Abstract
The viral protein VP35 of ebolavirus (EBOV) is implicated to have diverse roles in the viral life cycle. We employed a yeast two-hybrid screen to search for VP35 binding partners and identified the cytoplasmic dynein light chain (DLC8) as a protein that interacts with VP35. Mapping analysis unraveled a consensus motif, SQTQT, within VP35 through which VP35 binds to DLC8. The disruption of DLC8 binding does not affect the ability of VP35 to inhibit type I IFN production. Given that VP35 from various EBOV species interacts with DLC8, this interaction may have a role in regulating the EBOV life cycle.
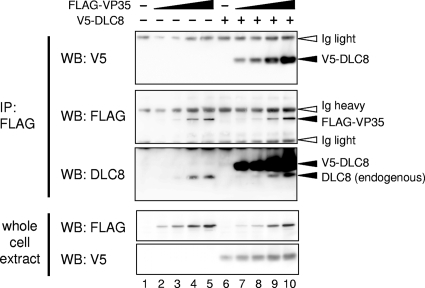
Ebolavirus (EBOV) belongs to the family Filoviridae of the order Mononegavirales. The EBOV virion contains an approximately 19-kb single-stranded RNA genome bearing seven genes. The genes encoding the nucleoprotein (NP), virion protein 35 (VP35), VP40, glycoprotein, VP30, VP24, and large (L) protein lie in this order from the 3′ to 5′ ends (33). VP35 is a multifunctional protein whose activities share some similarity with those of the P protein of other members of Mononegavirales, including the families Rhabdoviridae and Paramyxoviridae. VP35 forms the ribonucleoprotein complex that contains the NP, VP30, and polymerase L. Studies of reconstituted replication systems revealed that NP, VP35, VP30, and L are required for the transcription and replication of EBOV (28). In addition, VP35 inhibits type I interferon (IFN) production, thereby suppressing host innate immunity, the activity analogous to that of the P proteins of rabies virus and paramyxoviruses (2-4, 8, 13). VP35 also binds to double-stranded RNA, the activity associated with viral virulence (9, 16, 17). We have recently shown that VP35 interacts with the host cell machinery involved in the conjugation of small ubiquitin-like molecules and that this interaction is critical for inhibiting IFN transcription (T.-H. Chang, T. Kubota, M. Matsuoka, S. Jones, S. Bradfute, M. Bray, and K. Ozato, submitted for publication). In light of the multiple functions implicated for VP35, it is likely that it functions by interacting with various cellular partners. EBOV targets dendritic cells (DCs), which produce the highest amounts of IFNs in the body and confer resistance to many pathogens, for early infection (5, 7, 14, 25). To identify an interacting partner of EBOV VP35, we constructed a murine cDNA library from bone marrow-derived DCs stimulated with Newcastle disease virus in a pDEST22 vector using the CloneMiner cDNA library construction kit (Invitrogen). The cDNA library was screened with a full-length mouse-adapted VP35 as a bait using the ProQuest two-hybrid system (Invitrogen). VP35 from the mouse-adapted EBOV differs from that of the parental Zaire EBOV only in a single amino acid at position 12 (A → V). VP35 of both types potently inhibit IFN transcription in DCs (6) (Chang et al., submitted). Of 4.4 × 106 cDNA clones screened, 317 positive clones were isolated. Subsequent sequence analysis found that as many as 125 clones encode the 8-kDa dynein light chain (DLC8). DLC8 is a subunit of the cytoplasmic dynein motor complex involved in the microtubule-associated intracellular transport system (29, 33, 35). All 125 clones had a common DLC8 open reading frame, although 5′ untranslated regions were variably represented among these clones. To ascertain whether VP35 indeed interacts with DLC8 in the cells, pcDNA3.1 plasmids expressing FLAG-tagged VP35 (FLAG-VP35) and V5-tagged DLC8 (V5-DLC8) were transfected into 293T cells. Forty-eight hours later, the cells were harvested and the whole-cell lysates were subjected to immunoprecipitation with anti-FLAG agarose antibody beads and tested for V5-DLC8 binding by Western blot analysis (20). As shown in Fig. 1, FLAG-VP35 coprecipitated V5-DLC8 in a FLAG-VP35 dose-dependent manner (lanes 7 to 10 in the top panel). In addition, FLAG-VP35 coprecipitated endogenous DLC8 as detected with anti-DLC8 antibody (lanes 3 to 5 and 8 to 10 in the middle panel). The endogenous DLC8 was coprecipitated with FLAG-VP35 even in the absence of V5-DLC8. As expected, in the absence of FLAG-VP35, neither V5-DLC8 nor endogenous DLC8 was precipitated (lanes 1 and 6 in the middle panel). These results show that VP35 interacts with both endogenous and exogenous DLC8 in these cells.
