicates the position of Rep protein cleavage.
Compared to the database entry (GenBank accession number AJ427910), pUC19-APA9 shows five nucleotide substitutions: nucleotide 80 from A to T (resulting in amino acid substitution Q27L; nucleotide and amino acid numbering is according to AC1 sequence), nucleotides 407 and 408 from GC to CG (G136A), nucleotide 662 from C to A (A221D), and nucleotide 960 from A to G (silent substitution). All three amino acid substitutions found here are conserved in AC1 proteins of other ACMV strains. Additionally, DNA A (released from pCU19-APA9) together with DNA B (released from pUC19-AP3, kindly provided by Rob Briddon, Faisalabad, Pakistan) was infectious on Nicotiana benthamiana plants (K. Kittelmann and H. Jeske, unpublished results).
The plasmid pREP2:AC1mAC4 was derived from pREP2:AC1 by altering the ATG of the AC4 ORF to ACG by mutagenesis. pREP2:AC1 was amplified by PCR (95°C for 5 min, followed by 25 cycles of 95°C for 30 s, 69°C for 45 s, and 72°C for 20 min, with a final step at 72°C for 10 min) using a proofreading DNA polymerase (Fermentas, St. Leon-Roth, Germany) and oligonucleotides 3 and 4 (Table 1). The PCR fragment was gel purified and cloned, and the correctness of the construct was confirmed by sequencing.
Protein expression in S. pombe.
Plasmids pREP2, pREP2:AC1, and pREP2:AC1mAC4 were introduced into S. pombe HE621 (h−S leu1-32 ura4-D18) (kindly provided by Henning Schmidt, TU Braunschweig, Germany) by a lithium acetate-polyethylene glycol method of transformation according to the protocol of Clontech Matchmaker Two-Hybrid System 2 (catalogue number PT1030-1; Clontech, Saint-Germain-en-Laye, France). Small-scale induction of protein expression was carried out as described in Stratagene's ESP Yeast Protein Expression and Purification System and ESP Yeast Protein Expression Vectors instruction manual (75). Briefly, single transformant colonies were incubated overnight in YES medium (yeast extracts with supplements) at 30°C and 250 rpm, and 10 ml of YES medium was inoculated with cells from the overnight culture to an optical density at 600 nm of 0.2 to 0.4 and incubated for approximately 5 h to an optical density at 600 nm of 0.7 to 1.0 at 30°C and 250 rpm. The culture was split into two halves, harvested (by centrifugation for 5 min at room temperature at 1,000 × g), and washed twice with water. Cells were either resuspended in 10 ml of EMM+L (Edinburgh minimal medium supplemented with 0.01% leucine) to induce protein expression or in 10 ml of EMM+L+T (EMM+L medium supplemented with 5 μM thiamine) medium to repress it and incubated at 30°C and 250 rpm. Aliquots of cultures were withdrawn at different time points after induction and further processed for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), microscopy, and fluorescence-activated cell sorting analysis.
Preparation of cell extracts, SDS-PAGE, and Western blotting.
Cell pellets of 1-ml cultures were resuspended in 100 μl of ice cold PBST-PI buffer (phosphate-buffered saline [PBS] containing 1% Trition X-100, 1 mM phenylmethylsulfonyl fluoride, 1 μg/ml chymostatin, 100 μM leupeptin, 5 μg/ml antipain, 1 μM pepstatin A, 3.4 μg/ml bestain). Cells were broken with glass beads using a Fast Prep 24 device (MP Biomedicals, Illkirch, France) (10 cycles at 6 m/s and 15 s with intermittent cooling on ice). Cell extracts were separated into a supernatant and a pellet by centrifugation (5 min at 12,000 × g and 4°C). The pellet was resuspended in the same volume of PBST-PI buffer as the volume of the corresponding supernatant.
Protein samples were subjected to 10% SDS-PAGE according to Laemmli (44). Gels were blotted semidry (8) on nitrocellulose membranes (Protran nitrocellulose transfer membrane; Whatman Schleicher & Schuell, Dassel, Germany). Rep protein was detected using a polyclonal rabbit antiserum raised against TYLCSV Rep protein (11), alkaline phosphatase-conjugated goat anti-rabbit antibodies (Rockland Immunochemicals, Inc., Gilbertsville, PA), and nitroblue tetrazolium choride-5-bromo-4-choro-3′-indolylphosphate.
Clonal analysis of transformed cells.
S. pombe HE621 cells were transformed with the constructs as described above. For each construct, half of the transformed cells were plated on EMM+L medium for induction of protein expression, and the other half was plated on EMM+L+T medium to repress expression. Agar plates were incubated at 30°C, and the appearance of colonies was followed each day. No differences were detected by visual inspection. Nine days after transformation, the numbers of colonies were counted (Table 2). Light microscopical analysis of different colonies that were induced for AC1 protein expression (pREP2:AC1 and pREP2:AC1mAC4 on EMM+L agar plates) always revealed a limited number of phenotypically elongated cells (1 in 1,000 to 10,000 cells), whereas repressed as well as pREP2 (empty vector)- or pREP2:AC4-transformed colonies, irrespective of whether they were repressed or induced, were free of them.
TABLE 2.
Colony numbers for cells transformed with the indicated constructs on selective plates that induce or repress the target protein expression
| Construct | Inductiona | No. of colonies (%)b |
|---|---|---|
| pREP2 (empty vector): | + | 1,235 (111) |
| − | 1,111 (100) | |
| pREP2:AC1: | + | 1,862 (88) |
| − | 2,128 (100) | |
| pREP2:AC1mAC4: | + | 1,117 (83) |
| − | 1,346 (100) |
+, induction of protein expression; −, repression of protein expression.
For each construct, colony numbers on plates containing thiamine (protein expression repressed) were set to 100%.
For each construct (pREP2, pREP2:AC1, and pREP2:ACmAC4) and agar plate variant (EMM+L and EMM+L+T media), five colonies were picked and reincubated on EMM+L agar plates for induction of protein expression for 3 days at 30°C. Equal amounts of cells were scraped off the plates, resuspended in 80 μl of sample buffer, and disrupted after the addition of glass beads (45 mg) by two cycles of boiling at 95°C for 5 min and vortexing for 1 min. Four out of five colonies for pREP2:AC1 and three out of five colonies for pREP2:AC1mAC4 revealed the same trend of reduced Rep expression upon induction, as analyzed by Western blotting, whereas the others exhibited no differences in their expression levels.
In vitro cleavage assay.
Cell extract supernatants (250 to 500 μg of total protein) of cells harboring either pREP2, pREP2:AC1, or pREP2:AC1mAC4 were incubated in a total volume of 10 μl with or without 100 pmol of oligonucleotide 5 (Table 1) for 30 min at 37°C in cleavage buffer (25 mM Tris-HCl, pH 7.5, 75 mM NaCl, 5 mM MgCl2, 2.5 mM dithiothreitol, 0.5 mM EDTA) according to Laufs et al. (47). The reaction was stopped by the addition of 10 μl of 2× SDS gel loading buffer (100 mM Tris-HCl, pH 6.8, 4% SDS, 20% glycerin, 0.2% bromophenol blue, 200 mM dithiothreitol). Samples were heated to 95°C for 5 min and analyzed by SDS-PAGE and Western blotting.
Microscopy.
