Abstract
Herpesviruses account for 134 out of the 140 virus-encoded microRNAs (miRNAs) known today. Here we report the identification of 11 novel miRNAs encoded by herpesvirus of turkey (HVT), a virus used as a live vaccine in poultry against the highly oncogenic Marek's disease virus type 1. Ten of these miRNAs were clustered together within the repeat long region of the viral genome, demonstrating some degree of positional conservation with other mardiviruses. Close sequence and phylogenetic relationships of some miRNAs in this cluster indicate evolution by duplication. HVT miRNAs represent the first example of virus-encoded miRNAs that show evolution by duplication.
MicroRNAs (miRNAs) are increasingly recognized as major regulators of gene expression in several multicellular organisms and viruses. Herpesviridae, a large family of viruses associated with a number of diseases including cancer in humans and animals, account for most of the virus-encoded miRNAs known today (7, 8). Considering the distinct biological requirements of herpesviruses, such as long latency periods that require the avoidance of the host immune responses, it is perhaps not very surprising that herpesviruses make extensive use of this highly effective regulatory mechanism of gene expression (7, 9, 21).
Marek's disease (MD) is a highly contagious rapid-onset T-cell lymphoma of poultry caused by MD virus type 1 (MDV-1), an alphaherpesvirus of the genus Mardivirus (11), which also includes MDV-2 and the herpesvirus of turkeys (HVT). Originally isolated from domestic turkeys in the late 1960s (12, 24), HVT is widely used as a live vaccine against MD because of its antigenic relatedness to MDV-1 (1). HVT is estimated to have separated from the MDV-1/MDV-2 lineage only about 38 million years ago (19). This relatedness is further evident from the overall similarities in the genome structures and sequences of these viruses (1, 14). We and others have previously reported the characteristics of several miRNAs encoded by MDV-1 and MDV-2 (5, 6, 25, 26). In the present study, we extended these investigations to identify HVT-encoded miRNAs. For this, we size selected the small RNA (∼19- to 24-nucleotide [nt]) population from chicken embryo fibroblast (CEF) cultures infected with HVT by using procedures described previously (25). Sequence analysis of ∼480 clones using vector-specific primers identified a total of 1,346 high-quality reads containing small RNA sequences with both the 5′ and 3′ adapters used in the cloning. BLAST homology searches (2) of the sequences against the HVT genome (AF291866) identified a total of 406 sequences (30%) representing 11 candidate miRNAs. The complex steps involved in miRNA biogenesis, including the criteria for strand selection for incorporation in the RNA-induced silencing complex, have been described previously (4, 13). Although it is the miRNA strand that is predominantly incorporated into the RNA-induced silencing complex, the non-miRNA strand could also be processed less efficiently than miRNAs (23, 25, 26). In the present study, mature forms representing both strands of the duplex were demonstrated either by cloning or by Northern blotting for 8 of the 11 HVT miRNAs. The miRNAs derived from the 5′ and 3′ arms of the stem-loop precursors were designated with a -5p or -3p suffix, respectively (Fig. 1A).
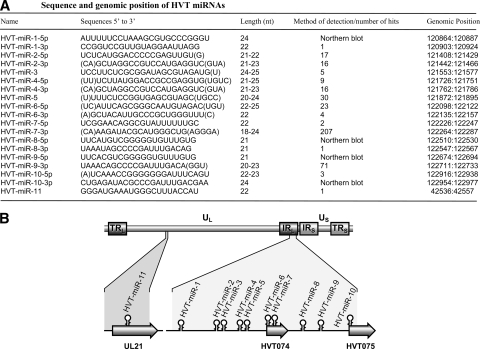
FIG. 1.
Cloning of HVT miRNAs. (A) Sequences of cloned candidate miRNA species, with nucleotide positions based on the published sequence (GenBank accession number AF291866). The methods of detection of the miRNAs or their cloning frequencies in the library are indicated. Sequence variations surrounding the recovered HVT miRNAs are indicated by parentheses. (B) The schematic diagram showing the positions of the miRNAs (stem-loops) in the HVT genome. The internal and terminal repeat long (TRL/IRL) and short (TRS/IRS) regions flanking the unique long (UL) and short (US) regions of the genome are shown. Genomic positions and orientations of HVT open reading frames UL21, HVT074, and HVT075 are shown.