FIG. 1.
Interaction of VP35 with DLC8. 293T cells were transfected with increasing amounts of pcDNA3.1 plasmid encoding FLAG-VP35 and a constant amount of pcDNA3.1 for V5-DLC8. Twenty-four hours posttransfection, the whole-cell extracts were immunoprecipitated (IP) with anti-FLAG antibody agarose beads, and the immunoprecipitates and whole-cell extracts were analyzed by Western blotting (WB) with antibody indicated at the left.
We next investigated a region within VP35 that interacts with DLC8. Plasmids encoding various truncated VP35 shown in Fig. 2A were tested for their ability to bind to DLC8. The immunoprecipitation experiments for which the results are depicted in Fig. 2B showed that deletions of 40 and 80 aa from the C terminus, ΔC/1-300 and ΔC/1-250, did not affect DLC8 binding (lanes 6 and 8 in the top panel). However, deletions of 80 or 120 aa from the N terminus, ΔN/80-340 and ΔN/120-340, abrogated DLC8 binding (lanes 12 and 14 in the top panel), although the deletion of the N-terminal 40 aa was without effect (lane 10 in the top panel). Since these data indicated that an internal N-terminal region is important for binding, we tested another deletion from positions 40 to 79 and found that this deletion completely eliminated DLC8 binding (Fig. 2B, Δ40-79, lane 16 in the top panel). Thus, DLC8 binding activity is mapped to a region from aa 40 to 79 of VP35 (see a summary of DLC binding in the legend to Fig. 2A). These observations were supported by the fact that all VP35 deletions were expressed at comparable levels with the expected molecular masses (Fig. 2B, bottom panel).
FIG. 2.
Mapping of the DLC8 binding site in VP35. (A) Schematic diagram of VP35 truncations tested for DLC8 binding. On the right is a summary of the binding assays below. (B) 293T cells were transfected with FLAG and full-length VP35 or the indicated truncations with or without V5-DLC8. Extracts were immunoprecipitated with anti-FLAG agarose beads, and the precipitates (IP) and whole-cell extracts were tested by Western blotting (WB) with the indicated antibodies (top and bottom panels, respectively). (C) Putative DLC8 binding motif in aa 71 to 75 of VP35 and those with indicated substitutions. (D) 293T cells were transfected with FLAG-VP35 mutants (S71A to T75A) and V5-DLC8, and binding to DCL8 was tested as described above.
A large number of DLC8-interacting partners, both cellular and viral proteins, have been reported (12, 15, 21). Based on these studies, a DLC8 binding consensus motif, K/SxTQT, has been proposed (24). An inspection of the sequence from aa 40 to 79 revealed a matching motif, SQTQT (Fig. 2C). To investigate whether this motif represents the DLC8 binding site, point mutations were introduced into each amino acid within the motif by alanine scan mutagenesis (Fig. 2C, S71A through T75A). The mutants were tested for DLC8 binding by a coimmunoprecipitation assay. As shown in Fig. 2D, while the substitution of first two amino acids retained DLC8 binding, the mutation in the third position greatly reduced DLC8 binding (lanes 3, 4, and 6 in the top panel). Moreover, the mutation in the last two amino acids completely abolished DLC8 binding (lanes 7 and 8). The expression levels of these mutants were comparable (Fig. 2D, bottom panels). These results indicate that this motif represents a binding surface through which VP35 interacts with DLC8 and that the last three amino acids are obligatory for DLC8 binding activity.