Aliquots of 100 μl of growing cultures at different time points were harvested (by centrifugation for 5 min at room temperature at 1,000 × g) and fixed by adding 100 μl of 70% ethanol. Cells were harvested as before and rehydrated in 20 μl of 50 mM sodium citrate, pH 7.0. Five microliters was applied to poly-l-lysine (Sigma, Taufkirchen, Germany)-coated glass slides and stained with 3 to 4 μl of 4′,6 diamidino-2-phenylindole (DAPI; 1 μg/ml in H2O). Samples were analyzed with an Axiovert 200 M microscope (Zeiss, Oberkochen, Germany). Differential interference contrast as well as DAPI fluorescence (filter set 49, catalog number 488049-0000-000; Zeiss, Oberkochen, Germany) images were taken with an AxioCam MR3 camera (Zeiss, Oberkochen, Germany) and processed with Axiovision software (version 4.6.3; Zeiss, Oberkochen, Germany).
Flow cytometry.
For flow cytometry, 1 to 2 ml of growing cell cultures was harvested at different time point (by centrifugation for 5 min at room temperature at 1,000 × g), washed once with 1 ml of water, pelleted as before, fixed in 1 ml of 70% ethanol, and stored at 4°C until further processing. A total of 1 × 107 to 3 × 107 cells were harvested as before, washed once in 1 ml of 50 mM sodium citrate, pH 7.0, pelleted, and treated with 0.1 mg/ml RNase A in 500 μl of 50 mM sodium citrate, pH 7.0, for at least 3 h at 37°C. Propidium iodide (Sigma, Taufkirchen, Germany) in 500 μl of 50 mM sodium citrate, pH 7.0, was added to a final concentration of 4 μg/ml. Cells were analyzed in a Cytomics FC500 (Beckman Coulter, Krefeld, Germany) after sonification for 45 s. Data were processed with WinMDI software (version 2.9; http://facs.scripps.edu/software.html). For sorting, cells were processed as described for flow cytometry. Sorting of 300,000 to 1,000,000 ethanol-fixed cells was performed in a FACS Vantage SE instrument (Becton Dickinson, Heidelberg, Germany). Sorted cells were harvested (by centrifugation for 5 min at room temperature at 2,700 × g) and subjected to SDS-PAGE after boiling in 15 μl of 1× cracking buffer (8 M urea, 5% SDS, 40 mM Tris-HCl, pH 6.8, 0.1 mM EDTA, 0.2% bromophenol blue) at 95°C for 7 min.
RESULTS
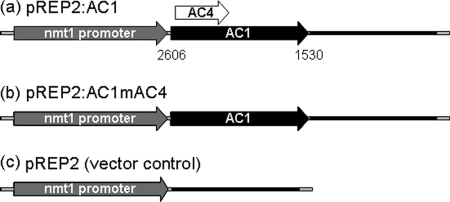
In order to analyze the effects of ACMV Rep protein in fission yeast, its ORF (AC1) was expressed under the control of the nmt1 promoter (50). Although the annotation for the viral ORFs of this ACMV strain (GenBank accession number AJ427910) implies that the start codon for AC4 is upstream of the start codon for AC1, comparison with the type strain of ACMV (27, 66) revealed a start codon for AC4 that is located within the AC1 ORF. In order to exclude any possible translation of AC4 from the nmt1 promoter in the plasmid pREP2:AC1 (Fig. 1a), a modified construct pREP2:AC1mAC4 (Fig. 1b) was generated in which the AC4 start codon was changed to ACG, a change which does not affect the amino acid sequence of Rep.
FIG. 1.
Schematic representation of expression constructs used in this study. The AC1 ORF (black arrow) includes the AC4 ORF (white arrow) in a different frame of the plasmid pREP2:AC1 (a). The AC4 start codon was destroyed in the plasmid pREP2:AC1mAC4 (b), preventing a potential expression of AC4 from this construct. As a control, the vector pREP2 (c) was used. First and last nucleotides of the AC1 ORF according to GenBank accession number AJ427910 are given below the arrow in panel a. Gray arrows and black bars denote the nmt1 promoter and nmt1 terminator, respectively. All features are drawn to scale. The backbone of the vector is not shown.
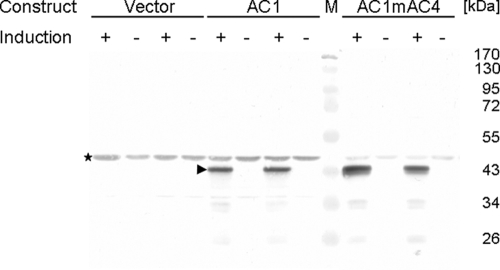
Under inducing conditions (medium lacking thiamine), Rep was detected in cell extract supernatants (Fig. 2) as well as in the corresponding pellets (data not shown) of cells harboring plasmids pREP2:AC1 or pREP2:AC1mAC4. Although Rep protein was already present in small amounts in cells analyzed at 6.5 h postinduction (hpi), full expression was obtained at 9 hpi and remained over the time period analyzed (up to 18 hpi) (data not shown). No AC1 expression was observed in cells grown under repressive conditions or in control cells harboring plasmid pREP2 (empty vector), irrespective of the growth conditions (Fig. 2).
FIG. 2.
Western blot analysis of AC1 protein expression. Cell extract supernatants were prepared 12 h after cells were shifted to minimal medium without (+ induction) or with (− induction) thiamine. For each of the constructs pREP2 (Vector), pREP2:AC1 (AC1), and pREP2:AC1mAC4 (AC1mAC4), the proteins of two independent clones are shown. The position of the AC1 protein is marked by an arrowhead; the asterisk indicates the position of a cross-reacting S. pombe protein. Marker proteins (M) with their molecular masses (kDa) are shown.
In order to examine the activity of ectopically expressed Rep, an in vitro cleavage assay was applied using an oligonucleotide comprising the conserved nonanucleotide sequence of geminiviruses. If Rep cleaves this substrate, an adduct containing the protein bound to the 5′ end of the cleaved oligonucleotide is expected (46). Extracts of cells expressing Rep from plasmid pREP2:AC1 or pREP2:AC1mAC4 as well as control cells harboring pREP2 were incubated with oligonucleotide 5 (Table 1). Reaction products were separated by SDS-PAGE and analyzed by Western blotting (11). In addition to the specific protein band present in all extracts of cells expressing Rep (Fig. 3), an additional band with higher molecular mass was observed after incubation with the oligonucleotide (Fig. 3). The observed shift matched well the expected size for the protein-DNA adduct (+1.5 kDa). Although adduct formation increased with increasing amounts of oligonucleotide (data not shown), complete conversion was not observed, possibly due to the reversal of the adduct formation by the nucleophilic attack of the phosphotyrosyl bond by the 3′ OH of the Rep-bound 5′ part of the substrate oligonucleotide (47).
FIG. 3.
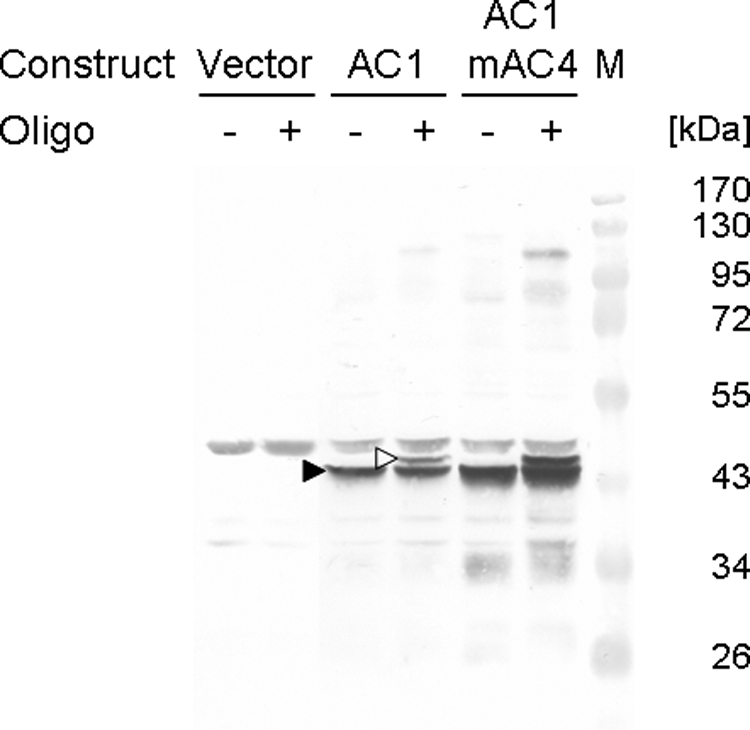
Western blot analysis of the reaction products after in vitro cleavage. Extracts of cells with either plasmid pREP2 (Vector), pREP2:AC1 (AC1), or pREP2:AC1mAC4 (AC1mAC4) were prepared 12 h after induction of protein expression and incubated with (+) or without (−) oligonucleotide 5 (Table 1) comprising a cleavage substrate for the Rep protein. Arrowheads mark the positions of AC1 protein (filled) and generated adduct (open). Marker proteins (M) with their molecular masses (kDa) are shown.