Examination of the genomic locations of the 11 candidate miRNAs showed that 10 of these miRNAs were clustered together in the same orientation in a 2.1-kb region (positions 120864 to 122977 in the TRL/IRL region) in the HVT genome, overlapping with two putative open reading frames, HVT074 and HVT075 (Fig. 1B). These included miRNA-6 and miRNA-7 embedded in HVT074 and miRNA-10 in HVT075, respectively, in the same orientations. The only HVT miRNA outside this cluster, miRNA-11, was located in the UL region in the same orientation to the coding region of tegument protein UL21. The clustering of the 10 miRNAs in the same orientation in the 2.1-kb region of the TRL/IRL suggests that these miRNAs could be derived from a single transcript. However, the cloning frequencies of the miRNAs from this cluster showed variations, with miRNA-7-3p being the most abundant, with 207 hits representing more than half of the HVT miRNA population. In contrast, miRNAs such as miRNA-1-3p, miRNA-8-3p, and miRNA-11 were represented only once (Fig. 1A). Such differences in the abundance of the miRNAs have also been observed with MDV-1 and MDV-2 miRNAs (25, 26) and are thought to be due to differences in their processing efficiency or stability.
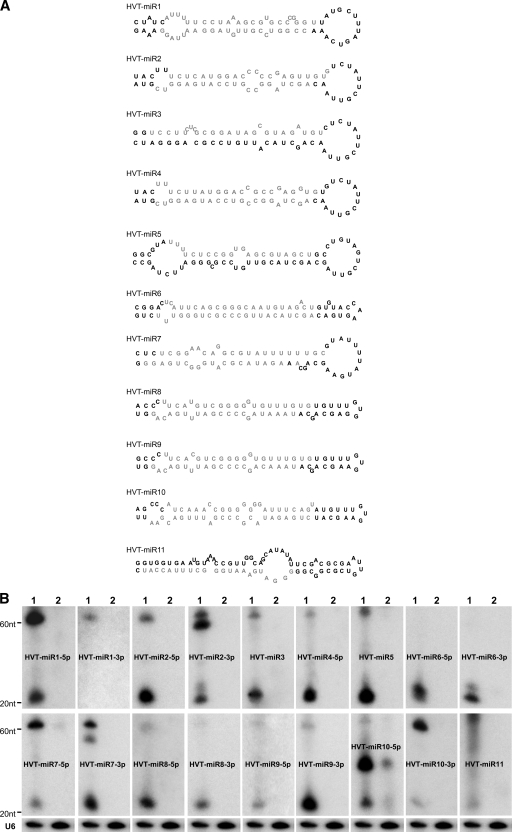
For validation of the authenticity of the candidate miRNAs, we used the standard criterion of miRNA annotation (3). First, we examined the potential precursor RNA hairpin structures of each of the 11 putative HVT miRNA candidates by using the 60- to 80-nt surrounding HVT genome sequence by MFOLD calculation (27). Secondary structures drawn using RNADRAW software showed that the miRNA precursors with average lengths of approximately 70 nt each were able to form characteristic hairpin structures (Fig. 2A), supporting the view that the cloned sequences represent novel HVT-encoded miRNAs. We next examined the expression of the mature HVT miRNAs by Northern blot analysis. For this, the total RNA isolated from HVT-infected CEFs was hybridized with individual miRNA probes. RNA from uninfected CEFs was included as a negative control. All of the cloned sequences are detectable in HVT-infected CEFs by Northern blotting, except for miR-1-3p, which appeared only once in the library (Fig. 2B). The other strand of miR-1-3p precursor is detectable, suggesting that miR-1-5p could be the miRNA strand and that miR-1-3p could serve as the non-miRNA strand. No miRNAs were detected with RNA extracted from the uninfected CEFs, although weak signals were observed with the miR-10-5p probe, which also gave a strong band between the precursor and mature forms in the HVT-infected cells. This band, as well as the mature band, also appears in the uninfected CEFs with much lower intensity. The reasons behind the detection of these signals in uninfected cells are not clear. Although BLAST searches did not reveal any similar sequences in the chicken genome, this cannot be ruled out, especially because of the incomplete annotation of the chicken genome sequence. Northern blotting detected the miRNA as well as the non-miRNA strands in several cases, although the latter generally showed lower expression levels based on the signal intensities (Fig. 2B). This is also reflected by the lower ratio of the mature non-miRNA strand to the pre-miRNA.
FIG. 2.
Identification of cloned HVT miRNAs. (A) Secondary structures of HVT pre-miRNAs predicted using the MFOLD algorithm. The mature miRNA strands are indicated in light gray. (B) Northern blot analysis demonstrating the expression of HVT miRNAs. Total RNAs from HVT-infected CEFs (lanes 1) and uninfected CEFs (lanes 2) were separated on a 15% denaturing polyacrylamide gel and probed with [γ-32P]ATP-radiolabeled antisense oligonucleotides to the indicated miRNAs. Size markers indicate the positions of the pre-miRNA and the mature miRNA. The cellular U6 small nuclear RNA served as the loading control. A representative blot of this set is shown.