Interestingly, the Sudan and Reston EBOVs carry a VP35 that differs from that of the Zaire species in the first two amino acids in the motif (Fig. 3A). We next examined whether the VP35s from these EBOVs were capable of binding to DLC8. As shown in Fig. 3B, the VP35s of all three EBOV species bound to DLC8 to comparable degrees and in a VP35 dose-dependent manner (lanes 3 to 5 versus lanes 6 to 8 or 9 to 11 in the top panel). These results indicate that VP35s from all three EBOV species avidly bind to DLC8 and that the last three amino acids of the motif are more critical for DLC8 binding than the first two. Given the conservation of DLC8 binding among different EBOV species, this binding property may be important for the EBOV life cycle.
FIG. 3.
Binding of DLC8 to other viral proteins. (A) Comparison of the DLC8 binding motif of other EBOV species. (B) FLAG-VP35 from the Zaire, Sudan, or Reston species was transfected in 293T cells, and the interaction with V5-DLC8 was tested by coimmunoprecipitation assays as described in the legends to Fig. 1 and 2. (C) Plasmids for FLAG-VP35, rabies virus P protein, Sendai virus P and V proteins, and mumps virus P and V proteins were transfected into 293T cells, and the interaction with V5-DLC8 was tested by coimmunoprecipitation assays as described in the legends to Fig. 1 and 2.
Previously, P proteins of rabies virus and Mokola virus have been shown to bind to DLC8 (18, 31, 32). The rabies virus P protein contains the KSTQT motif within its DCL8 binding region (18). These viruses are members of the Lyssavirus genus belonging to the Mononegavirales order, and similarly to VP35 of EBOV, the P proteins are the second viral gene products. We examined whether DLC8 binding is a common property among viruses belonging to Mononegavirales. The P and V proteins of mumps virus and Sendai virus, members of the Paramyxoviridae, were tested for binding to V5-DLC8 by coimmunoprecipitation analysis. As shown in Fig. 3C, the P and V proteins of both viruses failed to bind to DLC8, while the EBOV VP35 and the rabies virus P protein bound to DLC8 (Fig. 3C, top panel). In line with these results, a canonical DLC8 binding motif was not found in the P and V proteins of mumps virus and Sendai virus. These data indicate that DLC8 binding activity is conserved in two families, Rhabdoviridae and Filoviridae, but not in other viruses of Mononegavirales.
Since VP35 has been shown to interfere with host innate immunity by inhibiting type I IFN production (3, 4), it was of interest to test whether the VP35-DLC8 interaction contributes to the inhibition of type I IFN transcription. VP35 mutants lacking DLC8 binding were transfected into murine L and NIH 3T3 cells along with the dual IFN-β promoter-luciferase reporter plasmids. Cells were infected with Sendai virus for 16 h, and luciferase reporter activity was measured. As shown in Fig. 4, Sendai virus infection enhanced IFN-β promoter activity by about 30-fold more than that of the mock infection in the absence of VP35 (vector alone). This enhancement was abrogated when wild-type VP35 was coexpressed both in L and NIH 3T3 cells. Significantly, three VP35 mutants lacking DLC8 binding activity also abolished IFN-β promoter activation (Fig. 4, S73A, Q74A, and T75A). However, the R312A mutant tested as a control did not inhibit IFN-β promoter activation, consistent with the previous report (9). Similar results were obtained from reporter analyses in 293T cells (data not shown). These results indicate that the binding of VP35 to DLC8 does not play a role in the inhibition of virus-induced IFN-β transcription.
FIG. 4.
VP35 inhibits type I IFN transcription without requiring DLC8 binding. Murine L and NIH 3T3 cells were transfected with wild-type or mutant VP35 along with the pGL4 reporter driven by the IFN-β promoter and pRL-TK reporters. Eight hours posttransfection, cells were infected with Sendai virus for 16 h. Luciferase activities were normalized by Renilla luciferase activity, and reporter activities in infected cells are expressed relative to those in mock-infected cells. Values represent the average of three determinations ± standard deviation.