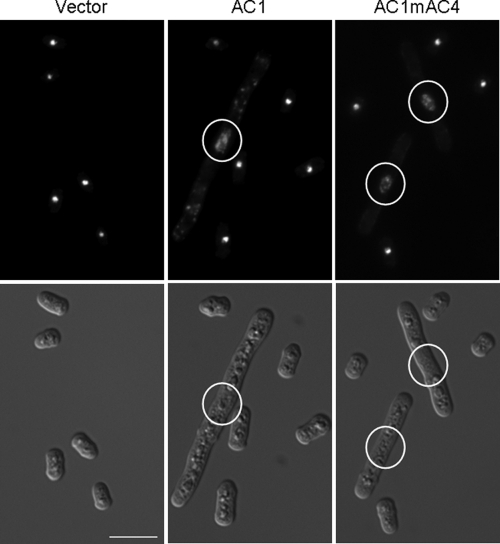
Cells harboring the different constructs were further analyzed by microscopy under inducing (Fig. 4) or noninducing (data not shown) conditions. At 18 h after the shift to minimal medium, cells harboring plasmid pREP2 appeared similar in cultures grown in medium with or without thiamine (Fig. 4). The same morphology of cells was observed for repressed cultures harboring pREP2:AC1 or pREP2:AC1mAC4 (data not shown). In contrast, induced pREP2:AC1 or pREP2:AC1mAC4 cultures displayed a conspicuous phenotype for some of the cells (Fig. 4). About 10% of the cells were elongated up to seven times the normal cell length. On average, these elongated cells had a mean length of 23 ± 6 μm, whereas repressed cells measured 8 ± 1 μm. DAPI staining of the cells revealed a single nucleus in elongated cells, ruling out the possibility that these cells have undergone normal cell division with incomplete separation of the daughter cells. In contrast to the sharp and round appearance of nuclei of normal cells, these nuclei were enlarged and less compact (Fig. 4).
FIG. 4.
Influence of Rep protein on cell morphology. Induced yeast cells carrying either plasmid pREP2, pREP2:AC1, or pREP2:AC1mAC4 were analyzed by light microscopy. Cells were fixed and stained with DAPI. DAPI images (upper row) show the same area as differential interference contrast images (lower row). Enlarged nuclei of elongated cells are encircled. Bar, 10 μm.
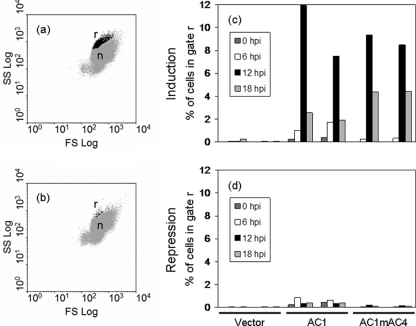
On account of the altered morphology in Rep-expressing cultures, we analyzed cells harboring either pREP2, pREP2:AC1, or pREP2:AC1mAC4 by flow cytometry. Compared to control samples with vector-containing yeasts, cultures induced for AC1 or AC1mAC4 expression displayed an additional distinct cell population in forward and side scatter plots (Fig. 5a) that appeared at time points when AC1 was detectable on Western blots. In order to quantify this population, the cells were gated, and the number of gated cells was quantified as a percentage of the total number of cells (Fig. 5c and d). At 6 hpi, few pREP2:AC1- or pREP2:AC1mAC4-carrying cells were located within this gate (Fig. 5c), and only small amounts of Rep protein were detectable on Western blots (data not shown). This result corresponds to the general finding that full derepression of the nmt1 promoter is achieved after 12 h in thiamine-free medium (50). At this time point, 8 to 12% of pREP2:AC1- or pREP2:AC1mAC4-harboring cells were counted in the gate (Fig. 5c). Thereafter, the percentage of gated cells dropped again to below 5% (Fig. 5c, 18 hpi). Expressing cells with altered morphology had probably stopped division and were overgrown by nonexpressing cells. Induced control cultures harboring pREP2 revealed less than 0.25% of the cells in the gate at any time point analyzed. Similarly, the percentages of gated cells were low for noninduced cultures (Fig. 5d), with less than 0.1% for pREP2-harboring cells and 0.9% for pREP2:AC1- or pREP2:AC1mAC4-carrying cells. A slightly higher number of gated counts for AC1 protein-expressing cells was expected due to the known leakiness of the nmt1 promoter.
FIG. 5.
Flow cytometric analysis of cells expressing Rep protein. Induced (a) and noninduced (b) cells harboring plasmid pREP2:AC1 were fixed at 12 hpi, stained with propidium iodide, and analyzed by flow cytometry. A distinct subpopulation of cells (black dots; r) with increased forward scatter (FS Log) and side scatter (SS Log) properties compared to normal cells (gray dots; n) was gated for further analysis. Induced (c) and noninduced (d) cells harboring either empty plasmid (Vector), pREP2:AC1 (AC1), or pREP2:AC1mAC4 (AC1mAC4) were analyzed at different time points. Aliquots of cells were withdrawn after 0, 6, 12, and 18 h; treated as described before; and analyzed by flow cytometry. Cells of the subpopulation present in cultures expressing the Rep protein (r) were gated to determine the fraction of cells in this gate for the different cultures. For each construct, the results of two independent clones are shown.
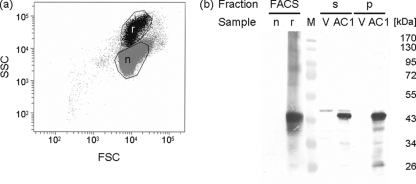
The light microscopy and flow cytometry results let us reason that only a subpopulation of the cells actually expressed Rep protein and that these cells were elongated with altered nuclei. In order to test this assumption, cells of the Rep-expressing subpopulation (subpopulation r) (Fig. 6a) and of the subpopulation of normal cell (subpopulation n) (Fig. 6a) from a culture induced for Rep protein expression were separated using fluorescence-activated cell sorting and analyzed by Western blotting (Fig. 6b). As expected, Rep protein was exclusively detected in subpopulation r (Fig. 6).
FIG. 6.
(a) AC1 protein is detected solely in cells forming a distinct subpopulation with altered properties in flow cytometry. Induced cells harboring pREP2:AC1 were fixed at 18 hpi. Two samples were obtained by sorting cells of the Rep-expressing subpopulation (black dots; r) and the normal population (gray dots; n) and analyzed by SDS-PAGE and Western blotting. (b) Cells of the same culture used for sorting were harvested at 18 hpi (AC1) as well as control cells with plasmid pREP2 (V). Cell lysates of these samples were separated in a supernatant (s) and a pellet (p) fraction by low-speed centrifugation and used as controls for presence or absence of Rep protein. Marker proteins (M) with their molecular masses (kDa) are shown. SSC, side scatter; FSC, forward scatter.