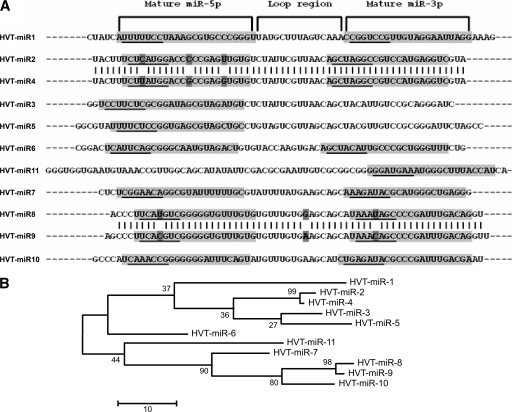
The discovery of 11 novel HVT-encoded miRNAs reported here together with the previously identified 14 in MDV-1 (5, 6, 20, 26) and 17 in MDV-2 (25) makes the genus Mardivirus a major source of virus-encoded miRNAs. Closer examination of some of the HVT miRNA sequences showed striking similarities, suggesting their generation by duplication. Gene duplication has long been recognized as a major route for evolution of genes including miRNAs in several species (10, 16-18). Duplications of miRNAs are generally observed in large miRNA clusters, suggesting that the expansion of the miRNA clusters is a major mode of miRNA evolution (10). In order to obtain evidence of miRNA duplication in the HVT miRNA cluster, we carried out multiple sequence alignment of the precursors of these miRNAs. As shown in Fig. 3A, the sequence of the precursors of miR-2 and miR-4 showed high sequence homology (95.3%) with identical sequences in the loop and the mature -3p regions, while the -5p mature miRNA region showed three substitutions, including the one in the seed region. Similarly, the precursors of miR-8 and miR-9 are highly homologous with only a single nucleotide difference in both -5p and -3p mature miRNA sequences as well as in the loop regions. The close sequence homology of the HVT miRNAs is also evident from the phylogenetic analysis which showed the branching of the closely related miRNAs (Fig. 3B). Because they are antisense regulators, alteration to the miRNA sequences of duplicated miRNAs can have a major impact on their targeting capabilities and capacities for acquiring novel functions, particularly for changes in the seed regions. The duplicated HVT miRNAs miR-8 and miR-9 had identical sequences in both -5p and -3p mature miRNA sequences, except for a single point mutation in the seed sequence (Fig. 3A). Since any transcript is just 1 nt away from being an miRNA target, such point mutations in the seed sequence could result in the loss or gain of new targets for these miRNAs, allowing “neofunctionalization” (22) of these duplicated miRNAs. Similarly, the three nucleotide substitutions including the one in the seed region of the -5p mature sequences of miR-2 and miR-4 will also have the potential to affect the expression of new targets. Although further work is needed to examine the targets of these novel miRNAs, HVT-encoded miRNAs represent the first clear example of evolution of miRNAs by duplication among viruses.
FIG. 3.
Phylogeny of HVT miRNAs. (A) Sequence alignment of HVT pre-miRNAs showing the positions of the mature -5p and -3p miRNAs (shaded areas) and the loop region. The seed regions of the mature miRNAs are underlined. (B) Phylogenetic tree (with the bootstrap values of the branches) showing the evolutionary relationships of the HVT pre-miRNAs by maximum parsimony analysis using the MEGA 4.0 package (15).
Acknowledgments
We thank Nick Knowles, Institute for Animal Health, Pirbright, for assistance with the phylogenetic analysis of the miRNAs.
This work was partly funded by the BBSRC, United Kingdom.
Footnotes
Published ahead of print on 29 April 2009.
REFERENCES
- 1.Afonso, C. L., E. R. Tulman, Z. Lu, L. Zsak, D. L. Rock, and G. F. Kutish. 2001. The genome of turkey herpesvirus. J. Virol. 75971-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215403-410. [DOI] [PubMed] [Google Scholar]
- 3.Ambros, V., B. Bartel, D. P. Bartel, C. B. Burge, J. C. Carrington, X. Chen, G. Dreyfuss, S. R. Eddy, S. Griffiths-Jones, M. Marshall, M. Matzke, G. Ruvkun, and T. Tuschl. 2003. A uniform system for microRNA annotation. RNA 9277-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartel, D. P. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116281-297. [DOI] [PubMed] [Google Scholar]
- 5.Burnside, J., E. Bernberg, A. Anderson, C. Lu, B. C. Meyers, P. J. Green, N. Jain, G. Isaacs, and R. W. Morgan. 2006. Marek's disease virus encodes microRNAs that map to meq and the latency-associated transcript. J. Virol. 808778-8786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burnside, J., M. Ouyang, A. Anderson, E. Bernberg, C. Lu, B. C. Meyers, P. J. Green, M. Markis, G. Isaacs, E. Huang, and R. W. Morgan. 2008. Deep sequencing of chicken microRNAs. BMC Genomics 9185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cullen, B. R. 2009. Viral and cellular messenger RNA targets of viral microRNAs. Nature 457421-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gottwein, E., and B. R. Cullen. 2008. Viral and cellular microRNAs as determinants of viral pathogenesis and immunity. Cell Host Microbe 3375-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grey, F., L. Hook, and J. Nelson. 2008. The functions of herpesvirus-encoded microRNAs. Med. Microbiol. Immunol. 197261-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hertel, J., M. Lindemeyer, K. Missal, C. Fried, A. Tanzer, C. Flamm, I. L. Hofacker, and P. F. Stadler. 2006. The expansion of the metazoan microRNA repertoire. BMC Genomics 725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.ICTV. 2006. 00.031.1.03 Mardivirus. In C. Búchen-Osmond (ed.), ICTVdB—the universal virus database, version 4. Columbia University, New York, NY.