We have shown here that the VP35 of EBOV binds to the DLC8, thus providing another example after that for Lyssavirus, in which the product of the second gene in the negative-strand RNA viruses interacts with this cellular factor. While this interaction is clearly based on the recognition of the common DLC binding motif SKTQT, the functional significance of the interaction is far from clear. The interaction of the rabies virus P protein with DLC8 was proposed to be of critical importance for the transport of the virus from the peripheral neurons to the central nervous system (CNS), which is pertinent to the fact that DLC8 is a subunit of the dynein motor complex involved in retrograde cargo transport (15, 18, 29-32). However, recent reports question the role of this interaction in viral transport, since the recombinant rabies virus P protein that lacks the DLC8 binding motif also moves to the brain and since the recombinant lentivirus pseudotyped with the rabies virus G protein confers retrograde transport within the brain network (26, 27). Further, it has been shown that the rabies virus P protein without the DLC8 binding domain impairs primary viral transcription, leading to reduced viral replication in the CNS, even though it allows viral entry into the CNS (34). The activity of the rabies virus P protein on viral transcription is closely linked to the formation of a complex with NP and polymerase L (10). Similarly, EBOV VP35 is shown to form a complex with NP and L and is essential for transcription and replication in the artificial minigenome systems (28). It is thus possible that VP35-DLC8 interaction plays a role in EBOV transcription and viral replication. Further, DLC8, by interacting with a variety of cellular factors, affects cellular functions. For example, DLC8 binds to the p35-binding protein and takes part in the regulation of apoptosis (23). It also binds to IκBα and is thought to regulate the activity of NFκB, a transcription factor involved in inflammation and apoptosis (11). Furthermore, DLC8 binds to nitric oxide synthase and inhibits its activity (19). Nitric oxide is a potent antimicrobial factor that regulates viral pathogenesis and host cell immunity (1). It can be envisaged that the interaction of VP35 with DLC8 influences multiple cellular functions, potentially to promote increased viral growth in EBOV-infected cells. In light of the evidence that fruit bats serve as a reservoir for EBOV, this interaction may be relevant to EBOV infection in these species as well (22). It will be of interest to study a recombinant EBOV without DLC8 binding for their transcription, replication, and host pathogenesis in an animal model.
Acknowledgments
We thank Kinjiroh Morimoto of Yasuda Women's University and Tadashi Fujita of Kyoto University for the kind gifts of plasmids for rabies virus P protein and the IFN-β promoter-luciferase reporter, respectively.
This research was supported in part by the Intramural Research Program of the NIH, National Institute of Child Health and Human Development, and the Trans NIH Biodefense Program.
Footnotes
Published ahead of print on 29 April 2009.
REFERENCES
- 1.Akaike, T., and H. Maeda. 2000. Nitric oxide and virus infection. Immunology 101300-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrejeva, J., K. S. Childs, D. F. Young, T. S. Carlos, N. Stock, S. Goodbourn, and R. E. Randall. 2004. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proc. Natl. Acad. Sci. USA 10117264-17269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Basler, C. F., A. Mikulasova, L. Martinez-Sobrido, J. Paragas, E. Muhlberger, M. Bray, H. D. Klenk, P. Palese, and A. Garcia-Sastre. 2003. The Ebola virus VP35 protein inhibits activation of interferon regulatory factor 3. J. Virol. 777945-7956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Basler, C. F., X. Wang, E. Muhlberger, V. Volchkov, J. Paragas, H. D. Klenk, A. Garcia-Sastre, and P. Palese. 2000. The Ebola virus VP35 protein functions as a type I IFN antagonist. Proc. Natl. Acad. Sci. USA 9712289-12294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bosio, C. M., M. J. Aman, C. Grogan, R. Hogan, G. Ruthel, D. Negley, M. Mohamadzadeh, S. Bavari, and A. Schmaljohn. 2003. Ebola and Marburg viruses replicate in monocyte-derived dendritic cells without inducing the production of cytokines and full maturation. J. Infect. Dis. 1881630-1638. [DOI] [PubMed] [Google Scholar]