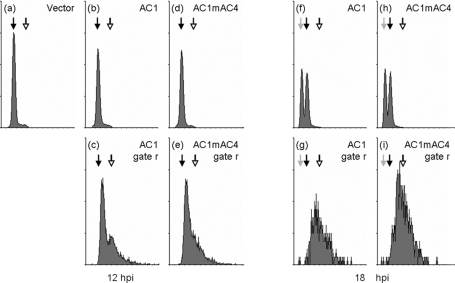
Normal and Rep-expressing cells were further investigated by fluorescence cytometry to quantify their DNA contents (Fig. 7). At 12 hpi (Fig. 7a to e), histograms revealed the typical appearance observed for fission yeasts in logarithmic growth phase, with a predominant peak for cells with 2C DNA content (Fig. 7a, b, and d) and a small, broad peak for binucleated cells during S phase (Fig. 7, DNA contents between 2C and 4C). There was no difference between the histograms of cells with either plasmid cultivated in thiamine-free medium if the normal cells were analyzed (Fig. 7a, b, and d). In contrast, cells of the Rep protein-expressing subpopulation r displayed additional DNA contents with higher values than 2C (Fig. 7c and e), reminiscent of rereplicating cells. Since the elongated Rep protein-expressing cells contained only one nucleus (Fig. 4), the higher DNA content indicates ongoing replication in single cells without subsequent cell division. At 18 h in minimal medium (Fig. 7f to i), the histograms showed two peaks for 1C and 2C DNA contents (Fig. 7), implying that cells had entered stationary phase and started starving. Again, no differences for normal cells (subpopulation n) with either plasmid were observed (Fig. 7f and h and data not shown), whereas Rep-expressing cells (subpopulation r) contained higher than 2C DNA contents (Fig. 7g and i). The mean values at 18 hpi were even higher than data at 12 hpi (Fig. 7c and e [12 h] and g and i [18 h]), with DNA contents of about 4C at the later time point. In order to quantify the changes, the histogram peaks of one sample were subdivided into three classes with DNA contents of either 1C, 2C, or >2C. Table 3 shows representative data for one set of constructs, and Table 4 shows the corresponding numbers of events for the different DNA contents. Whereas the majority of normal cells (78%) had a DNA content of 2C at 12 hpi, only smaller proportions of cells (6.7% and 16%, respectively) had DNA contents of 1C and beyond 2C. At 18 hpi, most cells had either a 1C (39%) or 2C (51%) DNA content. Rep protein-expressing cells revealed a shift toward higher DNA contents that increased over time. At 12 hpi, 43% displayed 2C DNA content; the rest of the subpopulation had even more DNA. Interestingly, more than 90% of the Rep-expressing cells accumulated more than 2C DNA content at 18 hpi. Almost no cells with 1C DNA content were detected in the subpopulation.
FIG. 7.
DNA contents of cells expressing Rep protein. Cells transformed with either plasmid pREP2 (Vector; a), pREP2:AC1 (AC1; b and f), or pREP2:AC1mAC4 (AC1mAC4; d and h) were grown under inducing conditions for protein expression. Cells were fixed, stained with propidium iodide, and measured for DNA contents by flow cytometry at 12 hpi (a to e) and 18 hpi (f to i). The subpopulation of cells present in cultures expressing the Rep protein, as shown in Fig. 5 (black dots), was analyzed separately (c and g, AC1 gate r; e and i, AC1mAC4 gate r). Gray, black, and open arrows in the histograms indicate 1C, 2C, and 4C DNA contents, respectively. The x axis shows increasing DNA contents as measured by fluorescence. The y axis is scaled to 4,096 counts in a, b, c, e, g, and i; to 128 counts in d and f; and to 16 counts in h and j. The experiment was repeated three times with similar results (data not shown).
TABLE 3.
Cell cycle analysis by flow cytometry of cells harboring the indicated plasmids at 12 and 18 hpi
| Cell population (gate) and plasmid | DNA content (%) at:a
|
|||||
|---|---|---|---|---|---|---|
| 12 hpi
|
18 hpi
|
|||||
| 1C | 2C | >2C | 1C | 2C | >2C | |
| Normal (n) | ||||||
| pREP2 | 3 | 82 | 15 | 36 | 52 | 12 |
| pREP2:AC1 | 4 | 79 | 18 | 41 | 50 | 9 |
| pREP2:AC1mAC4 | 6 | 80 | 14 | 43 | 49 | 9 |
| Avg | 4 | 80 | 16 | 40 | 50 | 10 |
| Rep-expressing (r) | ||||||
| pREP2:AC1 | 0 | 45 | 56 | 1 | 6 | 93 |
| pREP2:AC1mAC4 | 0 | 42 | 58 | 1 | 7 | 92 |
| Avg | 0 | 44 | 57 | 1 | 7 | 93 |
For cells with plasmids pREP2:AC1 and pREP2:AC1mAC4, the subpopulation was evaluated separately. Values for DNA content are rounded. The corresponding numbers of events are given in Table 4. Avg, arithmetic mean of DNA content for the group of cells.
TABLE 4.
Cell cycle analysis by flow cytometry of cells harboring the indicated plasmids at 12 and 18 hpi
| Cell population (gate) and plasmid | No. of events for the indicated DNA content at:a
|
|||||
|---|---|---|---|---|---|---|
| 12 hpi
|
18 hpi
|
|||||
| 1C | 2C | >2C | 1C | 2C | >2C | |
| Normal (n) | ||||||
| pREP2 | 2,792 | 77,606 | 14,232 | 34,311 | 50,043 | 11,157 |
| pREP2:AC1 | 3,160 | 64,557 | 14,574 | 38,570 | 47,309 | 8,701 |
| pREP2:AC1mAC4 | 5,173 | 64,722 | 11,435 | 39,489 | 45,384 | 8,108 |
| Rep-expressing (r) | ||||||
| pREP2:AC1 | 1 | 3,446 | 4,303 | 4 | 33 | 523 |
| pREP2:AC1mAC4 | 4 | 3,445 | 4,751 | 6 | 73 | 931 |
For cells harboring plasmids pREP2:AC1 and pREP2:AC1mAC4, the subpopulation was evaluated separately.
In summary, the results show that the geminiviral Rep protein is able to induce an accumulation of DNA to a certain extent in S. pombe, which is accompanied by morphological changes of the cells because cell division is probably blocked.
DISCUSSION
Several studies have described functions of geminiviral Rep protein in modifying the cell cycle of plant cells upon infection (reviewed in references 31 and 33). Nevertheless, open questions remain concerning the details of geminiviral replication such as the exact mechanism of Rep interference with the cell cycle. As S. pombe is a well-established model organism for cell cycle studies (24), we chose this organism to investigate the influence of a begomoviral Rep protein, controlling its expression by induction in thiamine-free medium (50).