- 12.Kawamura, H., D. J. King, Jr., and D. P. Anderson. 1969. A herpesvirus isolated from kidney cell culture of normal turkeys. Avian Dis. 13853-863. [PubMed] [Google Scholar]
- 13.Kim, V. N., J. Han, and M. C. Siomi. 2009. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 10126-139. [DOI] [PubMed] [Google Scholar]
- 14.Kingham, B. F., V. Zelnik, J. Kopacek, V. Majerciak, E. Ney, and C. J. Schmidt. 2001. The genome of herpesvirus of turkeys: comparative analysis with Marek's disease viruses. J. Gen. Virol. 821123-1135. [DOI] [PubMed] [Google Scholar]
- 15.Kumar, S., M. Nei, J. Dudley, and K. Tamura. 2008. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief. Bioinform. 9299-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu, N., K. Okamura, D. M. Tyler, M. D. Phillips, W. J. Chung, and E. C. Lai. 2008. The evolution and functional diversification of animal microRNA genes. Cell Res. 18985-996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu, J., Y. Fu, S. Kumar, Y. Shen, K. Zeng, A. Xu, R. Carthew, and C. I. Wu. 2008. Adaptive evolution of newly emerged micro-RNA genes in Drosophila. Mol. Biol. Evol. 25929-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu, J., Y. Shen, Q. Wu, S. Kumar, B. He, S. Shi, R. W. Carthew, S. M. Wang, and C. I. Wu. 2008. The birth and death of microRNA genes in Drosophila. Nat. Genet. 40351-355. [DOI] [PubMed] [Google Scholar]
- 19.McGeoch, D. J., F. J. Rixon, and A. J. Davison. 2006. Topics in herpesvirus genomics and evolution. Virus Res. 11790-104. [DOI] [PubMed] [Google Scholar]
- 20.Morgan, R., A. Anderson, E. Bernberg, S. Kamboj, E. Huang, G. Lagasse, G. Isaacs, M. Parcells, B. C. Meyers, P. J. Green, and J. Burnside. 2008. Sequence conservation and differential expression of Marek's disease virus microRNAs. J. Virol. 8212213-12220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murphy, E., J. Vanicek, H. Robins, T. Shenk, and A. J. Levine. 2008. Suppression of immediate-early viral gene expression by herpesvirus-coded microRNAs: implications for latency. Proc. Natl. Acad. Sci. USA 1055453-5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rastogi, S., and D. A. Liberles. 2005. Subfunctionalization of duplicated genes as a transition state to neofunctionalization. BMC Evol. Biol. 528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seo, G. J., L. H. Fink, B. O'Hara, W. J. Atwood, and C. S. Sullivan. 2008. Evolutionarily conserved function of a viral microRNA. J. Virol. 829823-9828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Witter, R. L., K. Nazerian, H. G. Purchase, and G. H. Burgoyne. 1970. Isolation from turkeys of a cell-associated herpesvirus antigenically related to Marek's disease virus. Am. J. Vet. Res. 31525-538. [PubMed] [Google Scholar]
- 25.Yao, Y., Y. Zhao, H. Xu, L. P. Smith, C. H. Lawrie, A. Sewer, M. Zavolan, and V. Nair. 2007. Marek's disease virus type 2 (MDV-2)-encoded microRNAs show no sequence conservation with those encoded by MDV-1. J. Virol. 817164-7170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yao, Y., Y. Zhao, H. Xu, L. P. Smith, C. H. Lawrie, M. Watson, and V. Nair. 2008. MicroRNA profile of Marek's disease virus-transformed T-cell line MSB-1: predominance of virus-encoded microRNAs. J. Virol. 824007-4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 313406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]