- 6.Bray, M., K. Davis, T. Geisbert, C. Schmaljohn, and J. Huggins. 1998. A mouse model for evaluation of prophylaxis and therapy of Ebola hemorrhagic fever. J. Infect. Dis. 178651-661. [DOI] [PubMed] [Google Scholar]
- 7.Bray, M., and T. W. Geisbert. 2005. Ebola virus: the role of macrophages and dendritic cells in the pathogenesis of Ebola hemorrhagic fever. Int. J. Biochem. Cell Biol. 371560-1566. [DOI] [PubMed] [Google Scholar]
- 8.Brzozka, K., S. Finke, and K. K. Conzelmann. 2005. Identification of the rabies virus alpha/beta interferon antagonist: phosphoprotein P interferes with phosphorylation of interferon regulatory factor 3. J. Virol. 797673-7681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cardenas, W. B., Y. M. Loo, M. Gale, Jr., A. L. Hartman, C. R. Kimberlin, L. Martinez-Sobrido, E. O. Saphire, and C. F. Basler. 2006. Ebola virus VP35 protein binds double-stranded RNA and inhibits alpha/beta interferon production induced by RIG-I signaling. J. Virol. 805168-5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chenik, M., M. Schnell, K. K. Conzelmann, and D. Blondel. 1998. Mapping the interacting domains between the rabies virus polymerase and phosphoprotein. J. Virol. 721925-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crépieux, P., H. Kwon, N. Leclerc, W. Spencer, S. Richard, R. Lin, and J. Hiscott. 1997. IκBα physically interacts with a cytoskeleton-associated protein through its signal response domain. Mol. Cell. Biol. 177375-7385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dohner, K., C. H. Nagel, and B. Sodeik. 2005. Viral stop-and-go along microtubules: taking a ride with dynein and kinesins. Trends Microbiol. 13320-327. [DOI] [PubMed] [Google Scholar]
- 13.Enterlein, S., K. L. Warfield, D. L. Swenson, D. A. Stein, J. L. Smith, C. S. Gamble, A. D. Kroeker, P. L. Iversen, S. Bavari, and E. Muhlberger. 2006. VP35 knockdown inhibits Ebola virus amplification and protects against lethal infection in mice. Antimicrob. Agents Chemother. 50984-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geisbert, T. W., L. E. Hensley, T. Larsen, H. A. Young, D. S. Reed, J. B. Geisbert, D. P. Scott, E. Kagan, P. B. Jahrling, and K. J. Davis. 2003. Pathogenesis of Ebola hemorrhagic fever in cynomolgus macaques: evidence that dendritic cells are early and sustained targets of infection. Am. J. Pathol. 1632347-2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greber, U. F., and M. Way. 2006. A superhighway to virus infection. Cell 124741-754. [DOI] [PubMed] [Google Scholar]
- 16.Hartman, A. L., B. H. Bird, J. S. Towner, Z. A. Antoniadou, S. R. Zaki, and S. T. Nichol. 2008. Inhibition of IRF-3 activation by VP35 is critical for the high level of virulence of Ebola virus. J. Virol. 822699-2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hartman, A. L., L. Ling, S. T. Nichol, and M. L. Hibberd. 2008. Whole-genome expression profiling reveals that inhibition of host innate immune response pathways by Ebola virus can be reversed by a single amino acid change in the VP35 protein. J. Virol. 825348-5358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jacob, Y., H. Badrane, P. E. Ceccaldi, and N. Tordo. 2000. Cytoplasmic dynein LC8 interacts with lyssavirus phosphoprotein. J. Virol. 7410217-10222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jaffrey, S. R., and S. H. Snyder. 1996. PIN: an associated protein inhibitor of neuronal nitric oxide synthase. Science 274774-777. [DOI] [PubMed] [Google Scholar]