The Rep protein expressed in S. pombe was able to specifically cleave an oligonucleotide comprising the conserved nonanucleotide sequence, indicating that its DNA cleavage and nucleotidyl transfer activities were not impaired. Remarkably, it was possible to show the cleavage activity of Rep directly in the soluble protein fraction without purification. Moreover, a hallmark of Rep function, interference with host cell cycle control, was probably mimicked in fission yeast. Upon Rep expression, about 10% of cells in the culture displayed a completely altered phenotype, with cell elongation up to sevenfold for single cells and enlarged and less compact nuclei than in noninduced or control cells. After transformation, cells were selected for the presence of the plasmid by complementation of a Ura deficiency, and cells in the analyzed cultures induced or repressed for protein expression were clonal. Therefore, it seems likely that a functional Rep protein is deleterious for the cell. In fact, if transformed cells are directly grown under inducing conditions, only very few cells displayed the elongation phenotype (Table 2). Given that colonies on the transformation plates are visible only after 5 or more days of growth, cells that do not express the Rep protein will probably overgrow nonexpressing cells, as was also observed in the time course analysis in liquid cultures (Fig. 5). Furthermore, if colonies (i.e., populations) of cells induced for Rep protein expression for 9 days directly after transformation were restreaked under inducing conditions for three more days, they tended to show a lower level of Rep protein expression than populations of corresponding cells repressed directly after transformation and restreaked under inducing conditions (data not shown). This indicates that the longer the cells are induced for Rep protein expression, the larger becomes the fraction of cells with a more or less “normal” morphology. This may reflect the downregulation of Rep expression by whatever mechanism or occurrence of mutations counteracting the deleterious effects on the cell of an active Rep protein.
It is a general experimental experience that only a subpopulation of cells in transformed S. pombe colonies express the ectopic target proteins (1, 3, 25, 26). This reduction is independent of the particular protein and may rely on genetic as well as epigenetic variations. Under similar laboratory conditions as described in this paper, e.g., only 26 of 131 (19.8%) transformed cells showed green fluorescent protein (A. Kadri, unpublished data).
To exclude any influence of the AC4 protein, the ORF of which overlaps AC1 and could potentially be expressed from pREP2:AC1, a plasmid was constructed in which the start codon of AC4 was mutated (pREP2:AC1mAC4). Introduction of pREP2:AC1mAC4 into the cells resulted in the same phenotype as observed for pREP2:AC1, indicating that the AC1-encoded Rep protein is responsible for the changes in cell shape and nucleus appearance. For the type strain of ACMV (ACMV-[KE], a Kenyan isolate), no definite function has been assigned to the AC4 protein, and disruption of its ORF within the viral context did not change infectivity or symptom development (20, 35). Likewise, the equivalent of AC4 in TGMV (AL4) was not required for infection of several host plant species (18, 62), which is in contrast to the well-established role of C4 for monopartite geminiviruses like TYLCV and Beet curly top virus (42, 61, 65). Recent reports assigned a silencing suppression function to AC4 for some bipartite geminiviruses including the Cameroon strain of ACMV (22, 79), which may explain roles in cell-to-cell movement and pathogenicity.
The cell elongation observed in this study upon ACMV Rep expression resembles cdc (cell division cycle) phenotypes of fission yeasts, which occur when growth continues in the absence of cell division. They generally indicate a cell cycle block. In order to determine whether the ACMV Rep-induced block occurred at G1 or G2, cells were analyzed by flow cytometry. Cells that did not express Rep accumulated predominantly in G2, as is generally observed for fission yeast cells. This difference in cell cycle phase distribution from that which occurs in budding yeast is caused by an extremely short G1 phase compared to G2 for an asynchronous, exponentially growing population of S. pombe cells (53).
Interestingly, cells of the Rep-expressing subpopulation possessed increased DNA contents and did not accumulate discrete 1C or 2C DNA complements characteristic for a cell cycle block in G1 or G2, respectively. The elevated DNA levels cannot be attributed to mitochondrial DNA although we observed an increase in mitochondria in elongated cells by DAPI staining as well (Fig. 4, pREP2:AC1). Correspondingly, Sazer and Sherwood (67) showed no increase in DNA content as analyzed by flow cytometry if cells were stained with propidium iodide in a cdc mutant that replicated mitochondrial DNA in the absence of nuclear DNA synthesis. Thus, ACMV Rep seems to promote reinitiation of nuclear DNA replication during the fission yeast cell cycle. The histograms for DNA content of Rep-expressing cells resemble those of cells undergoing rereplication due to deregulation of factors controlling initiation of S phase (37, 53). This is in contrast to the histograms for DNA content of fission yeast cells overexpressing ccs52, a protein activating the anaphase-promoting complex (13). This plant protein induces endoreduplication, resulting in discrete ploidy peaks in the histograms rather than a less distinct increase in DNA content observed for rereplication.
The results show that Rep action is highly conserved and also functions in the unicellular eukaryotic organism S. pombe. Similar findings were reported for S. cerevisiae, where DNA A of mung bean yellow mosaic India virus could replace the yeast autonomously replicating sequence in plasmid YCp50 and drive a geminivirus Rep-dependent replication of the respective recombinant plasmid (64). This particular Rep-driven plasmid replication also revealed a potential influence of AC4 on the process (64). Nevertheless, no altered phenotypes for yeast cells harboring constructs that supported replication of the viral DNA were described. The effect of expressing the viral Rep protein on DNA content of S. cerevisiae was not analyzed, and it appears quite conceivable that also in S. cerevisiae rereplication of cellular DNA was triggered. It is an open question whether a rereplication state of the yeast cells, as opposed to true endoreduplication as observed in infected plant cells (6, 45), allows geminivirus DNA replication.
Here, we show that the Rep protein as the only viral factor is sufficient to induce S-phase conditions in fission yeast. Since there is no viral origin of replication present in the plasmids used in yeast cells here, it is probable that ACMV Rep has induced yeast chromosomal DNA rereplication. Nagar et al. (55) observed that besides viral DNA, host plant DNA replication also was induced by TGMV infection and suggested that this might lead to endoreduplication. A recent study (6) found that genes that are necessary for a replication-competent environment tended to be upregulated during cabbage leaf curl virus infection, whereas genes coupled to mitosis tended to be downregulated. In fact, the authors showed an increase in ploidy, explained as a consequence of endoreduplication. In plants, Rep action on the cell cycle may occur via interaction with pRBR and the E2F/DP transcription factors (29, 33, 72). However, maize streak virus Rep mutants with altered retinoblastoma protein (RB) binding motifs were still infectious but changed the tissue tropism of the virus (51, 69, 70). Additionally, Rep mutants in the LXCXE motif of bean yellow dwarf virus, a mastrevirus infecting dicotyledonous plants, were able to systemically infect N. benthamiana and bean albeit RB binding efficiency was reduced, as measured in yeast two-hybrid assays (48). Remarkably, no RB homologue has been identified in fission yeast to date. It would therefore be interesting to know whether an alternative pathway for S phase induction by Rep exists. Alternatively, a yet unidentified RB or a protein performing its function in fission yeast might be the target for Rep action on the cell cycle. Clarifying the nature of such a Rep target awaits further investigation. In that context, it may be interesting to analyze the effect of Clink, a nanovirus cell cycle regulator protein, in S. pombe. Clink binds plant RB via its LXCXE motif, and a mutant with alterations in this motif fails to interact with RB and is not able to influence the cell cycle (5). It has been shown that Clink induces transcription of PCNA and CDC6 (Cdc18 in S. pombe), both of which are E2F-regulated genes, and activation of expression was dependent on the authentic LXCXE motif (45). Moreover, discrete increments in ploidy in Arabidopsis were observed only upon expression of Clink but not of a Clink variant mutated in the LXCXE motif (45).
If there is no RB homologue in fission yeast, Rep might interact directly or indirectly with one of the factors that is important for DNA replication in S. pombe. Initiation of DNA synthesis relies on the activation of preformed complexes at the onset of S phase. In S. pombe, Cdc18 (Cdc6 in S. cerevisiae and other eukaryotes) and Cdt1 are required for the assembly of prereplication complexes at origins of DNA replication (7, 57). At the beginning of S phase, inactivation of Cdc18 by phosphorylation by cyclin-dependent kinase is followed by degradation (37, 38), and rereplication is prevented until cells enter the next cell cycle and synthesize Cdc18 and Cdt1 again. Rereplication due to successive S phases in the absence of mitosis is induced by high-level expression of Cdc18 (54, 58) and is even enhanced by additionally overexpressing Cdt1 (57). Other conditions that are known to cause rereplication are represented by a bypass of M phase. This type of polyploidization occurs when Cdc13/Cdc2 are inactivated either by complete disruption of the cdc13+ gene or cdc2/cdc13 mutants or by overexpression of the mitotic cyclin/cyclin-dependent kinase inhibitor Rum1 (10, 34, 53). Rum1 normally negatively regulates Cdc13/Cdc2, which in turn is necessary for inactivation of Cdc18 (37, 38). It could be anticipated that Rep interacts with one or several of these factors to induce resumption of DNA replication. Additional investigation is required to analyze if expression of Rep results in either elevated levels of Cdc18/Cdt1 or inactivation of Cdc13/Cdc2 or stabilization of Rum1. These experiments might provide evidence for an alternative pathway of S phase induction by Rep in fission yeast.
Acknowledgments
We thank Rob Briddon for the plasmid pUC19:APA-9 and Henning Schmidt for the plasmid pREP2 and S. pombe strain HE621. We are grateful to Peter Scheurich and Jessica Tepperink for help with flow cytometry and to Martin Pfannkuchen and Michael Schweikert for microscopy advice. We also thank Dieter Hülser for helpful discussion and Anan Kadri and Fania Geiger for critical reading of the manuscript.
This work was supported by DFG (Je 116/11-1).
Footnotes
Published ahead of print on 15 April 2009.
REFERENCES
- 1.Aberle, H. J., M. L. Rütz, M. Karayavuz, S. Frischmuth, C. Wege, D. Hülser, and H. Jeske. 2002. Localizing BC1 movement proteins of Abutilon mosaic geminivirus in yeasts by subcellular fractionation and freeze-fracture immunolabelling. Arch. Virol. 147103-107. [DOI] [PubMed] [Google Scholar]
- 2.Ach, R. A., T. Durfee, A. B. Miller, P. Taranto, L. Hanley-Bowdoin, P. C. Zambryski, and W. Gruissem. 1997. RRB1 and RRB2 encode maize retinoblastoma-related proteins that interact with a plant D-type cyclin and geminivirus replication protein. Mol. Cell. Biol. 175077-5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alfa, C., P. Fantes, J. Hyams, M. McLeod, and E. Warbrick. 1993. Experiments with fission yeast: a laboratory course manual. Cold Spring Harbor Press, Cold Spring Harbor, NY.
- 4.Arguello-Astorga, G., L. Lopez-Ochoa, L. J. Kong, B. M. Orozco, S. B. Settlage, and L. Hanley-Bowdoin. 2004. A novel motif in geminivirus replication proteins interacts with the plant retinoblastoma-related protein. J. Virol. 784817-4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aronson, M. N., A. D. Meyer, J. Györgyey, L. Katul, H. J. Vetten, B. Gronenborn, and T. Timchenko. 2000. Clink, a nanovirus-encoded protein, binds both pRB and SKP1. J. Virol. 742967-2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ascencio-Ibanez, J. T., R. Sozzani, T.-J. Lee, T.-M. Chu, R. D. Wolfinger, R. Cella, and L. Hanley-Bowdoin. 2008. Global analysis of Arabidopsis gene expression uncovers a complex array of changes impacting pathogen response and cell cycle during geminivirus infection. Plant Physiol. 148436-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baum, B., H. Nishitani, S. Yanow, and P. Nurse. 1998. Cdc18 transcription and proteolysis couple S phase to passage through mitosis. EMBO J. 175689-5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bjerrum, O. J., and Schafer-Nielsen, C. 1986. Buffer systems and transfer parameters for semidry electroblotting with a horizontal apparatus, p. 315-327. In M. J. Dunn (ed.), Proceedings of the fifth meeting of the International Electrophoresis Society. VHC, Weinheim, Germany.
- 9.Böttcher, B., S. Unseld, H. Ceulemans, R. B. Russell, and H. Jeske. 2004. Geminate structures of African cassava mosaic virus. J. Virol. 786758-6765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Broek, D., R. Bartlett, K. Crawford, and P. Nurse. 1991. Involvement of p34cdc2 in establishing the dependency of S phase on mitosis. Nature 349388-393. [DOI] [PubMed] [Google Scholar]
- 11.Campos-Olivas, R., J. M. Louis, D. Clérot, B. Gronenborn, and A. M. Gronenborn. 2002. The structure of a replication initiator unites diverse aspects of nucleic acid metabolism. Proc. Natl. Acad. Sci. USA 9910310-10315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Castillo, A. G., D. Collinet, S. Deret, A. Kashoggi, and E. R. Bejarano. 2003. Dual interaction of plant PCNA with geminivirus replication accessory protein (Ren) and viral replication protein (Rep). Virology 312381-394. [DOI] [PubMed] [Google Scholar]
- 13.Cebolla, A., J. M. Vinardell, E. Kiss, B. Olah, F. Roudier, A. Kondorosi, and E. Kondorosi. 1999. The mitotic inhibitor css52 is required for endoreduplication and ploidy-dependent cell enlargement in plants. EMBO J. 184476-4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choudhury, N. R., P. S. Malik, D. K. Singh, M. N. Islam, K. Kaliappan, and S. K. Mukherjee. 2006. The oligomeric Rep protein of mungbean yellow mosaic India virus (MYMIV) is a likely replicative helicase. Nucleic Acids Res. 346362-6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clérot, D., and F. Bernardi. 2006. DNA helicase activity is associated with the replication initiator protein Rep of tomato yellow leaf curl geminivirus. J. Virol. 8011322-11330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davies, J. W., and J. Stanley. 1989. Geminivirus genes and vectors. Trends Genet. 577-81. [DOI] [PubMed] [Google Scholar]
- 17.Desbiez, C., C. David, A. Mettouchi, J. Laufs, and B. Gronenborn. 1995. Rep protein of tomato yellow leaf curl geminivirus has an ATPase activity required for viral DNA replication. Proc. Natl. Acad. Sci. USA 925640-5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elmer, J. S., L. Brand, G. Sunter, W. Gardiner, D. M. Bisaro, and S. G. Rogers. 1988. Genetic analysis of tomato golden mosaic virus. II. The conserved AL1 ORF product is essential for replication. Nucleic Acids Res. 167043-7060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Etessami, P., R. Callis, S. Ellwood, and J. Stanley. 1988. Delimitation of essential genes of cassava latent virus DNA 2. Nucleic Acids Res. 164811-4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Etessami, P., K. Saunders, J. Watts, and J. Stanley. 1991. Mutational analysis of complementary-sense genes of African cassava mosaic DNA A. J. Gen. Virol. 721005-1012. [DOI] [PubMed] [Google Scholar]
- 21.Felsani, A., A. M. Mileo, and M. G. Paggi. 2006. Retinoblastoma family proteins as key targets of the small DNA virus oncoproteins. Oncogene 255277-5285. [DOI] [PubMed] [Google Scholar]