- 20.Kubota, T., M. Matsuoka, T. H. Chang, P. Tailor, T. Sasaki, M. Tashiro, A. Kato, and K. Ozato. 2008. Virus infection triggers SUMOylation of IRF3 and IRF7, leading to the negative regulation of type I interferon gene expression. J. Biol. Chem. 28325660-25670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leopold, P. L., and K. K. Pfister. 2006. Viral strategies for intracellular trafficking: motors and microtubules. Traffic 7516-523. [DOI] [PubMed] [Google Scholar]
- 22.Leroy, E. M., B. Kumulungui, X. Pourrut, P. Rouquet, A. Hassanin, P. Yaba, A. Delicat, J. T. Paweska, J. P. Gonzalez, and R. Swanepoel. 2005. Fruit bats as reservoirs of Ebola virus. Nature 438575-576. [DOI] [PubMed] [Google Scholar]
- 23.Lo, K. W., H. M. Kan, L. N. Chan, W. G. Xu, K. P. Wang, Z. Wu, M. Sheng, and M. Zhang. 2005. The 8-kDa dynein light chain binds to p53-binding protein 1 and mediates DNA damage-induced p53 nuclear accumulation. J. Biol. Chem. 2808172-8179. [DOI] [PubMed] [Google Scholar]
- 24.Lo, K. W., S. Naisbitt, J. S. Fan, M. Sheng, and M. Zhang. 2001. The 8-kDa dynein light chain binds to its targets via a conserved (K/R)XTQT motif. J. Biol. Chem. 27614059-14066. [DOI] [PubMed] [Google Scholar]
- 25.Mahanty, S., and M. Bray. 2004. Pathogenesis of filoviral haemorrhagic fevers. Lancet Infect. Dis. 4487-498. [DOI] [PubMed] [Google Scholar]
- 26.Mazarakis, N. D., M. Azzouz, J. B. Rohll, F. M. Ellard, F. J. Wilkes, A. L. Olsen, E. E. Carter, R. D. Barber, D. F. Baban, S. M. Kingsman, A. J. Kingsman, K. O'Malley, and K. A. Mitrophanous. 2001. Rabies virus glycoprotein pseudotyping of lentiviral vectors enables retrograde axonal transport and access to the nervous system after peripheral delivery. Hum. Mol. Genet. 102109-2121. [DOI] [PubMed] [Google Scholar]
- 27.Mebatsion, T. 2001. Extensive attenuation of rabies virus by simultaneously modifying the dynein light chain binding site in the P protein and replacing Arg333 in the G protein. J. Virol. 7511496-11502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muhlberger, E., M. Weik, V. E. Volchkov, H. D. Klenk, and S. Becker. 1999. Comparison of the transcription and replication strategies of Marburg virus and Ebola virus by using artificial replication systems. J. Virol. 732333-2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pfister, K. K. 2005. Dynein cargo gets its groove back. Structure 13172-173. [DOI] [PubMed] [Google Scholar]
- 30.Poisson, N., E. Real, Y. Gaudin, M. C. Vaney, S. King, Y. Jacob, N. Tordo, and D. Blondel. 2001. Molecular basis for the interaction between rabies virus phosphoprotein P and the dynein light chain LC8: dissociation of dynein-binding properties and transcriptional functionality of P. J. Gen. Virol. 822691-2696. [DOI] [PubMed] [Google Scholar]
- 31.Rasalingam, P., J. P. Rossiter, T. Mebatsion, and A. C. Jackson. 2005. Comparative pathogenesis of the SAD-L16 strain of rabies virus and a mutant modifying the dynein light chain binding site of the rabies virus phosphoprotein in young mice. Virus Res. 11155-60. [DOI] [PubMed] [Google Scholar]
- 32.Raux, H., A. Flamand, and D. Blondel. 2000. Interaction of the rabies virus P protein with the LC8 dynein light chain. J. Virol. 7410212-10216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanchez, A., M. P. Kiley, B. P. Holloway, and D. D. Auperin. 1993. Sequence analysis of the Ebola virus genome: organization, genetic elements, and comparison with the genome of Marburg virus. Virus Res. 29215-240. [DOI] [PubMed] [Google Scholar]
- 34.Tan, G. S., M. A. Preuss, J. C. Williams, and M. J. Schnell. 2007. The dynein light chain 8 binding motif of rabies virus phosphoprotein promotes efficient viral transcription. Proc. Natl. Acad. Sci. USA 1047229-7234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu, H., M. W. Maciejewski, S. Takebe, and S. M. King. 2005. Solution structure of the Tctex1 dimer reveals a mechanism for dynein-cargo interactions. Structure 13213-223. [DOI] [PubMed] [Google Scholar]