- 22.Fondong, V. N., R. V. C. Reddy, C. Lu, B. Hankoua, C. Felton, K. Czymmek, and F. Achenjang. 2007. The consensus N-myristoylation motif of a geminivirus AC4 protein is required for membrane binding and pathogenicity. Mol. Plant Microbe Interact. 20380-391. [DOI] [PubMed] [Google Scholar]
- 23.Fontes, E. P. B., V. A. Luckow, and L. Hanley-Bowdoin. 1992. A geminivirus replication protein is a sequence-specific DNA binding protein. Plant Cell 4597-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Forsburg, S. L. 1999. The best yeast? Trends Genet. 15340-344. [DOI] [PubMed] [Google Scholar]
- 25.Frischmuth, S., T. Kleinow, H.-J. Aberle, C. Wege, D. Hülser, and H. Jeske. 2004. Yeast two-hybrid systems confirm the membrane-association and oligomerization of BC1 but do not detect an interaction of the movement proteins BC1 and BV1 of Abutilon mosaic geminivirus. Arch. Virol. 1492349-2364. [DOI] [PubMed] [Google Scholar]
- 26.Frischmuth, S., C. Wege, D. Hülser, and H. Jeske. 2007. The movement protein BC1 promotes redirection of the nuclear shuttle protein BV1 of Abutilon mosaic geminivirus to the plasma membrane in fission yeast. Protoplasma 230117-123. [DOI] [PubMed] [Google Scholar]
- 27.Frischmuth, T., and J. Stanley. 1998. Recombination between viral DNA and the transgenic coat protein gene of African cassava mosaic geminivirus. J. Gen. Virol. 791265-1271. [DOI] [PubMed] [Google Scholar]
- 28.Grafi, G., R. J. Burnett, T. Helentjaris, B. A. Larkins, J. A. DeCaprio, W. R. Sellers, and W. G. Kaelin. 1996. A maize cDNA encoding a member of the retinoblastoma protein family: involvement in endoreduplication. Proc. Natl. Acad. Sci. USA 938962-8967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gronenborn, B. 2004. Nanoviruses: genome organisation and protein function. Vet. Microbiol. 98103-109. [DOI] [PubMed] [Google Scholar]
- 30.Gutierrez, C. 2000. DNA replication and cell cycle in plants: learning from geminiviruses. EMBO J. 19792-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gutierrez, C., E. Ramirez-Parra, M. Mar Castellano, A. P. Sanz-Burgos, A. Luque, and R. Missich. 2004. Geminivirus DNA replication and cell cycle interactions. Vet. Microbiol. 98111-119. [DOI] [PubMed] [Google Scholar]
- 32.Hanley-Bowdoin, L., J. S. Elmer, and S. G. Rogers. 1990. Expression of functional replication protein from tomato golden mosaic virus in transgenic tobacco plants. Proc. Natl. Acad. Sci. USA 871446-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hanley-Bowdoin, L., S. B. Settlage, and D. Robertson. 2004. Reprogramming plant gene expression: a prerequisite to geminivirus DNA replication. Mol. Plant Pathol. 5149-156. [DOI] [PubMed] [Google Scholar]
- 34.Hayles, J., D. Fisher, A. Woollard, and P. Nurse. 1994. Temporal order of S phase and mitosis in fission yeast is determined by the state of the p34cdc2-mitotic B cyclin complex. Cell 78813-822. [DOI] [PubMed] [Google Scholar]
- 35.Hong, Y., and J. Stanley. 1995. Regulation of African cassava mosaic virus complementary-sense gene expression by N-terminal sequences of the replication-associated protein AC1. J. Gen. Virol. 762415-2422. [DOI] [PubMed] [Google Scholar]
- 36.Horns, T., and H. Jeske. 1991. Localization of Abutilon mosaic virus DNA within leaf tissue by in-situ hybridization. Virology 181580-588. [DOI] [PubMed] [Google Scholar]
- 37.Jallepalli, P. V., G. W. Brown, M. Muzi-Falconi, D. Tien, and J. T. Kelly. 1997. Regulation of the replication initiator protein p65cdc18 by CDK phosphorylation. Genes Dev. 112767-2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jallepalli, P. V., D. Tien, and T. J. Kelly. 1998. sud1+ targets cyclin-dependent kinase-phosphorylated Cdc18 and Rum1 proteins for degradation and stops unwanted diploidization in fission yeast. Proc. Natl. Acad. Sci. USA 958159-8164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jeske, H. 2009. Geminiviruses. Curr. Top. Microbiol. Immunol. 331185-226. [DOI] [PubMed] [Google Scholar]
- 40.Jeske, H. 2007. Replication of geminiviruses and the use of rolling circle amplification for their diagnosis, p. 141-156. In H. Czosnek (ed.), Tomato yellow leaf curl virus disease. Springer, Dordrecht, The Netherlands.
- 41.Jeske, H., M. Lütgemeier, and W. Preiss. 2001. Distinct DNA forms indicate rolling circle and recombination-dependent replication of Abutilon mosaic geminivirus. EMBO J. 206158-6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jupin, I., F. De Kouchkovsky, F. Jouanneau, and B. Gronenborn. 1994. Movement of tomato yellow leaf curl geminivirus (TYLCV): involvement of the protein encoded by ORF C4. Virology 20482-90. [DOI] [PubMed] [Google Scholar]
- 43.Kong, L. J., B. M. Orozco, J. L. Roe, S. Nagar, S. Ou, H. S. Feiler, T. Durfee, A. B. Miller, W. Gruissem, D. Robertson, and L. Hanley-Bowdoin. 2000. A geminivirus replication protein interacts with the retinoblastoma protein through a novel domain to determine symptoms and tissue specificity of infection in plants. EMBO J. 193485-3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227680-685. [DOI] [PubMed] [Google Scholar]
- 45.Lageix, S., O. Catrice, J.-M. Deragon, B. Gronenborn, T. Pélissier, and B. C. Ramírez. 2007. The nanovirus-encoded Clink protein affects plant cell cycle regulation through interaction with the retinoblastoma-related protein. J. Virol. 814177-4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Laufs, J., S. Schumacher, N. Geisler, I. Jupin, and B. Gronenborn. 1995. Identification of the nicking tyrosine of geminivirus Rep. protein. FEBS Lett. 377258-262. [DOI] [PubMed] [Google Scholar]
- 47.Laufs, J., W. Traut, F. Heyraud, V. Matzeit, S. G. Rogers, J. Schell, and B. Gronenborn. 1995. In vitro cleavage and joining at the viral origin of replication by the replication initiator protein of tomato yellow leaf curl virus. Proc. Natl. Acad. Sci. USA 923879-3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu, L., K. Saunders, C. L. Thomas, J. W. Davies, and J. Stanley. 1999. Bean yellow dwarf virus RepA, but not Rep, binds to maize retinoblastoma protein, and the virus tolerates mutations in the consensus binding motif. Virology 256270-279. [DOI] [PubMed] [Google Scholar]
- 49.Lucy, A. P., M. I. Boulton, J. W. Davies, and A. J. Maule. 1996. Tissue specificity of Zea mays infection by maize streak virus. Mol. Plant Microbe Interact. 922-31. [Google Scholar]
- 50.Maundrell, K. 1990. nmt1 of fission yeast. J. Biol. Chem. 26510857-10864. [PubMed] [Google Scholar]
- 51.McGivern, D. R., K. C. Findlay, N. P. Montague, and M. I. Boulton. 2005. An intact RBR-binding motif is not required for infectivity of maize streak virus in cereals, but is required for invasion of mesophyll cells. J. Gen. Virol. 86797-801. [DOI] [PubMed] [Google Scholar]
- 52.Moffat, A. 1999. Geminiviruses emerge as serious crop threat. Science 2861835. [Google Scholar]
- 53.Moreno, S., and P. Nurse. 1994. Regulation of progression through the G1 phase of the cell cycle by the rum1+ gene. Nature 367236-242. [DOI] [PubMed] [Google Scholar]
- 54.Muzi Falconi, M., G. W. Brown, and T. J. Kelly. 1996. cdc18+ regulates initiation of DNA replication in Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. USA 931566-1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nagar, S., L. Hanley-Bowdoin, and D. Robertson. 2002. Host DNA replication is induced by geminivirus infection of differentiated plant cells. Plant Cell 142995-3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nagar, S., T. J. Pedersen, K. M. Carrick, L. Hanley-Bowdoin, and D. Robertson. 1995. A geminivirus induces expression of a host DNA synthesis protein in terminally differentiated plant cells. Plant Cell 7705-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nishitani, H., Z. Lygerou, T. Nishimoto, and P. Nurse. 2000. The Cdt1 protein is required to license DNA for replication in fission yeast. Nature 404625-628. [DOI] [PubMed] [Google Scholar]
- 58.Nishitani, H., and P. Nurse. 1995. p65cdc18 plays a major role controlling the initiation of DNA replication in fission yeast. Cell 83397-405. [DOI] [PubMed] [Google Scholar]
- 59.Orozco, B. M., and L. Hanley-Bowdoin. 1998. Conserved sequence and structural motifs contribute to the DNA binding and cleavage activities of a geminivirus replication protein. J. Biol. Chem. 27324448-24456. [DOI] [PubMed] [Google Scholar]
- 60.Papouli, E., S. Chen, A. A. Davies, D. Huttner, L. Krejci, P. Sung, and H. D. Ulrich. 2005. Crosstalk between SUMO and ubiquitin on PCNA is mediated by recruitment of the helicase Srs2p. Mol. Cell 19123-133. [DOI] [PubMed] [Google Scholar]
- 61.Piroux, N., K. Saunders, A. Page, and J. Stanley. 2007. Geminivirus pathogenicity protein C4 interacts with Arabidopsis thaliana shaggy-related protein kinase AtSKeta, a component of the brassinosteroid signalling pathway. Virology 362428-440. [DOI] [PubMed] [Google Scholar]
- 62.Pooma, W., and I. T. D. Petty. 1996. Tomato golden mosaic virus open reading frame AL4 is genetically distinct from its C4 analogue in monopartite geminiviruses. J. Gen. Virol. 771947-1951. [DOI] [PubMed] [Google Scholar]
- 63.Prakash, S., R. E. Johnson, and L. Prakash. 2005. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu. Rev. Biochem. 74317-353. [DOI] [PubMed] [Google Scholar]
- 64.Raghavan, V., P. S. Malik, N. R. Choudhury, and S. K. Mukherjee. 2004. The DNA-A component of a plant geminivirus (Indian mung bean yellow mosaic virus) replicates in budding yeast cells. J. Virol. 782405-2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rojas, M. R., H. Jiang, R. Salati, B. Xoconostle-Cazares, M. R. Sudarshana, W. J. Lucas, and R. L. Gilbertson. 2001. Functional analysis of proteins involved in movement of the monopartite begomovirus, Tomato yellow leaf curl virus. Virology 291110-125. [DOI] [PubMed] [Google Scholar]
- 66.Saunders, K., A. Lucy, and J. Stanley. 1992. RNA-primed complementary-sense DNA synthesis of the geminivirus African cassava mosaic virus. Nucleic Acids Res. 206311-6315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sazer, S., and S. W. Sherwood. 1990. Mitochondrial growth and DNA synthesis occur in the absence of nuclear DNA replication in fission yeast. J. Cell Sci. 97509-516. [DOI] [PubMed] [Google Scholar]
- 68.Settlage, S. B., A. B. Miller, and L. Hanley-Bowdoin. 1996. Interactions between geminivirus replication proteins. J. Virol. 706790-6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shepherd, D. N., D. P. Martin, D. R. McGivern, M. I. Boulton, J. A. Thomson, and E. P. Rybicki. 2005. A three-nucleotide mutation altering the Maize streak virus Rep pRBR-interaction motif reduces symptom severity in maize and partially reverts at high frequency without restoring pRBR-Rep binding. J. Gen. Virol. 86803-813. [DOI] [PubMed] [Google Scholar]
- 70.Shepherd, D. N., D. P. Martin, A. Varsani, J. A. Thomson, E. P. Rybicki, and H. H. Klump. 2006. Restoration of native folding of single-stranded DNA sequences through reverse mutations: an indication of a new epigenetic mechanism. Arch. Biochem. Biophys. 453106-120. [DOI] [PubMed] [Google Scholar]
- 71.Stanley, J. 1995. Analysis of African cassava mosaic virus recombinants suggests strand nicking occurs within the conserved nonanucleotide motif during the initiation of rolling circle DNA replication. Virology 206707-712. [DOI] [PubMed] [Google Scholar]
- 72.Stanley, J. 2004. Subviral DNAs associated with geminivirus disease complexes. Vet. Microbiol. 98121-129. [DOI] [PubMed] [Google Scholar]
- 73.Stanley, J., D. M. Bisaro, R. W. Briddon, J. K. Brown, C. M. Fauquet, B. D. Harrison, E. P. Rybicki, and D. A. Stenger. 2005. Geminiviridae, p. 301-326. In C. M. Fauquet, M. A. Mayo, J. Maniloff, U. Desselberger, and L. A. Ball (ed.), Virus taxonomy. Eighth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, San Diego, CA.
- 74.Stanley, J., and M. R. Gay. 1983. Nucleotide sequence of cassava latent virus DNA. Nature 301260-262. [Google Scholar]
- 75.Stratagene. 2002. ESP yeast protein expression and purification system and ESP yeast protein expression vectors instruction manual. Stratagene, Heidelberg, Germany.
- 76.Sunter, G., and D. M. Bisaro. 1992. Transactivation of geminivirus AR1 and BR1 gene expression by the viral AL2 gene product occurs at the level of transcription. Plant Cell 41321-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Townsend, R., J. Stanley, S. J. Curson, and M. N. Short. 1985. Major polyadenylated transcripts of cassava latent virus and location of the gene encoding coat protein. EMBO J. 433-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Townsend, R., J. Watts, and J. Stanley. 1986. Synthesis of viral DNA forms in Nicotiana plumbaginifolia protoplasts inoculated with cassava latent virus (CLV); evidence for the independent replication of one component of the CLV genome. Nucleic Acids Res. 141253-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vanitharani, R., P. Chellappan, J. S. Pita, and C. M. Fauquet. 2004. Differential roles of AC2 and AC4 of cassava geminiviruses in mediating synergism and suppression of posttranscriptional gene silencing. J. Virol. 789487-9498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Waseem, N. H., K. Labib, P. Nurse, and D. P. Lane. 1992. Isolation and analysis of the fission yeast gene encoding polymerase δ accessory protein PCNA. EMBO J. 115111-5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Watts, F. Z. 2006. Sumoylation of PCNA: Wrestling with recombination at stalled replication forks. DNA Repair 5399-403. [DOI] [PubMed] [Google Scholar]
- 82.Wege, C. 2007. Movement and localization of tomato yellow leaf curl viruses in the infected plant, p. 185-206. In H. Czosnek (ed.), Tomato yellow leaf curl virus disease. Springer, Dordrecht, The Netherlands.
- 83.Xie, Q., A. P. Sanz-Burgos, G. J. Hannon, and C. Gutierrez. 1996. Plant cells contain a novel member of the retinoblastoma family of growth regulatory proteins. EMBO J. 154900-4908. [PMC free article] [PubMed] [Google Scholar]
- 84.Xie, Q., P. Suarez-Lopez, and C. Gutierrez. 1995. Identification and analysis of a retinoblastoma binding motif in the replication protein of a plant DNA virus: requirement for efficient viral DNA replication. EMBO J. 144073-4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhang, W., N. H. Olson, T. S. Baker, L. Faulkner, M. Agbandje-McKenna, M. Boulton, J. W. Davies, and R. McKenna. 2001. Structure of the Maize streak virus geminate particle. Virology 279471-477. [DOI] [PubMed] [Google Scholar]